Abstract
We investigated the regulation of the activin/nodal-inducible distal element (DE) of the Xenopus goosecoid (gsc) promoter. On the basis of its interaction with the DE, we isolated a Xenopus homolog of the human Williams-Beuren syndrome critical region 11 (XWBSCR11), and further, show that it interacts with pathway-specific Smad2 and Smad3 in a ligand-dependent manner. Interestingly, we also find that XWBSCR11 functions cooperatively with FoxH1 (Fast-1) to stimulate DE-dependent transcription. We propose a mechanism in which FoxH1 functions together with Smads as a cofactor for the recruitment of transcription factors like XWBSCR11 in the process of activin/nodal-mediated gsc-specific induction. This mechanism provides considerable opportunities for modulation of transcription across a variety of activin/nodal-inducible genes, increasing diversity in promoter selection, thus leading to the differential induction of activin/nodal target genes.
Keywords: Williams-Beuren syndrome, goosecoid, Smad, transforming growth factor, FoxH1, Fast1
Vertebrate embryogenesis proceeds through a series of inductive events in which signaling molecules produced by one cell population influence the developmental fate and morphogenesis of neighboring cells. Identifying these signaling molecules and understanding how they function is a primary goal of modern embryology. Through the use of amphibians as model systems, significant advances have been made in the understanding of mesoderm induction, one of the earliest inductive interactions in vertebrate development. From previous studies, we have learned that mesoderm induction requires the activity of the maternal transcription factor, VegT (Zhang et al. 1998). Depletion of maternal VegT results in diminished expression of members of the TGF-β superfamily, which, in turn, affects mesodermal patterning (Kofron et al. 1999). Activin and nodal family members of the TGF-β superfamily have been shown to initiate the formation of mesoderm in naïve, isolated animal cap explants, thereby mimicking the function of Spemann's organizer (Asashima et al. 1990; Thomsen et al. 1990; Jones et al. 1995). TGF-β superfamily members with this activity include Xnr1, Xnr2, Xnr4, Xnr5, and Xnr6, derriere, Vg1, and activin itself (Thomsen and Melton 1993; Jones et al. 1995; Joseph and Melton 1997; Takahashi et al. 2000). Questions have remained regarding how these activin-like molecules function in vivo to regulate mesoderm formation.
Activin and nodals (Xnrs) signal through a transmembrane serine/threonine kinase receptor complex composed of a type II receptor, ActRIIA or ActRIIB, and a type I receptor ActRIB (ALK4; for review, see Heldin et al. 1997; Massague and Chen 2000; Reissman et al. 2001). Signaling begins when a ligand binds to its cognate type II receptor, which then heterodimerizes and phosphorylates a distinct type I receptor. The activated type I receptor, in turn, phosphorylates receptor-regulated Smads, Smad2 or Smad3. The activated Smad forms a complex with Smad4 and translocates to the nucleus to regulate transcription. Although Smads can bind to DNA directly through a conserved region known as the MH1 domain and can activate gene transcription via the region known as the MH2 domain, interaction of Smads with DNA is relatively weak. Thus, existing evidence favors a model in which nuclear cofactors assemble with Smads to form a high-affinity complex with cognate DNA to affect target gene expression.
FoxH1 (also known as FAST-1), a member of the winged helix family of transcription factors, is the first transcription factor shown to interact with Smad2 and Smad4 to mediate activin signaling (Chen et al. 1996, 1997). FoxH1 was identified initially by its ability to bind to a 50-bp activin-response element located within the promoter region of the Xenopus Mix2 gene, a mesendoderm-specific gene (Huang et al. 1995; Chen et al. 1996). Further analyses with a dominant-negative form of FoxH1 or antibodies that specifically block FoxH1 function, suggest that FoxH1 is a central regulator of activin/nodal signals and is required for general mesoderm specification (Watanabe and Whitman 1999). Consistent with this notion, FoxH1-binding sites have been identified in the promoter region of several activin/nodal target genes such as goosecoid (gsc), Mix 2 and nodal-1 (Chen et al. 1996; Labbe et al. 1998; Osada et al. 2000). A dominant-negative FoxH1 was shown to block activin-mediated induction of gsc, brachyury, and Mix2, presumably by binding to the FoxH1 sites located within their promoters (Watanabe and Whitman 1999). Recent analysis of FoxH1 mutant mice has also indicated that FoxH1 is the major transcriptional inducer of nodal signaling in early mouse development. (Hoodless et al. 2001; Yamamoto et al. 2001).
Gsc is an organizer-specific gene whose transcriptional regulation has been well characterized (Watabe et al. 1997; Laurent et al. 1997; Germain et al. 2000). Gsc expression is initially detected in the organizer, and its transcription continues until neurula stage in the prechordal plate mesoderm. The transcriptional regulatory region of gsc contains both distal (DE) and proximal (PE) control elements whose importance for proper expression of gsc within the organizer has been demonstrated (Watabe et al. 1995) and has led to the suggestion that the regulation of gsc transcription in many ways reflects the regulation of mesoderm induction. The DE consists of a 29-bp response element that is activated specifically by activin or activin-like signals in the presence of cyclohexamide (Watabe et al. 1995). The DE responds not only to endogenous activin-like signals in Xenopus, but also to nodals and BVg1 (Watabe et al. 1995; M. Hashimoto and K.W.Y. Cho, unpubl.). The DE is highly conserved among the mouse, Xenopus, and zebrafish gsc promoters (Watabe et al. 1995; Danilov et al. 1998; McKendry et al. 1998). This observation suggests that the activin/nodal signaling pathways utilized in the formation and patterning of mesoderm have been conserved widely across the vertebrate order and underscores the importance of understanding the nature of gsc regulation as a means to elucidate the molecular mechanisms involved during mesoderm induction. Interestingly, sequence comparison of the 29-bp gsc DE and the 50-bp Mix2 activin-responsive element does not reveal any significant identity between these two elements despite the fact that both respond directly to activin (Watabe et al. 1995; Chen et al. 1996). Dissimilarity of the activin-responsive elements between gsc and Mix2 begs the question of the nature of the underlying molecular mechanisms governing activin/nodal-mediated gene activation.
Previous studies suggested that DE-mediated gsc regulation might be controlled by two different transcription factors, a maternal protein that interacts with DE until the midblastula transition and a second protein that binds from the onset of gastrulation (McKendry et al. 1998). According to this model, gsc is initially induced by a maternal transcription factor and subsequently the expression is maintained via Mixer/Milk in response to zygotic activin/nodal signal (McKendry et al. 1998; Germain et al. 2000).
Here we identify a Xenopus homolog of the human gene Williams-Beuren syndrome (WBS) critical region 11 (WBSCR11), as a critical factor in the phenomenon of DE-mediated activin signaling (Franke et al. 1999; Osborne et al. 1999). WBS is a complex, multisystemic developmental disorder characterized by distinctive facial features, mental disability, and growth retardation (Franke 1999; Osborne et al. 1999). The WBSCR11 gene has been mapped to the breakpoint region known to cause WBS. The Xenopus homolog of WBSCR11 (XWBSCR11) is expressed during early Xenopus embryogenesis, at the right time and place during development to affect gsc transcription. Here, we demonstrate that XWBSCR11 forms a ligand-dependent complex with Smad2 or Smad3, and selectively affects the transcription of gsc as revealed by both gain-of-function and loss-of-function analyses. Additionally, we have found that XWBSCR11 can interact directly with FoxH1, thereby influencing the transcription of the DE-mediated activin/nodal signaling in a cooperative manner. These observations led us to propose a model for activin/nodal-mediated signaling in which differential recruitment of cofactors such as XWBSCR11 together with Smads and FoxH1 modulates the transcriptional activities and specificities of a variety of activin/nodal target genes.
Results
Cloning of a Xenopus gsc activin-response element (DE) binding protein
Study of the regulation of gsc gene expression has provided a valuable tool in the analysis of the mechanisms of TGF-β signaling by activin/nodal-related factors (Watabe et al. 1995; Labbe et al. 1998; Germain et al. 2000). The DE of the gsc gene has emerged as a key regulatory element in the transduction of activin/nodal signals. We have made use of the yeast one-hybrid system to identify a transcription factor mediating the regulation of the DE. A Xenopus oocyte cDNA library fused to the GAL4 transcriptional activation domain was transformed into a yeast strain bearing a multimerized form of the DE upstream of HIS3 and lacZ reporter genes. A total of 11 positive clones were identified, among which eight clones showed strong homology to the human gene WBSCR11 also identified as GTF2IRD1 (Franke et al. 1999; Osborne et al. 1999). A full-length cDNA with an ∼3-kb ORF was subsequently isolated from a Xenopus oocyte library. XWBSCR11 is a 994-amino-acid protein that contains five highly conserved helix–loop–helix (HLH) containing repeats, and a carboxy-terminal serine rich region (Fig. 1). WBSCR11 is thought to be a transcription factor, which may contribute to the spectrum of developmental symptoms found in WBS (Osborne et al. 1999; Franke et al. 1999). WBS is a multisystemic developmental disorder caused by the hemizygous deletion of an unspecified number of genes on chromosome 7q11.23. The HLH repeats of both XWBSCR11 and WBSCR11 bear homology to those found in the human protein TFII-I, suggesting they are members of a family of proteins (Grueneberg et al. 1997; Osborne et al. 1997; Roy et al. 1997; Bayarsaihan and Ruddle 2000).
Figure 1.
