Abstract
Sister chromatid separation at the metaphase-to-anaphase transition is induced by the proteolytic cleavage of one of the cohesin complex subunits. This process is mediated by a conserved protease called separase. Separase is associated with its inhibitor, securin, until the time of anaphase initiation, when securin is degraded in an anaphase-promoting complex/cyclosome (APC/C)-dependent manner. In budding yeast securin/Pds1 not only inhibits separase/Esp1, but also promotes its nuclear localization. The molecular mechanism and regulation of this nuclear targeting are presently unknown. Here we show that Pds1 is a substrate of the cyclin-dependent kinase Cdc28. Phosphorylation of Pds1 by Cdc28 is important for efficient binding of Pds1 to Esp1 and for promoting the nuclear localization of Esp1. Our results uncover a previously unknown mechanism for regulating the Pds1–Esp1 interaction and shed light on a novel role for Cdc28 in promoting the metaphase-to-anaphase transition in budding yeast.
Keywords: Cell cycle regulation, Pds1p, Esp1p, Cdc28p, mitosis
In all eukaryotic cells, cyclin-dependent kinases (CDKs) are essential for driving cell cycle progression. Periodic activation of different cyclin–CDK complexes is largely responsible for the regulation of various cell cycle processes, such as passage through START, initiation of DNA replication, and entry into mitosis. It is still not clear how cyclin–CDK activity is translated into this complex interplay of events, mainly because many of the critical cyclin–CDK substrates are unknown.
A central step in mitosis is the separation of sister chromatids, which takes place at the metaphase-to-anaphase transition. The proper timing of sister chromatid separation is key for accurate chromosome segregation: both premature separation, prior to the formation of bipolar spindle attachments between the chromatids and the spindle microtubules, and a prolonged delay in separation will result in extensive chromosome missegregation. Sister chromatid separation is triggered by the ubiquitin-dependent degradation of an anaphase inhibitor known as securin. Prior to anaphase, securin associates with and inhibits an anaphase activator known as separase, a protease that cleaves a subunit of the cohesin complex that mediates the association between the two sister chromatids (Ciosk et al. 1998; Uhlmann et al. 1999; Nasmyth et al. 2000; Cohen-Fix 2001). At the onset of anaphase securin is degraded in a process that involves the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, acting in conjunction with the Cdc20/Fizzy protein (Visintin et al. 1997; Zachariae and Nasmyth 1999). This, in turn, allows separase to promote anaphase initiation. The activity of the APC/C is also required for mitotic exit, where it acts in concert with the Cdh1 protein to promote the degradation of several proteins, including the mitotic cyclins (Schwab et al. 1997; Visintin et al. 1997). The APC/C is subjected to regulation by the spindle checkpoint pathway (Li et al. 1997; Fang et al. 1998; Alexandru et al. 1999), which inhibits APC/C activation until all chromosomes have formed stable bipolar spindle attachments, thereby preventing the premature dissociation of sister chromatids.
The budding yeast securin is called Pds1. Pds1 is a nuclear protein that is present in cells from late G1/early S until the onset of anaphase, when it is degraded in an APC/CCdc20-dependent manner (Cohen-Fix et al. 1996; Visintin et al. 1997). Pds1 physically associates with Esp1, the budding yeast separase, and inhibits its activity (Uhlmann et al. 1999). Although Pds1 is not essential for viability, cells lacking Pds1 are temperature-sensitive for growth and are unable to arrest mitotic progression in response to DNA or spindle damage (Yamamoto et al. 1996a). In the presence of DNA damage, Pds1 is required for inhibiting both anaphase initiation and mitotic exit (Yamamoto et al. 1996b). The latter is likely to be mediated by the ability of Pds1 to inhibit mitotic cyclin degradation (Cohen-Fix and Koshland 1999; Tinker-Kulberg and Morgan 1999). The presence of DNA damage leads to Pds1 phosphorylation via the DNA damage checkpoint pathway in a Mec1- and Chk1-dependent manner (Cohen-Fix and Koshland 1997; Sanchez et al. 1999). This phosphorylation is essential for Pds1's ability to inhibit cell cycle progression in the presence of DNA damage, but is not observed in undamaged cells, nor is it required for the response to other types of cellular impairments, such as spindle damage (Wang et al. 2001).
Pds1 plays a dual role in regulating the activity of Esp1. In addition to inhibiting Esp1, Pds1 is also required for the activation of Esp1, by promoting its efficient nuclear localization (Jensen et al. 2001). Cells lacking Pds1 are viable but do not accumulate Esp1 in the nucleus, resulting in the failure to initiate anaphase at elevated temperatures (Yamamoto et al. 1996a; Jensen et al. 2001). It is presently unknown how Pds1 targets Esp1 to the nucleus or how this process is regulated. The requirement for securin in separase's nuclear localization was first observed in fission yeast, where the nuclear localization of the separase/Cut1 was dependent on securin/Cut2 (Kumada et al. 1998). Interestingly, the requirement for securin in separase's activation is not limited to organisms that undergo a closed mitosis (i.e., no nuclear envelope breakdown). In Drosophila, cells defective in securin function (pim mutants) fail to separate sister chromatids (Leismann et al. 2000), and the absence of the human securin, known as pituitary tumor transforming gene (PTTG), results in reduced levels of separase (Zou et al. 1999; Jallepalli et al. 2001). Thus, the dual role for securins in regulating the activity of separase appears to be conserved throughout evolution.
The central role played by Pds1 in regulating mitosis prompted us to examine the upstream pathways that regulate the activity of Pds1. In this study we describe a previously unknown modification of Pds1. We show that Pds1 is phosphorylated by the cyclin-dependent kinase Cdc28, a key regulator of cell cycle progression in budding yeast, and that this phosphorylation is distinct from the previously known DNA damage-induced phosphorylation. We provide evidence that phosphorylation of Pds1 by Cdc28 is important for its efficient binding to Esp1 and for promoting the nuclear localization of Esp1. Our results uncover a previously unknown target of Cdc28 and reveal a novel mechanism for regulating the Pds1–Esp1 interaction.
Results
Pds1 is phosphorylated in vitro in a DNA damage-independent manner
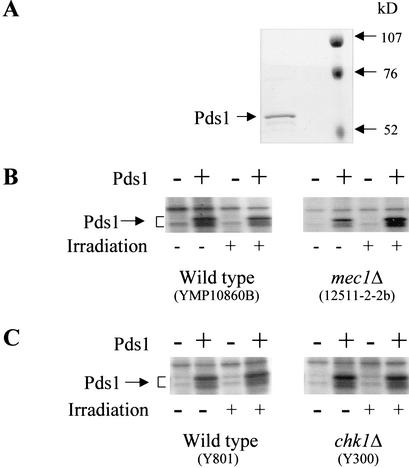
In response to DNA damage, Pds1 is phosphorylated in a Mec1- and Chk1-dependent manner (Cohen-Fix and Koshland 1997; Sanchez et al. 1999). In an attempt to reconstitute this pathway in vitro, we examined whether recombinant Pds1 could be phosphorylated when incubated in the presence of a protein extract prepared from wild-type cells that were exposed to γ radiation. We purified bacterially expressed Pds1, the majority of which migrated on SDS-PAGE as a single band with a minor degradation product of slightly faster mobility (Fig. 1A). Analysis by mass spectrometry confirmed that the purified protein was Pds1 (data not shown). Following incubation with [γ-32P]ATP and protein extracts from either irradiated or nonirradiated wild-type cells, a radiolabeled doublet specific to Pds1 was observed in all reactions containing purified Pds1, even those that contained extracts from nonirradiated cells (Fig. 1B,C, wild type). It was formally possible that DNA fragments generated during the extraction procedure activated the DNA damage checkpoint pathway and led to Pds1 phosphorylation. To test this, we used extract from strains in which either the MEC1 or CHK1 gene was deleted. As seen in Figure 1, B and C, Pds1 was still phosphorylated in the absence of Mec1 or Chk1. Thus, unexpectedly, our results showed that there is a DNA damage-independent pathway for Pds1 phosphorylation.
