Abstract
Triacylglycerol (TAG) is known to be synthesized in a reaction that uses acyl-CoA as acyl donor and diacylglycerol (DAG) as acceptor, and which is catalyzed by the enzyme acyl-CoA:diacylglycerol acyltransferase. We have found that some plants and yeast also have an acyl-CoA-independent mechanism for TAG synthesis, which uses phospholipids as acyl donors and DAG as acceptor. This reaction is catalyzed by an enzyme that we call phospholipid:diacylglycerol acyltransferase, or PDAT. PDAT was characterized in microsomal preparations from three different oil seeds: sunflower, castor bean, and Crepis palaestina. We found that the specificity of the enzyme for the acyl group in the phospholipid varies between these species. Thus, C. palaestina PDAT preferentially incorporates vernoloyl groups into TAG, whereas PDAT from castor bean incorporates both ricinoleoyl and vernoloyl groups. We further found that PDAT activity also is present in yeast microsomes. The substrate specificity of this PDAT depends on the head group of the acyl donor, the acyl group transferred, and the acyl chains of the acceptor DAG. The gene encoding the enzyme was identified. The encoded PDAT protein is related to lecithin:cholesterol acyltransferase, which catalyzes the acyl-CoA-independent synthesis of cholesterol esters. However, budding yeast PDAT and its relatives in fission yeast and Arabidopsis form a distinct branch within this protein superfamily, indicating that a separate PDAT enzyme arose at an early point in evolution.
Keywords: oil seeds, phospholipids
Triacylglycerol (TAG) is the most common lipid-based energy reserve in nature. The main pathway for synthesis of TAG is believed to involve three sequential acyl-transfers from acyl-CoA to a glycerol backbone (1, 2). For many years, acyl-CoA:diacylglycerol acyltransferase (DAGAT), which catalyzes the third acyl transfer reaction, was thought to be the only enzyme specifically involved in TAG synthesis. It acts by diverting diacylglycerol (DAG) from membrane lipid synthesis into TAG (2). Genes encoding this enzyme were recently identified both in mice (3) and in plants (4–6), and the encoded proteins were shown to be homologous to acyl-CoA:cholesterol acyltransferase (ACAT). It was also recently reported that another DAGAT exists in the oleaginous fungus Mortierella ramanniana, which is unrelated to the mouse DAGAT, the ACAT gene family, or to any other known gene (K. Lardizabal, personal communication). In addition to these acyl-CoA-dependent enzymes, recent work has shown that, in microsomal preparations from oil seeds, TAG synthesis can also occur in the absence of acyl-CoA (7, 8). However, the enzyme involved in this acyl-CoA-independent synthesis of TAG has not yet been identified in any organism.
In this paper, we characterize the acyl-CoA-independent synthesis of TAG in plants, and we conclude that it is mediated by an enzyme that we call phospholipid:diacylglycerol acyltransferase (PDAT). This enzyme is proposed to be involved in the accumulation of high levels of hydroxylated fatty acid (ricinoleic acid) and epoxidated fatty acid (vernolic acid) in TAG in castor bean (Ricinus communis) and the hawk's-beard Crepis palaestina, respectively. Furthermore, a similar enzyme is shown to be present in the yeast Saccharomyces cerevisiae, and the gene encoding this enzyme, YNR008w, is identified.
Materials and Methods
Yeast Strains and Plasmids.
The wild-type yeast strains used were either FY1679 (MATα his3-Δ200 leu2-Δ1 trp1-Δ6 ura3-52) (9) or W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) (10). The YNR008w∷KanMX2 disruption strain 10280B, which is congenic to FY1679, was obtained from the Euroscarf collection (Frankfurt). A 2751-bp fragment containing the YNR008w gene with 583 bp of 5′ and 183 bp of 3′ flanking DNA was amplified from W303-1A genomic DNA by using Taq DNA polymerase with 5′-TCTCCATCTTCTGCAAAACCT-3′ and 5′-CCTGTCAAAAACCTTCTCCTC-3′ as primers. The resulting PCR product was purified by agarose gel electrophoresis and cloned into the EcoRV site of pBluescript. For complementation experiments, the cloned fragment was released from pBluescript by HindIII–SacI digestion and then cloned between the HindIII and SacI sites of pFL39 (11), thus generating pUS1. For overexpression of the PDAT gene, a 2202-bp EcoRI fragment from the pBluescript plasmid, which contains only 24 bp of 5′ flanking DNA, was cloned into the BamHI site of the GAL1 expression vector pJN92 (12), thus generating pUS4.
Microsomal Preparations.
Microsomes from developing seeds of sunflower (Helianthus annuus), R. communis, and C. palaestina were prepared by the procedure of Stobart and Stymne (13). To obtain yeast microsomes, 1 g of yeast cells (fresh weight) was resuspended in 8 ml of ice-cold buffer [20 mM Tris⋅Cl, pH 7.9/10 mM MgCl2/1 mM EDTA/5% (vol/vol) glycerol/1 mM DTT/0.3 M ammonium sulfate] in a 12-ml glass tube. To this tube, 4 ml of glass beads (diameter 0.45–0.5 mm) was added, and the tube was then heavily shaken (three time for 60 s each) in an MSK cell homogenizer (B. Braun Melsungen, Germany). The homogenized suspension was centrifuged at 1,500 × g for 15 min at 6°C, and the resulting supernatant was again centrifuged, at 100,000 × g for 1.5 h at 6°C. The 100,000 × g pellet was resuspended in 0.1 M potassium phosphate (pH 7.2), and stored at −80°C. It is subsequently referred to as the crude yeast microsomal fraction.