Structural analysis of XWBSCR11. (Top) Amino acid alignments between Xenopus and human WBSCR11. These repeats show strong homology to domains found in a family of transcription factors related to TFII-I, especially the human protein WBSCR11. The five HLH domains are underlined in gray, and the carboxy-terminal serine-rich region is double underlined. (Bottom) The five HLH domain repeats are highlighted as hatched boxes. A comparison of WBSCR11 and XWBSCR11 is shown with percent similarity between domains indicated. (Dark hatched boxes) Hydrophobic zipper-like domains are also conserved between XWBSCR11 and TFII-I related family members. The GenBank accession no. for XWBSCR11 is 446344.
Expression analysis of XWBSCR11
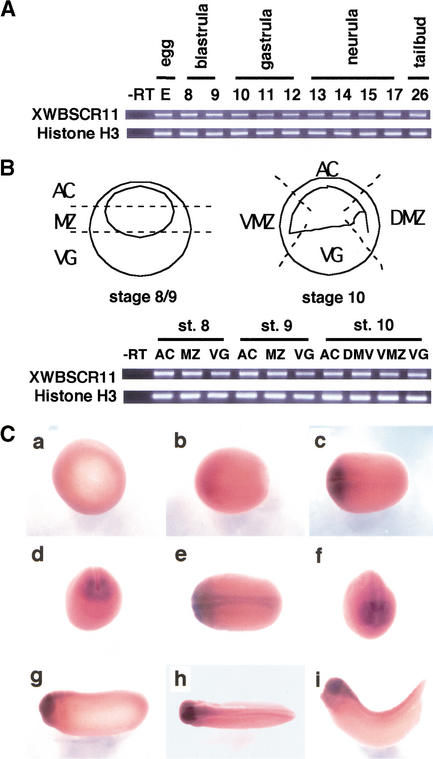
Developmental RT–PCR analyses demonstrate that XWBSCR11 is expressed both maternally and zygotically at relatively consistent levels throughout Xenopus development (Fig. 2A). XWBSCR11 is expressed uniformly throughout the embryo in pre- and early gastrula stages, as revealed by RT–PCR analysis of tissue samples (Fig. 2B). Although the early ubiquitous expression of XWBSCR11 is not apparent by whole mount in situ hybridization analysis, XWBSCR11 expression can be detected in the early neurula through tailbud stages and is enriched in the head region (Fig. 2C). These expression patterns are consistent with a potential role of XWBSCR11 in the early and late expression of gsc in vertebrates (Blum et al. 1992; Gaunt et al. 1993; Stachel et al. 1993; Thisse et al. 1994).
Figure 2.
Expression analysis of XWBSCR11 during Xenopus development. (A) RT–PCR analysis with RNA isolated from whole embryos demonstrates the expression of XWBSCR11 throughout Xenopus embryogenesis. (B) RT–PCR of tissue explants from blastula (stage 8/9) and early gastrula (stage 10) embryos. The schematic diagram shows the location of the dissection for the various tissue explants. XWBSCR11 does not appear to be differentially expressed between animal cap (AC), marginal zone (MZ), or vegetal pole (VG) tissues in blastula and gastrula embryos, nor between dorsal (DMZ) and ventral marginal zone (VMZ) in gastrula embryos. RT–PCR of histone H3 is shown as a loading control in both A and B. (C) Whole mount in situ analysis of XWBSCR11 expression. (a) Gastrula stage; (b–g) neurula stages; (h–i) tailbud stages. In a–c, e, and g–i, anterior is left.
XWBSCR11 binds to the gsc DE in vitro
To confirm the DNA binding specificity of XWBSCR11 for the gsc DE, a series of GST-XWBSCR11 fusion proteins was generated and used for electrophoretic mobility shift analysis (EMSA) along with radiolabeled oligonucleotides for both wild-type and a mutant form of the DE. The XWBSCR11 protein was subdivided into various fragments and fused to GST. GST fusion proteins containing either repeats 1–2 (GST-XWBSCR11 R1–2), repeats 2–3 (GST-XWBSCR11 R2–3), or repeats 4–5 (GST-XWBSCR11 R4–5) were purified following expression in bacteria (data not shown). For EMSA, three wild-type oligonucleotide probes corresponding to the full-length 29-bp DE (D29), a 17-bp (5′ portion, D17), and a 12-bp (3′ portion, D12) of the DE were used as well as a mutated version of D17 (M4) (Fig. 3). In our study, mutation of the 17-bp region within the DE (corresponding to the M4 mutant) was shown to abolish activin responsiveness in reporter gene assays (T. Watabe and K.W.Y. Cho, unpubl.). In the case of all three GST-XWBSCR11 fusion proteins, mobility of the DNA probe was observed only with the D29 and D17 oligonucleotides, and not with the D12 and M4 mutant forms, demonstrating the specificity of XWBSCR11 for the DE, and suggesting further that the binding of XWBSCR11 to the DE occurs in the 17-bp region. Specific interaction of XWBSCR11 with D17 was also confirmed in the yeast one-hybrid assay using a multimerized form of the D17 fused to the HIS3 and lacZ reporter genes (data not shown).
Figure 3.
Formation of a complex between bacterially expressed XWBSCR11 protein and the gsc DE (activin response element) revealed by EMSA. Truncated GST-XWBSCR11 fusion proteins containing repeats 1 and 2 (GST-XWBSCR11 R1–2), repeats 2 and 3 (GST-XWBSCR11 R2–3), or repeats 4 and 5 (GST-XWBSCR11 R4–5) were generated and used for EMSA with the full-length, 29-bp gsc DE (D29), as well as a 17-bp 5′ version (D17), a 12-bp 3′ version (D12), and a mutant form of D17 (M4). In the case of all three GST-XWBSCR11 fusion proteins, mobility retardation of the DNA probe was observed only with the D29 (DE) and D17 oligonucleotides and not with the D12 and M4 mutant forms, demonstrating the specificity of XWBSCR11 for the DE and suggesting further that the binding of XWBSCR11 to the DE occurs in the 17-bp region. (Bottom) Sequences of the gsc DE oligonucleotides used in this study. Mutations in the 17-bp portion of the DE used to generate the M4 oligonucleotide, which abolished activin responsiveness in reporter assays, are represented as lower case characters. M4 mutant oligos compete poorly for XWBSCR11 fusion protein binding to the DE (data not shown).
Interestingly, these data also suggest that the carboxy-terminal region of the XWBSCR11 protein possesses stronger binding to the DE, as GST-XWBSCR11 R4–5 as compared with GST-XWBSCR11 R1–2 and GSTXWBSCR11 R2–3 shifted significantly more oligonucleotides in the EMSA, despite the fact that an equivalent amount of fusion proteins is used for each experiment. This result is interesting, as it suggests a mode whereby XWBSCR11 interacts with the DE of the gsc gene and that each of the HLH repeats may have different activities.
A constitutively active form of XWBSCR11 activates a DE-containing reporter gene whereas a repressive form of XWBSCR11 attenuates activin-dependent activation
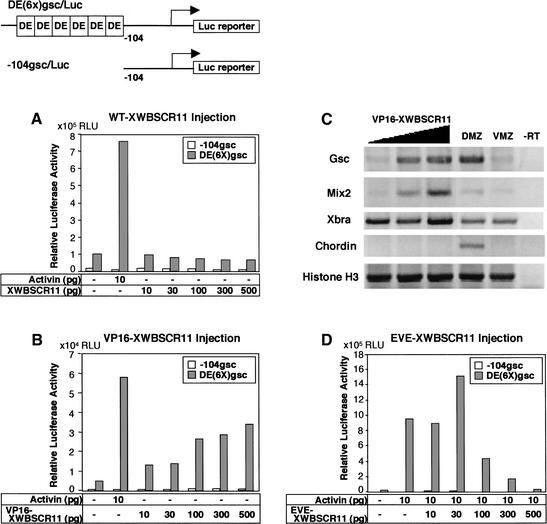
To examine the effect of XWBSCR11 on gsc transcription via the DE, the animal pole (prospective ectoderm) of two-cell stage Xenopus embryos was microinjected with mRNA encoding wild-type XWBSCR11 (WTXWBSCR11), a VP16-fusion protein of XWBSCR11 (VP16-XWBSCR11), or an even-skipped fusion protein of XWBSCR11 (EVE-XWBSCR11), with a reporter gene containing a multimerized form of the DE [DE(6×)gsc/Luc]. The VP16 fusion protein is a constitutively active form, generated by the fusion of the full-length XWBSCR11 protein to the VP16 transcriptional activator domain from herpes simplex virus (Sadowski et al. 1988). We assayed the ability of this construct to activate the reporter gene in animal cap ectoderm. Whereas WT-XWBSCR11 failed to induce the DE(6×)gsc/Luc reporter construct (Fig. 4A), VP16-XWBSCR11 activated DE(6×)gsc/Luc in an activin-independent, dose-dependent manner (Fig. 4B). This activation is DE dependent as a control reporter construct (−104gsc/Luc containing only a 104-bp promoter fragment of gsc lacking the DE, remains unaffected by the VP16-XWBSCR11 fusion protein (Fig. 4B). These data suggest that XWBSCR11 binding to the DE results in activation of the gsc gene, consistent with its role as a potential activator of gsc transcription. Furthermore, the lack of transcriptional activation of the DE by simple overexpression of WT-XWBSCR11 suggests that pathway activation is required for DE responsiveness, presumably by either eliciting post-translational modification of the XWBSCR11 protein, or perhaps interaction with an activated Smad member, or interaction with another factor. The VP16 fusion is able to bypass this requirement. To examine the specificity of VP16-XWBSCR11 on endogenous gene expression, we overexpressed the protein in the ventral marginal zone region and examined the expression of various dorsal marker genes. VP16-XWBSCR11 was able to induce the expression of gsc and mix2, but the expression of Xbra and histone H3 was unaffected, and chordin was not induced, suggesting that VP16-XWBSCR11 is capable of specifically activating a subset of organizer-specific genes (Fig. 4C).
