Abstract
Activation of gene transcription in mammalian cells requires several classes of coactivators that participate in different steps of the activation cascade. Using conventional and affinity chromatography, we have isolated a human coactivator complex that interacts directly with the C-terminal domain (CTD) of RNA polymerase II (Pol II). The CTD-binding complex is structurally and functionally indistinguishable from our previously isolated CRSP coactivator complex. The closely related, but transcriptionally inactive, ARC-L complex failed to interact with the CTD, indicating a significant biochemical difference between CRSP and ARC-L that may, in part, explain their functional divergence. Electron microscopy and three-dimensional single-particle reconstruction reveals a conformation for CTD–CRSP that is structurally distinct from unliganded CRSP or CRSP bound to SREBP-1a, but highly similar to CRSP bound to the VP16 activator. Together, our findings suggest that the human CRSP coactivator functions, at least in part, by mediating activator-dependent recruitment of RNA Pol II via the CTD.
Keywords: CRSP, Mediator, CTD, structure, transcription
Regulation of eukaryotic RNA polymerase II (Pol II) transcription by sequence-specific enhancer and promoter-binding proteins is dependent on several different classes of cofactors and coactivators (Lemon and Tjian 2000; Malik and Roeder 2000; Peterson and Workman 2000; Näär 2001). Some of these coactivators are recruited to enhancer/promoter DNA by transcriptional activators to facilitate various steps in the gene activation process. For example, certain chromatin-directed activities, such as ATP-dependent nucleosome remodeling factors and histone acetyltransferases, assist enhancer and promoter-binding proteins and general transcription factors in accessing their target sequences in chromatin-packaged DNA (Lemon and Tjian 2000; Näär et al. 2001; Roth et al. 2001). Other classes of coregulators, such as TFIID, are more closely integrated with the transcriptional machinery and have been proposed to act at steps subsequent to chromatin remodeling to enhance activator-dependent recruitment of the transcriptional apparatus to the promoter (Albright and Tjian 2000; Näär et al. 2001). The TFIID complex, composed of TBP and associated TAFs, recognizes the TATA box and downstream promoter sequences and can be recruited to the promoter by activators.
A different class of cofactors, including yeast Mediator, do not directly bind promoter sequences, but can be recruited by activators. In addition, yeast Mediator can associate with RNA Pol II via the C-terminal domain (CTD) of the large RNA Pol II subunit and has been proposed to act as a bridge between activators and the transcriptional machinery (Myers and Kornberg 2000). The RNA Pol II CTD is composed largely of tandem repeats of the YSPTSPS consensus amino acid sequence; yeast CTD contains 26 repeats, whereas mammalian CTD harbors 52 repeats. The CTD appears to serve multiple functions in the transcription initiation and elongation process. Recently, the CTD has also been shown to play a role in coupling gene transcription to mRNA processing events, such as 5′-capping, splicing, RNA cleavage, and polyadenylation (Gerber et al. 1995; Cho et al. 1997; Corden and Patturajan 1997; McCracken et al. 1997a,b; Tanner et al. 1997; Yue et al. 1997; Hirose et al. 1999; Otero et al. 1999; Conaway et al. 2000).
A family of human cofactor complexes distantly related to yeast Mediator have been isolated recently (Fondell et al. 1996; Jiang et al. 1998; Sun et al. 1998; Boyer et al. 1999; Gu et al. 1999; Näär et al. 1999; Rachez et al. 1999; Ryu et al. 1999). Unlike yeast Mediator, however, these human cofactor complexes (which include ARC/DRIP, TRAP/SMCC, NAT, CRSP, and PC2) have not been shown to interact with the CTD of RNA Pol II.
