Abstract
Genome-wide location analysis indicates that the yeast nucleosome-remodeling complex RSC has ∼700 physiological targets and that the Rsc1 and Rsc2 isoforms of the complex behave indistinguishably. RSC is associated with numerous tRNA promoters, suggesting that the complex is recruited by the RNA polymerase III transcription machinery. At RNA polymerase II promoters, RSC specifically targets several gene classes, including histones, small nucleolar RNAs, the nitrogen discrimination pathway, nonfermentative carbohydrate metabolism, and mitochondrial function. At the histone HTA1/HTB1 promoter, RSC recruitment requires the Hir1 and Hir2 corepressors, and it is associated with transcriptional inactivity. In contrast, RSC binds to promoters involved in carbohydrate metabolism in response to transcriptional activation, but prior to association of the Pol II machinery. Therefore, the RSC complex is generally recruited to Pol III promoters and it is specifically recruited to Pol II promoters by transcriptional activators and repressors.
Keywords: Chromatin, transcription, RNA polymerase III, nucleosome-remodeling, gene expression, microarray analysis
Chromatin plays a crucial role in the control of nuclear processes including transcription, recombination, DNA replication, and repair. Two general types of chromatin-modifying complexes have been described. One type covalently modifies histones by acetylation, methylation, or phosphorylation of the histone N-terminal tails protruding from the nucleosome core (Wu and Grunstein 2000; Jenuwein and Allis 2001). The second class is the ATP-dependent nucleosome-remodeling complexes that noncovalently modify and reposition nucleosomes in chromatin (Kingston and Narlikar 1999; Kornberg and Lorch 1999; Vignali et al. 2000; Urnov and Wolffe 2001). The yeast SWI/SNF complex, which comprises an Swi2/Snf2 catalytic subunit and ∼10 associated factors, was the first nucleosome-remodeling complex to be described, and it remains the best studied (Peterson and Workman 2000; Sudarsanam and Winston 2000). Five other nucleosome-remodeling complexes containing a Swi2-like catalytic subunit have been purified from yeast, namely, the RSC, INO80, ISW1, ISW2, and CHD1 complexes (Cairns et al. 1996; Tsukiyama et al. 1999; Shen et al. 2000; Tran et al. 2000). Mutations in subunits of these complexes affect expression of specific genes, indicating that the complexes have distinct transcriptional roles in vivo.
In principle, there are three basic mechanisms by which nucleosome-remodeling complexes may interact with the genome and affect transcription (Struhl 1998; Peterson and Workman 2000; Hassan et al. 2001). First, nucleosome-remodeling complexes could interact nonspecifically and globally with the genome. Such untargeted action might differently affect gene expression, depending on the positions and/or conformation of nucleosomes in individual promoter regions. Second, nucleosome-remodeling complexes could be generally targeted to promoters by associating with components of the basic transcription machinery. SWI/SNF can interact with the Pol II holoenzyme under certain conditions in vitro (Wilson et al. 1996), although it is unclear whether this interaction is physiologically relevant. Third, nucleosome-remodeling complexes can be recruited to specific promoters via interactions with DNA-binding transcriptional activators and repressors. In vivo, SWI/SNF is recruited by the Swi5 activator to the HO promoter at a certain stage of the cell cycle (Cosma et al. 1999), and it is also recruited by the Gcn4 and Gal4 activators (J. Deckert and K. Struhl, in prep.). SWI/SNF recruitment to the histone HTA1 promoter requires both Hir1 and Hir2 corepressors, although SWI/SNF contributes positively to transcription in this situation (Dimova et al. 1999). The ISW2 complex is recruited to promoters by the Ume6 repressor, and it is important for repression of target genes (Goldmark et al. 2000; Fazzio et al. 2001; Kent et al. 2001). These models are not mutually exclusive, and, indeed, histone acetylases and deacetylases have both promoter-specific and genome-wide activities (Kuo et al. 2000; Reid et al. 2000; Vogelauer et al. 2000).
RSC is an abundant nucleosome-remodeling complex in yeast cells, and it is the only such complex that is essential for growth (Cairns et al. 1996). RSC is closely related to the SWI/SNF complex (Cairns et al. 1996; Cao et al. 1997; Treich and Carlson 1997), and the two complexes contain some common subunits (Cairns et al. 1998). Sth1, a homolog of Swi2, is the catalytic subunit of the RSC complex (Du et al. 1998). Biochemical studies suggest the existence of distinct RSC complexes. Rsc1 and Rsc2 are related proteins that associate with the other RSC subunits, but in a mutually exclusive manner (Cairns et al. 1999). Unlike other Rsc subunits, loss of either Rsc1 or Rsc2 does not significantly affect cell growth, although the resulting strains show common and distinct phenotypes. Loss of both Rsc1 and Rsc2 causes lethality, suggesting that there are Rsc1 and Rsc2 isoforms of the RSC complex that have related, though nonidentical functions (Cairns et al. 1999). More recently, the RSCa complex, which lacks the Rsc3 and Rsc30 subunits, has been purified. Rsc3 and Rsc30 form a heterodimer within the RSC complex, and transcriptional microarray experiments suggest that they have both cooperative and opposite functions (Angus-Hill et al. 2001).
Mutations in several RSC subunits show a typical G2/M arrest characterized by large budded cells containing 2N or 4N chromosomes (Cao et al. 1997; Tsuchiya et al. 1998; Angus-Hill et al. 2001). The basis for this G2/M arrest is unknown, but it depends on the spindle body checkpoint. Whole-genome analysis of gene expression in rsc3 and rsc30 mutants indicates that RSC affects the expression of ribosomal protein and cell wall genes (Angus-Hill et al. 2001). However, it is unclear whether these transcriptional effects are directly or indirectly mediated by RSC. Inactivation of the Sth1 and Rsc8, but not the Sfh1 component of RSC leads to inappropriate expression of the CHA1 gene under noninducing conditions, suggesting that RSC might negatively regulate CHA1 expression (Moreira and Holmberg 1999).
Understanding the biological function of nucleosome-remodeling complexes requires the knowledge of their direct physiological targets. Many investigators have used whole-genome microarrays to identify genes whose expression is affected by mutations in transcription factors, but such experiments have limitations for defining direct targets of these transcription factors. First, genome-wide expression analyses performed with mutants cannot easily distinguish between direct and indirect effects at individual promoters. Second, yeast cells contain at least five nucleosome-remodeling complexes that might have partially redundant functions that will not be uncovered by a single mutation. Third, the use of deletion mutants to measure gene expression provides a steady-state measurement of cells that have adapted to the mutations. Fourth, conditional alleles often cause partial loss of function, and the analysis is complicated by the loss of viability or cell cycle arrest under nonpermissive conditions.