Lipid Substrates.
Radio-labeled ricinoleic (12-hydroxy-9-octadecenoic) and vernolic (12,13-epoxy-9-octadecenoic) acids were synthesized enzymatically from [1-14C]oleic acid and [1-14C]linoleic acid, respectively, by incubation with microsomal preparations from seeds of R. communis and C. palaestina, respectively (14). The synthesis of phosphatidylcholines (PC) or phosphatidylethanolamines (PE) with 14C-labeled acyl groups in the sn-2 position was performed by using either enzymatic (15), or synthetic (16) acylation of [14C]oleic, [14C]ricinoleic, or [14C]vernolic acid. Dioleoyl-PC that was labeled in the sn-1 position was synthesized from sn-1-[14C]oleoyl-lyso-PC and unlabeled oleic acid as described in ref. 16. sn-1-Oleoyl-sn-2-[14C]ricinoleoyl-DAG was synthesized from PC by the action of phospholipase C type XI from Bacillus cereus (Sigma) as described in ref. 17. Monovernoloyl- and divernoleoyl-DAG were synthesized from TAG extracted from seeds of Euphorbia lagascae, using the TAG-lipase (Rizhopus arrhizus; Sigma) as previously described (7). Monoricinoleoyl-DAG was synthesized according to the same method, using TAG extracted from castor bean.
Lipid Analysis.
Total lipid composition of yeast was determined from cells harvested from a 40-ml liquid culture, broken in a glass-bead shaker, and extracted into chloroform as described by Bligh and Dyer (18), and then separated by TLC in hexane/diethyl ether/acetic acid (80:20:1) on precoated silica gel 60 plates (Merck). The lipid areas were located by brief exposure to I2 vapors and identified by means of appropriate standards. Polar lipids, sterol esters, and triacylglycerols, as well as the remaining minor lipid classes, referred to as other lipids, were excised from the plates. Fatty acid methyl esters were prepared by heating the dry excised material at 85°C for 60 min in 2% (vol/vol) sulfuric acid in dry methanol. The methyl esters were extracted with hexane and analyzed by GLC through a 50 m × 0.32 mm CP-Wax58-CB fused-silica column (Chrompack), with methylheptadecanoic acid as an internal standard. The fatty acid content of each fraction was quantified and used to calculate the relative amount of each lipid class. To determine the total lipid content, 3-ml aliquots from yeast cultures were harvested by centrifugation, and the resulting pellets were washed with distilled water and lyophilized. The weight of the dried cells was determined, and the fatty acid content was quantified by GLC analyses after conversion to methyl esters as described above. The lipid content was then calculated as nmol of fatty acid (FA) per mg dry weight yeast.
Enzyme Assays.
Aliquots of crude microsomal fractions (corresponding to 10 nmol of microsomal PC) from developing plant seeds or yeast cells were lyophilized overnight. 14C-labeled substrate lipids dissolved in benzene were then added to the dried microsomes. The benzene was evaporated under a stream of N2, leaving the lipids in direct contact with the membranes, and 0.1 ml of 50 mM potassium phosphate (pH 7.2) was added. The suspension was thoroughly mixed and incubated at 30°C for the time period indicated, up to 90 min. Lipids were extracted from the reaction mixture into chloroform (18) and separated by TLC in hexane/diethyl ether/acetic acid (35:70:1.5) on silica gel 60 plates (Merck). The radioactive lipids were visualized and quantified on the plates by electronic autoradiography (Instant Imager; Packard).
Yeast Cultivation.
Yeast cells were grown at 28°C on a rotary shaker in liquid YPD medium (1% yeast extract/2% peptone/2% glucose). In the experiments described in Fig. 2, cells were precultured for 20 h in liquid YPD medium, harvested, and resuspended in an equal volume of minimal medium (19) containing 16 g/liter glycerol. The cells were then grown for an additional 24 h before being harvested. Selection for the plasmid in transformed cells was maintained by growing cells in synthetic medium (20) lacking uracil and supplemented with 2% (vol/vol) glycerol and 2% (vol/vol) ethanol. Transformed cells with genes overexpressed from the GAL1 promoter were induced after 2 or 25 h of growth by the addition of 2% final concentration (wt/vol) galactose. The cells were then incubated for another 22 h before being harvested.
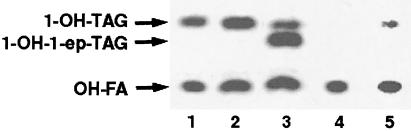
Figure 2.