Figure 1.
Pds1 is phosphorylated in vitro in a Mec1- and Chk1-independent manner. (A) Purification of Pds1. Pds1 was expressed in Escherichia coli and purified as described under Materials and Methods. The purified protein was resolved by 10% SDS-PAGE and visualized by Coommassie blue staining. (B,C) Modification of Pds1 in vitro. In vitro kinase reactions were carried out as described under Materials and Methods in the absence or presence of purified Pds1, as indicated. The protein extract was made from the following strains that were either irradiated or left untreated: (B) wild type and mec1Δ (YMP10860b and 12511-2-2b, respectively); (C) wild type and chk1Δ (Y801 and Y300, respectively). Radiolabeled proteins were resolved by 10% SDS-PAGE and detected by autoradiography.
Pds1 can be phosphorylated by Cdc28 in vitro
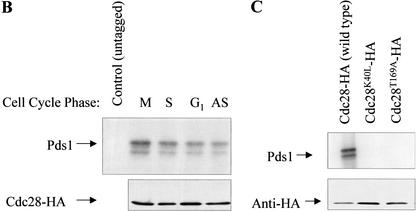
In search of a kinase that phosphorylates Pds1 in a DNA-damage-independent manner, we analyzed the protein sequence of Pds1 and found five putative cyclin-dependent kinase (CDK) phosphorylation sites (Fig. 2A). CDKs phosphorylate serine and threonine residues found within the consensus S/TPXK/R (single-letter amino acid code), although the K/R at position +3 relative to the phosphoacceptor site is not absolutely necessary (Songyang et al. 1994; Zhang et al. 1994; Srinivasan et al. 1995; Holmes and Solomon 1996). To test whether Pds1 is a substrate for the budding yeast CDK Cdc28 in vitro, cyclin–Cdc28 complexes were immunoprecipitated from either asynchronously growing cells or from cells arrested in different phases of the cell cycle, and the immunoprecipitates were used in in vitro phosphorylation reactions containing purified Pds1. As seen in Figure 2B, Pds1 could be phosphorylated by HA-tagged Cdc28 immunoprecipitated from different cell cycle stages but not by immunoprecipitates from control cells that express untagged Cdc28. The phosphorylation by Cdc28 purified from G1-arrested cells was reproducibly less efficient. This was not a result of less Cdc28 in this reaction as the levels of immunoprecipitated Cdc28 were the same in all reactions (Fig. 2B, lower panel).
Figure 2.
Pds1 is a Cdc28 substrate in vitro. (A) The assignment of the putative Cdc28 phosphorylation sites was done according to Songyang et al. (1994), Zhang et al. (1994), and Srinivasan et al. (1995). (B) Pds1 is phosphorylated in vitro by Cdc28 isolated from cells arrested in different cell cycle phases. HA-tagged Cdc28 was immunoprecipitated from protein extracts made from cells that were either grown to mid-log phase (asynchronous, AS), or arrested in G1 (with α-factor, 200 nM final concentration), in S phase (with hydroxyurea, 0.2 M final concentration), or in mitosis, M, (with nocodazole, 15 μg/mL final concentration). Asynchronous cells expressing untagged Cdc28 were used as a control. Immunoprecipitated Cdc28 was used in the in vitro phosphorylation reaction as described under Materials and Methods. The reaction products were resolved by 10% SDS-PAGE followed by autoradiography to detect 32P-labeled Pds1 (upper panel) or Western blot analysis with anti-HA antibody to show the relative amounts of the Cdc28 protein in each reaction (lower panel). (C) Pds1 is phosphorylated by wild-type but not by mutant Cdc28. Cells carrying plasmids expressing Cdc28-HA, Cdc28T169A-HA, or Cdc28K40L-HA were grown to mid-log phase. Protein extraction, immunoprecipitation, and in vitro kinase reactions were carried out and analyzed as described for panel B.
To establish that the phosphorylation activity was specific to Cdc28 and not caused by a copurifying kinase, we used two Cdc28 mutants that were previously shown to be defective in their kinase activity: Cdc28K40L, in which the ability to bind ATP is abolished, and Cdc28T169A, in which activation by CAK is eliminated. Unlike immunoprecipitates containing wild-type Cdc28, those containing either of the two mutant forms were unable to promote Pds1 phosphorylation (Fig. 2C, top panel), although the levels of the different Cdc28 derivatives were the same in all reactions (Fig. 2C, lower panel). Thus Pds1 appears to be a Cdc28 substrate. This conclusion is also consistent with an observation made by David Morgan and Jeff Ubersax (UCSF), who identified Pds1 in a large-scale screen for Cdc28 substrates. In this screen, a series of GST-fused proteins were tested for their ability to undergo in vitro phosphorylation by an altered Cdc28, Cdc28-as1, the ATP-binding site of which was modified to accommodate bulky ATP analogs (see below; Bishop et al. 2000). The labeling reactions were carried out in the presence of a radioactive bulky ATP analog that could be used by no kinase other than Cdc28-as1 (D. Morgan and J. Ubersax, unpubl.). Taken together, these results suggest that Pds1 is specifically phosphorylated by Cdc28 in vitro.
Pds1 is phosphorylated in vivo in a Cdc28-dependent manner
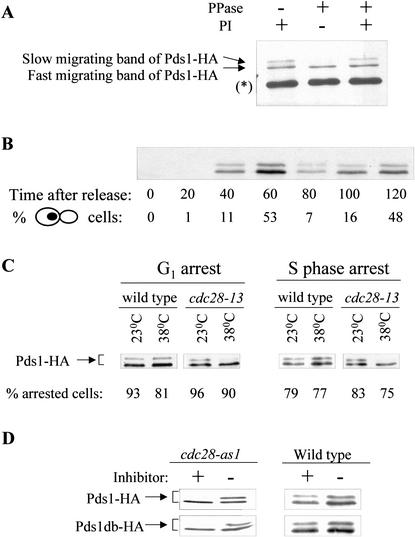
The observation that Pds1 can be phosphorylated in vitro by Cdc28 prompted us to examine whether Pds1 is phosphorylated in vivo in a Cdc28-dependent manner. Overexpressed Pds1 and endogenous HA-tagged Pds1 have been shown to migrate as a doublet by SDS-PAGE (Cohen-Fix et al. 1996), but phosphatase treatment of Pds1 in crude extracts preparation did not yield a change in this electrophoretic mobility (Cohen-Fix and Koshland 1997). We decided to reexamine the in vivo phosphorylation state of Pds1 using a more purified preparation. Pds1-HA was immunoprecipitated from asynchronously growing cells, and the immunoprecipitates were treated with alkaline phosphatase. As can be seen in Figure 3A, the slow-migrating band of Pds1 disappeared upon phosphatase treatment. Moreover, the presence of phosphatase inhibitors prevented this change, indicating that the slower-migrating Pds1 form was caused by phosphorylation. We therefore concluded that Pds1 is phosphorylated in vivo and that the electrophoretic mobility of Pds1 can be used as an indicator for its phosphorylation state. To examine when in the cell cycle Pds1 is phosphorylated, cells expressing Pds1-HA under the native PDS1 promoter were arrested in G1 with α-factor mating pheromone followed by a synchronous release from the arrest. As observed previously, the levels of Pds1 peak prior to anaphase (Fig. 3B; Cohen-Fix et al. 1996), and judging by the mobility shift Pds1 is phosphorylated for as long as it is present in the cell. Using densitometry scanning we estimate that the fraction of the slow-migrating Pds1 form in cycling cells is ∼20% of the total Pds1, but under these separation conditions some phospho-Pds1 forms may not show a shift in protein mobility (see below).