To define physiologically relevant targets of DNA-binding proteins in a wild-type cell rather than observing the results of genetic alterations, we and others have combined the technique of chromatin immunoprecipitation with DNA microarray technology to identify the location of specific DNA-binding proteins over the entire genome (Ren et al. 2000; Iyer et al. 2001; Lieb et al. 2001; Simon et al. 2001; Wyrick et al. 2001). However, such genome-wide location analysis has never been applied to a nucleosome-remodeling complex. Here we use genome-wide location analysis to identify the physiological targets of the RSC complex. Our results indicate that the Rsc1 and Rsc2 isoforms of the RSC complex associate with the same genes, that RSC is generally targeted to Pol III promoters, and that RSC is specifically recruited to specific Pol II promoters in response to transcriptional activation or repression.
Results
Genome-wide location of the RSC complex
Genome-wide location analysis, a process that combines chromatin immunoprecipitation followed by hybridization on microarrays, has been successfully used to identify physiological targets for many DNA-binding proteins (Ren et al. 2000; Iyer et al. 2001; Lieb et al. 2001; Simon et al. 2001; Wyrick et al. 2001). However, the application of this technique to RSC involves overcoming several technical challenges that do not apply to proteins that bind directly to specific DNA sequences. As RSC is an abundant complex that may associate with a large fraction of the genome, the conventional approach of normalization to the median signal might be problematic. Conventional normalization works well when a small fraction of the 6000 yeast genes are bound, because the majority of signals provide an internal control. In addition, our preliminary studies indicate that the cross-linking efficiency of RSC is ∼10-fold less than that of a DNA-binding protein. Thus, signal-to-noise ratios are smaller than they are for DNA-binding proteins. To address those problems, we analyzed myc-tagged versions of five different RSC subunits, namely, Rsc1, Rsc2, Rsc3, Rsc8, and Sth1; and each subunit was profiled in at least three independent experiments. The large number of data sets allowed us to apply statistical methods to increase our confidence in bona fide signals. Furthermore, the experimental data were normalized using two different approaches to more fully evaluate the binding behavior of RSC across the genome.
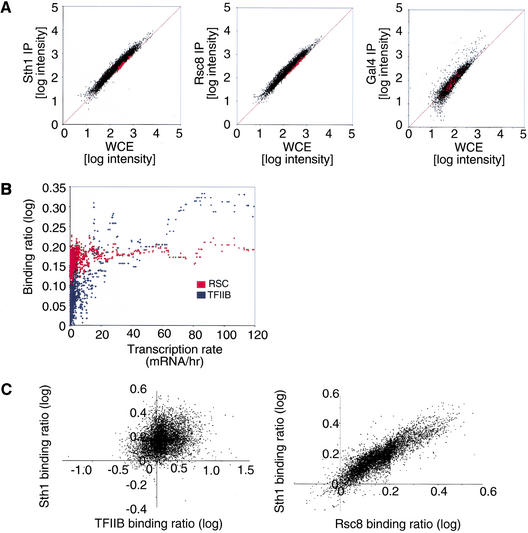
We first addressed whether RSC has a preference for intergenic regions or protein-coding regions (ORFs) by analyzing Rsc8 and Sth1 on intergenic microarrays containing 64 additional ORF features, which were used to normalize the data. As shown in Figure 1A, the binding ratios for both Rsc8 and Sth1 were slightly higher at most intergenic regions than at ORFs, a result confirmed by quantitative analysis on randomly selected promoters (RSC occupancy at the ACT1, CLB2, PGK1, and SDL1 promoters is about 1.2- to 1.4-fold higher than that at the POL1 coding region; http://web.wi.mit.edu/young/RSC). Quantitative analysis indicates that RSC association with telomeres, centromeres, and four ORF-less regions occurs at the level of the POL1 ORF. On a genome-wide level, RSC occupancy does not correlate with Pol II transcription rates of individual genes (Fig. 1B), which was determined previously by microarray experiments (Holstege et al. 1998). As expected (Kuras et al. 2000), occupancy of the general transcription factor TFIIB does correlate with transcription rate. In addition, the occupancy of TFIIB does not correlate with either Sth1 or Rsc8 (Fig. 1C; data not shown). Therefore, RSC has a slight genome-wide preference for promoter regions, but it is not generally recruited to Pol II promoters in a manner that correlates with TFIIB recruitment or transcription rate.
Figure 1.
Genome-wide location analysis of the RSC complex. (A) Preferential association of RSC with promoter regions versus ORF sequences. The ORF-normalized log intensity of the IP DNA and the whole cell extract DNA (WCE) is shown for a Sth1 (left), Rsc8 (middle), and Gal4 (right) experiment. The ORF control features are shown in red. (B) RSC occupancy correlates poorly with Pol II transcription rate. The binding ratio of Sth1 and Rsc8 was averaged, to generate the RSC binding ratio (red). The moving median (window size = 20) of the RSC (red) and TFIIB (blue) binding ratio is plotted as a function of the expression level as determined by microarray (Holstege et al. 1998). Intergenic regions assigned to more than one gene were not considered in the analysis. (C) RSC occupancy correlates poorly with TFIIB occupancy. The weighted average log binding ratio of Sth1 versus TFIIB (left) or Sth1 versus Rsc8 (right) is plotted, with the signal from the ORF features used to normalize the data.
Rsc1 and Rsc2 isoforms of the RSC complex have indistinguishable binding profiles
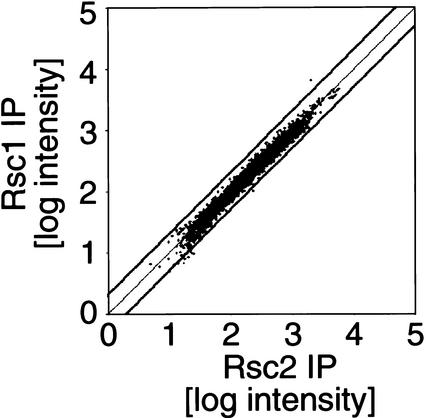
Rsc1 and Rsc2 are related proteins that are present in different isoforms of the RSC complex (Cairns et al. 1999). To examine whether Rsc1 and Rsc2 have distinct binding profiles, Rsc1- and Rsc2-immunoprecipitated DNAs were labeled with different fluorophores and hybridized to the same intergenic DNA microarray. This experimental design eliminates most of the error associated with comparisons between two independent microarray experiments. As shown in Figure 2, Rsc1 and Rsc2 have virtually identical genomic binding profiles. Although a few promoters such as EGT2 show a slightly skewed Rsc1:Rsc2 binding ratio, quantitative PCR analysis of EGT2 and several other potential Rsc1- or Rsc2-specific targets reveals no significant (variation <50%) difference in Rsc1 or Rsc2 binding. Thus, our results suggest that the Rsc1 and Rsc2 isoforms of the RSC complex have indistinguishable binding profiles, although we cannot completely exclude the possibility of a small number of Rsc1- and/or Rsc2-specific genes.