PDAT activity in yeast microsomes, as visualized by autoradiogram of neutral lipid products separated on TLC. Microsomal membranes from the wild-type yeast strain FY1679 (lanes 1–3), a congenic yeast strain [FVKT004–04C(AL)] that is disrupted for YNR008w (lane 4), or the same disruption strain transformed with the plasmid pUS1, containing the YNR008w gene behind its native promoter (lane 5), were assayed for PDAT activity. As substrates, we used 2 nmol of sn-1-oleoyl-sn-2-[14C]ricinoleoyl-PC together with either 5 nmol of dioleoyl-DAG (lanes 2, 4, and 5) or rac-oleoyl-vernoloyl-DAG (lane 3). The enzymatic assay and lipid analysis were performed as described in Materials and Methods. Abbreviations: 1-OH-TAG, monoricinoleoyl-TAG; 1-OH-1-ep-TAG, monoricinoleoyl-monovernoloyl-TAG; OH-FA, unesterified ricinoleic acid.
Results
Acyl-CoA-Independent Synthesis of TAG by Oil Seed Microsomes.
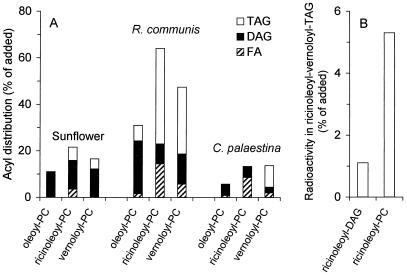
A large number of unusual fatty acids can be found in oil seeds (21). Many of these fatty acids, such as ricinoleic (22) and vernolic acids (23), are synthesized by using PC with oleoyl or linoleoyl groups esterified to the sn-2 position, respectively, as the immediate precursor. However, even though PC can be a substrate for unusual fatty acid synthesis and is the major membrane lipid in seeds, unusual fatty acids are rarely found in the membranes. Instead, they are mainly incorporated into TAG. A mechanism for efficient and selective transfer of these unusual acyl groups from PC into TAG must therefore exist in oil seeds that accumulate such unusual fatty acids. This transfer reaction was biochemically characterized in seeds from castor bean (R. communis) and C. palaestina, plants that accumulate high levels of ricinoleic and vernolic acid, respectively, and sunflower (H. annuus), a plant that has only common fatty acids in its seed oil. Thus, we incubated crude microsomal fractions from developing seeds with PC having 14C-labeled oleoyl, ricinoleoyl (12-hydroxy-9-octadecenoyl), or vernoloyl (12,13-epoxy-9-octadecenoyl) groups at the sn-2 position. After the incubation, lipids were extracted and analyzed by TLC. We found that the amount of radioactivity that was incorporated into the neutral lipid fraction increased linearly over a period of 4 h (data not shown). The distribution of [14C]acyl groups within the neutral lipid fraction was analyzed after 80 min (Fig. 1A). Interestingly, the amount and distribution of radioactivity between different neutral lipids were strongly dependent both on the plant species and on the type of [14C]acyl chain. Thus, sunflower microsomes incorporated most of the label into DAG, regardless of the type of [14C]acyl group. In contrast, R. communis microsomes preferentially incorporated [14C]ricinoleoyl and [14C]vernoloyl groups into TAG, whereas [14C]oleoyl groups mostly were found in DAG. C. palaestina microsomes, finally, incorporated only [14C]vernoloyl groups into TAG, with [14C]ricinoleoyl groups being found mostly as free fatty acids, and [14C]oleoyl groups in DAG. From these studies, we conclude that the high in vivo levels of ricinoleic acid and vernolic acid in the TAG pool of R. communis and C. palaestina, respectively, may be explained by an efficient and selective transfer of the corresponding acyl groups from PC to TAG in these organisms.
Figure 1.
Metabolism of 14C-labeled PC into the neutral lipid fraction by plant microsomes. (A) Microsomes from developing seeds of sunflower, R. communis, and C. palaestina were incubated for 80 min at 30°C with PC (8 nmol) having oleic acid in its sn-1 position, and 14C-labeled oleic, ricinoleic, or vernolic acid in its sn-2 position. Radioactivity incorporated into TAG (open bars), DAG (filled bars), and unesterified fatty acids (hatched bars) is shown as percentage of added labeled substrate. (B) Synthesis in vitro of TAG carrying two vernoloyl and one [14C]ricinoleoyl group by microsomes from R. communis. The substrates added were unlabeled divernoloyl-DAG (5 nmol), together with either sn-1-oleoyl-sn-2-[14C]ricinoleoyl-DAG (0.4 nmol, 7,700 dpm/nmol) or sn-1-oleoyl-sn-2-[14C]ricinoleoyl-PC (0.4 nmol, 7,700 dpm/nmol). The microsomes were incubated with the substrates for 30 min at 30°C as described in Materials and Methods. The data shown are the average of two experiments.
PDAT: An Enzyme That Catalyzes Acyl-CoA-Independent Synthesis of TAG.
It has previously been suggested that an acyl-CoA-independent synthesis of TAG could be catalyzed by a DAG:DAG acyltransferase (7, 8). It is also known that acyl chains can be transferred from PC to DAG by the reversed action of a CDP-choline:choline phosphotransferase (24). We therefore investigated whether DAG could serve as both acyl donor and acyl acceptor in the reactions catalyzed by the oil seed microsomes. To this end, we incubated unlabeled divernoloyl-DAG with either sn-1-oleoyl-sn-2-[14C]ricinoleoyl-DAG or sn-1-oleoyl-sn-2-[14C]ricinoleoyl-PC in the presence of R. communis microsomes. We found that the synthesis of TAG molecules containing both [14C]ricinoleoyl and vernoloyl groups was 5-fold higher when [14C]ricinoleoyl-PC served as acyl donor as compared with [14C]ricinoleoyl-DAG (Fig. 1B). These data strongly suggest that PC is the immediate acyl donor and DAG the acyl acceptor in the acyl-CoA-independent formation of TAG by oil seed microsomes. We therefore propose that this reaction is catalyzed by an enzyme that we call phospholipid:diacylglycerol acyltransferase (PDAT).