Figure 4.
Injection of full-length wild-type (WT-XWBSCR11), as well as constitutively activating (VP16-XWBSCR11) and repressing (EVE-XWBSCR11) forms of XWBSCR11 with the DE(6×)gsc/Luc promoter reporter constructs. (A) WT-XWBSCR11 is unable to activate a gsc ARE reporter construct [DE(6×)gsc/Luc], whereas activin stimulates this reporter. A control reporter construct (−104gsc/Luc) lacking the ARE does not respond to activin or WT-XWBSCR11. (B) A fusion protein containing the VP16 activation domain of the herpes simplex virus fused to XWBSCR11 (VP16-XWBSCR11) is able to activate DE(6×)gsc/Luc in an activin-independent, dose-dependent manner, whereas the −104gsc/Luc control reporter remains unaffected. VP16 alone did not activate DE(6×)gsc/Luc (data not shown). (C) RT–PCR analysis of endogenous genes that are activated by VP16-XWBSCR11. Note that whereas VP16-XWBSCR11 can activate both gsc and Mix2 genes, it is unable to affect the transcription of Xbra, chordin, and histone H3. (D) A fusion protein containing the transcriptional repressor domain from Drosophila eve was fused to XWBSCR11 (EVE-XWBSCR11). EVE-XWBSCR11 repressed the activin-dependent induction of DE(6×)gsc/Luc, whereas the −104gsc/Luc control reporter remained unaffected. Eve alone did not repress the induction of DE(6×)gsc/Luc (data not shown). RNAs were injected during the two-cell stage at the given concentrations. Animal caps were harvested at stage 8 (blastula), and assayed at stage 10.5 (early gastrula). In all cases, promoter constructs were coinjected at a concentration of 160 pg/embryo.
The even-skipped fusion protein is a transcriptionally repressive form, generated by the fusion of the full-length XWBSCR11 protein to the transcriptional repressor domain from Drosophila eve (Onichtchouk et al. 1996). Expression of EVE-XWBSCR11 alone had no effect on the activity of the DE(6×)gsc/Luc reporter (data not shown). However, EVE-XWBSCR11 reduced the activation of the DE(6×)gsc/Luc reporter gene by activin in a dose-dependent manner (Fig. 4D). Eve-XWBSCR11 had no effect on the −104gsc/Luc control reporter (Fig. 4D). This result lends further support to the hypothesis that XWBSCR11 binds to the DE and positively affects gsc transcription.
XWBSCR11 morpholinos block endogenous gsc gene expression
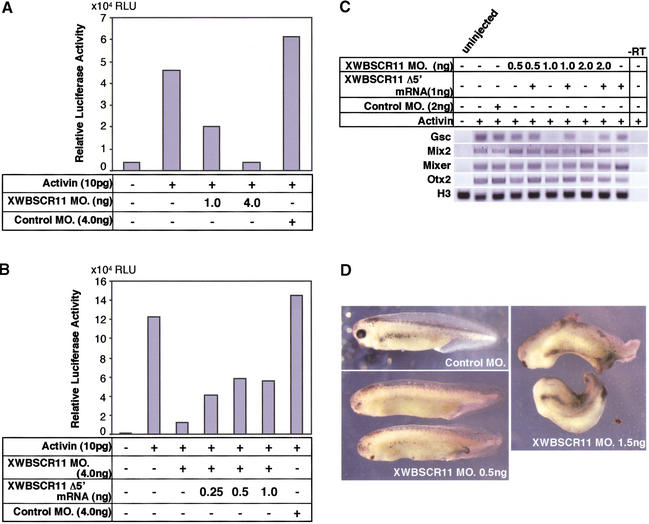
We wanted to determine the effect of blocking XWBSCR11 protein expression on the activin/nodal-dependent activation of the DE(6×)gsc/Luc reporter construct, as well as on the expression of the endogenous gsc gene. We made use of antisense morpholino oligonucleotides, which act by preventing translation of protein (Summerton and Weller 1997; Heasman et al. 2000), directed against a specific 25-bp region of the 5′ end of the XWBSCR11 cDNA (see Materials and Methods), as an alternative loss-of-function approach to examine the effect of blocking endogenous XWBSCR11 expression. XWBSCR11 morpholino oligonucleotides were coinjected with activin mRNA into the animal pole region of Xenopus embryos as above. Injection of XWBSCR11 morpholinos abolished activin responsiveness of the DE(6×)gsc/Luc reporter gene in a dose-dependent manner, whereas injection of a control morpholino oligonucleotide had no effect (Fig. 5A). Further, this effect could be partially rescued by the injection of a wild-type form of XWBSCR11 lacking a portion of the 5′ sequence (Δ5′-XWBSCR11) corresponding to the region recognized by the morpholino oligonucleotide (Fig. 5B). The fact that the rescue was incomplete may be due to the reduced activity of the Δ5'-XWBSCR11 missing 21 amino acids at the amino-terminal region. We interpret these results as demonstrating that blocking endogenous XWBSCR11 expression results in failure to stimulate the activin-dependent DE(6×)gsc/Luc reporter gene activation, which is in agreement with the overexpression study using the EVE-XWBSCR11 repressor fusion.
Figure 5.
Effects of XWBSCR11 morpholinos on reporter gene activity and endogenous gene expression. (A) XWBSCR11 morpholinos were coinjected with activin (4 pg/embryo) and the DE(6×)gsc/Luc reporter gene into animal caps at the two- to four-cell stage, then assayed for luciferase activity at stage 10.5. The XWBSCR11 morpholino specifically abolished activin responsiveness of the reporter gene, as a control morpholino similarly injected had no effect. (B) This effect was rescued by the injection of a wild-type form of XWBSCR11 lacking a portion of the 5′ sequence corresponding to the region bound by the morpholino. (C) XWBSCR11 morpholino (0.5, 1, and 2 ng/embryo) was coinjected with activin mRNA (4 pg/embryo) in animal caps and assayed for gene expression. gsc expression decreased with increasing XWBSCR11 morpholino concentration, whereas levels of histone H3 appeared constant and Mix2, Mixer and Otx2 expression was unaffected. However, injection of XWBSCR11 morpholino oligonucleotides at concentrations >8 ng per embryo inhibited Mix2 expression (data not shown). A control morpholino had no effect. (D) WXBSCR11 morpholino-injected (0.5 ng/embryo) embryos had disrupted anterior structures whereas they appeared to go through normal gastrulation movements. At higher concentrations (1.5 ng), head truncation was more prominent and gastrulation movements were affected. Control morpholino-injected embryos were normal.
Injection of XWBSCR11 morpholino oligonucleotide was also used to examine the effects on endogenous gsc expression by RT–PCR analysis. Increasing amounts of XWBSCR11 morpholino oligonucleotides were coinjected with activin mRNA into the animal pole region of two-cell stage Xenopus embryos. RNA samples were isolated from animal caps at the blastula stage, and subjected to RT–PCR analysis. We found a concomitant decrease in endogenous gsc expression, whereas levels of Mix2, Otx2, Mixer, and Histone H3 appeared relatively constant (Fig. 5C). Furthermore, the inhibition of endogenous gsc was rescued by coinjection of Δ5'-XWBSCR11 lacking the morpholino annealing site. These data support and extend the observation that XWBSCR11 serves to bind to the DE to positively affect gsc, but not Mix2, transcription in vivo. Interestingly, when XWBSCR11 morpholino injection was targeted to the organizer, we observed only partial inhibition of endogenous gsc expression (data not shown). The failure of the XWBSCR11 morpholino to completely block organizer gsc expression while inhibiting activin-induced gsc expression in animal caps may be due to the fact that the organizer gsc activity is not only regulated by activin-like signals, but also by maternal Wnt-like signals and zygotic transcription factors such as Xlim, Mixer, and Milk (Germain et al. 2000; Karavanov et al. 2000).
We also examined the phenotypes of XWBSCR11 morpholino-injected embryos. XWBSCR11 morpholino was injected into the marginal zone of four-cell stage embryo. The injected embryos appeared to go through normal gastrulation movements, but displayed defects in the head region at later stage (Fig. 5D). Interestingly, the rest of the axial structures and mesoderm patterning was not affected at lower concentrations. The head defects observed in XWBSCR11 morpholino-injected embryos were not caused by general defects in gastrulation movements, which one often observes in various overexpression assays, but were specific XWBSCR11 knockdown phenotypes. First, XWBSCR11 is expressed in the head region of neurula stage embryos, thus, depletion of XWBSCR11 protein in embryos is expected to cause defects in the head. Second, the phenotypes were obtained with as little as 0.5 ng of XWBSCR11 morpholinos, a concentration believed to be nontoxic to embryos. From these experiments, we conclude that XWBSCR11 is an endogenous transcription factor involved in activin/nodal-mediated gsc regulation.