In a recent study, we discovered that the human activator recruited cofactor fraction (ARC) consists of two distinct complexes, ARC-L and CRSP (Taatjes et al. 2002). Both are highly related, but display contrasting cofactor properties. ARC-L is somewhat larger and contains additional subunits (ARC240, ARC250, cdk8, cyclin C) not present in CRSP, whereas CRSP contains a 70-kD subunit (CRSP70) not present in ARC-L. On the basis of subunit composition and in vitro transcription assays, ARC-L most closely resembles the NAT and SMCC cofactor complexes (Sun et al. 1998; Gu et al. 1999; Taatjes et al. 2002). Previous studies with NAT, SMCC, and ARC/DRIP revealed weak interactions with RNA Pol II, but direct and specific binding to the CTD was not observed (Sun et al. 1998; Gu et al. 1999; Näär et al. 1999; Chiba et al. 2000). Here, we show that the human CRSP coactivator complex, but not ARC-L, interacts strongly with the CTD of RNA Pol II. CTD-affinity chromatography specifically isolated a large, multisubunit complex indistinguishable from the previously identified CRSP coactivator. Both complexes possess highly similar or identical subunit composition and display indistinguishable coactivator function in vitro. Further, structural analysis of the CTD-binding complex by electron microscopy (EM) and single particle reconstruction reveals a structure very similar to a specific activator-bound form of the CRSP complex.
Results and Discussion
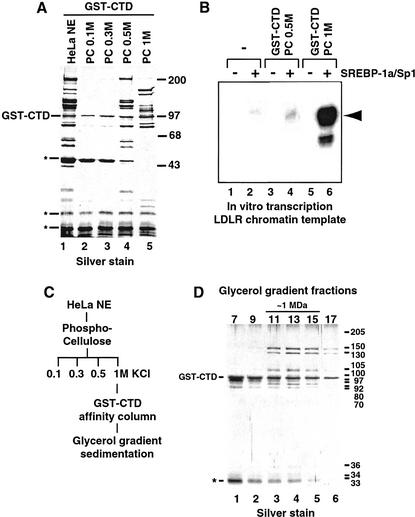
To identify putative human cofactors that interact selectively with the RNA Pol II CTD, we screened HeLa nuclear extract (NE) using an affinity resin composed of the human RNA Pol II CTD (52 heptad repeats) fused to glutathione-S-transferase (GST–CTD). More than 30 polypeptides from HeLa NE were specifically retained on the GST–CTD column as compared with control resins (Fig. 1A, lane 1; data not shown). Fractionation of the HeLa NE over a phosphocellulose (PC) column prior to CTD affinity purification revealed that the polypeptides bound to the CTD column could be separated into two populations eluting at 0.5 M KCl (PC 0.5M) and 1 M KCl (PC 1M), respectively (Fig. 1A, lanes 4 and 5). Intriguingly, the polypeptide pattern from the PC 1M fraction closely resembled that of the CRSP coactivator identified previously in our laboratory (Ryu et al. 1999). To investigate a possible relationship between the PC 1M-derived CTD-binding polypeptides and CRSP, we examined whether the CTD-binding fractions (from PC 0.5M and PC 1M) could substitute for CRSP in a chromatin-based in vitro transcription assay. This assay utilized a LDLR-derived chromatin template driven by the SREBP-1a and Sp1 activators that require the CRSP coactivator component of ARC for transcriptional activation (Näär et al. 1999; Taatjes et al. 2002). The CTD-binding polypeptides purified from the PC 0.5M phosphocellulose fraction were largely inactive (Fig. 1B, cf. lanes 2 and 4); however, the PC 1M-derived polypeptides strongly potentiated (>100-fold) SREBP-1a/Sp1-dependent activation (Fig. 1B, cf. lanes 2 and 6), suggesting that this class of polypeptides harbors a functional CTD-binding coactivator. Further purification of the PC 1M-derived CTD-associated polypeptides using glycerol gradient sedimentation (Fig. 1C) confirmed that they were components of a large ∼1-MD multiprotein complex (Fig. 1D, lanes 3–5). The transcriptionally inactive PC 0.5M-derived CTD-binding polypeptides also purify as a large, multisubunit complex. Peptide microsequence analysis of individual subunits indicates that this complex is composed of novel gene products that are unrelated to subunits of known transcriptional coactivator complexes and may not be directly involved in regulation of transcription initiation (A.M. Näär, unpubl.). We have not pursued the characterization of this CTD-binding complex further.