PDAT Activity Is Also Present in Yeast Microsomes.
In an attempt to identify the gene(s) coding for this enzyme, we considered the possibility that it might be present in other oil-accumulating organisms. It is well known that yeast cells can accumulate substantial amounts of TAG, especially during the late exponential and stationary phase, or after exhaustion of certain growth medium components (25). Therefore, we investigated whether budding yeast (S. cerevisiae) contains a PDAT activity that can contribute to TAG synthesis in vivo. Wild-type yeast cells were cultivated under conditions where TAG synthesis is induced. Microsomal membranes were prepared from these cells and incubated with sn-2-[14C]ricinoleoyl-PC and DAG, and the 14C-labeled products formed were analyzed. We found that the PC-derived [14C]ricinoleoyl groups within the neutral lipid fraction mainly were found in free fatty acids or TAG, and also that the amount of TAG synthesized depended on the amount of DAG that was added to the reaction (Fig. 2). The in vitro synthesis of TAG containing both ricinoleoyl and vernoloyl groups, a TAG species not present in vivo, from exogenous added sn-2-[14C]ricinoleoyl-PC and unlabeled vernoloyl-DAG (Fig. 2, lane 3) clearly demonstrates the existence of an acyl-CoA-independent synthesis of TAG involving PC and DAG as substrates in yeast microsomal membranes. We conclude from these experiments that TAG synthesis in yeast can be catalyzed by an enzyme similar to the PDAT found in plants.
Identification of the PDAT-Encoding Gene in Yeast.
In mammalian cells, the synthesis of cholesterol esters is catalyzed mainly by the enzyme acyl-CoA:cholesterol acyltransferases (ACAT), which is related to DAGAT. However, the synthesis of cholesterol esters can also occur in an acyl-CoA-independent reaction where an acyl group is transferred from PC to cholesterol. This reaction is catalyzed by lecithin:cholesterol acyltransferase (LCAT), an enzyme that is found in mammalian blood (26). The sequence homologies between LCAT and DAGAT enzymes, which use cholesterol and DAG as acyl acceptors, respectively, raised the possibility that an LCAT-related enzyme could be responsible for the PDAT activity found in yeast and plant microsomes.
A homology search revealed that there is one gene in the yeast genome, YNR008w, that shows a significant sequence similarity to LCAT. We therefore considered the possibility that YNR008w might encode the PDAT activity. Nothing is known about the function of YNR008w, except that the gene is not essential for growth under normal circumstances. We obtained a strain from the Euroscarf collection (9) in which this gene had been disrupted and found that the disruption has no apparent effect on the sensitivity to temperature, salt, or oxidative stress, or nitrogen starvation, nor does it affect growth on different carbon sources. To test whether YNR008w might encode a PDAT enzyme, we prepared microsomal membranes from this strain and assayed them for PDAT activity, using PC labeled at the sn-2 position with radioactive fatty acids. The activity was completely absent in the disruption strain (Fig. 2, lane 4). Significantly, the activity could be partially restored by the presence of YNR008w on a single-copy plasmid (Fig. 2, lane 5). Moreover, we found that acyl groups of PE also were efficiently incorporated into TAG by microsomes from the wild-type strain, whereas no incorporation occurred from this substrate in the mutant strain (data not shown). Taken together, these findings show that YNR008w encodes a yeast PDAT, which catalyzes the transfer of an acyl group from the sn-2 position of phospholipids to DAG, thus forming TAG. It should be noted that no cholesterol esters were formed from radioactive PC even in incubations with added ergosterol, nor were the amounts of radioactive free fatty acids formed from PC affected by disruption of the YNR008w gene (data not shown). This observation demonstrates that the yeast PDAT does not have cholesterol ester-synthesizing or phospholipase activities.
The expression of the PDAT gene in wild-type yeast was analyzed by Northern blots with RNA isolated from yeast cells grown in rich medium and harvested at different time points ranging from mid-exponential to late-stationary phase (data not shown). We found that the level of expression was generally low, but slightly higher during exponential growth as compared with cells in the stationary phase, where the mRNA was barely detectable. Thus, the expression of the PDAT gene does not coincide with the major stage for TAG accumulation, which occurs during the late stationary phase in yeast.
Lipid analyses were performed on both wild-type and mutant yeast cells to determine the effect of the YNR008w disruption on the lipid composition. We found that the mutant as compared with the wild-type strain showed only minor differences in the amounts of major polar lipids (PC, PE, and phosphatidylinositol), sterol esters, and TAGs, regardless of whether the cells were harvested in exponential or stationary phase (data not shown). From these experiments, we conclude that, whereas the PDAT activity encoded by YNR008w may contribute to TAG synthesis in vivo, other pathways are able to fully compensate for the loss of PDAT activity in the mutant strain. The data also suggest that the PDAT enzyme is not the major contributor to the high levels of TAG accumulated in yeast at late stationary phase.