Figure 3.
Pds1 is phosphorylated in vivo in a Cdc28-dependent manner. (A) Pds1-HA was immunoprecipitated from mid-log-phase wild-type OCF1522 cells using anti-HA antibodies. The immunoprecipitates were subjected to treatment with alkaline phosphatase (PPase) in the presence and absence of phosphatase inhibitors (PI). Pds1 mobility was analyzed by 10% SDS-PAGE followed by Western blot analysis using anti-HA antibodies. The asterisk in brackets indicates the heavy chain of the antibody used for immunoprecipitation. (B) Wild-type cells (OCF1522) expressing Pds1-HA from its native promoter were grown at 30°C, arrested in G1 with α-factor mating pheromone and then released into media lacking pheromone. Samples were taken at the indicated time points, processed for Western blot analysis, and scored for cell morphology by DIC and DAPI staining. The percent of large budded cells with a single nucleus, indicative of a post-S phase/pre-anaphase state, is shown. (C) cdc28-13 and wild-type strains (RA2817-8a and RA2817-1d, respectively), both carrying a plasmid encoding for PDS1-HA expressed from a galactose-inducible promoter (pOC42), were grown to mid-log phase at 23°C, arrested in G1 with α factor (5 μM) or in S phase with hydroxyurea (200 mM), and then shifted to 38°C for 3.5 h. Following the temperature shift, the expression of Pds1-HA was induced for 1 h (still at 38°C), after which protein extracts were prepared and analyzed by Western blot analyses. The percent of cells in G1 or S phase is indicated. (D) Centromeric plasmids encoding for Pds1-HA or Pds1db-HA (Pds1 lacking a destruction box; Cohen-Fix et al. 1996) expressed from a galactose-inducible promoter (pOC42 and pOC57-HA, respectively) were introduced into either a wild-type strain or a cdc28-as1 strain (Bishop et al. 2000; see Materials and Methods). Following an arrest in G1 with α factor, cells were either mock-treated or treated with 10 μM of PP1 (analog 9) for 4.5 h. The expression of Pds1-HA or Pds1-db-HA was induced for 1 h, after which protein extracts were prepared and analyzed by Western blot analysis.
To test whether in vivo Pds1 is phosphorylated in a Cdc28-dependent manner, we used a strain carrying a temperature-sensitive allele of CDC28, cdc28-13, that is inactive for its kinase activity at the nonpermissive temperature of 38°C. Wild-type and cdc28-13 strains, both expressing HA-tagged Pds1 from a galactose-inducible promoter, were arrested in G1 with the α-factor mating pheromone (Fig. 3C, left) or in S phase with hydroxyurea (Fig. 3C, right) and then shifted to the nonpermissive temperature for 3.5 h. The arrest prior to the temperature shift was done to ensure that Pds1 phosphorylation was examined while both strains were in the same cell cycle phase. Following the temperature shift the expression of Pds1-HA was induced for 1 h, and the phosphorylation state of Pds1 was monitored by SDS-PAGE (Fig. 3C). At the permissive temperature, under both arresting conditions, Pds1-HA appeared as a doublet in both the wild-type and cdc28-13 strains. However, in the cdc28-13 strain after the shift to the nonpermissive temperature, there was a significant reduction in the relative amount of the slow-migrating Pds1 form, both in G1 and in S phase, indicating that this Pds1 form is a result of Cdc28-dependent phosphorylation. Parenthetically, it should be noted that despite the pheromone treatment, which induces the Cdc28-inhibitor Far1 and markedly reduces the levels of Cln1 and Cln2 (Peter and Herskowitz 1994; Gartner et al. 1998), we could still detect Cdc28-dependent phosphorylation of Pds1 in G1. These results suggest that in the presence of pheromone, Cdc28 may only be partially inactive, and that the G1 arrest is achieved by keeping the Cdc28 kinase levels below a certain threshold.
We also examined the involvement of Cdc28 in the phosphorylation of Pds1 by using the cdc28-as1 allele that encodes for a Cdc28 derivative with an altered ATP-binding site that is inactive in the presence of a synthetic inhibitor, PP1 (Bishop et al. 2000; see Materials and Methods). Wild-type and cdc28-as1 cells carrying plasmids encoding for galactose-inducible PDS1-HA or PDS1-db-HA (mutated in the Pds1 destruction box; Cohen-Fix et al. 1996) were arrested in G1 with the α-factor mating pheromone, and induced with galactose in the presence or absence of the PP1 inhibitor. As can be seen in Figure 3D, in the absence of the inhibitor Pds1-HA and Pds1-db-HA appeared as a doublet in both the wild-type strain and the cdc28-as1 mutant strain. In contrast, in the presence of the inhibitor most of the Pds1 in the cdc28-as1 strain was in the fast-migrating form. Taken together, these results show that in vivo Pds1 is phosphorylated in a Cdc28-dependent manner.
The ability of Pds1 to interact efficiently with Esp1 depends on the phosphorylation of Pds1 by Cdc28
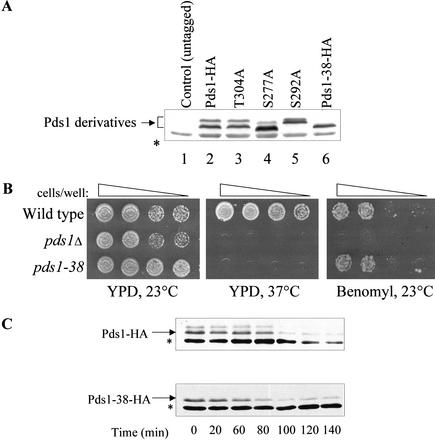
To determine the role of the Cdc28-dependent phosphorylation of Pds1, we wished to generate Pds1 mutants that could not undergo this type of phosphorylation. To this end we searched for the Pds1 residues that when mutated abolished the mobility shift observed by SDS-PAGE. As mentioned earlier, Pds1 has five consensus sites for Cdc28 (Fig. 2A). Three of these sites reside in the C terminus of the protein, a deletion of which severely compromises the ability of Pds1 to act as a mitotic regulator (R. Agarwal and O. Cohen-Fix, unpubl.). We therefore substituted the serines or threonines of these three sites in PDS1-HA with alanines, either individually or in combination, and examined the effect on protein mobility by SDS-PAGE and the growth phenotype conferred when expressed from the native PDS1 promoter. Pds1 is essential for growth at elevated temperatures and under conditions that induce a spindle checkpoint response. Thus, the functionality of pds1 alleles can be assessed by examining their ability to promote growth at 37°C or in the presence of spindle poisons such as benomyl. The S277A and S292A amino acid substitution led to changes in the migration of the two Pds1 forms, whereas the T304A mutant form migrated as wild-type Pds1 (Fig. 4A, cf. lanes 3, 4, and 5 to lane 2). None alone or in a pairwise combination conferred a growth phenotype that was distinguishable from that of the wild-type control strain (data not shown). However, when all three single amino acid substitutions were combined to yield the pds1-38-HA allele, the resultant protein behaved by SDS-PAGE as the fast-migrating Pds1 form, consistent with its inability to undergo Cdc28-dependent phosphorylation (Fig. 4A, lane 6), and a strain carrying this allele as its sole source of Pds1 was temperature-sensitive but not benomyl-sensitive (Fig. 4B). The fact that the growth phenotype on benomyl of the pds1-38 strain was distinct from that of the pds1Δ strain suggested that the three substitutions affected a specific Pds1 function rather than completely eliminated all of its activity. The phenotype conferred by a pds1 allele in which the serines or threonines of all five Cdc28 consensus sites were substituted with alanines was indistinguishable from that conferred by the pds1-38 allele (data not shown). Owing to the lack of a detectable change in mobility in the T304A mutated protein we currently do not know whether threonine 304 is phosphorylated in vivo, but it should be noted that not all protein phosphorylations result in an altered mobility (e.g., see Ross et al. 2000).