XWBSCR11 binds to Smad2 and Smad3 in an activin-dependent manner
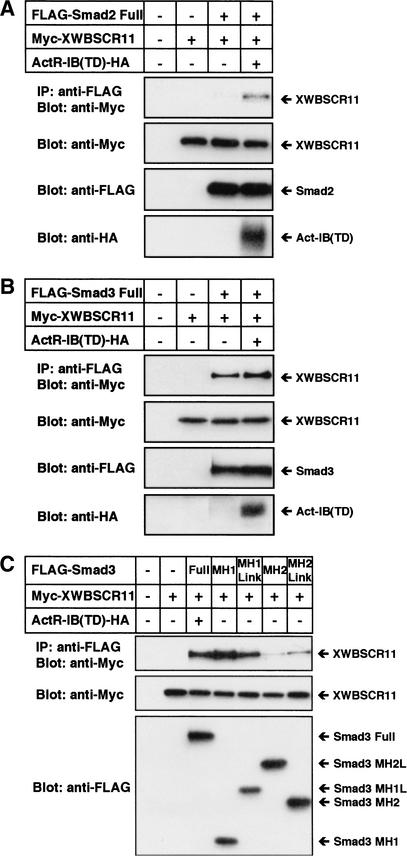
Activin/nodal-type TGF-β signals are transduced from activated receptors to the nucleus via a complex formed by both Smad2 and Smad4. In the case of the Mix2 gene, FoxH1 is able to form a complex with Smad2/Smad4 by interacting with Smad2 via its carboxy-terminal Smad-interaction domain (Chen et al. 1996, 1997). We examined whether an analogous interaction occurs between XWBSCR11 and Smads. Cos7 cells were transfected in the presence and absence of an epitope-tagged form of a constitutively active activin receptor [ActR-IB(TD)–HA], together with either mouse Smad2 or Smad3 tagged with the Flag epitope, and full-length, Myc-tagged XWBSCR11. Extracts isolated from the cells were immunoprecipitated with anti-Flag antibodies, followed by Western blot analysis with anti-Myc antibodies. XWBSCR11 was able to interact with Smad3 but not Smad2 in the absence of receptor, although the interaction appeared to be weak (Fig. 6A,B). In the presence of activated ActR-IB receptor, XWBSCR11 interacted strongly with Smad3. Interaction of XWBSCR11 and Smad2 was only detected when activated ActR-IB was present. Next, we determined which domain of Smad3 interacts with XWBSCR11. As shown in Figure 6C, the MH1 domain of Smad3 preferentially interacts with XWBSCR11. Similarly the MH1 domain of Smad2 interacts with XWBSCR11 (data not shown). These data support a model whereby an activated pathway-specific Smad interacts with a transcriptional partner XWBSCR11 in a ligand-dependent manner, to affect transcription of gsc. It should be noted that XWBSCR11 does not contain a Smad-interaction motif that was shown to be critical for interaction between Smads and Mixer, Milk, and FoxH1 transcription factors (Germain et al. 2000).
Figure 6.
XWBSCR11 binds to Smad2 and Smad3 in an activin-dependent manner. (A) Cos7 cells were transiently transfected in the presence and absence of an HA-tagged, constitutively active activin receptor [ActR-IB(TD)–HA] with Flag-tagged Smad2 and full-length Myc-tagged XWBSCR11. Following immunoprecipitation (IP) with anti-Flag antibodies, and Western analysis with anti-Myc antibodies, XWBSCR11 showed interaction with Smad2 in the presence of activated receptor. (B) Cos7 cells were transfected as in A, but with Flag-tagged Smad3 rather than Smad2. Following IP and Western analysis, XWBSCR11 showed interaction with Smad3 in the presence and absence of activated receptor. The interaction was stronger in the presence of the activated receptor. (C) Cos7 cells were transfected with various combinations of ActR-IB(TD)–HA and Myc-tagged XWBSCR11 in the presence of Smad3 deletion constructs. The MH1 domain of Smad3 interacts with XWBSCR11.
XWBSCR11 interacts physically with Xenopus FoxH1
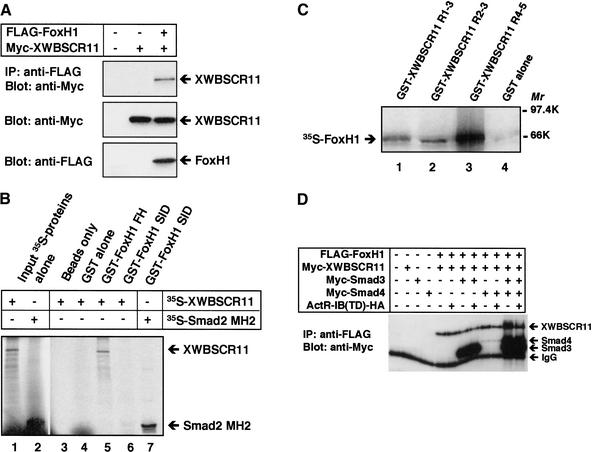
Several lines of evidence suggest that FoxH1 transcription factors, members of the forkhead-containing family, play a role in the induction of not only the Mix.2 gene, but also other activin/nodal response genes, including gsc and nodal (Labbe et al. 1998; Watanabe and Whitman, 1999; Osada et al. 2000). To understand the mechanisms whereby XWBSCR11 could interact with Smads, and possibly FoxH1 to affect activin/nodal signaling, we sought to determine whether a physical interaction between FoxH1 and XWBSCR11 could occur. Lysates from Cos7 cells transfected with full-length, Myc-tagged XWBSCR11 together with Flag-tagged FoxH1 were immunoprecipitated with anti-Flag antibodies. These experiments revealed that FoxH1 can interact physically with XWBSCR11 (Fig. 7A).
Figure 7.
XWBSCR11 interacts physically with Xenopus FoxH1 (Fast1). (A) Immunoprecipitation with anti-Flag antibodies of Cos7 cells transiently transfected with full-length, Myc-tagged XWBSCR11 together with Flag-tagged FoxH1 reveals the activin-independent interaction of XWBSCR11 and FoxH1. (B) GST-FoxH1 FH or GST-FoxH1 SID were used for immunoprecipitation of full-length in vitro translated 35S-labeled XWBSCR11. XWBSCR11 interacts preferentially with the FH domain of FoxH1 in vitro. 35S-labeled Smad2 MH2 is capable of interacting with GST-FoxH1 SID as expected, demonstrating that this protein remains active. (C) GST-XWBSCR11 R1–2, GST-XWBSCR11 R2–3, and GST-XWBSCR11 R4–5 were used for IP of full-length, in vitro-translated 35S-labeled FoxH1. All three GST-XWBSCR11 fusion proteins were able to interact with FoxH1, but the interaction appeared particularly robust between FoxH1 and GST-XWBSCR11 R4–5. (D) Interaction of WBSCR11 and Fox1. Cos7 cells were transfected with the indicated constructs. Cell lysates were subjected to an anti-Flag immunoprecipitation, followed by immunoblotting with anti-Myc antibodies.
Previous reports have indicated that FoxH1 interacts with Smad2 via a carboxy-terminal Smad-interacting domain (SID; Chen et al. 1997; Germain et al. 2000). We wondered which particular subdomain of FoxH1 might interact with XWBSCR11. The forkhead and the SID domains of Xenopus FoxH1 were expressed in bacteria as GST fusion proteins and used for immunoprecipitation of in vitro-translated full-length 35S-labeled XWBSCR11. We found that the forkhead, but not the SID domain of FoxH1 was able to interact with XWBSCR11 in vitro (Fig. 7B).
The reciprocal experiment was also performed to determine the region of XWBSCR11 that interacts with full-length 35S-labeled FoxH1. XWBSCR11 contains five highly conserved HLH repeats proposed to function in DNA binding. We performed coimmunoprecipitation analysis using the GST fusion proteins (GST-XWBSCR11 R1–2, GST-XWBSCR11 R2–3, and GST-XWBSCR11 R4–5) that were used previously for gel shift analysis (Fig. 7C). Although all three fusion proteins were able to interact physically with in vitro translated full-length 35S-labeled FoxH1 in this assay, the interaction with GST-XWBSCR11 R4–5 was particularly robust. Coincidentally, this region was also identified as the domain that had the strongest binding to the DE by EMSA (Fig. 3).
Then, we examined whether Smad2 or Smad3 can affect the complex formation between XWBSCR11 and FoxH1. Cos7 cells were cotransfected with various combinations of Flag–FoxH1, Myc–XWBSCR11, Myc–Smad3, Myc–Smad4, and ActR-IB(TD). Cell lysates were subjected to an anti-Flag immunoprecipitation, and coprecipitating XWBSCR11 was detected by immunoblotting with anti-Myc antibodies. The amount of precipitating XWBSCR11 did not change significantly throughout the experiment suggesting that Smad3 does not affect the complex formation between XWBSCR11 and Fox1 (Fig. 7D). Similar experiments were carried out for Smad2, and essentially identical results were obtained (data not shown). Together these results suggest a potential mechanism whereby a complex containing XWBSCR11, Smads, and FoxH1 interact to affect DE-mediated transcription of gsc.