Figure 2.
Rsc1 and Rsc2 have indistinguishable binding profiles. Amplified DNA from Myc–Rsc1 and Myc–Rsc2 immunoprecipitations was labeled with Cy3 and Cy5, respectively, and hybridized to the same intergenic-DNA microarray. The weighted average intensity (three independent experiments) obtained for Rsc2-IP versus Rsc1-IP is shown for all the features on the array. The thick black lines represent a twofold cutoff.
Mutant cells lacking Rsc1 or Rsc2 have similar, but nonidentical, phenotypes (Cairns et al. 1999). Our results suggest that the distinct phenotypes are not caused by differential interaction of Rsc1 and Rsc2 isoforms with specific promoters. The distinct phenotypes could be explained by Rsc1- and Rsc2-containing complexes having slightly different functions at common target genes. However, because Rsc1 and Rsc2 are homologous proteins, with Rsc2 being more abundant than Rsc1, we favor the idea that some genes are more sensitive to a dosage decrease in RSC, and therefore more sensitive to an rsc2 deletion, than an rsc1 deletion.
RSC is targeted to ∼700 targets in the yeast genome
Although RSC shows a small preference for intergenic regions generally, some intergenic regions show substantially greater RSC occupancy than others, suggesting that RSC is targeted specifically to some genes. To identify promoters bound by RSC with high confidence, we performed genome-wide location analysis on five different Rsc subunits (Rsc1, Rsc2, Rsc3, Rsc8, and Sth1). Each experiment was done at least in triplicate and analyzed using a single-array error model combined with a weighted average method as described previously (Ren et al. 2000). Because the five subunits profiled components of a common RSC complex or, based on our profiles of Rsc1 and Rsc2, part of isoforms that bind to the same promoters, it is appropriate to combine P values of individual experiments (Fisher 1954). The combined P value minimizes the effects of experimental error in individual experiments, and allows us to determine the promoters that are consistently bound by all the different Rsc subunits profiled. Although the evidence strongly supports the view that RSC comes in Rsc1 and Rsc2 forms of a common complex, we cannot exclude the possibility that partial RSC complexes (i.e., lacking one or more Rsc subunits) might associate with a small subset of promoters. The approach of combining P values cannot distinguish between partial and complete RSC complexes, although it will tend to eliminate targets of partial RSC complexes from consideration.
The combined P values indicate that, of ∼6400 intergenic regions tested, RSC binds to 267 regions with P values <10−4, 429 regions with P values <0.001, 671 regions with P values <0.01, and 1006 regions with P values <0.05 (http://web.wi.mit.edu/young/RSC). By subtracting the number of intergenic regions expected to pass these P-value boundaries by chance, we estimate that RSC has ∼700 physiological targets that it occupies to a greater degree than most intergenic regions. This corresponds to ∼11% of the intergenic regions.
To further investigate RSC targets and functions, we restricted our analysis to the 429 intergenic regions that passed a P-value cutoff of 0.001. We tested 43 such regions for RSC binding by conventional PCR analysis, and RSC binding was verified for all 43 regions (http://web.wi.mit.edu/young/RSC). None of these confirmed targets were enriched in a control immunoprecipitation performed on the untagged parental strain.
RSC binds to numerous promoters transcribed by RNA polymerase III
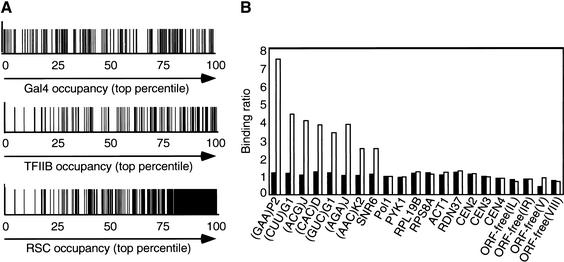
Surprisingly, 112 of the 429 RSC targets contain a tRNA promoter, a number that is far beyond that expected by chance (P value = 10−65). Furthermore, 38% of the 407 tRNA genes have P values <0.01, and 87% are found in the top fourth of RSC occupancy values (Fig. 3A). In addition, other Pol III-transcribed genes, such as RPR1 (Lee et al. 1991) and SCR1 (Felici et al. 1989), are clear RSC targets. Quantitative PCR analysis confirms that RSC occupancies at all six tRNA promoters tested and the U6 promoter are three- to sevenfold above the background level (Fig. 3B). In fact, one of these tRNA promoters, (AAC)K2, was among the 13% of tRNAs whose RSC occupancy values were not in the top quartile. Therefore, RSC is associated with numerous, and probably the majority of, Pol III promoters.
Figure 3.
RSC is generally recruited to RNA polymerase III promoters. (A) For each intergenic region that contains a tRNA gene (indicated by a vertical line), binding of TFIIB (defined by increasing binding ratio) and RSC (defined by decreasing combined P value) is shown in percentiles. (B) Rsc8 occupancies at the indicated promoter and control regions in myc-tagged (open bars) and untagged (solid bars) strains are defined by the ratio between the amount of PCR product obtained with the Rsc8-immunoprecipitated DNA and input DNAs, with the value for the POL1 structural gene being arbitrarily set to 1.
The striking association of RSC with Pol III promoters is specific, and not simply owing to RSC binding to transcriptionally active regions, because RSC occupancy does not correlate with TFIIB occupancy or Pol II transcription rates (Fig. 1B,C). In addition, RSC associates with some repressed promoters (see below), and it does not associate with some highly activated promoters such as ACT1, PYK1, RPS8A, or RPL19B (Fig. 3B). Finally, RSC does not associate with the rDNA promoter (RDN37 in Fig. 3B), which is transcribed by RNA polymerase I. Taken together, these observations suggest that RSC is recruited to promoters by the Pol III transcription machinery (see Discussion).
RSC is targeted to Pol II promoters involved in specific cellular functions
We determined whether RSC targets are overrepresented in gene categories defined by the Munich Information Center for Protein Sequences (http://web.wi.mit.edu/young/RSC). Functional categories that significantly overlap (P ≤ 0.05 from a cumulative hypergeometric distribution test) with the RSC targets are shown in Table 1. These overlaps are specific, because they do not occur when the RSC data are randomly permutated or when a mock immunoprecipitation is analyzed (data not shown). RSC binds to many genes involved in mitochondrial function, nitrogen and carbohydrate metabolism, and transcriptional regulation. It also binds to promoters expressing histones, small nucleolar RNAs, two S-phase cyclins, and two subunits of CAC, the chromatin assembly complex. Interestingly, genes involved in the spindle pole body formation, cell wall integrity, cell cycle control (other than the histones and the two S-phase cyclins), and ribosomal proteins are not enriched among RSC targets, despite the fact that expression of these genes is affected in rsc mutants (Angus-Hill et al. 2001). Given that RSC is targeted to genes involved in specific cellular functions, we then addressed the mechanism of RSC association at some of those genes.