Figure 4.
The effect of alanine substitutions at putative Cdc28 phosphorylation sites on the electrophoretic mobility of Pds1. (A) Protein extracts from wild type (lane 2) or strains carrying mutations in PDS1 as indicated (lanes 3–6) were prepared from logarithmically growing cells, resolved by 10% SDS-PAGE, and analyzed by Western blot analysis using anti-HA antibodies. The Pds1-38-HA protein (lane 6) contains all three mutations (S277A, S292A, and T304A). (B) The pds1-38 strain is temperature-sensitive but not benomyl-sensitive. Wild-type, pds1Δ, and pds1-38 strains were grown to log phase and spotted by serial dilutions onto YPD plates incubated at 23°C or 37°C and onto YPD plates containing benomyl (12.5 μg/mL). (C) Pds1-38 stability at 37°C. Cells expressing Pds1-HA or Pds1-38-HA were synchronized in S phase using hydroxyurea, temperature-shifted for 1 h after a full arrest was achieved, and then released into prewarmed YPD (37°C) containing α factor to arrest the cells at the subsequent G1. Protein extracts were prepared every 20 min and analyzed by Western blot analysis. The asterisk represents an anti-HA cross-reacting band in the extracts.
One reason for the temperature sensitivity of the pds1-38-HA strain could have been that the Pds1-38-HA protein is unstable at the elevated temperature. To examine this possibility, wild-type and pds1-38-HA strains were released synchronously from an S-phase arrest into media containing the α-factor mating pheromone at 37°C, and the Pds1 levels were monitored over time. In both the strains, the Pds1 protein levels started to decline after 80–100 min following the release (Fig. 4C). Thus, at 37°C, Pds1-38-HA is not unstable nor is there a defect in its degradation. We therefore conclude that the temperature sensitivity of the pds1-38-HA strain was due to a partial defect in Pds1 function and that Cdc28 phosphorylation plays an important role in regulating the activity of Pds1.
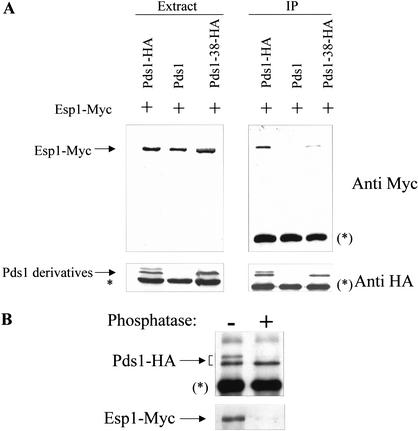
Earlier work has shown that the Pds1–Esp1 interaction is required for two purposes: (1) to inhibit the activity of Esp1 (Ciosk et al. 1998) and (2) to promote the nuclear localization of Esp1 (Kumada et al. 1998; Jensen et al. 2001). To examine how the pds1-38 mutations affect the ability of this protein to interact with Esp1, coimmunoprecipitation experiments were conducted. Cells expressing Esp1-Myc, with Pds1-HA, Pds1-38-HA, or untagged Pds1 were grown at 23°C and subjected to coimmunoprecipitation as described under Materials and Methods. As shown in Figure 5A, the ability of the HA-tagged Pds1-38 to associate with Esp1 was significantly reduced compared with HA-tagged wild-type Pds1. To further examine the effect of Pds1 phosphorylation on its ability to interact with Esp1, Pds1-HA immunoprecipitated from wild-type cells was either mock treated or subjected to phosphatase treatment, and then mixed with a protein extract prepared from a pds1Δ ESP1-Myc strain. This strain was chosen as a source for Esp1-Myc to ensure that Esp1-Myc was not already associated with endogenous Pds1. The mixing reactions were carried out in the absence of ATP to prevent rephosphorylation of the treated Pds1-HA. The ability of both phosphatase-treated and mock-treated Pds1 to interact with Esp1-Myc was then examined. As can be seen in Figure 5B, the phosphatase treatment significantly reduced the ability of Pds1 to interact with Esp1. Taken together our results strongly suggest that the phosphorylation of Pds1 by Cdc28 is required for the Pds1–Esp1 interaction.
Figure 5.
Pds1 phosphorylation is required for its efficient binding to Esp1. (A) Pds1-38 has reduced affinity for Esp1. Protein extracts were prepared from strains expressing Esp1-Myc and untagged Pds1 (OCF1548-6D), Pds1-HA (OCF1548-5C), or Pds1-38-HA (RA2806-2b), all grown to mid-log phase at 23°C. The extracts were subjected to immunoprecipitation using anti-HA antibodies. Total extracts (left panel) and immunoprecipitates (IP, right panel) were separated by SDS-PAGE followed by Western blot analyses using anti-HA antibodies (bottom panel) or anti-myc antibodies (top panel). Densitometric scans showed that the ratio of the immunoprecipitated Pds1-HA to Pds1-38-HA was ∼1.2, whereas the ratio of the Esp1-Myc that coimmunoprecipitated with Pds1-HA to the Esp1-Myc that coimmunoprecipitated with Pds1-38-HA was ∼6.8. The asterisk indicates an anti-HA cross-reacting band in the extracts, whereas an asterisk in brackets represents the cross-reacting band of the heavy chain of the antibody used for immunoprecipitation. (B) Nonphosphorylated Pds1 binds weakly to Esp1 in vitro. Pds1-HA was immunoprecipitated from extracts made from mid-log phase wild-type cells expressing Pds1-HA (OCF1522), using anti-HA antibodies. The immunoprecipitates were split in two and were either treated with alkaline phosphatase or mock-treated, after which they were mixed with an extract prepared from the pds1Δ ESP1-Myc strain (RA2804-25c). The immunoprecipitates were then washed extensively, and the bound proteins were eluted using SDS sample buffer. The proteins were resolved by SDS-PAGE followed by Western blot analyses of Pds1-HA (top panel) and Esp1-Myc (bottom panel).