XWBSCR11 and Fast-1-binding sites map to distinct locations on the Xenopus gscpromoter
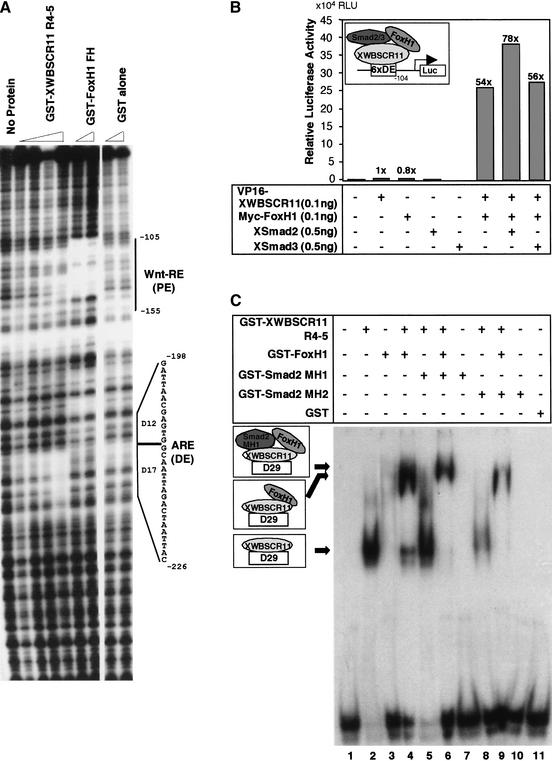
XWBSCR11 was able to physically interact with FoxH1 in Xenopus, and FoxH1 has been shown to affect the transcription of mouse gsc via an element downstream of the mouse DE (Labbe et al. 1998). Therefore, we thought to determine the precise location of XWBSCR11 and FoxH1 binding sites in the Xenopus gsc promoter using DNase I footprint analysis. GST fusion proteins containing the FoxH1 forkhead domain and XWBSCR11 repeats 4–5 were purified from bacteria and used for footprint analysis of the −309 Xenopus gsc promoter.As shown in Figure 8A, XWBSCR11 binds to the DE primarily in the 17-bp region, and the FoxH1 forkhead domain binds to a region corresponding to the 3′ end of the previously identified proximal element (PE), shown to respond to Xtwin/Xsia-mediated Wnt signals (Laurent et al. 1997). These results demonstrate that XWBSCR11 and FoxH1 bind two distinct regions within the gsc promoter to regulate the expression of gsc.
Figure 8.
Binding of XWBSCR11 and FoxH1 to the gsc promoter. (A) Bacterially expressed GST-XWBSCR11 R4–5 and GST-FoxH1 FH were used for footprint analysis of the −309 gsc promoter. XWBSCR11 R4–5 bound to the promoter primarily in the 17-bp DE, and FoxH1 FH bound to the 3′ end of the previously identified proximal element (PE). The PE has been shown previously to respond to Xtwin/Xsia-mediated Wnt signals. (B) XWBSCR11 and Xenopus FoxH1 synergistically activate transcription of the DE(6×)gsc/Luc reporter. Injection of either VP16-XWBSCR11 or a Myc-tagged Fast1 alone did not activate the DE(6x)gsc/Luc reporter. However, coexpression of Myc-tagged FoxH1 together with VP16-XWBSCR11 resulted in significant reporter gene activation, reaching a maximum induction level of 54-fold. The synergistic effect of FoxH1 and XWBSCR11 was not affected by the presence of Smad2 or Smad3. (C) Bacterially expressed GST-XWBSCR11-R4-5, GST-FoxH1, and various GST-Smads were used to perform an EMSA with DE oligonucleotides. (Arrows) Presumed ternary complexes among these proteins and DNA.
Interaction of XWBSCR11 and Xenopus Fast-1 cooperatively activates transcription of the DE(6×)gsc/Luc reporter gene
Previous reports have indicated that a VP16-FoxH1 fusion protein activates gsc expression in vivo (Watanabe and Whitman 1999). Because our DNase I footprint analysis indicates that the FoxH1-binding site in the gsc promoter is independent of that of XWBSCR11, we sought to understand the nature of the activation of gsc by XWBSCR11 and FoxH1, using the DE(6×)gsc/Luc reporter as a model. Injection of either VP16-XWBSCR11 or a Myc-tagged FoxH1 alone does not activate the DE(6×)gsc/Luc reporter gene (Fig. 8B). However, when Myc-tagged FoxH1 is coexpressed together with VP16-XWBSCR11, an increase in reporter activation was observed, reaching a maximum induction level of 54-fold. The results suggest that FoxH1 can synergistically cooperate with XWBSCR11 to activate DE-mediated activin signaling in vivo. Coinjection of Smad2 or Smad3 together with FoxH1 and VP16-XWBSCR11 did not significantly affect the activity of DE-mediated activin signaling. This result is interesting as work by Labbe et al. (1998) had shown previously that Smad3 can negatively affect gsc transcription. This discrepancy may be due to the fact that, in our experiments, the promoter construct [DE(6×)gsc/Luc] used does not contain a FoxH1-binding site, whereas in the context of the entire gsc promoter, including the FoxH1-binding site, differential regulation by Smad2 and Smad3 may occur. Together, the data demonstrate that XWBSCR11 and FoxH1 bind to separate regions of the Xenopus gsc promoter, and further, that they are able to interact physically and suggest that, in so doing, synergistically cooperate to activate gsc transcription.
To support this notion further, we examined whether FoxH1 and XWBCR11 can form a complex on the DE using an electrophoretic mobility shift assay (EMSA). As shown in Figure 8C, full-length XWBSCR11 alone binds to DE and the XWBSCR11–DE complex is further shifted in the presence of the GST-FoxH1 fusion protein (cf. lanes 2 and 4) to generate a XWBSCR11–FoxH1–DE complex. The XWBSCR11–FoxH1–DE complex is further shifted in the presence of Smad2–MH1 fusion protein (lane 6) indicating that these proteins are capable of forming a ternary complex. As Smad2–MH1 alone is incapable of binding to DE (lane 7), we suggest that Smad2 binding to FoxH1–XWBSCR11–DE is mediated via XWBSCR11.
Discussion
WBS is caused by the heterozygous deletion of genes on chromosome 7q11.23. Human WBSCR11 was identified as a gene that resides within the WBS deletion and is thought to contribute to the spectrum of developmental symptoms found in WBS (Franke et al. 1999; Osborne et al. 1999). Both human WBSCR11 and its Xenopus ortholog XWBSCR11, identified here, contain HLH repeats that bear similarity to those found in the transcription factor, TFII-I, leading to the suggestion that XWBSCR11 functions as a transcription factor. Here, we show that XWBSCR11 functions in DNA binding via selective HLH repeats and interacts with the transcription factor FoxH1 and the signal transduction molecules Smad2 and Smad3 to affect activin/nodal-mediated induction of the DE of the gsc gene.
XWBSCR11 was identified by a yeast one-hybrid screen for proteins that were capable of binding to and activating the activin/nodal response element (DE) of the gsc gene. Characterization of the XWBSCR11 protein demonstrated that it was the Xenopus ortholog of human WBSCR11 and mouse BEN, a transcription factor thought to be involved in the regulation of HoxC8 (Bayarsaihan and Ruddle 2000). The HLH repeat domains of XWBSCR11 bear similarity to those found in the pyrimidine-rich initiator (INR)-binding protein TFII-I (Roy et al. 1997), suggesting that these are members of a family of HLH repeat-containing transcription factors with a broad range of target genes. XWBSCR11 is ubiquitously expressed maternally and shows a more restricted zygotic expression pattern, making it a potential candidate both temporally and spatially as a regulator of transcription of the gsc gene and potentially other mesoderm response genes. This expression is important, as it has been reported that Mix/Mixer family proteins regulate the expression of DE-mediated gsc expression during mesoderm formation by forming a complex with Smad2 and Smad4 in response to activin-like signals (Germain et al. 2000). Although Mix/Mixer proteins bind to the DE in early Xenopus embryos, these transcription factors are not expressed maternally. The broad maternal distribution of XWBSCR11 suggests that it may be able to interact with widely distributed factors in the early embryo and may be capable of integrating signals from multiple pathways to affect transcription of target genes.
Binding of XWBSCR11 to the DE of the gsc gene was confirmed by gel shift analysis with GST fusion proteins containing repeat pairs 1 and 2, 2 and 3, and 4 and 5, with the GST fusion protein containing repeats 4 and 5 exhibiting the most robust binding to the DE of the gsc gene. Interestingly, of the five HLH repeat regions, the highest degree of homology (96%) was found in repeat 4 between Xenopus and human WBSCR11, implicating a conserved function of this region. As mentioned previously, WBSCR11 is thought to function as a transcription factor, although its exact role in humans is unknown, and it is not known to what extent it contributes to the symptoms observed in WBS (Franke et al. 1999; Osborne et al. 1999). One interesting possibility is that WBSCR11 functions similarly in humans in the regulation of gsc transcription, and it is the deregulation of gsc that contributes to some of the traits found in WBS, such as the craniofacial abnormalities. This possibility is consistent with mouse studies in which mice homozygous for a deletion of the gsc gene exhibit mandibular defects as a result of malformations in branchial arch-derived structures (Rivera-Perez et al. 1995, 1999; Yamada et al. 1995).
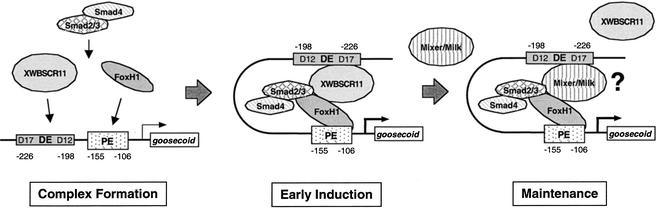
In addition to its ability to bind to the DE, we have shown that XWBSCR11 is a key regulator of gsc expression via the DE. Overexpression of a constitutively active XWBSCR11 protein is sufficient to activate endogenous gsc expression, and a dominant-suppressive version of XWBSCR11 (EVE-XWBSCR11) specifically inhibits gsc induction mediated via the DE. Loss-of-function experiments using XWBSCR11 morpholinos in animal caps also showed the requirement of XWBSCR11 in gsc induction. The fact that homologs of XWBSCR11 are present in both mouse and human suggests a conserved function of this protein in embryos. We propose that XWBSCR11 is an important factor regulating DE-mediated gsc induction (see Fig. 9).