Figure 1.
(A) Silver-stained SDS-polyacrylamide gel of GST–CTD-purified material from HeLa cell nuclear extract (HeLa NE, lane 1) and Phosphocellulose (PC) fractions eluting at 0.1 M (lane 2), 0.3 M (lane 3), 0.5 M (lane 4), and 1 M KCl (lane 5). The GST–CTD coeluting with associated proteins is indicated at left. (*) Nonspecific polypeptides. The migration of molecular weight standards (in kilodaltons) is indicated at right. (B) In vitro transcription analysis of coactivator activity associated with GST–CTD-bound polypeptides isolated from the PC 0.5M (lanes 3,4) and PC 1M (lanes 5,6) fractions. Transcriptional activity from the Low Density Lipoprotein Receptor (LDLR)-derived chromatin-assembled DNA template was assessed in the absence (lanes 1,3,5) or presence (lanes 2,4,6) of activators (SREBP-1a/Sp1). The expected location of the primer extension product is indicated by the arrowhead at right. (C) Purification scheme of the CTD-binding coactivator. HeLa NE was separated by PC chromatography. The 1 M fraction was incubated with GST–CTD resin and eluted with glutathione after extensive washing. Eluted material was separated on a 2-mL 15%–40% glycerol gradient. (D) SDS-PAGE and silver-stain analysis of glycerol gradient-purified CTD–CRSP. Estimated subunit sizes are shown at right (in kilodaltons) and the migration of GST–CTD and a nonspecific protein (*) are indicated at left.
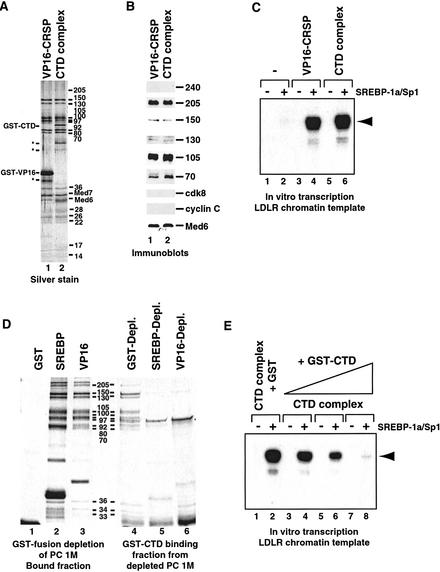
Direct comparison of SDS–polyacrylamide silver-stained gels of the PC 1M-derived CTD-binding complex and CRSP confirms that these two coactivator complexes are highly related or identical (Fig. 2A, cf. lanes 1 and 2). Immunoblotting confirmed the identity of several of the polypeptides found in the CTD-binding complex as bona fide CRSP subunits (Fig. 2B, cf. lanes 1 and 2). Interestingly, no ARC-L-specific polypeptides (ARC240, ARC250, cdk8, cyclin C) appeared to bind the CTD affinity column, despite their presence in the PC 1M fraction. This suggests that the ARC-L complex, in contrast to CRSP, is unable to interact with the CTD of RNA Pol II. This result is consistent with previous reports indicating that SMCC and NAT, which are highly related to ARC-L, are also unable to associate directly with the RNA Pol II CTD (Sun et al. 1998; Gu et al. 1999). Because ARC-L lacks the CRSP70 subunit, it is possible that the CRSP–CTD interaction is mediated by CRSP70. However, it is notable that a CRSP70 homolog is absent in yeast Mediator. Alternately, we propose that the additional subunits in ARC-L may occlude a CTD-specific binding surface on the CRSP complex. Some additional protein density is present near the CTD-binding region in ARC-L (Taatjes et al. 2002).