The nuclear localization of Esp1 is dependent on the Cdc28-dependent phosphorylation of Pds1
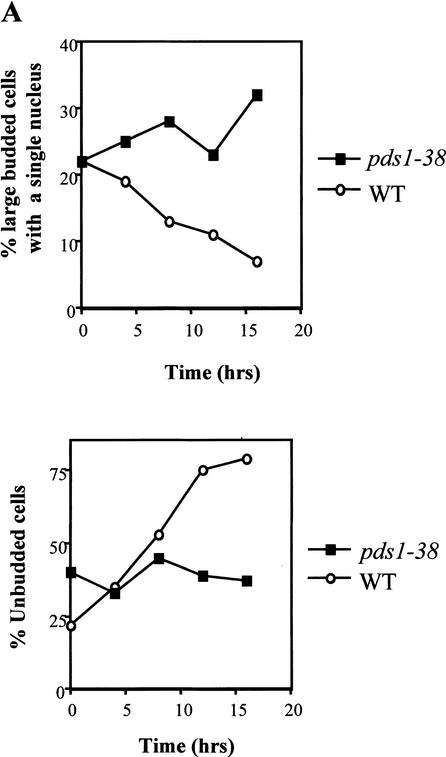
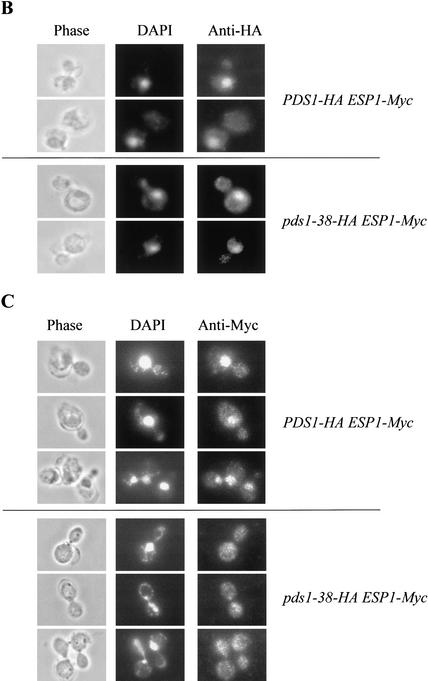
The picture that emerges from the data presented above is that mutating the Cdc28 consensus sites in Pds1 results in the reduced binding of Pds1 to Esp1. The fact that pds1-38 mutants are not benomyl-sensitive suggested that the absence, or partial absence, of Cdc28-dependent phosphorylation did not severely affect the ability of Pds1 to act as a mitotic inhibitor. When grown at 37°C for several hours, pds1Δ cells show a cut phenotype (i.e., an unequal distribution of the DNA mass between the two daughter cells; Yamamoto et al. 1996a) and wild-type cells accumulate in G1 as they enter stationary phase (Fig. 6A). In contrast, the pds1-38 strain grown at 37°C for several hours accumulated as large budded cells with a single nucleus and a 2N DNA content (Fig. 6A; data not shown), suggesting that these cells have difficulties in carrying out anaphase. Such a phenotype would be expected from a mutant strain that is defective in promoting Esp1 activity but has retained the ability to inhibit mitotic progression. Given the known role of Pds1 in the activation of Esp1, we examined the ability of Pds1-38 to promote the nuclear localization of Esp1. In wild-type cells, Pds1 is concentrated in the nucleus for as long as it is present, namely, from late G1/early S phase to the time of anaphase initiation. Esp1, on the other hand, is present throughout the cell in G1 and is first seen in the nucleus only as cells enter mitosis (Jensen et al. 2001). To monitor the localization of Pds1 and Esp1, strains expressing Esp1-Myc and either Pds1-HA or Pds1-38-HA were used. Both strains were grown at 37°C for 4 h, and the localization of Pds1 and Esp1 was monitored by indirect immunofluorescence. Western blot analysis revealed that there was no difference between the wild-type and pds1-38 strains in the Esp1-Myc levels (data not shown). Pds1-38 itself showed a normal nuclear localization at both permissive and nonpermissive temperatures (Fig. 6B; data not shown). However, in the pds1-38 strain Esp1 failed to concentrate in the nucleus at 37°C (Fig. 6C), much like the phenotype observed in a pds1Δ strain (Jensen et al. 2001). This suggests that Cdc28-mediated phosphorylation of Pds1 is needed for efficient recruitment of Esp1 into the nucleus. In a pds1Δ strain the failure to accumulate Esp1 in the nucleus was observed at 23°C as well (Jensen et al. 2001; data not shown). The pds1-38 mutant strain has no detectable defect in the nuclear accumulation of Esp1 at 23°C, although even at this temperature the ability of Pds1-38 to bind Esp1 is significantly reduced (Fig. 5A). These observations suggest that the low-affinity interaction between Esp1 and Pds1-38 may be sufficient to promote the nuclear localization of Esp1 at 23°C but not at 37°C, possibly because of the slower growth rate at 23°C, allowing more time for Esp1 to accumulate (also see Discussion). This further suggests that other factors contribute to the low-affinity Pds1–Esp1 interaction.
Figure 6.
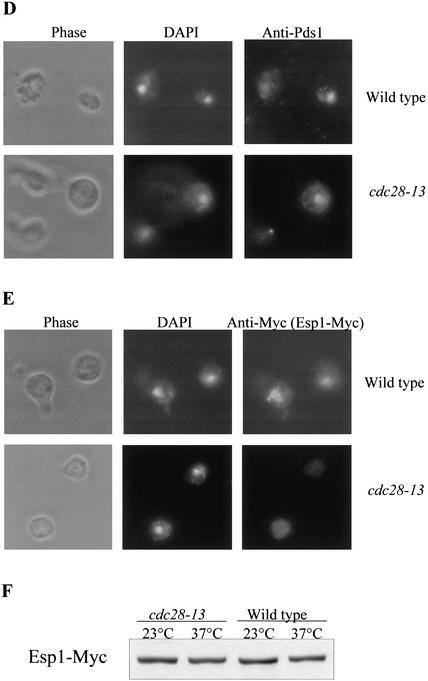
Cdc28-dependent phosphorylation of Pds1 is required for Esp1 accumulation in the nucleus. (A) The distribution of cell types in a wild-type versus a pds1-38 strain grown at 37°C. Wild-type (OCF1522) and pds1-38 mutant (RA2806-2b) strains were grown to log phase at 23°C and then shifted to 37°C. Samples were collected every 4 h and fixed in 70% ethanol; cell morphology was analyzed by microscopy. The percentage of large budded cells with a single nucleus (upper panel) and unbudded cells (lower panel) in wild-type cells (circles) and pds1-38 mutant cells (squares) is shown. (B,C) Esp1 does not accumulate in the nucleus in the pds1-38 strain. An HA-tagged Pds1 strain (OCF1548-5C, upper panels) and an HA-tagged Pds1-38 strain (RA2806-2b, lower panels), both of which also carried Myc-tagged Esp1 expressed from its own promoter, were grown to early-log phase and then shifted to 37°C for 4 h. Cells were fixed with 4% formaldehyde and processed for indirect immunofluorescence as described under Materials and Methods using anti-HA antibodies for detecting Pds1-HA or Pds1-38-HA (B) and anti-Myc antibodies for detecting Esp1-Myc (C). In each case several typical examples are shown. (D,E) The nuclear localization of Esp1 is Cdc28-dependent. Wild-type (RA2817-8a, upper panel) or cdc28-13 (RA2817-1d, lower panel) cells carrying Myc-tagged Esp1 expressed from its own promoter and Pds1-HA expressed from a galactose-inducible promoter were grown to early log phase at 23°C in media containing raffinose, after which they were arrested in G1 with α factor (5 μM). After a complete arrest the cultures were shifted to 37°C for 3.5 h. Pds1 expression was then induced from a galactose promoter for 2 h. Cells were fixed and processed for indirect immunofluorescence as described under Materials and Methods using anti-Pds1 antibodies to detect Pds1 (D) and anti-Myc antibodies to detect Esp1 (E). (F) The levels of Esp1-Myc in wild-type and cdc28-13 cells at 23°C and 37°C.