Figure 9.
Model of activation of gsc transcription via the DE and PE. DE-mediated gsc regulation is regulated by two phases. During the early inductive phase, XWBSCR11 acts through the DE to stimulate gsc transcription. FoxH1 binds to the PE and cooperates to activate transcription, perhaps via the bending of DNA bringing the DE and PE in close proximity, thereby forming a large transcription complex among these various factors. During the late maintenance phase, Mixer/Milk homeoproteins bind to the DE, and Xtwn and Xsia homeoproteins bind to the PE to affect gsc transcription. It is unclear at present how the interplay among these transcription factors modulates the activity of gsc.
We find that overexpression of a constitutively active form of XWBSCR11 (VP16-XWBSCR11) was able to induce the expression of both gsc and mix2 (Fig. 4C), raising the possibility that mix2 could be regulated by XWBSCR11. However, at present, there is no in vivo evidence supporting the notion that XWBSCR11 is involved in mix2 regulation, as the activin-mediated induction of mix2 was unaffected by XWBSCR11-specific morpholinos. Furthermore, there are no XWBSCR11 transcription factor binding sites within the promoter region of mix2. We reason that the activation of mix2 by VP16-XWBSCR11 occurs when VP16-XWBSCR11 is overexpressed, causing fortuitous interaction with endogenous FoxH1 as XWBSCR11 is capable of interacting with FoxH1 in vitro.
Activin/nodal-type TGF-β signals are transduced from activated receptors to the nucleus via a complex formed by both Smad2 and Smad4. We find that although XWBSCR11 can interact with both Smad2 and Smad3, XWBSCR11 interaction with Smad3 can occur independent of activated receptor expression. The in vivo relevance of this preferential binding of XWBSCR11 to Smad3 is unclear at present. Xenopus Smad2 and Smad3 are over 82.5% identical at the amino acid level (Howell et al. 2000), and they have both been shown to mediate TGF-β signaling. However, these related activin/nodal intracellular transducing molecules may have distinct functions. In Xenopus, we and others found that Smad3 is expressed both maternally and zygotically, and its transcripts are detected ubiquitously in embryos (Howell et al. 2001; C. Ring and K.W.Y. Cho, unpubl.). In mouse embryos, Smad3 is ubiquitously expressed together with Smad2 mRNA in the embryo proper, but in the presumptive endoderm, only Smad2 mRNA is expressed (Waldrip et al. 1998; Tremblay et al. 2000). Mouse embryos lacking Smad2 fail to generate the appropriate proximo-distal polarity, and the entire epiblast adopts an extraembryonic mesodermal fate (Waldrip et al. 1998; Heyer et al. 1999), whereas Smad3-deficient mice are viable and survive to adulthood with minor defects (Zhu et al. 1998; Datto et al. 1999; Yang et al. 1999). Furthermore, it has been shown that Smad3 suppresses transcriptional activity from a FoxH1-dependent site present in the mouse gsc promoter, whereas Smad2 enhanced activation (Labbe et al. 1998). Although the physiological relevance of such an interaction is still unclear, differential effects of the related Smad2 and Smad3 suggest a mechanism whereby diverse responses to activin/nodal ligands could be generated.
Although FoxH1 is unable to bind to the DE, surprisingly, it is capable of interacting synergistically with XWBSCR11 to stimulate DE-mediated induction of transcription. This result is somewhat unexpected as previous findings indicate that ectopic expression of FoxH1 alone is unable to activate FoxH1 target genes, including gsc (Chen et al. 1997; Watatnabe et al. 1999). However, in this case, we were able to observe strong synergistic induction of DE(6×)gsc/Luc. Perhaps under normal circumstances, endogenous gsc expression is controlled via formation of an XWBSCR11–Smad2/Smad4 complex on the DE, but its expression is enhanced further via a physical interaction with FoxH1 and the concomitant binding of FoxH1 to the downstream enhancer site. The occurrence of these higher order interactions in vivo remains to be shown.
The fact that XWBSCR11 and FoxH1 can interact directly to form a complex is consistent with this notion. Recruitment of promoter-specific transcription factors such as XWBSCR11–Smad via a FoxH1 or FoxH1–Smad complex may constitute an effective strategy for increasing range and potential for modulation of promoter selectivity across activin/nodal target genes. Furthermore, the synergistic effects of FoxH1 and XWBSCR11 on the DE of gsc demonstrate how drastically the expression levels of a given gene can be affected by changes in the availability of various transcription factors. Thus, expression of specific transcription factors, potentially dictated by the developmental history of individual cells, may create profoundly different responses toward the same activin/nodal ligand signal in those cells. In the case of gsc, activin/nodal-mediated DE activation occurs via XWBSCR11, and perhaps maximal activation of gsc requires FoxH1. In addition to the involvement of these transcription factors, other factors such as Mix/Mixer and Xtwn/Xsia homeoproteins are known to bind to DE and PE, respectively, adding further complexity in the regulation of gsc (Laurent et al. 1997; Germain et al. 2000). As the list of transcription factors that bind to the promoter region of gsc to affect transcription grows, the following questions might be interesting in terms of gsc regulation: As both Mixer and XWBSCR11 can bind to the DE, does the zygotic expression of Mixer displace XWBSCR11 (Fig. 9)? Is the expression of gsc via the DE mediated by different transcription factors expressed at distinct times in development? Given the binding of FoxH1 to the PE of the gsc promoter, what is the interaction of FoxH1 with the Wnt signaling cascade, shown previously to regulate this region of the gsc promoter? Further characterization of the transcriptional regulation of gsc will continue to provide insights into the molecular mechanisms underlying the signaling cascades that govern mesoderm induction.
Materials and methods
Embryo handling and microinjection assays
Embryo manipulations and microinjections were as described previously (Cho et al. 1991; Watabe et al. 1995).
One-hybrid screen and cDNA identification
A yeast one-hybrid assay was used to identify proteins capable of interacting with a multimerized gsc-ARE. Briefly, yeast bearing six tandem copies of the 17-bp DE (Watabe et al. 1995) upstream of HIS3 and lacZ reporter genes were transformed with a Xenopus oocyte cDNA library in the pGAD10 fusion vector (Clontech), containing an amino-terminal GAL4 transcriptional transactivation domain. To confirm binding specificity for the gsc-ARE, plasmid DNA from positive colonies was then individually retransformed into yeast containing a wild-type or mutant DE upstream of lacZ. Of the 4 × 106 clones screened, 11 independent clones that bound to both DE-HIS3 and DE-lacZ strains were identified. Of these, XWBSCR11 was identified eight times. Sequence analysis of several of the independently isolated XWBSCR11 pGAD10 cDNAs revealed that they contained full-length coding sequence fused in-frame with the GAL4 activation domain. DNA sequences were compiled and aligned by use of the programs DNA Strider and MacAlign.
RNA isolation, whole mount in situ hybridization, and RT–PCR analysis
RT–PCR assays were performed as described in Hawley et al. (1995) as follows: XWBSCR11, 58°C annealing, 30 cycles (forward primer, 5′-AGGCAGCCCTGGAACTCTAGG-3′; reverse primer, 5′-ATCCTCATCATCAGGTCTTGGG-3′); Xsmad3, 54°C annealing, 35 cycles (forward primer, 5′-AGTAGAGAGAGAG GAAGAAG-3′; reverse primer, 5′-TATGAGCAGGAGACAT AGAG-3′), Xsmad2, 55°C annealing, 35 cycles (forward primer, 5′-CAGCAACAGACTCAATACAGC-3′; reverse primer, 5′-CAAGGTGCTTTTCGTTGATGG-3′); Gsc, 52°C annealing, 33 cycles (forward primer, 5′-AGAGTTCATCTCAGAGAG-3′; reverse primer, 5′-TCTTATTCCAGAGGAACC-3′); Mix2, 55°C annealing, 33 cycles (forward primer 5′-TGCCCCTAACAGCT CCTCATACCAA-3′; reverse primer, 5′-TGGCCGGCAAACA AACTCAT-3′); Otx2, 55°C annealing, 33 cycles (forward primer, 5′-GGAGGCCAAAACAAAGTG-3′; reverse primer, 5′-TCATGGGGTAGGTCCTCT-3′); Mixer, 55°C annealing, 33 cycles (forward primer, 5′-CACCAGCCCAGCACTTAAC-3′; reverse primer, 5′-CCAATGTCACATCAACTGAAG-3′). Whole mount in situ hybridization was performed as described (Harland 1991), using an antisense probe synthesized with T7 RNA polymerase after the digestion of the full-length XWBSCR11 cDNA with SacII.