Table 1.
Gene categories overlapping with the RSC targets
| Categories
|
Number of genes in category
|
Number of genes in overlap
|
Overlap P value
|
|---|---|---|---|
| Mitochondrial organization | 334 | 52 | 501E-06 |
| Metabolism | 1022 | 113 | 0.00050119 |
| Cellular organization | 2131 | 207 | 0.00251189 |
| Amino acid biosynthesis | 116 | 19 | 0.00316228 |
| Nitrogen and sulfur metabolism | 75 | 14 | 0.00316228 |
| Transcriptional control | 313 | 40 | 0.00398107 |
| Amino acid metabolism | 202 | 28 | 0.00501187 |
| Organization of chromosome structure | 40 | 9 | 0.00501187 |
| Regulation of nitrogen and sulfur utilization | 29 | 7 | 0.00794328 |
| mRNA synthesis | 400 | 47 | 0.00794328 |
| Nutritional response pathway | 22 | 6 | 0.00794328 |
| Metabolism of energy reserves | 37 | 8 | 0.01 |
| Organization of cytoplasm | 562 | 62 | 0.01 |
| Other transport facilitators | 55 | 10 | 0.01258925 |
| Other nucleotide-metabolism activities | 7 | 3 | 0.01584893 |
| Other intracellular-transport activities | 25 | 6 | 0.01584893 |
| Regulation of carbohydrate utilization | 119 | 17 | 0.01995262 |
| G-proteins | 3 | 2 | 0.01995262 |
| mRNA transcription | 519 | 56 | 0.02511886 |
| Detoxification | 96 | 14 | 0.02511886 |
| Carbohydrate metabolism | 412 | 45 | 0.03162278 |
| Pentose-phosphate pathway | 9 | 3 | 0.03162278 |
| Cell rescue, defense, cell death, and aging | 341 | 38 | 0.03981072 |
RSC targets are defined as described in the text. The categories are from MIPS (http://www.mips.biochem.mpg.de). Only the categories with an overlap with a P ≤ 0.05 (from a cumulative hypergeometric distribution test) are shown. A complete list of the overlap values is available at http://web.wi.mit.edu/young/RSC.
Specific recruitment of RSC to the HTA1 promoter depends on a negative regulatory element
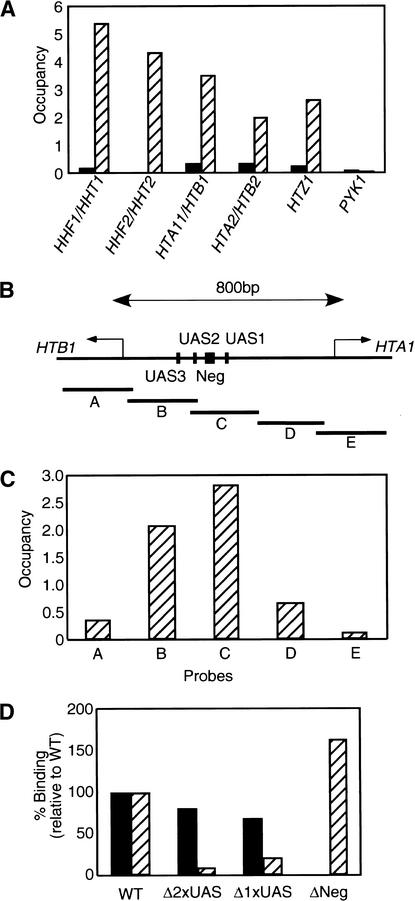
In accordance with the microarray experiments, quantitative PCR analysis indicates that all five Rsc subunits tested are associated with all loci encoding the core histone promoters (Fig. 4A; data not shown). The HTA1/HTB1 promoter region (Fig. 4B) contains binding sites for many transcriptional regulators implicated in both the activation and repression of the divergently transcribed HTA1 and HTB1 genes (Osley et al. 1986; Spector et al. 1997; Iyer et al. 2001; Simon et al. 2001). RSC binds to regions containing the various upstream activating sequences and the negative regulatory element, but RSC occupancy drops in the flanking regions and is at near-background levels in the protein coding regions (Fig. 4C). The association of RSC with a particular region of the promoter strongly suggests that RSC is specifically recruited to the HTA1/HTB1 promoter.
Figure 4.
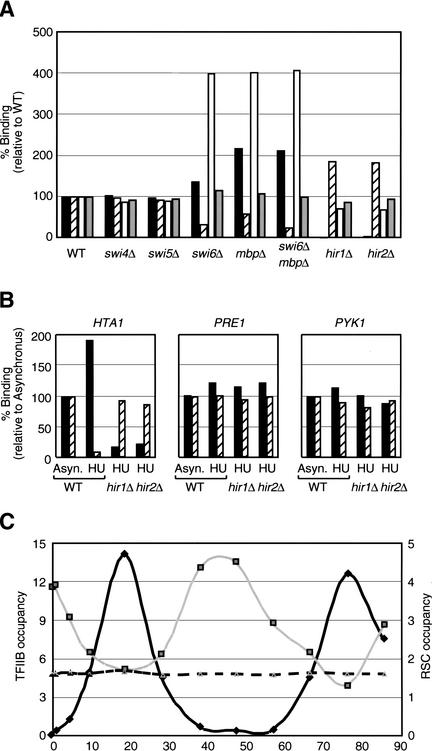
RSC recruitment to the HTA1/HTB1 promoter requires the negative regulatory element. (A) Rsc8 occupancies at the indicated histone promoters and the PYK1 control in tagged (striped bars) and untagged (solid bars) strains are defined as described in Materials and Methods. (B) Schematic representation of the HTA1/HTB1 locus as well as the position of the PCR products (A–E) used for quantitative analysis of RSC occupancy. (C) Rsc8 occupancy at the indicated regions (UAS, upstream activating sequence; Neg, negative regulatory element) of the HTA1/HTB1 locus. Similar results were also obtained for Rsc1. (D) Relative binding of Rsc8 (solid bars) and TFIIB (striped bars) to various deletions of the HTA1/HTB1 locus. (WT) Wild type; (Δ2xUAS) lacks both UAS2 and UAS3; (Δ1xUAS) lacks UAS1; (ΔNeg) lacks the negative regulatory element. For each deletion allele, Rsc8 and TFIIB binding are defined relative to that of the endogenous HTA1/HTB1 promoter.