To eliminate the possibility that the Pds1-38 defect is unrelated to its inability to undergo Cdc28-dependent phosphorylation, we also examined the ability of overexpressed Pds1 to promote the nuclear localization of Esp1 in wild-type or cdc28-13 cells. It was shown previously that in G1, Esp1 does not concentrate in the nucleus but that ectopic expression of Pds1 can induce its nuclear localization (Jensen et al. 2001). If the phosphorylation of Pds1 by Cdc28 is required for this process, one would predict that cdc28-13 cells would be impaired in Esp1's nuclear accumulation when assayed at the nonpermissive temperature even in the presence of high Pds1 levels. To test this, wild-type and cdc28-13 strains, both carrying ESP1-Myc and galactose-inducible PDS1-HA, were arrested in G1 and shifted to 38°C for 3.5 h, followed by the induction of Pds1-HA expression for 2 h. In both strains 50% of the G1 cells showed nuclear Pds1-HA staining, again showing that Cdc28 activity is not required for nuclear localization of Pds1 (Fig. 6D). Whereas >30% of the wild-type G1 cells showed the accumulation of Esp1 in the nucleus, this was observed in <10% of the cdc28-13 G1 cells (Fig. 6E). Under these conditions both strains expressed similar levels of Esp1-Myc (Fig. 6F). Taken together, our data strongly suggest the Cdc28-dependent phosphorylation of Pds1 is required for Pds1 to promote the nuclear localization of Esp1.
Discussion
Cell cycle progression is the net result of regulatory mechanisms that drive the cell cycle and those that exert an inhibitory effect. The latter are often required to ensure that late cell cycle events are initiated only after early ones have been successfully completed. During mitosis, it is imperative that sister chromatids separate only after all duplicated chromosomes have formed bipolar spindle attachments. Because sister chromatid separation is driven by the proteolytic cleavage of one of the cohesin complex subunits, it is conceivable that the protease that drives this process, separase, would be subjected to multiple levels of regulation. In budding yeast, the activity of the Esp1/separase is inhibited by Pds1/securin, making Pds1 a target for numerous regulatory mechanisms that influence the timing of anaphase initiation. These include the spindle checkpoint pathway, acting via the inhibition of APC/C activity (Fang et al. 1998; Wassmann and Benezra 1998), and the DNA damage checkpoint pathway, stabilizing Pds1 through Chk1-mediated phosphorylation (Wang et al. 2001). Pds1 not only inhibits Esp1 but also acts as an Esp1 activator by driving its nuclear accumulation (Jensen et al. 2001). Despite the diverse roles played by Pds1 in mitotic regulation, little is known about the molecular mechanism of its action.
Mitotic CDKs are known to be required for progression through mitosis. In budding yeast, Clb–Cdc28 complexes are necessary for spindle formation and regulation of the APC/C (Richardson et al. 1992; Irniger et al. 1995; Amon 1997). In this study we present several lines of evidence suggesting that Pds1 is a target of Cdc28: (1) Pds1 was phosphorylated in vitro by immunopurified wild-type Cdc28. This phosphorylation was specific to Cdc28, because mutant forms of Cdc28 impaired in their catalytic activity did not promote Pds1 phosphorylation. (2) The phosphorylation of Pds1 in vivo was abolished by mutations or conditions that inactivated Cdc28. These included a temperature-sensitive allele of CDC28, cdc28-13, and a chemically repressible Cdc28 derivative, Cdc28-as1. (3) Mutagenesis of three of the Cdc28 consensus sites in Pds1 altered the mobility of Pds1 as detected by SDS-PAGE and abolished the slow-migrating Pds1 form, similar to the change in mobility observed after phosphatase treatment. Taken together, our data suggest that Pds1 is a bona fide Cdc28 substrate.
The Cdc28-dependent phosphorylation of Pds1 occurred at any phase of the cell cycle in which Pds1 was present. Given that not all phosphorylations may result in a mobility shift, the fraction of Pds1 molecules that is phosphorylated by Cdc28 at any given time and the number of Cdc28 phosphorylation sites occupied per molecule are currently unknown. Nonetheless, Cdc28-mediated phosphorylation appears to be critical for the ability to interact efficiently with Esp1 and to promote the nuclear localization of Esp1. This conclusion is based on the following observations: (1) Pds1-38, in which three Cdc28 consensus phosphorylation sites at the C terminus of Pds1 were converted to alanines, had a significantly reduced affinity for Esp1 at the permissive temperature and was unable to promote the nuclear localization of Esp1 at elevated temperatures. (2) Dephosphorylation of in vivo phosphorylated Pds1 reduced its ability to interact with Esp1 in vitro. (3) The ability of high levels of Pds1 to drive Esp1 into the nucleus in G1 was dependent on Cdc28 activity. Our results also show that phosphorylation by Cdc28 is not required for the nuclear localization of Pds1 itself. Moreover, phosphorylation at Ser 277, Ser 292, and possibly Thr 304 is not essential for the role of Pds1 as a mitotic inhibitor, as the pds1-38 mutant strain, unlike the pds1Δ strain, was not sensitive to microtubule depolymerizing drugs. The ability of Pds1-38 to inhibit mitotic progression suggests that it is still capable of binding to Esp1, albeit at much a lower affinity. The difference between the ability to promote the nuclear localization of Esp1 and to inhibit mitotic progression may lie in the different cellular compartments in which these functions take place. Inhibition of mitotic progression is likely to take place in the nucleus, where the levels of Pds1-38, or wild-type Pds1, are high and therefore low-affinity interactions may be compensated by the overall high Pds1-38 concentration. In contrast, the Pds1–Esp1 interaction that is required to promote the nuclear localization of Esp1 may take place in the cytoplasm, where Pds1-38 levels are exceedingly low, and low-affinity interactions may be insufficient to drive Esp1 into the nucleus. Moreover, Pds1 is likely to reside in the cytoplasm for only a short period of time, whereas in the nucleus there is more opportunity to maintain a Pds1–Esp1 interaction. At present we do not know why the defect in Esp1 nuclear localization in the pds1-38 strain is detected only at elevated temperatures. It is possible that the affinity between Pds1-38 and Esp1 is further reduced at the elevated temperature and that the low-affinity state, detected by the immunoprecipitation assay at 23°C, is sufficient to promote nuclear accumulation at low but not high temperatures.
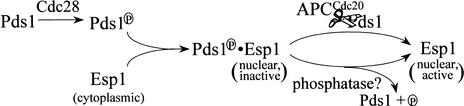
Our results suggest that following Pds1 phosphorylation by Cdc28, Pds1 associates with Esp1 and promotes its nuclear accumulation (Fig. 7). This mechanism ensures that prior to anaphase, nuclear Esp1 will be inactive because of its association with Pds1, and only after Pds1 inactivation, either by APC/C-mediated degradation or by the activity of a putative phosphatase (see below), will Esp1 become active. Interestingly, the phosphorylation of securins by mitotic CDKs appears to be conserved, as the human securin homolog of Pds1, PTTG, has been shown to be phosphorylated by Cdc2 (Ramos-Morales et al. 2000). In this case the CDK-dependent phosphorylation of securin is unlikely to be required for the nuclear localization of separase due to nuclear envelope break-down upon mitotic entry, but a requirement for targeting to a subnuclear structure (e.g., chromatin) is still possible. In both humans and Drosophila, the loss of securin function is accompanied by reduced separase activity, suggesting that the involvement of securin in separase activation is conserved. Whether the CDK-dependent phosphorylation is required for the positive function of securin in Drosophila and humans remains to be determined.
Figure 7.