Figure 2.
Comparative analysis of VP16-purified CRSP and the PC 1M-derived CTD-binding complex. (A) Silver stain of SDS-PAGE-separated VP16–CRSP (lane 1) and the CTD-binding complex (lane 2). The bands corresponding to GST–CTD and GST–VP16 are indicated at left, along with nonspecific proteins (*). The molecular weights of the subunits are indicated at right (in kilodaltons). (B) Immunoblot analysis of VP16–CRSP (lane 1) and the PC 1M-derived CTD-binding complex (lane 2). The ARC-L/CRSP subunits analyzed are indicated at right. (C) Direct comparison of coactivator activity associated with equal quantities of VP16-purified CRSP and CTD-purified complex. SREBP-1a/Sp1-dependent activation of the LDLR-derived chromatin template (lanes 1,3,5, no activator; lanes 2,4,6, SREBP-1a and Sp1) was analyzed in the absence of added protein (lanes 1,2), in the presence of 0.5 nM CTD-binding complex (lanes 5,6), or in the presence of 0.5 nM VP16-purified CRSP (lanes 3,4). The primer extension product is indicated by the arrowhead at right. (D) Analysis of presence of CTD-binding complex in activator- or control-depleted PC 1M. Silver-stain analysis shows depletion of CTD-complex from PC 1M by CRSP-targeting activation domains. The PC 1M fraction was depleted (see Materials and Methods) using resins containing GST (lane 1), GST–SREBP1a (lane 2), or GST–VP16 (lane 3). Lanes 1–3 show bound material after first depletion. After depletion, the PC 1M fractions were incubated with GST–CTD resin and bound material was then analyzed by SDS-PAGE and silver staining (lanes 4–6). (E) Addition of exogenous CTD inhibits gene activation dependent on the CTD-purified complex. SREBP-1a/Sp1-dependent activation was analyzed as in C after the addition of 5 pmole (lanes 3,4), 15 pmole (lanes 5,6), or 50 pmole (lanes 7,8) of GST–CTD, or 50 pmole of GST alone (lanes 1,2). The primer extension product is indicated at right by an arrowhead.
Although the CTD-binding complex was found to be a potent coactivator in our in vitro transcription assays and exhibited a subunit composition similar to CRSP, we nevertheless wished to directly compare their coactivator activities in our chromatin-based transcription system. As shown in Figure 2C, the CTD-binding complex and CRSP exhibit similar specific activities in this assay (cf. lanes 4 and 6 for single-point analysis, titrations not shown). These results further suggest that the CTD-binding complex and CRSP are functionally related. Accordingly, we will refer to the CTD-binding complex (derived from the PC 1M fraction) as CTD–CRSP throughout the rest of the text.
Other biochemical similarities between CTD–CRSP and CRSP were established by specific activator-binding experiments. We documented previously the ability of SREBP-1a and VP16 activation domains to bind CRSP (Taatjes et al. 2002). Here, we find that affinity resins bearing the SREBP-1a or VP16 activation domains efficiently deplete CTD–CRSP from the PC 1M fraction (Fig. 2D, cf. lanes 2 and 3 with 5 and 6). In contrast, GST control resins failed to bind CRSP from this same fraction (Fig. 2D, cf. lanes 1 and 4). Conversely, prior depletion of PC 1M with GST–CTD significantly reduced the amount of CRSP bound to GST–SREBP-1a and VP16 activation domain resins (data not shown).