Increased TAG Content in Yeast Cells That Overexpress PDAT.
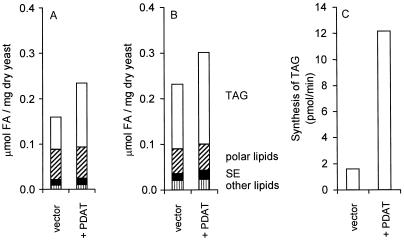
The effect of overexpressing the PDAT-encoding gene was studied by transforming a wild-type yeast strain with the pUS4 plasmid, in which the gene is expressed from the galactose-induced GAL1 promoter. Cells containing the empty expression vector were used as a control. The cells were grown in synthetic glycerol/ethanol medium, and expression of the gene was induced after either 2 h (early logarithmic phase) or 25 h (stationary phase) by the addition of galactose. The cells were then incubated for another 21 h, after which they were harvested and assays were performed. We found that overexpression of PDAT had no significant effect on the growth rate as determined by the optical density. However, the total lipid content, measured as total μmol of fatty acids per mg of yeast dry weight, was 47% (logarithmic phase) or 29% (stationary phase) higher in the PDAT-overexpressing strain than in the control. Furthermore, we found that the polar lipid and sterol ester contents were unaffected by overexpression of PDAT. Instead, the elevated lipid content in these cells is entirely because of an increased TAG content (Fig. 3 A and B). Thus, the amount of TAG was increased by 2-fold in PDAT-overexpressing early logarithmic phase cells and by 40% in stationary phase cells. It is interesting to note that a significant increase in the TAG content was achieved by overexpressing PDAT even under conditions (i.e., in stationary phase) where DAGAT is induced and thus contributes significantly to TAG synthesis. In vitro PDAT activity assayed in microsomes from the PDAT-overexpressing strain was 7-fold higher than in the control strain (Fig. 3C), a finding that is consistent with the increased levels of TAG that we observed in vivo. These results clearly demonstrate the potential use of the PDAT gene in increasing the oil content in transgenic organisms.
Figure 3.
Lipid content (A and B) and PDAT activity (C) in PDAT-overexpressing yeast cells. The PDAT gene in the plasmid pUS4 was overexpressed from the galactose-induced GAL1 promoter in the wild-type strain W303-1A (10). Its expression was induced after 2 h (A) or 25 h (B) of growth as described in Materials and Methods. The amount of TAG (open bar), polar lipids (hatched bar), sterol esters (filled bar), and other lipids (striped bar) of these cells is presented as μmol of fatty acids per mg of dry weight. The data shown are the mean values of results with three independent yeast cultures. (C) In vitro synthesis of TAG by microsomes prepared from yeast cells, cultivated as in A, containing either the empty vector (vector) or the PDAT plasmid (+PDAT). The substrate lipids dioleoyl-DAG (2.5 nmol) and sn-1-oleoyl-sn-2-[14C]oleoyl-PC (2 nmol) were added to aliquots of microsomes, which were then incubated for 10 min at 30°C. The results shown are the mean values of two experiments.
Substrate Specificity of Yeast PDAT.
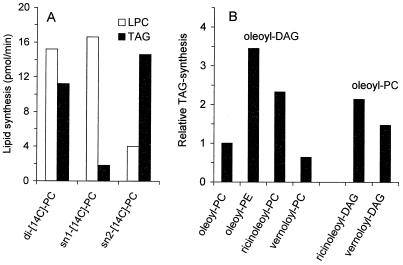
The enzymatic activity of LCAT has been thoroughly characterized and shown to have a preference for transfer of the sn-2 acyl group of PC (27). The substrate specificity of yeast PDAT was analyzed by using microsomes prepared from the PDAT-overexpressing strain (Fig. 4). We found that the rate of TAG synthesis, under conditions given in Fig. 4 with dioleoyl-PC as the acyl-donor, was 0.15 nmol per min per mg of protein. With both oleoyl groups of PC labeled, we were able, under the given assay conditions, to detect the transfer of 11 pmol/min of [14C]oleoyl chain into TAG and the formation of 15 pmol/min of lyso-PC. In microsomes from the PDAT-deficient strain, no TAG at all and only trace amounts of lyso-PC were detected, strongly suggesting that yeast PDAT catalyzes the formation of equimolar amounts of TAG and lyso-PC when supplied with PC and DAG as substrates. The fact that somewhat more lyso-PC than TAG is formed can be explained by the presence of a phospholipase in yeast microsomes, which produces lyso-PC and unesterified fatty acids from PC (data not shown).
Figure 4.