To address the basis of RSC recruitment to the HTA1/HTB1 promoter, deleted versions of this promoter lacking the UASs or the negative regulatory element were analyzed on a centromeric plasmid. As histone H2A and H2B protein levels may potentially regulate the HTA1/HTB1 promoter through a feedback mechanism (Moran et al. 1990), the introduced plasmids did not contain the intact protein-coding sequences. We distinguished the endogenous HTA1/HTB1 locus from the plasmid harboring the deletion alleles by using primers that yield PCR products of two different sizes. The deletion alleles appear to be functional, because TFIIB is present when the negative element is deleted and absent when UASs are removed (Fig. 4D). Importantly, RSC occupancy is abolished upon removal of the negative regulatory element, but is barely affected upon deletion of various UASs. Therefore, the negative regulatory element is required to target RSC to the HTA1/HTB1 promoter region.
RSC binding to the HTA1/HTB1 promoter requires the Hir1 and Hir2 corepressors and correlates with transcription inactivity
In vivo, the HTA1/HTB1 promoter is bound by the cell cycle transcriptional activators such as MBF, a heterodimer composed of Mbp1 and Swi6 (Iyer et al. 2001; Simon et al. 2001), presumably via consensus DNA sequences in the UASs. In addition, this promoter is repressed outside the S phase of the cell cycle by the Hir1 and Hir2 corepressors, which function through the negative regulatory element (Spector et al. 1997). In accordance with their effects on transcription, deletion of the Swi6 and/or Mbp1 activators causes a decrease in TFIIB occupancy, whereas deletion of Hir1 or Hir2 results in increased TFIIB association (Fig. 5A). Loss of Swi6 and/or Mbp1 does not completely abolish TFIIB occupancy, indicating that there is also some MBF-independent activation of the HTA1/HTB1 promoter.
Figure 5.
Association of RSC with the HTA1/HTB1 locus correlates with transcriptional inactivity. (A) Rsc8 occupancy at the HTA1/HTB1 (black bars), HTA2/HTB2 (open bars), and SOD1 (gray bars) promoters and TFIIB occupancy at the HTA1/HTB1 locus (striped bars) in the indicated mutant strains is presented relative to binding in the wild-type strain. (B) Binding of Rsc8 (black bars) and TFIIB (striped bars) in cells treated with 0.2 M hydroxyurea for 40 min (HU) relative to untreated cells (Asyn.) at the HTA1, PRE1, and PYK1 promoters. (C) TFIIB (black line) and Rsc8 association with the HTA1 (gray line) and PRE1 (dashed line) promoters in cells synchronized with α factor. The zero time point represents removal of α factor and the release of cell cycle arrest at Start.
Strikingly, deletion of either Hir1 or Hir2 completely abolishes RSC association with the HTA1/HTB1 locus, but does not significantly affect other RSC target genes such as SOD1 and CLB5 (Fig. 5A; data not shown). In contrast, RSC occupancy is increased in strains lacking Swi6 and/or Mbp1, and it is unaffected in strains lacking unrelated cell cycle regulators Swi4 or Swi5. Unlike the other histone promoters, HTA2/HTB2 is not regulated by the Hir proteins (Spector et al. 1997), and RSC occupancy at this promoter is not significantly affected by the deletion of Hir1 or Hir2 (Fig. 5A). Inhibition of DNA synthesis by hydroxyurea leads to Hir-dependent repression of HTA1/HTB1 (Osley and Lycan 1987; Spector et al. 1997). Upon incubation with hydroxyurea for 40 min, RSC binding at the HTA1/HTB1 promoter increases 1.6- to 2-fold in a manner that depends on either Hir1 or Hir2, and TFIIB occupancy decreases (Fig. 5B). These results indicate that RSC association with the HTA1/HTB1 promoter depends on the Hir1 and Hir2 corepressors and is correlated with transcriptional repression.
To investigate the role of RSC in the cell cycle regulation of histone gene expression, we determined TFIIB and RSC occupancy at the HTA1/HTB1 locus as cells progress through the cell cycle (Fig. 5C). In agreement with the results in mutant and hydroxyurea-treated cells, TFIIB and RSC occupancies are negatively correlated. During S phase, when histone genes are highly transcribed, TFIIB binding peaks, whereas RSC binding reaches a trough. However, RSC occupancy at the HTA1/HTB1 promoter during S phase does not reach background level. This might be caused by imperfect synchronization of the cells coupled with a slow off-rate of RSC binding, although other explanations are possible. RSC occupancy at the PRE1 promoter is unaffected by cell cycle progression, indicating that regulation occurs at the level of RSC recruitment, rather than RSC activity per se. Taken together, these experiments suggest that RSC assists Hir1 and Hir2 in repressing HTA1/HTB1 transcription outside of the S phase.
Regulated recruitment of RSC prior to transcriptional activation
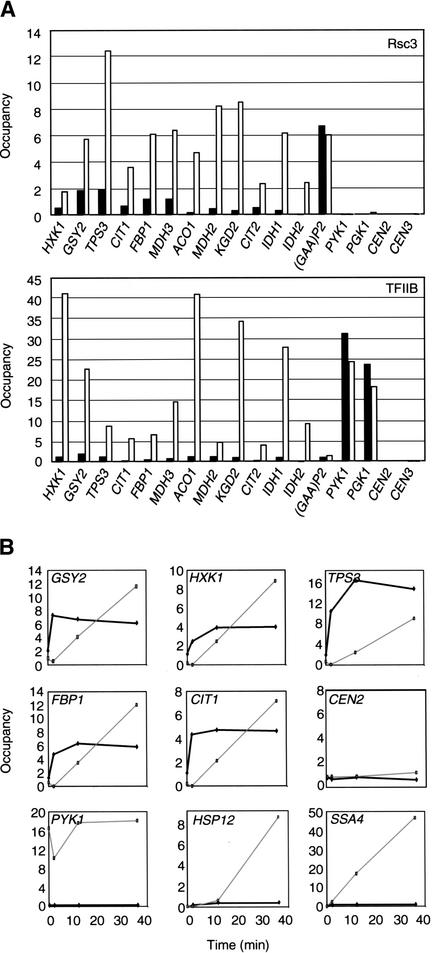
A significant number of RSC target genes are involved in nonfermentative carbohydrate metabolism. These genes are repressed when cells are grown in rich glucose-containing medium, but are strongly induced upon depletion of a fermentable carbon source or in response to other unfavorable environmental conditions (DeRisi et al. 1997; Holstege et al. 1998; Posas et al. 2000; Alexandre et al. 2001; Causton et al. 2001). By analogy with Hir-dependent repression of HTA1/HTB1, we imagined that RSC might be associated with the transcriptionally repressed state of genes involved in carbohydrate metabolism. In contrast to this expectation, RSC and TFIIB occupancies at several target promoters (HXK1, GSY2, TPS3, CIT1, FBP1, and MDH3) are significantly increased when glucose-grown cells are shifted for 30 min to medium containing 3% ethanol (Fig. 6A). We also analyzed RSC occupancy at other promoters that are involved in nonfermentative carbohydrate metabolism, but that did not appear as targets in the microarray experiments, which were performed in glucose medium. In many cases (ACO1, MDH2, KGD2, CIT2, IDH1, and IDH2), RSC occupancy is very low in glucose medium, but dramatically increased upon shift to ethanol medium. Therefore, regulated association of RSC is correlated with transcriptional activation of genes involved in nonfermentative carbohydrate metabolism.