Plasmid constructs
XWBSCR11 fragments containing the first two repeats (XWBSCR11 R1–2), the first three repeats (XWBSCR11 R1–3), the middle two repeats (XWBSCR11 R2–3), and the last two repeats without the carboxy-terminal serine-rich stretch (XWBSCR11 R4–5Δ3′) were generated from the full-length XWBSCR11 cDNA. The primers used for the PCR to generate these XWBSCR11 fragments were: XWBSCR11 wt-EcoRI, 5′-CCGGAATT CGATGGCTCTGGCAGGAAAGCAG-3′; XWBSCR11dR1-EcoRI, 5′-CCGGAATTCGGTCCTTGCATCAAAGCATCTC-3′; XWBSCR11 4(UP), 5′-CCGGAATTCGAGTTTTGATGCC AGACTTTCC-3′; XWBSCR11 3d3–5-NotI, 5′-ATAGTTTA GCGGCCGCTTCGCTACAAGCCTCTTCTCC-3′; XWBSCR11 3d4–5-EcoRI, 5′-CCCGAATTCCTGCCTACAACACAGTCT TCC- 3′; XWBSCR11 R5RV/X, 5′-GGGCTCGAGCTTGGGAC CAGTGTTACC-3′. The combinations of the primers to generate the XWBSCR11 fragments mentioned above were: wt-EcoRI and XWBSCR11 3d3–5-NotI for XWBSCR11 R1–2; XWBSCR11WT-EcoRI and XWBSCR113d4–5-EcoRI for XWBSCR11R1–3; XWBSCR11dR1-EcoRI and XWBSCR113d4–5-EcoRI for XWBSCR11 R2–3; and XWBSCR11 R4(UP) and XWBSCR11 R5RV/X for XWBSCR11 R4–5Δ3′. The XWBSCR11 R1–2 fragment was digested by the restriction enzymes EcoRI and NotI, subcloned into pGEX4T3 digested with EcoRI and NotI, and sequenced. XWBSCR11 R1–3 and XWBSCR11 R2–3 fragments were digested with EcoRI, subcloned into pGEX4T3 digested with EcoRI, and sequenced. The XWBSCR11 R4–5Δ3′ fragment was digested with EcoRI and XhoI and subcloned into pGEX4T3 digested with EcoRI and XhoI. The Xenopus GST-FoxH1 FH, GST-FoxH1 SID constructs are described in Chen et al. (1997), and GST-Smad2 MH1 and GST-Smad2 MH2 are in Nishita et al. (2000).
Morpholino constructs
Antisense morpholino oligos were generated by Gene Tools using the following sequences: XWBSCR11, 5′-CATACTGCT TTCCTGCCAGAGCCAT-3′; FoxH1, 5′-AGTACAGACTGG AGGGGTCTCTCAT-3′. As a control, a random morpholino oligo was used. The Δ-5′-XWBSCR11 construct was generated by PCR amplification of XWBSCR11 cDNA with the following two primers:forward primer (76–98), 5′-CGGAATTC CGCCATGGGTGGGAAGAATGAGATTATTAC-3′; reverese primer, 5′-ATAGTTTAGCGGCCGCGTAGTTTATAGGGCC AG-3′.
Expression and purification of GST-XWBSCR11 fusion proteins
Plasmid constructs were transfected into BL21(DE3) bacteria. Single colonies were picked and grown overnight at 37°C in 50 mL of TBM9+Amp (ampicillin) media (Roy et al. 1997). The overnight culture was reinoculated, grown at 30°C to an OD600 of ∼0.7 and induced for 2 h with 0.1 mM isopropyl-β-D-thiogalactopylanoside (IPTG). The cells were harvested by centrifugation, and the bacterial pellet was resuspended in 10 mL of lysis/sonication buffer (50 mM sodium phosphate at pH 8.0, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 1.5% sarcosyl, and 1 mM PMSF). After addition of 10 mg of lysozyme (Sigma) into the suspension, the mixture was incubated on ice for 30–45 min. The lysate was sonicated and centrifuged. The supernatant was transferred to a conical tube, and 600 μL of a 50% slurry of glutathione–sepharose 4B beads (Amersham Pharmacia) was allowed to bind the GST fusion protein. Then, beads were centrifuged and washed three times with 10 mL of wash buffer (20 mM sodium phosphate at pH 8.0, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.5% Tween-20, 1 mM DTT, and 1 mM PMSF). The resulting bead-coupled proteins were used directly for GST pull-down experiments. For gel shift and DNase I footprint assays, the GST fusion protein was then eluted for 15 min at room temperature in 0.5 mL of elution buffer (50 mM sodium phosphate at pH 8.0, 300 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% Triton X-100, 1.0% sarcosyl, 1 mM PMSF, and 10 mM reduced glutathione).
Gel shift assay
Approximately 0.5 micrograms of the eluted GST-XWBSCR11 fusion protein was mixed with the annealed oligonucleotides. The sequence of the oligonucleotides were: D29-F, 5′-GA TCCATTAATCAGATTAACGGTGAGCAATTAG-3′; D29-R, 5‘-CTAATTGCTCACCGTTAATCTGATTAATG-3‘; D17-F, 5′-GATCCATTAATCAGATTAACG-3′; D17-R, 5′- CGTTAA TCTGATTAATG-3′; D12-F, 5′-GATCGTGAGCAATTAG-3′; D12-R, 5′-CTAATTGCTCAC-3′; M4-F, 5′-GATCCAgTcAg CAGcTgAcCG-3′ (lowercase indicates the site of point mutation); M4-R, 5′-CGgTcAgCTGcTgAcTG-3′. Each pair of the D29, D17, D12, and M4 oligos was annealed at room temperature, end-labeled with [32P]dATP by use of the exo(−) Klenow fragment (Stratagene), and purified by a Sephadex G-50 column. The purified GST-XWBSCR11, GST-FoxH1, and/or GST-Smad2 fusion proteins were pre-incubated on ice for 10 min in a buffer containing 20 mM HEPES-KOH (pH 7.9), 7.5 mM MgCl2, 2 mM EDTA, 2 mM DTT, 50 μg/mL poly[d(I-C)], and 10% glycerol. Then, labeled probe was added to the mixture, and allowed to form a complex on ice. Samples were separated by native PAGE, and analyzed by autoradiography.
DNase I footprint assay
A fragment corresponding to the region from +4 to −309 of the gsc promoter was subcloned into the pBluescriptII KS+ plasmid (Stratagene). The plasmid was digested with HindIII, and end-labeled with [32P]dATP. The labeled plasmid was then digested with BamHI to generate the single-end labeled probe. The probe was mixed with various combination of GST fusion proteins on ice for 30 min in binding buffer (20 mM HEPES-KOH at pH 7.9, 7.5 mM MgCl2, 2 mM DTT, 15 μg/mL poly[d(I-C)], and 10% glycerol). The reaction mixture was incubated with DNaseI, and separated by 5% Urea-PAGE for autoradiography.
GST pull-down assay
A GST-XWBSCR11 fusion bead slurry (20 μL) was mixed with 35S-labeled full-length FoxH11, FoxH11 FH, or FoxH11 SID in a 1.5-mL tube. The binding reaction was carried out in 0.2 mL of PBST buffer (20 mM sodium phosphate at pH 8.0, 150 mM NaCl, 0.5% Tween-20, and 0.1% BSA) for 50 min at room temperature. The mixture was centrifuged briefly, and washed three times with 1 mL of PBST without BSA. Subsequently, the beads were boiled in 15 μL of SDS-PAGE loading buffer and analyzed after SDS-PAGE.
Coimmunoprecipitation assay
In vivo coimmunoprecipitation assays were performed with transfected Cos7 cells as described in Kawabata et al. (1998). 35S-labeled proteins were generated by use of an in vitro translation kit (Promega) and used for in vitro coimmunoprecipitation assay as described previously (Nishita et al. 2000). Full-length XWBSCR11, XWBSCR11 lacking repeat 1 (XWBSCR11 ΔR1), XWBSCR11 lacking repeats 1–2 (XWBSCR11 ΔR1–2), XWBSCR11 lacking repeats 4–5 (XWBSCR11 ΔR4–5), and XWBSCR11 lacking repeats 3–5 (XWBSCR11 ΔR3–5) were generated by PCR and engineered to contain a T7 promoter at the 5′ end to generate sense transcripts. The following primer pairs were use to generate the desired fragments: full-length XWBSCR11 (forward primer, 5′-GTAATACGACTCACTATAGGGCCGC CACCATGGCTCTGGCAGGAAAGCAG-3′; reverse primer, 5′-TCTAGTAGTTTATAGGGCCAGG-3′); XWBSCR11 ΔR1 (forward primer, 5′-GTAATACGACTCACTATAGGGCCGCCA CCATGGTCCTTGCATCAAAGCATCTC-3′; reverse primer, 5′- TCTAGTAGTTTATAGGGCCAGG-3′); XWBSCR11ΔR1–2 (forward primer, 5′-GTAATACGACTCACTATAGGG CCGCC ACCATGGTTTGCGATGATAGACCCTGTG-3′; reverse primer primer, 5′- TCTAGTAGTTTATAGGGCCAGG-3′); XWBSCR11 ΔR4–5 (forward primer, 5′-GTAATACGACTCACTATAGGGC CGCCACCATGGCTCTGGCAGGAAAGCAG-3′; reverse primer, 5′-CTGCCTACAACACAGTCTTCC-3′); XWBSCR11 ΔR3–5 (forward primer, 5′-GTAATACGACTCACTATAGGGCCGC CACCATGGCTCTGGCAGGAAAGCAG-3′; reverse primer, 5′-TTCGCTACAAGCCTCTTCTCC-3′). PCR fragments were purified and used to generate RNA transcripts by use of a T7 mMessage Machine kit (Ambion). The RNA was subjected to in vitro translation with a TNT kit (Promega). GST fusion proteins containing the Xenopus FoxH1 FH, FoxH1 SID (Chen et al. 1997), and Smad2 MH2 domains were used in a coimmunoprecipitation analysis (Nishita et al. 2000).
Acknowledgments
We are grateful to Dr. Malcolm Whitman for various Xenopus FoxH1 constructs and to Dr. Tetsuro Watabe for his technical assistance in the yeast one-hybrid screen. The authors also thank Drs. Masahiro Kawabata and Minako Hashimoto for helpful comments. This work was supported by National Institutes of Health Grants HD29507 (to K.W.Y.C.) and by Grant-in-Aid for Scientific Research (12219217) of the Ministry of Education, Culture, Sport, Science, and Technology of Japan (to K.M.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kwcho@uci.edu; FAX (949) 824-4709.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.963802.