Substrate specificity of yeast PDAT. (A) sn-position specificity of yeast PDAT regarding the acyl donor substrate. Dioleoyl-DAG (2.5 nmol) together with 4 nmol of sn-1-[14C]oleoyl-sn-2-[14C]oleoyl-PC (di-[14C]-PC), sn-1-[14C]oleoyl-sn-2-oleoyl-PC (sn1-[14C]-PC), or sn-1-oleoyl-sn-2-[14C]oleoyl-PC (sn2-[14C]-PC) was used as substrate. (B) Specificity of yeast PDAT regarding the phospholipid headgroup and the acyl composition of the phospholipid as well as the DAG. Dioleoyl-DAG (2.5 nmol) together with 4 nmol of sn-1-oleoyl-sn-2-[14C]oleoyl-PC (oleoyl-PC), sn-1-oleoyl-sn-2-[14C]oleoyl-PE (oleoyl-PE), sn-1-oleoyl-sn-2-[14C]ricinoleoyl-PC (ricinoleoyl-PC), or sn-1-oleoyl-sn-2-[14C]vernoloyl-PC (vernoloyl-PC) was used as substrate. In the experiments presented in the two bars to the far right, 2.5 nmol of monoricinoleoyl-DAG (ricinoleoyl-DAG) or monovernoloyl-DAG (vernoloyl-DAG) were used together with 4 nmol of sn-1-oleoyl-sn-2-[14C]oleoyl-PC. Microsomes from W303-1A cells overexpressing the PDAT gene, as described in Fig. 3A, were incubated at 30°C for 10 min (A) or 90 min (B). The synthesis of radiolabeled TAG (solid bars) and lyso-PC (LPC, open bars) are the mean values of two experiments.
The specificity of yeast PDAT for different acyl group positions was investigated by incubating the microsomes with dioleoyl-PC carrying a [14C]acyl group at either the sn-1 position (Fig. 4A, bar 2) or the sn-2 position (Fig. 4A, bar 3). We found that the major 14C-labeled product formed in the former case was lyso-PC, and, in the latter case, TAG. We conclude that yeast PDAT has specificity for the transfer of acyl groups from the sn-2 position of the phospholipid to DAG, thus forming sn-1-lyso-PC and TAG. Under the given assay conditions, trace amounts of 14C-labeled DAG are formed from the sn-1-labeled PC by the reversible action of a CDP-choline:choline phosphotransferase (data not shown). This labeled DAG can then be further converted into TAG by the PDAT activity. It is therefore not possible to distinguish whether the minor amounts of labeled TAG that are formed in the presence of dioleoyl-PC carrying a [14C]acyl group in the sn-1 position are synthesized directly from the sn-1-labeled PC by a PDAT that also can act on the sn-1 position, or if it is first converted to sn-1-labeled DAG and then acylated by a PDAT with strict selectivity for the transfer of acyl groups at the sn-2 position of PC. Taken together, our experiments suggest that the PDAT encoded by YNR008w catalyses an acyl transfer from the sn-2 position of PC to DAG, thus causing the formation of TAG and lyso-PC.
The substrate specificity of yeast PDAT was further analyzed with respect to the head group of the acyl donor, the acyl group transferred, and the acyl chains of the acceptor DAG molecule. The two major membrane lipids of S. cerevisiae are PC and PE, and as shown in Fig. 4B (bars 1 and 2), dioleoyl-PE is nearly 4-fold more efficient than dioleoyl-PC as acyl donor in the PDAT-catalyzed reaction. Moreover, the rate of acyl transfer is strongly dependent on the type of acyl group that is transferred. Thus, a ricinoleoyl group at the sn-2 position of PC is 2.5 times more efficiently transferred into TAG than an oleoyl group in the same position (Fig. 4B, bars 1 and 3). In contrast, yeast PDAT has no preference for the transfer of vernoloyl groups over oleoyl groups (Fig. 4B, bars 1 and 4). The acyl chain of the acceptor DAG molecule also affects the efficiency of the reaction. Thus, DAG with a ricinoleoyl or a vernoloyl group is a more efficient acyl acceptor than dioleoyl-DAG (Fig. 4B, bars 1, 5, and 6). Taken together, these results clearly show that the efficiency of the PDAT-catalyzed acyl transfer is strongly dependent on the properties of the substrate lipids.
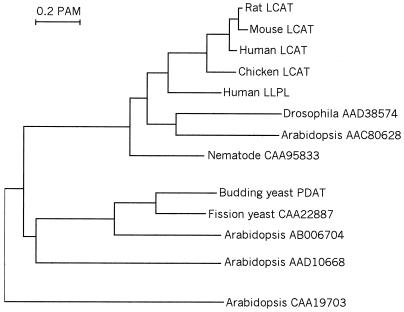
Three PDAT-Related Proteins Form a Separate Branch Within the LCAT Protein Superfamily.
To identify possible PDAT orthologs in other organisms, we searched the databases for related sequences. A large number of protein sequences from different eukaryotes show similarity to both LCAT and PDAT. To further analyze how they are related to each other, we used the most conserved part of the sequences, where all proteins can be unambiguously aligned, to compute a dendrogram (Fig. 5). This conserved region comprises the N-terminal third of the proteins and includes the putative lipase active site motif, HS(M/L)G. Interestingly, whereas most of the available sequences show a strong similarity to human and mouse LCAT, two sequences (one from fission yeast and one from Arabidopsis thaliana) are clearly more closely related to the budding yeast PDAT than to any of the LCATs. It is therefore likely that these two enzymes also possess PDAT activity. The presence of PDAT-related proteins in both fungi and plants suggests that a distinct branch encoding enzymes with this activity arose at an early point in evolution.
Figure 5.