Together, these findings establish that the activator-targeted human CRSP coactivator can interact with the CTD of RNA Pol II. By virtue of this interaction with the CTD, CRSP (but not ARC-L) may help recruit RNA Pol II to the promoter. We observed previously that CRSP and ARC-L possess contrasting transcriptional properties in vitro; CRSP displayed potent, coactivator activity, whereas ARC-L was inactive (Taatjes et al. 2002). Given that the RNA Pol II CTD has been implicated in the activation of transcription (Gerber et al. 1995), we speculated that the CRSP-specific interaction with the CTD may predicate its coactivator function. To substantiate this, we examined whether disruption of this interaction would inhibit CRSP-dependent transcriptional activation. An excess of free GST–CTD was added to the transcription reactions, which potently inhibited CRSP-dependent transcriptional activation in a dose-dependent manner (Fig. 2E, lanes 3–8). In contrast, addition of GST alone had no significant effect (Fig. 2C, cf. lanes 5 and 6 with E, lanes 1 and 2). These results further show the functional importance of the CTD in potentiating transcript initiation. However, given the essential role of the CTD in multiple aspects of the transcription process, including transcript elongation, we cannot exclude the possibility that the exogenously added CTD may also titrate other activities required for transcription.
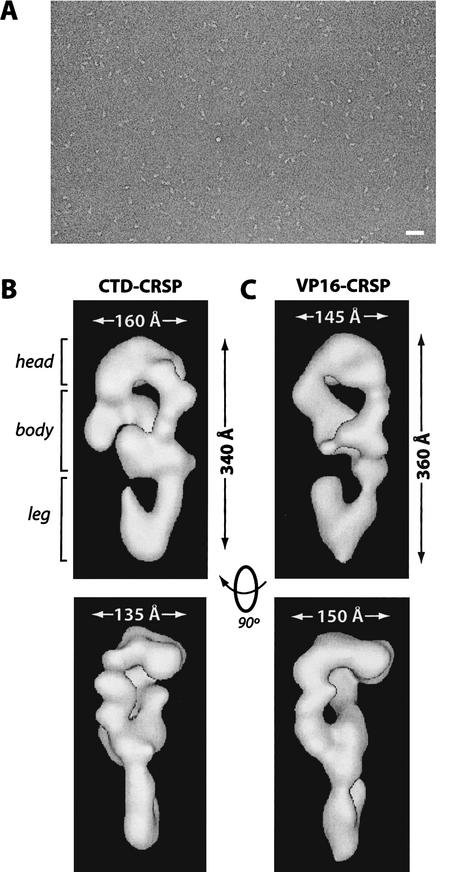
In addition to the biochemical characterization of CTD–CRSP, we determined its structural characteristics using EM and single particle reconstruction techniques (see Materials and Methods). A micrograph of a typical negatively stained CTD–CRSP sample is shown in Figure 3A. The three-dimensional structure of CTD–CRSP, reconstructed from 3662 single particle images, following multiple rounds of angular refinement, is shown in Figure 3B. The complex is somewhat elongated and possesses three distinct regions, the head region, which contacts the protein-dense body in two areas to form a lobular density with a central cavity, and a hook-like leg domain that contacts the body from the opposite side. The relatively large size of the complex (340 Å × 160 Å × 135 Å) suggests it may be capable of mediating many protein–protein interactions at the promoter. TFIID, for example, is considerably smaller by comparison (200 Å × 135 Å × 110 Å) (Andel et al. 1999).
Figure 3.
CTD–CRSP and VP16–CRSP are structurally similar. (A) Negatively stained electron micrograph of CTD–CRSP sample. Bar, 800 Å. (B,C) Three-dimensional reconstruction of CTD–CRSP and VP16–CRSP at 32 Å resolution. Complexes are rendered to 1.25 MD, their approximate predicted molecular weight. Dimensions shown. Rotation of the volumes 90° gives the second side view of the coactivator.