Figure 6.
RSC occupancy at several promoters involved in carbohydrate metabolism correlates with, and precedes, transcriptional activation. (A) Rsc3 and TFIIB occupancies at the indicated promoters upon a 30-min shift from medium containing 2% glucose (black) to medium containing 3% ethanol (white). Similar results were also obtained for Rsc1. (B) Rsc3 (thick black line) and TFIIB (thin gray line) occupancies at the indicated promoters in glucose-grown cells that were treated with 7.5% ethanol for the indicated times.
To address the kinetics of RSC association, cells growing in rich glucose media were stressed by the addition of a high concentration of ethanol (7.5% final). In agreement with the results above, RSC occupancy is dramatically and rapidly increased at the GSY2, HXK1, TPS3, FBP1, and CIT1 promoters (Fig. 6B). Interestingly, this dramatic increase in RSC occupancy precedes the increase in TFIIB occupancy. RSC is not recruited to all promoters that are strongly activated by ethanol stress. CTT1, HSP12, and SSA4 are strongly induced under these conditions (here confirmed by a large increase in TFIIB occupancy), but RSC binding to these promoters is not observed (Fig. 6B; data not shown). RSC is therefore targeted to specific classes of genes involved in carbohydrate metabolism, and it is highly likely that specific activators are involved in the regulated recruitment of RSC. Furthermore, these observations strongly suggest that transcriptional activators can recruit RSC to target promoters prior to assembly of the preinitiation complex and hence transcriptional initiation.
Association of RSC with genes involved in the nitrogen discrimination pathway
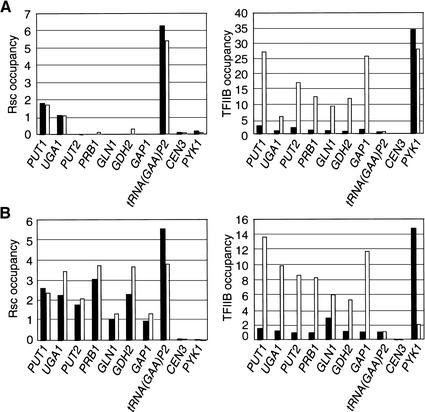
RSC targets also include many genes involved in the nitrogen discrimination pathway (NDP), which is responsible for the utilization of poor nitrogen sources when good nitrogen sources such as ammonium and glutamine are not available (Hofman-Bang 1999). The NDP genes are also regulated by the TOR signaling pathway (Hardwick et al. 1999; Shamji et al. 2000). Ure2, a component of the TOR signaling cascade, sequesters transcriptional activators such as Gln3 and Gat1 in the cytoplasm, leading to low expression of nitrogen source-regulated genes (Beck and Hall 1999; Bertram et al. 2000). Strains lacking Ure2 specifically cripple the communication between TOR and the NDP genes, without affecting other TOR-dependent pathways. In the conditions used in the microarray experiments (YPD medium), the NDP genes are poorly expressed, again suggesting the possibility that RSC association is correlated with transcriptional repression.
We analyzed RSC occupancy in wild-type cells grown in synthetic medium containing good (ammonium) or poor (proline) nitrogen sources, as well as the effect of a ure2 mutation in YPD-grown cells. We examined NDP promoters that either were (PUT1 and UGA1) or were not (PUT2, PRB1, GLN1, GDH2, and GAP1) identified as RSC targets in the microarray experiments. As expected from their transcriptional properties, all NDP promoters showed much higher levels of TFIIB occupancy in proline-grown or ure2 mutant cells. In accordance with the microarray experiments, only PUT1 and UGA1 are bound by RSC in wild-type cells grown in YPD medium (Fig. 7A). Ure2 does not affect RSC association with these promoters, nor does it confer RSC occupancy at the other NDP promoters tested. Interestingly, RSC associates with all NDP promoters tested in synthetic medium, but the levels of RSC occupancy in synthetic medium are not significantly different in ammonium-grown or proline-grown cells (Fig. 7B). Thus, unlike the case of the histone or carbohydrate metabolism genes, RSC occupancy in response to nitrogen source and Ure2 occurs constitutively during both the induced and the noninduced state. Furthermore, these results suggest that nitrogen-regulated activators such as Put3, Gln3, and Gat1 are not involved in the recruitment of RSC to NDP promoters. The difference between YPD and synthetic medium suggests that RSC occupancy is regulated in a novel manner that appears unrelated to nitrogen source.
Figure 7.
Constitutive RSC occupancy at promoters involved in the nitrogen discrimination pathway. (A) Rsc8 and TFIIB occupancies at the indicated promoters in wild-type (solid bars) and ure2 mutant (open bars) strains grown in YPD medium. (B) Rsc8 and TFIIB occupancies at the indicated promoters in strains grown in minimal medium with 0.2% ammonium sulfate (solid bars) or 0.1% proline (open bars) as the sole nitrogen source.
Discussion
Rsc1 and Rsc2 forms of the RSC complex associate with common subsets of promoters
The physiological targets of several DNA-binding activators have been determined on a genome-wide level (Ren et al. 2000; Iyer et al. 2001; Lieb et al. 2001; Simon et al. 2001). In this report, we present the first genome-wide location analysis of a chromatin-modifying activity. As expected from experiments involving other chromatin-modifying activities (Cosma et al. 1999; Agalioti et al. 2000; Reid et al. 2000; Bhaumik and Green 2001; Larschan and Winston 2001), cross-linking of RSC subunits to promoters is significantly (5- to 10-fold) less efficient than cross-linking of specific DNA-binding proteins. This modest cross-linking efficiency and the relatively large size of the intergenic regions on the arrays make it difficult to distinguish bona fide RSC targets from false positives that arise from experimental error. We overcame this difficulty by independently analyzing five RSC subunits and then combining P values of individual experiments. Combining P values is appropriate when individual subunits associate with promoters as part of a common complex, and RSC subunits behave predominantly, and perhaps exclusively, in this fashion. Our results indicate that the Rsc1 and Rsc2 isoforms of RSC have indistinguishable binding profiles. In YPD medium, RSC has ∼700 targets, which represents ∼11% of the intergenic regions in the yeast genome.