Evolutionary dendrogram showing LCAT- and PDAT-related proteins from different eukaryotes. The dendrogram was calculated from aligned protein sequences corresponding to amino acid residues 174–335 in yeast PDAT. The clustalx multiple alignment program (28) was used with default settings to align the sequences and compute pairwise alignment scores. An unrooted tree was then obtained from these scores by using the neighbor-joining method (29), with correction for multiple substitutions and exclusion of gapped positions.
Discussion
During the past few years, the biosynthesis of TAG has received much attention because of its central role both in fat accumulation in animal cells and in the synthesis of storage lipids in oil seed plants. Until recently, the only enzyme thought to be directly involved in the synthesis of TAG was DAGAT, which catalyzes the acyl-CoA-dependent acylation of DAG into TAG. Genes encoding enzymes with DAGAT activity have recently been identified in the mouse (3), in plants (4–6), and in microbes (K. Lardizabal, personal communication). We have now shown that plant and yeast cells also can synthesize TAG by a reaction in which a phospholipid acts as the acyl donor. An acyl-CoA-independent enzyme, which we call phospholipid:diacylglycerol acyltransferase or PDAT, catalyzes this reaction.
It is clear from the results in Fig. 1 that the specificity of the PDAT enzyme with respect to the acyl group that is transferred from PC into TAG varies between different plant species. Thus, either the ricinoleoyl or the vernoloyl group of PC is specifically incorporated into TAG when microsomal fractions from castor bean and C. palaestina are used, respectively. This finding suggests that PDAT could play an important role in the specific channeling of bilayer-disturbing fatty acids (e.g., ricinoleic and vernolic acid) from PC into the TAG pool. The synthesis of ricinoleic and vernolic acid in transgenic A. thaliana has been reported (22, 23, 30), but only low to moderate levels of these fatty acids were observed in the seed oil. It is conceivable that the expression of a PDAT enzyme with specificity for the desired unusual fatty acid could help to achieve higher levels of ricinoleic and vernolic acids in the seed oil of these transgenic plants.
The physiological role of PDAT and its intracellular location remain to be determined. However, it should be noted that budding yeast PDAT and its putative orthologs in fission yeast and Arabidopsis all share an N-terminal extension of 80–140 amino acid residues, which is absent in the other LCAT-related enzymes. Moreover, this extension contains a predicted membrane-spanning region, which could serve to anchor the enzyme in a membrane. As for the in vivo function of PDAT, it is clear from our results that the in vivo accumulation of TAG in yeast is not dependent on PDAT. Despite the fact that a DAGAT-encoding gene has not yet been identified in yeast, it is clear from in vitro studies using microsomal preparations that a CoA-dependent synthesis of TAG exists in yeast. We have further shown that this activity remains in microsomes from yeast cells that lack PDAT (data not shown). This finding suggests that the CoA-dependent synthesis of TAG, which is most likely catalyzed by a yeast DAGAT, is sufficient to maintain adequate TAG levels under normal conditions. It is therefore conceivable that the yeast PDAT may have some other more specialized role in vivo. In this context, we note that PDAT also catalyzes a breakdown of the major membrane lipids (PC and PE). Possibly, yeast PDAT could be involved in the regulation of membrane lipid composition in response to different growth conditions. As shown in Fig. 4B, the two major membrane lipids in yeast, PC and PE, can both act as acyl donors in the synthesis of TAG by PDAT. However, the rate of synthesis is clearly dependent on the polar head group, the type of acyl group that is transferred, and the acyl groups of the acceptor DAG molecule. Taken together, these results suggest that the substrate specificity of PDAT may play an important role in determining the membrane lipid composition in vivo.
The dendrogram in Fig. 5 suggests that separate branches leading to the PDAT-related and LCAT-related enzymes arose at an early stage in eukaryotic evolution. Interestingly, whereas PDAT is the only member of the LCAT protein family that has so far been found in fungi, the opposite is true in animals, where only LCAT-related but no PDAT-related enzymes have been found. This raises the question whether the separation of PDAT-related and LCAT-related enzymes in fact reflects not an ancient gene duplication but rather the divergence of fungi from animals. According to this view, PDAT would be the true homolog of LCAT in fungi, although with a substrate specificity that differs considerably from the latter. However, an examination of the available plant sequences strongly suggests that this is not the case. Thus, A. thaliana has one clearly PDAT-related and one LCAT-related sequence, as well as two other more divergent members of the same protein superfamily, one of which is closer to PDAT than to LCAT within the dendrogram (Fig. 5). We conclude that separate enzymes encoding PDAT and LCAT most likely were present already in the common ancestor of plants, animals, and fungi, and that the apparent absence of LCAT in fungi and PDAT in animals probably reflects a more recent loss of these enzymes in the respective kingdoms.
In conclusion, we have found an enzyme that we call phospholipid:diacylglycerol acyltransferase or PDAT that is present in both plants and yeast. It catalyzes the transfer of acyl groups from the sn-2 position of the major phospholipids to DAGs, thus forming TAGs and lyso-phospholipids. The yeast gene encoding the PDAT enzyme was identified, and the encoded protein was found to be related to the human enzyme LCAT. The activity of yeast PDAT was shown to be dependent on the type of polar head-group of the donor lipid, the acyl group transferred, and the acyl chains of the acceptor molecule DAG. Although the in vivo function of PDAT still remains to be determined, it is conceivable that it could play a role in regulating the fatty acid composition of membrane lipids.