In a previous study, we used EM single-particle reconstruction techniques to identify structural characteristics of the CRSP coactivator bound to different activators (Taatjes et al. 2002). This study revealed that CRSP was conformationally flexible and capable of adopting multiple activator-dependent conformations. Specifically, CRSP assumed three distinct conformations when unliganded, or bound to VP16 or SREBP-1a. Interestingly, CRSP adopts a conformation very similar to VP16–CRSP (for comparison, see Fig. 3C ) when bound to the CTD, as evident by visual comparison and cross-correlation analysis (see Materials and Methods). The fact that CTD–CRSP adopts a conformation similar to VP16–CRSP suggests that this conformational state may represent a particular activated form of the CRSP complex that efficiently potentiates transcript initiation. Such structurally dynamic transitions may facilitate activation by allowing CRSP to associate with other ligands, such as specific activators or other components of the transcriptional apparatus.
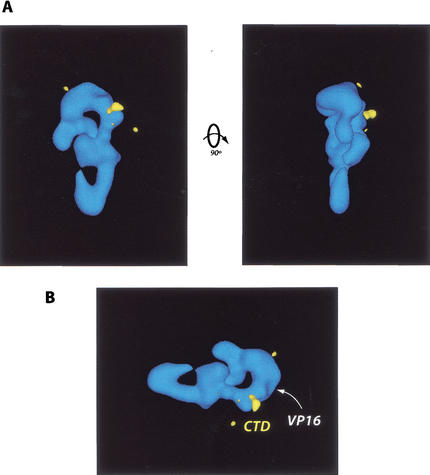
The CTD-binding site was localized on the CRSP complex using CTD–CRSP samples labeled with anti-GST antibodies (CTD is present as a GST fusion protein). Samples of CTD–CRSP were prepared as described (Fig. 1C), followed by addition of antibody in a fivefold excess. Subsequent three-dimensional reconstruction and difference mapping of antibody-labeled versus unlabeled samples localized the CTD-binding site to a relatively small region between the head and body of the complex (Fig. 4A). Both polyclonal and monoclonal antibodies against GST were used for this analysis and yielded similar results in independent experiments. Incidentally, VP16 binds a similar, but not identical, region on the CRSP complex (Fig. 4B; Taatjes et al. 2002). Thus, the CTD and VP16 bind proximal regions and induce similar conformations in the CRSP coactivator. This suggests that VP16 may be able to substitute for, but not compete with, a potential function of the CTD in activating transcription. By inducing a CTD-bound conformation in the CRSP coactivator, VP16 may circumvent CTD-dependent regulatory mechanisms that would otherwise moderate transcript initiation. Although it is likely that VP16 and the CTD target different peptide sequences in CRSP, these sequences may reside in the same subunit. VP16 is proposed to bind CRSP77 (TRAP80) (Ito et al. 1999); interestingly, the homolog of this subunit is essential for viability in Drosophila (Boube et al. 2000). Although the subunit that mediates CRSP interaction with the CTD is unknown, it also may be essential for viability given the likely importance of the CRSP–RNA Pol II interaction in activating gene expression.
Figure 4.
(A) Localization of the CTD binding site (yellow) on the CRSP coactivator. This site was identified via EM analysis and difference mapping of structures generated from CTD–CRSP samples incubated with anti-GST antibodies, which target the GST–CTD fusion protein bound to the CRSP complex. (B) VP16 and CTD bind similar regions on the CRSP complex. As in A, the CTD-binding site is shown in yellow. The VP16-binding site is indicated by the white arrowhead.
Our findings identify the CRSP complex as a probable link between RNA Pol II and human transcriptional activators, analogous to the suggested function of Mediator in yeast. Furthermore, yeast Mediator selectively binds the hypophosphorylated form of RNA Pol II CTD; phosphorylation appears to prevent CTD–Mediator association (Myers et al. 1998). This is consistent with our current results, insofar as we observe strong interaction between recombinant, unphosphorylated CTD and the human CRSP coactivator complex. Thus, despite substantial divergence in subunit composition and structure, some core biochemical functions of yeast Mediator and human CRSP, such as interaction with activators, RNA Pol II CTD binding, and transcriptional coactivation, appear to have been maintained over evolutionary time.