References
- Asashima M, Nakano H, Shimada K, Kinoshita K, Ishii K, Shibai H, Ueno N. Mesodermal induction in early amphibian embryos by activin A (erythroid differentiation factor) Roux’s Arch Dev Biol. 1990;198:330–335. doi: 10.1007/BF00383771. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D, Ruddle FH. Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix–loop–helix domains. Proc Natl Acad Sci. 2000;97:7342–7347. doi: 10.1073/pnas.97.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Gaunt SJ, Cho KW, Steinbeisser H, Blumberg B, Bittner D, De Robertis EM. Gastrulation in the mouse: The role of the homeobox gene goosecoid. Cell. 1992;69:1097–1106. doi: 10.1016/0092-8674(92)90632-m. [DOI] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcription partner for MAD proteins in TGF-beta signaling. Nature. 1996;383:691–616. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Cho KWY, Blumberg B, Steinbeisser H, De Robertis EM. Molecular nature of Spemann's organizer: The role of the Xenopus homeobox gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilov V, Blum M, Schweickert A, Campione M, Steinbeisser H. Negative autoregulation of the organizer-specific homeobox gene goosecoid. J Biol Chem. 1998;273:627–635. doi: 10.1074/jbc.273.1.627. [DOI] [PubMed] [Google Scholar]
- Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol Cell Biol. 1999;4:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke Y, Peoples RJ, Francke U. Identification of GTF2IRD1, a putative transcription factor within the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet. 1999;86:296–304. doi: 10.1159/000015322. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ, Blum M, De Robertis EM. Expression of the mouse goosecoid gene during mid-embryogenesis may mark mesenchymal cell lineages in the developing head, limbs and body wall. Development. 1993;117:769–778. doi: 10.1242/dev.117.2.769. [DOI] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes & Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- Grueneberg DA, Henry RW, Brauer A, Novina CD, Cheriyath V, Roy AL, Gilman M. A multifunctional DNA-binding protein that promotes the formation of serum response factor/homeodomain complexes: Identity to TFII-I. Genes & Dev. 1997;11:2482–2493. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: An improved whole mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hawley SH, Wunnenberg-Stapleton K, Hashimoto C, Laurent MN, Watabe T, Blumberg BW, Cho KWY. Disruption of BMP signals in embryonic Xenopus ectoderm leads to direct neural induction. Genes & Dev. 1995;9:2923–2935. doi: 10.1101/gad.9.23.2923. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. β-Catenin signaling activity dissected in the early Xenopus embryo: A novel antisense approach. Dev Biol. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Heldin C H, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Heyer J, Escalante-Alcalde D, Lia M, Boettinger E, Edelman W, Stewart CI, Kucherlapati R. Postgastrulation of Smad2-deficient embryos show defects in embryo turning anterior morphogenesis. Proc Natl Acad Sci. 1999;96:112595–12600. doi: 10.1073/pnas.96.22.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoodless PA, Pye M, Chazaud C, Labbe E, Attisano L, Rosant J, Wrana JL. FoxH1(Fast) functions to specify the anterior primitive streak in the mouse. Genes & Dev. 2001;15:1257–1271. doi: 10.1101/gad.881501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Mohun TJ, Hill CS. Xenopus Smad3 is specifically expressed in the chordoneural hinge, notochord and in the endocardium of the developing heart. Mech Dev. 2001;104:147–150. doi: 10.1016/s0925-4773(01)00365-3. [DOI] [PubMed] [Google Scholar]
- Huang HC, Muyrtaugh L, C, Vize PD, Whitman M. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 1995;14:5969–5973. doi: 10.1002/j.1460-2075.1995.tb00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CVE. Nodal-related signals induce axis mesoderm and dorsalize mesoderm during gastrulation. Development. 1995;121:3651–3662. doi: 10.1242/dev.121.11.3651. [DOI] [PubMed] [Google Scholar]
- Joseph EM, Melton DA. Xnr4: A Xenopus nodal-related gene expressed in the Spemann's organizer. Dev Biol. 1997;184:367–372. doi: 10.1006/dbio.1997.8510. [DOI] [PubMed] [Google Scholar]
- Karavanov A, Mochizuki T, Curtiss PE, Ault KT, Sugimoto N, Watabe T, Shiokawa K, Jamrich M, Cho KWY, Dawid IB, et al. Xlim-1 and LIM domain binding protein 1 cooperate with other transcription factors in the regulation of the goosecoid promoter. Dev Biol. 2000;224:470–485. doi: 10.1006/dbio.2000.9778. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Demel T, Xanthos J, Lohr J, Sun B, Sive H, Osada S, Wright C, Wylie C, Heasman J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGF-β growth factors. Development. 1999;126:5759–5770. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulates TGF β-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Laurent M, Hashimoto C, Blitz IL, Rothbacher U, Cho KWY. The Xenopus homeobox gene Twin mediates Wnt induction of goosecoid in establishment of Spemann's organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-β signaling. Genes & Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- McKendry R, Harland RM, Stachel SE. Actvin-induced factors maintain goosecoid transcription through a paired homeodomain binding site. Dev Biol. 1998;204:172–186. doi: 10.1006/dbio.1998.9065. [DOI] [PubMed] [Google Scholar]
- Nishita M, Hashimoto M K, Ogata S, Laurent MN, Ueno N, Shibuya H, Cho KW. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann's organizer. Nature. 2000;403:781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Gawantka V, Dosch R, Delius H, Hirschfeld K, Blumenstock C, Niehrs C. The Xvent-2 homeobox gene is part of the BMP-4 signalling pathway controlling dorsoventral patterning of Xenopus mesoderm. Development. 1996;122:3045–3053. doi: 10.1242/dev.122.10.3045. [DOI] [PubMed] [Google Scholar]
- Osada S, Saijoh Y, Frisch A, Yen CY, Adachi H, Watanabe M, Whitman M, Hamada H, Wright CVE. Activin/nodal responsiveness and symmetric expression in a Xenopus nodal-related gene converge on a FAST-regulated module in intron 1. Development. 2000;127:2503–2514. doi: 10.1242/dev.127.11.2503. [DOI] [PubMed] [Google Scholar]
- Osborne LR, Campbell T, Daradich A, Scherer SW, Tsui LC. Identification of a putative transcription factor gene (WBSCR11) that is commonly deleted in Williams-Beuren syndrome. Genomics. 1999;57:279–284. doi: 10.1006/geno.1999.5784. [DOI] [PubMed] [Google Scholar]
- Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes & Dev. 2001;15:2010–2022. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Wakamiya M, Behringer RR. Goosecoid acts cell autonomously in mesenchyme-derived tissues during craniofacial development. Development. 1999;126:3811–3821. doi: 10.1242/dev.126.17.3811. [DOI] [PubMed] [Google Scholar]
- Roy AL, Du H, Gregor PD, Novina CD, Martinez E, Roeder RG. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 1997;16:7091–7104. doi: 10.1093/emboj/16.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Grunwald DJ, Myers PZ. Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development. 1993;117:1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- Summerton J, Weller D. Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Yokota C, Takano K, Tanegashima K, Onuma Y, Goto JI, Asashima M. Two novel nodal-related genes initiate early inductive events in Xenopus Nieuwkoop center. Development. 2000;127:5319–5329. doi: 10.1242/dev.127.24.5319. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Halpern ME, Postlethwait JH. Goosecoid expression in neurectoderm and mesendoderm is disrupted in zebrafish cyclops gastrulas. Dev Biol. 1994;164:420–429. doi: 10.1006/dbio.1994.1212. [DOI] [PubMed] [Google Scholar]
- Thomsen G, Melton DA. Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell. 1993;74:433–441. doi: 10.1016/0092-8674(93)80045-g. [DOI] [PubMed] [Google Scholar]
- Thomsen G, Woolf T, Whitman M, Sokol S, Vaughan J, Vale W, Melton DA. Activins are expressed early in Xenopus embryogenesis and can induce axial mesoderm and anterior structures. Cell. 1990;63:485–493. doi: 10.1016/0092-8674(90)90445-k. [DOI] [PubMed] [Google Scholar]
- Tremblay K, Hoodless PA, Bikoff EK, Robertson EJ. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development. 2000;127:3079–3090. doi: 10.1242/dev.127.14.3079. [DOI] [PubMed] [Google Scholar]
- Waldrip W, Birkoff E, Hoodless P, Wrana J, Robertson E. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- Watabe T, Kim S, Candia A, Rothbacher U, Hashimoto C, Inoue K, Cho KWY. Molecular mechanisms of Spemann's organizer formation: conserved growth factor synergy between Xenopus and mouse. Genes & Dev. 1997;9:3038–3050. doi: 10.1101/gad.9.24.3038. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Whitman M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development. 1999;126:5621–5634. doi: 10.1242/dev.126.24.5621. [DOI] [PubMed] [Google Scholar]
- Yamada G, Mansouri A, Torres M, Stuart ET, Blum M, Schultz M, De Robertis EM, Gruss P. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Meno C, Sakai Y, Shiratori H, Mochida K, Ikawa Y, Saijoh Y, Hamada H. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes & Dev. 2001;15:1242–1256. doi: 10.1101/gad.883901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Letterio JJ, Lechleider RJ, Chen L, Hayman R, Gu H, Roberts AB, Deng C. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-β. EMBO J. 1999;18:1280–1291. doi: 10.1093/emboj/18.5.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, King ML, Houston D, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94:703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]