The RSC target genes cluster in specific cellular functions such as Pol III transcripts, genes encoding histones and small nucleolar RNAs, and genes involved in the nitrogen discrimination pathway, nonfermentative carbohydrate metabolism, and mitochondrial function; this clustering provides independent validation for the microarray analysis. Importantly, the relationship between RSC and genes involved in nitrogen regulation and nonfermentative carbohydrate metabolism was uncovered, despite the fact that the microarray experiments involved cells grown under “inappropriate” conditions (YPD medium). Nevertheless, the relevance of RSC to these classes of genes was firmly established by the observation that RSC associates with many more genes of these classes under more relevant growth conditions (e.g., nonfermentative carbon sources or synthetic medium). Genome-wide location analysis can therefore uncover potential biological functions, even when the experiments are performed under standard conditions that are not optimal for observing these biological functions.
Our genome-wide location analysis has implications for interpreting RSC functions inferred from transcriptional profiling and phenotypic analysis of rsc mutants. Although rsc3 and rsc30 mutations affect expression of most, if not all, ribosomal protein genes (Angus-Hill et al. 2001), RSC does not associate with a significant fraction of the ribosomal protein genes. This suggests that the effect of RSC on expression of ribosomal protein genes is indirect, particularly because expression of ribosomal protein genes is extremely sensitive to growth rate (Warner 1989). Similarly, RSC does not preferentially associate with promoters involved in cell wall integrity, despite the fact that many such genes are affected by rsc mutations (Angus-Hill et al. 2001). This emphasizes the importance of identifying targets of transcriptional regulators in a wild-type context, rather than by the use of mutations.
RSC mutants arrest at the G2/M stage of the cell cycle (Cao et al. 1997; Tsuchiya et al. 1998; Angus-Hill et al. 2001), but there is no significant overlap between RSC targets and genes involved in cell cycle control or the genes whose expression oscillates during the cell cycle. The few cell cycle-regulated genes that are bound by RSC are not biased toward any specific phase of the cell cycle. These observations suggest that RSC is not dedicated to cell cycle regulation, although we cannot exclude the possibility that G2/M arrest is caused by defective expression of a key G2/M-specific gene(s). Interestingly, mutations in histone tails or in other chromatin-modifying activities (e.g., histone acetylases), arrest cells at the G2/M transition or slow progression through G2/M (Megee et al. 1990; Morgan et al. 1991; Zhang et al. 1998; Clarke et al. 1999; Howe et al. 2001). We suggest that the G2/M phenotype of rsc mutants reflects the general importance of chromatin-modifying activities during mitosis (Krebs et al. 2000), a stage in which chromosomes are condensed.
Distinct modes of RSC association with promoters
In principle, interactions of chromatin-modifying activities with the genome can be untargeted, generally targeted to promoters, or specifically targeted to certain promoters (Struhl 1998; Peterson and Workman 2000). Our results strongly suggest that RSC association with the genome occurs by all three mechanisms. RSC is specifically targeted to certain promoters, and such recruitment is presumably mediated by activators and repressors (see below). In addition, RSC is likely to function in an untargeted genome-wide fashion, because it shows a slight preference for most intergenic regions over protein-coding regions. This preference is not correlated with transcriptional activity, indicating that RSC is not recruited by the Pol II machinery. Instead, the preference is likely to reflect the fact that noncoding regions are typically more accessible to nuclear proteins than are protein-coding regions (Kuo et al. 2000; Mai et al. 2000; Lieb et al. 2001), particularly because RSC nonspecifically interacts with DNA and nucleosomes in vitro (Lorch et al. 1999; Sengupta et al. 2001). Given that RSC is the most abundant nucleosome-remodeling activity in yeast (Cairns et al. 1996), its untargeted action is expected.
Unexpectedly, RSC appears to be generally targeted to promoters transcribed by RNA polymerase III. Specifically, RSC associates with numerous tRNA promoters as well as other Pol III promoters. In contrast, RSC association correlates poorly with TFIIB association and hence Pol II transcription, and RSC does not associate with the ribosomal RNA promoter that is transcribed by Pol I. Some putative Pol III promoters score negatively in the microarray experiments, but it is unclear whether individual promoters are transcriptionally active or inactive in vivo. In addition, fold-enrichment of RSC binding to targets (in comparison to control genomic regions) is typically lower when assayed on microarrays than by quantitative PCR; hence, Pol III promoters scoring negatively on the microarrays might show RSC occupancy when assayed by a more sensitive and quantitative method. The simplest mechanism to account for this striking RNA polymerase-specific association is that RSC is recruited to promoters by a component(s) of the Pol III transcription machinery. Such a mechanism would restrict RSC association to promoters in which the Pol III machinery is stably bound. Although not definitive, our results provide strong evidence for general promoter targeting of a chromatin-modifying activity, a phenomenon not previously described.
Constitutive and regulated recruitment of RSC in response to activation and repression
In vivo, nucleosome-remodeling complexes can be recruited to specific promoters in a regulated manner by activators and repressors, whereupon they positively or negatively affect transcriptional activity. SWI/SNF recruitment by the Swi5, Gcn4, and Gal4 activators contributes to transcription activation of the target genes (Cosma et al. 1999; J. Deckert and K. Struhl, in prep.), and recruitment of the ISW2 complex by the Ume6 repressor is important for transcriptional repression of target genes (Goldmark et al. 2000; Fazzio et al. 2001; Kent et al. 2001). However, the connection between activator- and repressor-dependent recruitment to transcriptional output is not always straightforward. SWI/SNF recruitment to the HTA1/HTB1 promoter depends on the Hir corepressors, but SWI/SNF plays a positive role in histone gene transcription (Dimova et al. 1999). SWI/SNF does negatively regulate transcription of some genes (Sudarsanam et al. 2000), but it is unknown whether this is through SWI/SNF recruitment to the repressed promoters. Mutational analysis has implicated RSC in repression of the CHA1 gene (Moreira and Holmberg 1999), but our microarray experiments and quantitative PCR analysis (data not shown) suggest that CHA1 is not a RSC target; hence, the effect of rsc mutations on CHA1 expression might be indirect or due to the global action of RSC.