A model for the regulation of the Pds1–Esp1 interaction by Cdc28. See text for details.
The dependence of the Pds1–Esp1 interaction on Cdc28-mediated phosphorylation suggests that the association between the two proteins could be dissolved not only by APC/C-mediated degradation of Pds1 but also by the action of an as yet unidentified phosphatase (Fig. 7). A possible phosphatase is the Cdc14 protein that was shown to remove Cdc28-mediated phosphorylation from the Cdh1, Sic1, and Swi5 proteins (Jaspersen et al. 1999), and overexpression of Cdc14 resulted in the disappearance of the slower-migrating Pds1 form (Visintin et al. 1998). At the time, the nature of this form was not known, but the results presented here suggest that this effect could be caused by the dephosphorylation of Pds1. Why would a cell need two mechanisms to counteract Pds1's inhibitory grip on Esp1? Having redundant mechanisms for regulating key cell cycle transitions is not uncommon. For example, Clb–Cdc28 inactivation at the exit from mitosis occurs via at least two pathways, the APC/CCdh1-dependent degradation of the mitotic cyclins and the Sic1-dependent inhibition of Clb–Cdc28 activity (Schwab et al. 1997; Visintin et al. 1998). In addition, there may be a pool of Esp1-bound phosphorylated Pds1 that is resistant to APC/CCdc20-mediated degradation and for which dephosphorylation may be the only way to release Esp1. Tinker-Kulberg and Morgan (1999) proposed a role for Esp1 at the exit of mitosis, and at that late cell cycle stage, when the nuclear Pds1 levels are low, dephosphorylation may be an effective way to free Esp1. Dephosphorylation may also render Pds1 more susceptible to degradation, either because dephosphorylated Pds1 is a better substrate for the APC/C or simply because by increasing the Pds1–Esp1 dissociation rate Pds1 may become more accessible to the ubiquitination machinery. Finally, it has been shown recently that Esp1 regulates the early release of Cdc14 from the nucleolus (Stegmeier et al. 2002). It is tempting to speculate that if Cdc14 is indeed the Pds1 phosphatase there may be a positive feedback loop where released Cdc14 dephosphorylates Pds1. As the nuclear concentrations of Pds1 drop owing to APC/C activity, dephosphorylation could play a significant role in dissociating the Esp1–Pds1 complex, thereby further activating Esp1 and allowing more Cdc14 to be released from the nucleolus. Thus, the Cdc28-mediated phosphorylation of Pds1 adds another layer of complexity to the regulation of mitotic progression.
Materials and methods
Plasmids and strains
The yeast strains used are described in Table 1. All strains were W303 derivatives except when indicated otherwise. The plasmids used were pOC57, pOC52 (Cohen-Fix et al. 1996), and pOC42 (CEN GAL1-PDS1-HA:URA3). PKB174 (CEN CDC28-HA TRP1), PKB173 (CEN cdc28K40L-HA TRP1), and PKB175 (CEN cdc28T169A-HA TRP1) were a generous gift from Philipp Kaldis (National Cancer Institute, Frederick, MD; originally plasmids CWB174, CWB173, and CWB186, respectively, from C. Wittenberg, The Scripps Research Institute, La Jolla, CA).
Table 1.
Strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| YMP10860a | MATαRNR1-LEU2 sml1d::KAN his3 leu2 trp1 ura3 | L. Hartwell |
| 12511-2-2ba | Same as YMP10860b but mec1Δ::TRP1 | L. Hartwell |
| Y300 | MATa can1-100 ade2 his3 leu2 trp1 ura3 | S. Elledge and Y. Sanchez |
| Y801 | Same as Y300 but chk1Δ::HIS3 | S. Elledge and Y. Sanchez |
| PKY 238 | MATa CDC28-HA::URA3 leu2 trp1 his3 ade2 | M. Solomon and P. Kaldis |
| OCF 1522 | MATa bar1 PDS1-HA::URA3 leu2 trp1 his3 ade2ura3 | (Cohen-Fix and Koshland 1997) |
| OCF 1525 | Same as 1522 but pds1Δ::LEU2 | (Cohen-Fix and Koshland 1997) |
| RA2817-1d | MATa cdc28-13 ESP1-Myc::TRP1 ura3 leu2 his3 | This study |
| RA2817-8a | MATa ESP1-Myc::TRP1 ura3 leu2 his3 | This study |
| cdc28-as1 | MATa bar1 cdc28d::cdc28-as1 ura3 trp1 ade2 can1-100 | David Morgan and Jeff Ubersax |
| RA2808 | Same as 1522 but pds1T304A-HA::URA3 | This study |
| RA2810 | Same as 1522 but pds1S277AHA::URA3 | This study |
| RA2811 | Same as 1522 but pds1S292A-HA::URA3 | This study |
| RA2815 | Same as 1522 pds1S277AS292T301A-HA::URA3 (also referred to as pds1-38) | This study |
| OCF1548-6D | MATa bar1 ESP1-Myc::TRP1 his3 leu2 ura3 ade2 | This study |
| OCF1548-5C | MATa PDS1-HA::URA3 ESP1-Myc::TRP1 his3 leu2 ade2 | This study |
| RA2806-2b | MATa pds1-38-HA::URA3 ESP1-Myc::TRP1 leu2 his3 ade2 | This study |
| RA2804-25c | MATa pds1Δ::LEU2 ESP1-Myc::TRP1 ura3 his3 ade2 | This study |
A derivative of the A364a background.
Media and reagents
YPD media contained 1% yeast extract, 2% bactopeptone, adenine (2.5 mg/L), and 2% glucose; YPGal and YPRaf contained 2% galactose or 2% raffinose instead of glucose, respectively. α-Factor mating pheromone (Sigma) was used at 0.1 μM for bar1 strains and 5 μM for BAR1+ strains. Hydroxyurea (Sigma) was used at 0.2 M. Nocodazole (Aldrich) was used at 15 μg/mL. [γ-32P]ATP was from NEN life science products (3000 Ci/mmole). Enterokinase was from Invitrogen. Calf intestine alkaline phosphatase was from Roche. Anti-HA antibodies (16B12), anti-Myc antibodies (9E10), and protein A sepharose linked to anti-HA antibodies were from Covance. Anti-Pds1 antibodies were a generous gift from Doug Koshland (The Carnegie Institution of Washington, DC).
Purification of Pds1
Pds1 was amplified by PCR using a 5′ primer that placed a BamHI site at the start codon and a 3′ primer that placed an XbaI site at the stop codon. The BamHI–XbaI fragment was cloned into a GST tagging vector (pGST, a generous gift from Peter Sheffield, University of Virginia, Charlottesville; Sheffield et al. 1999). GST-Pds1 was expressed in Escherichia coli strain BL21 codon plus (Stratagene). Bacteria transformed with the GST-Pds1 plasmid were grown to an OD600 of 0.5, and the expression of Pds1 was induced by IPTG (1 mM) for 16 h at 14°C. The cells were harvested, lysed in buffer A (50 mM Tris-HCl at pH 7.5 containing 0.15 M NaCl, and a protease inhibitors mix from Roche). After centrifugation, the supernatant was passed through a glutathione–sepharose column. The column was washed extensively with buffer A, and Pds1 was eluted by cleaving off the GST tag with enterokinase (dissolved in 50 mM Tris-HCl at pH 7.5 containing 1 mM DTT). Mass spectrometry was done by the Keck Foundation, Yale University (New Haven, CT).