Materials and methods
GST-pulldown assays
A total of 1 mL of HeLa NE or PC fractions was applied to 25 μL of GST–CTD beads (Peterson et al. 1992) and mixed at 4°C for 3 h. Beads were washed 7 × 1 mL with 0.5 M KCl HEGN (20 mM HEPES at pH 7.6, 0.1 mM EDTA, 10% glycerol, 0.1% NP-40, 1 mM DTT, 1 mM benzamidine, 0.25 mM PMSF, 2 μg/mL aprotinin) and 1 × 1 mL of 0.1 M KCl HEG + 0.02% NP40. The beads were then eluted with a Tris-buffered 20-mM glutathione solution (0.1 M KCl). For depletion experiments, 500 μL of PC 1M was mixed with 100 μL of GST, GST SREBP-1a (amino acids 1–50), or GST–VP16 (amino acids 413–490) beads for 2 h. The supernatant was then transferred to 100 μL of fresh beads and mixed for another 2 h at 4°C. The double-depleted fraction was then incubated with GST–CTD and analyzed as above. For elution of GST–CTD-associated proteins, 2 × 1 bead volumes of 0.1 M KCl HEGN with 0.25% sarkosyl or 20 mM glutathione was added and mixed at 4°C for 1 h each.
Purification of CTD–CRSP
HeLa NE was prepared as described (Dignam et al. 1983) and loaded onto a P11 PC column equilibrated in 0.1 M KCl HEG (20 mM HEPES at pH 7.6, 0.1 mM EDTA, 10% glycerol, 1 mM DTT, and protease inhibitors). Eluted fractions were dialyzed against 0.1 M KCl HEG. Typically, 10 mL of the PC 1M fraction was mixed over 200 μL GST–CTD beads at 4°C for 3 h. The beads were washed 7 × 10 mL with 0.5 M KCl HEGN and 1 × 10 mL 0.1M KCl HEGN. Immobilized proteins were then eluted with 20 mM glutathione at 4°C. The eluate was applied to a 2-mL glycerol gradient (15%–40% in 0.1 M KCl HEG), which ran at 4°C for 7 h at 55K RPM. Fractions were collected in 100-μL aliquots.
Immunoblotting
Immunoblotting was performed essentially as described (Näär et al. 1999) using the specified antibodies.
Chromatin-based in vitro transcription
The template plasmids were assembled into chromatin as described (Näär et al. 1998). Drosophila embryo cytosolic extract (S-190), purified Drosophila core histones, Mg/ATP, and an ATP-regenerating system were added to supercoiled DNA template, and assembly was performed for 4.5 h at 27°C. Transcription reactions were performed as described (Näär et al. 1998).
Electron microscopy and single particle reconstruction
Electron micrographs were obtained using a Tecnai 12 TEM at 30,000× magnification. Glycerol gradient-purified CTD–CRSP samples were applied to a glow-discharged carbon grid and negatively stained with a 4% uranyl acetate solution. Each sample was dialyzed versus a 5% trehalose solution (20 mM HEPES, 0.1 mM EDTA, 0.1M KCl) prior to analysis. Micrographs (38) were digitized with a scan step of 13.3 μm (4.4 Å/pixel). Image pairs of tilted (35°–45°) and untilted (0°) complexes were obtained and analyzed via random conical tilt (Radermacher et al. 1987) using the SPIDER and WEB software packages (Frank et al. 1996). Untilted images were subjected to reference-free alignment and merged into 24 distinct classes (indicative of their orientation on the grid) following in-plane shifts and rotations (Penczek et al. 1992). Three-dimensional structures for each class were calculated by back projection using corresponding tilted images. These three-dimensional structures were then correlated against each other to establish a homogeneous data set. Related classes (comprising 82% of the data set and having a correlation coefficient of 0.80 or higher) were subsequently merged to generate an initial three-dimensional reference structure. This structure was subjected to multiple rounds of angular refinement by use of the previously defined homogeneous data set (3662 particles). Experimental images were matched to reference projections. On the basis of highest cross-correlation; a refined volume was then calculated with the newly identified Euler angles (Penczek et al. 1994). This process was repeated multiple times until the angles did not change and the resolution of the reconstruction did not improve. Final angular refinement was performed by generating 798 reference projections with an angular step of 5°. The CTD–CRSP structure was reconstructed to a resolution of 32 Å, on the basis of the 0.5 Fourier shell correlation criteria (Harauz and van Heel 1986). At this resolution, no CTF correction was needed.