Acknowledgments
This work was supported by financial contributions from the Swedish Foundation for Strategic Research, the Swedish Farmers Research Foundation, Stiftelsen Svensk Oljeväxtforskning, and VL-stiftelsen.
Abbreviations
- TAG
triacylglycerol
- DAGAT
diacylglycerol acyltransferase
- DAG
diacylglycerol
- ACAT
acyl-CoA:cholesterol acyltransferase
- PDAT
phospholipid:diacylglycerol acyltransferase
- LCAT
lecithin:cholesterol acyltransferase
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120067297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120067297
References
- 1.Bell R M, Coleman R A. Annu Rev Biochem. 1980;49:459–487. doi: 10.1146/annurev.bi.49.070180.002331. [DOI] [PubMed] [Google Scholar]
- 2.Stymne S, Stobart K. In: The Biochemistry of Plants: A Comprehensive Treatise. Stumpf P K, editor. Vol. 9. New York: Academic; 1987. pp. 175–214. [Google Scholar]
- 3.Cases S, Smith S J, Zheng Y W, Myers H M, Lear S R, Sande E, Novak S, Collins C, Welch C B, Lusis A J, et al. Proc Natl Acad Sci USA. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobbs D H, Lu C, Hills M J. FEBS Lett. 1999;452:145–149. doi: 10.1016/s0014-5793(99)00646-8. [DOI] [PubMed] [Google Scholar]
- 5.Routaboul J M, Benning C, Bechtold N, Caboche M, Lepiniec L. Plant Physiol Biochem. 1999;37:831–840. doi: 10.1016/s0981-9428(99)00115-1. [DOI] [PubMed] [Google Scholar]
- 6.Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor D C. Plant J. 1999;19:645–653. doi: 10.1046/j.1365-313x.1999.00555.x. [DOI] [PubMed] [Google Scholar]
- 7.Stobart K, Mancha M, Lenman M, Dahlqvist A, Stymne S. Planta. 1997;203:58–66. [Google Scholar]
- 8.Dahlqvist A, Banas A, Stymne S. In: Advances in Plant Lipid Research. Sánches J, Cerdá-Olmedo E, Martinez-Force E, editors. Seville, Spain: Universidad de Sevilla; 1998. pp. 211–214. [Google Scholar]
- 9.Entian K-D, Kötter P. Methods Microbiol. 1998;26:431–449. [Google Scholar]
- 10.Thomas B J, Rothstein R. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 11.Kern L, de Montigny J, Jund R, Lacroute F. Gene. 1990;88:149–157. doi: 10.1016/0378-1119(90)90026-n. [DOI] [PubMed] [Google Scholar]
- 12.Ronne H, Carlberg M, Hu G-Z, Nehlin J O. Mol Cell Biol. 1991;11:4876–4884. doi: 10.1128/mcb.11.10.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stobart K, Stymne S. In: Methods in Plant Biochemistry. Harwood J L, Bowyer J R, editors. Vol. 4. London: Academic; 1990. pp. 19–46. [Google Scholar]
- 14.Bafor M, Smith M A, Jonsson L, Stobart A K, Stymne S. Biochem J. 1991;280:507–514. doi: 10.1042/bj2800507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banas A, Johansson I, Stymne S. Plant Sci. 1992;84:137–144. [Google Scholar]
- 16.Kanda P, Wells M A. J Lipid Res. 1981;22:877–879. [PubMed] [Google Scholar]
- 17.Ståhl U, Ek B, Stymne S. Plant Physiol. 1998;117:197–205. doi: 10.1104/pp.117.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 19.Meesters P A E P, Huijberts G N M, Eggink G. Appl Microbiol Biotechnol. 1996;45:575–579. [Google Scholar]
- 20.Sherman F, Fink G R, Hicks J B. Laboratory Course Manual for Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 21.van de Loo F J, Fox B G, Somerville C. In: Lipid Metabolism in Plants. Moore T S, editor. Boca Raton, FL: CRC; 1993. pp. 91–126. [Google Scholar]
- 22.van de Loo F J, Broun P, Turner S, Somerville C. Proc Natl Acad Sci USA. 1995;92:6743–6747. doi: 10.1073/pnas.92.15.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M, Lenman M, Banas A, Bafor M, Singh S, Schweizer M, Nilsson R, Liljenberg C, Dahlqvist A, Gummeson P-O, et al. Science. 1998;280:915–918. doi: 10.1126/science.280.5365.915. [DOI] [PubMed] [Google Scholar]
- 24.Slack C R, Roughan P G, Browse J A, Gardiner S E. Biochim Biophys Acta. 1985;833:438–448. [Google Scholar]
- 25.Rattray J B, Schibeci A, Kidby D K. Bacteriol Rev. 1975;39:197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glomset J A. J Lipid Res. 1968;9:155–167. [PubMed] [Google Scholar]
- 27.Veedamali M L, Subramanian S, Subbaiah P V. Biochemistry. 1998;37:13626–13633. doi: 10.1021/bi980351e. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 30.Broun P, Boddupalli S, Somerville C. Plant J. 1998;13:201–210. doi: 10.1046/j.1365-313x.1998.00023.x. [DOI] [PubMed] [Google Scholar]