Cell lysis and immunoprecipitation
Yeast cell lysates were prepared by breaking cells with glass beads buffer B (0.05 M Tris-HCl at pH 7.5, 0.15 M NaCl, 5 mM EGTA, 5 mM EDTA, 10 mM sodium pyrophosphate, and protease inhibitors mix from Roche). The extract (2 mg of protein) was incubated at 4°C for 2 h with anti-HA antibody prebound to protein A sepharose. The immunoprecipitates were washed four times with 1 mL of extraction buffer. Bound proteins were eluted by boiling in 2× SDS loading buffer. Proteins were separated by SDS-PAGE (10% gel) and analyzed by Western blot analysis as indicated.
Kinase assays
For radioactive kinase assays, 2.5 μg of purified Pds1 was mixed with 2.5 μL of yeast extract (4 μg of protein), 1 mM ATP, 50 μg/mL creatine kinase, 35 mM phosphocreatine, 1.3 μCi/μL [γ-32P]ATP, and 5 mM MgCl2 in buffer C (0.05 M Tris-HCl at pH 7.5, 0.1 M NaCl, 1.25 mM EGTA, 1.25 mM EDTA, protease inhibitors [Roche], and phosphatase inhibitors [Sigma]). The reaction was stopped by adding 2× SDS sample buffer. The yeast extracts used above were made from cells that were either left untreated or subjected to a radiation dose of 12 krad using a γ-irradiator.
Phosphatase treatment
Immunoprecipitates prepared as described above were resuspended in 50 μL of buffer (50 mM Tris-HCl at pH 7.5) containing 100 U of calf intestine phosphatase in the absence or presence of the phosphatase inhibitors mix (100× dilution of Cocktail 1 and 2 from Sigma). After incubation at 30°C for 30 min, the immunoprecipitated complexes were washed three times in lysis buffer B and analyzed by Western blot analysis.
Inhibition of the cdc28-as1 strain
Wild-type and cdc28-as1 strains (a generous gift from David Morgan and Jeff Ubersax, University of California, San Francisco) carrying plasmids encoding galactose-inducible Pds1-HA or Pds1db-HA were arrested in G1 at 23°C using α-factor mating pheromone. The cultures were then split into two parts; one was treated with the 10 μM final concentration of synthetic inhibitor PPI derivative (C 3-1-naphthylmethyl PPI [PPI derivative 9]; Bishop et al. 2000), and the other was mock-treated. The cultures were incubated at 23°C for 4.5 h followed by the galactose induction of Pds1-HA or Pds1db-HA for 1 h. Protein extracts were then prepared and examined by Western blot analysis using anti-HA antibodies.
Protein techniques
Protein extracts were prepared as described (Cohen-Fix et al. 1996).
Site-directed mutagenesis
The Quik-Change XL site-directed mutagenesis kit (Stratagene) was used to mutate serines/threonines in PDS1-HA to alanines, and the complete pds1 mutant allele was sequenced. The template used was pOC52. A ClaI–SacI fragment containing the mutagenized PDS1 with a neighboring URA3 marker was used to transform the pds1Δ strain, OCF1525.
Microscopy
Fluorescence and DIC microscopy were performed using an E800 Microscope (Nikon) linked to a cooled CCD camera. To visualize Pds1-HA or Esp1-Myc, cells were fixed for 20 min or 1 h with 4% formaldehyde at room temperature and processed as described (Cohen-Fix et al. 1996). The primary antibodies used were as described above. The nuclei were visualized by DAPI.
Acknowledgments
We thank Lee Hartwell, David Morgan, Jeff Ubersax, Mark Solomon, Steve Elledge, Philipp Kaldis, Yolanda Sanchez, and Peter Sheffield for providing us with yeast strains and plasmids. We are especially thankful to David Morgan and Jeff Ubersax for helpful advice, allowing us to cite their unpublished observations, and for sharing their very last bit of Cdc28-as1 inhibitor with us. We also thank April Robbins, William Jacoby, and Karen Ross for stimulating discussions during the course of the work; and April Robbins, Philipp Kaldis, Karen Ross, Joe Campbell, Melody Ng, Angelika Amon, David Morgan, Sue Biggins, and Steve Elledge for excellent comments on the manuscript. This work was funded by intramural NIDDK grant to O.C.F.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ornacf@helix.nih.gov; FAX (301) 402-0053.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.971402.
References
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A. Regulation of B-type cyclin proteolysis by Cdc28-associated kinases in budding yeast. EMBO J. 1997;16:2693–2702. doi: 10.1093/emboj/16.10.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O. Making and breaking sister chromatid cohesion. Cell. 2001;106:137–140. doi: 10.1016/s0092-8674(01)00439-1. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Pds1p of budding yeast has dual roles: Inhibition of anaphase initiation and regulation of mitotic exit. Genes & Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Jovanovic A, Jeoung DI, Bourlat S, Cross FR, Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28–Cln2 kinase, in vivo. Mol Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JK, Solomon MJ. Predictive scale for evaluating cyclin-dependent kinase substrates. A comparison of p34cdc2 and p33cdk2. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters J, Kinzler KW, Vogelstein B, Lengauer C. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jensen S, Segal M, Clarke DJ, Reed SI. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: Evidence that proper spindle association of Esp1 is regulated by Pds1. J Cell Biol. 2001;152:27–40. doi: 10.1083/jcb.152.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumada K, Nakamura T, Nagao K, Funabiki H, Nakagawa T, Yanagida M. Cut1 is loaded onto the spindle by binding to Cut2 and promotes anaphase spindle movement upon Cut2 proteolysis. Curr Biol. 1998;8:633–641. doi: 10.1016/s0960-9822(98)70250-7. [DOI] [PubMed] [Google Scholar]
- Leismann O, Herzig A, Heidmann S, Lehner CF. Degradation of Drosophila PIM regulates sister chromatid separation during mitosis. Genes & Dev. 2000;14:2192–2205. doi: 10.1101/gad.176700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gorbea C, Mahaffey D, Rechsteiner M, Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc Natl Acad Sci. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: Cutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28–Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- Ramos-Morales F, Dominguez A, Romero F, Luna R, Multon MC, Pintor-Toro JA, Tortolero M. Cell cycle regulated expression and phosphorylation of hpttg proto-oncogene product. Oncogene. 2000;19:403–409. doi: 10.1038/sj.onc.1203320. [DOI] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes & Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Ross KE, Kaldis P, Solomon MJ. Activating phosphorylation of the Saccharomyces cerevisiae cyclin-dependent kinase, cdc28p, precedes cyclin binding. Mol Biol Cell. 2000;11:1597–1609. doi: 10.1091/mbc.11.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piw-nica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- Srinivasan J, Koszelak M, Mendelow M, Kwon YG, Lawrence DS. The design of peptide-based substrates for the cdc2 protein kinase. Biochem J. 1995;309:927–931. doi: 10.1042/bj3090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and following DNA damage. Genes & Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Liu D, Wang Y, Qin J, Elledge SJ. Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes & Dev. 2001;15:1361–1372. doi: 10.1101/gad.893201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmann K, Benezra R. Mad2 transiently associates with an APC/p55Cdc complex during mitosis. Proc Natl Acad Sci. 1998;95:11193–11198. doi: 10.1073/pnas.95.19.11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996a;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996b;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: Cell division and the anaphase-promoting complex. Genes & Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sanchez RJ, Wang S, Guarnaccia C, Tossi A, Zahariev S, Pongor S. Substrate specificity of CDC2 kinase from human HeLa cells as determined with synthetic peptides and molecular modeling. Arch Biochem Biophys. 1994;315:415–424. doi: 10.1006/abbi.1994.1519. [DOI] [PubMed] [Google Scholar]
- Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]