Antibody labeling experiments
After eluting the CTD–CRSP complex from the affinity column, a fivefold excess of anti-GST antibodies were added and mixed for 1 h at 4°C. This sample was then run over a glycerol gradient to remove unbound antibody. Antibody-labeled CTD–CRSP samples were then analyzed by electron microscopy as described above, except that no tilted images were obtained, as the unlabeled CTD–CRSP structure was used as reference.
Cross-correlation analysis
All 24 classes within the CTD–CRSP data set had correlation coefficients between 0.78 and 0.88. Classes at the lower end of this range may represent degraded complexes, alternate conformers, or distorted complexes. Such classes were excluded from angular refinement because they reduced the quality (resolution) of the reconstruction. The average correlation coefficient of the classes used for angular refinement of CTD–CRSP was 0.83, which serves as a reference indicative of conformational similarity. Cross-correlation of CTD–CRSP and VP16–CRSP yielded a correlation coefficient of 0.88. For comparison, the correlation coefficient between conformationally distinct VP16–CRSP and SREBP–CRSP structures is 0.77.
Structural analysis of CTD–CRSP and VP16–CRSP was done completely independently (via random conical tilt) without reference bias. The dynamic nature of the CRSP coactivator suggests that, despite adopting specific and distinct conformational states, a degree of flexibility is maintained in each. Consequently, structures resolved by electron microscopy likely represent an average conformation about which the structure oscillates. For these reasons, it is likely that the structures of VP16–CRSP and CTD–CRSP are not 100% identical, although they clearly represent the same conformational state.
Acknowledgments
We thank R. Freiman, C. Kane, and Q. Zhou for critical reading of the manuscript. We also thank A. Ladurner for anti-CRSP70 antibodies, and Y. Nedialkov and S. Triezenberg for monoclonal antibodies against GST. D.J.T. is a postdoctoral fellow of the American Cancer Society (no. PF0007801GMC). This work was funded by grants from the NIH and Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jmlim@uclink4.Berkeley.edu; FAX (510) 643-9547.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.987602.
References
- Albright SR, Tjian R. TAFs revisited: More data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- Andel F, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. doi: 10.1126/science.286.5447.2153. [DOI] [PubMed] [Google Scholar]
- Boube M, Faucher C, Joulia L, Cribbs DL, Bourbon H. Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes & Dev. 2000;14:2906–2917. doi: 10.1101/gad.17900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer TG, Martin MED, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- Chiba N, Suldan Z, Freedman LP, Parvin JD. Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J Biol Chem. 2000;275:10719–10722. doi: 10.1074/jbc.275.15.10719. [DOI] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li YH, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West MA, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–153. [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon BD, Tjian R. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Myers LC, Kornberg RD. Mediator of transcriptional regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär AM, Beaurang PA, Robinson KM, Oliner JD, Avizonis D, Scheek S, Zwicker J, Kadonaga JT, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes & Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näär AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- Näär AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: New techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Peterson SR, Dvir A, Anderson CW, Dynan WS. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapeptide repeats. Genes & Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure random conical tilt series applied to the 50s ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Näär AM, Andel F, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Tanner S, Stagljar I, Georgiev O, Schaffner W, Bourquin JP. A novel SR-related protein specifically interacts with the carboxy-terminal domain (CTD) of RNA polymerase II through a conserved interaction domain. Biol Chem. 1997;378:565–751. doi: 10.1515/bchm.1997.378.6.565. [DOI] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]