Our results indicate that recruitment of RSC can be regulated in accord with transcriptional activation or repression, depending on the promoter. RSC is recruited to many promoters involved in nonfermentative carbohydrate metabolism in a manner that correlates with transcriptional activation. Moreover, the fact that RSC recruitment precedes association of the preinitiation complex strongly supports the argument that RSC does not directly bind these promoters, but rather is recruited by specific DNA-binding activators. Although we have not directly identified any activators that recruit RSC, the Hap2–5 complex and the Rtg1/Rtg3 heterodimer are good candidates. The activation-dependent association of RSC with promoters involved in carbohydrate metabolism is consistent with, and probably accounts for, the observation that conditional sth1 alleles show a growth defect on carbon sources such as ethanol and glycerol under semipermissive conditions (Du et al. 1998).
In contrast to promoters involved in carbohydrate metabolism, RSC recruitment to the HTA1/HTB1 promoter requires both the Hir1 and Hir2 corepressors, and recruitment is correlated with transcriptional inactivity. Again, the regulated and Hir-dependent recruitment to specific promoters makes an argument against direct binding of RSC. The Hir corepressors are also required for recruitment of SWI/SNF to the same promoter, but mutational analysis indicates that SWI/SNF plays a positive role in the expression of histone genes during S phase (Dimova et al. 1999). As our results strongly suggest that RSC is involved in repression of HTA1/HTB1 outside of the S phase, it appears that SWI/SNF and RSC have opposite functions at this promoter. However, HTA1 RNA levels are not significantly affected by mutational inactivation of Rsc8, Sth1, or Rsc3 (Angus-Hill et al. 2001; data not shown), suggesting that RSC is redundant with another function (perhaps even SWI/SNF) for transcriptional repression.
Unlike the histone and carbohydrate metabolism promoters, RSC occupancy at the NDP promoters occurs both under induced and repressed conditions. This constitutive association of RSC with the NDP promoters suggests the Put3, Gln3, and Gat1 activators are not involved in RSC recruitment. We presume that some other DNA-binding protein(s) interacting at NDP promoters plays a role in RSC recruitment; perhaps such a DNA-binding protein is affected by whether cells are grown in rich or synthetic medium. Alternatively, the Rsc3 and Rsc30 subunits, which contain a zinc cluster domain similar to a class of DNA-binding proteins (Angus-Hill et al. 2001), might recognize sequences in these NDP promoters, and hence might directly mediate association of the RSC complex. At present, however, there is no evidence that the Rsc3/Rsc30 heterodimer or the intact RSC complex has specific DNA-binding activity.
Taken together, our results suggest that RSC can be recruited to a promoter in an activator- or repressor-dependent fashion, but that RSC does not have any activating or repressing activity per se. These results support a model, developed for SWI/SNF, in which nucleosome-remodeling complexes increase chromatin fluidity, a process that can lead to either activation or repression, depending on the promoter context.
Materials and methods
Yeast strains and DNAs
All yeast strains were derived from FT4 (a ura3-52 trp1-Δ63 his3-Δ200 leu2::PET56; Tzamarias and Struhl 1994). Rsc1, Rsc2, Rsc3, Rsc8, and Sth1 proteins were tagged at their C termini with a 9myc::TRP1 cassette (Cosma et al. 1999). Correct targeting of genes was verified by PCR, and the proper expression was monitored by Western blot analysis with anti-myc (9e10) monoclonal antibody. The epitope-tagged strains grow indistinguishably from the wild-type parental strain, indicating that the tagged versions of the RSC subunits are functional in vivo. The swi4, swi5, swi6, mbp1, hir1, hir2, and ure2 mutant strains were constructed by deleting the respective ORF using hisG-based constructs (Alani et al. 1987) or PCR-based methods (Longtine et al. 1998). The following fragments of the HTA1/HTB1 promoter region and sequences encoding the first four residues of histones H2A and H2B were cloned between the HindIII and XbaI sites of Ycplac33: Δ2xUAS, 68-bp deletion lacking residues −522 to −590, relative to ATG of HTA1; Δ1xUAS, 51-bp deletion lacking residues −395 to −446; ΔNeg, 54-bp deletion lacking residues −512 to −458. More detailed descriptions are available at http://web.wi.mit.edu/young/RSC.
For the microarray experiments, strains were grown in YPD medium. For other experiments, strains were also grown in YPD medium except when otherwise specified. For determining the elements in the HTA1/HTB1 promoter that mediate RSC recruitment (Fig. 4D), cells were grown in glucose minimal medium containing casamino acids and lacking uracil. Hydroxyurea arrest (Fig. 5B) was performed by treating cells with 0.2 M hydroxyurea for 40 min. For cell cycle experiments (Fig. 5C), cells were arrested at Start with 5 μg/mL of α factor, and synchronization was monitored by microscopy, fluorescence-activated cell sorting, and TFIIB occupancy. To analyze regulated recruitment of RSC at promoters involved in nonfermentative carbohydrate metabolism (Fig. 6), YPD-grown cells were either shifted to YP medium containing 3% ethanol for 30 min or directly treated with 7.5% ethanol and incubated for varying times. To analyze RSC recruitment at NDP promoters as a function of nitrogen source (Fig. 7B), cells were grown in glucose minimal medium containing either 0.2% ammonium sulfate or 0.1% proline as the sole nitrogen source.
Genome-wide location analysis
Genome-wide location analysis of Myc-Rsc1, Myc-Rsc2, Myc-Rsc3, Myc-Rsc8, Myc-Sth1, and Myc-TFIIB was performed in microarrays containing ∼6400 intergenic regions as described previously (Ren et al. 2000). For each subunit, at least three independent chromatin preparations were immunoprecipitated with 9e10 monoclonal anti-myc antibodies (Santa Cruz). For some experiments, the microarrays contained an additional 64 features corresponding to the middle of large ORFs. The complete list of the RSC target genes, as well as downloadable data sets, are available at http://web.wi.mit.edu/young/RSC. To examine whether Rsc1 and Rsc2 have distinct binding profiles, Rsc1- and Rsc2-immunoprecipitated DNAs were labeled with different fluorophores and hybridized to the same intergenic DNA microarray. This experimental design eliminates most of the error associated with comparisons between two independent microarray experiments.
Quantitative analysis of RSC and TFIIB occupancy
For analysis of individual promoter regions, chromatin immunoprecipitation and quantitative PCR analysis were carried out as described previously (Kuras and Struhl 1999) using 9e10 monoclonal anti-myc or rabbit polyclonal anti-TFIIB antibodies. Sequences of oligonucleotide primers used for quantitative PCR are available at http://web.wi.mit.edu/young/RSC. All experiments were repeated with at least two RSC subunits, and the results were similar. For most experiments, RSC and TFIIB occupancies at an individual genomic region were calculated by determining the apparent IP efficiency (the amount of PCR product from the immunoprecipitated sample divided by the amount of PCR product in the input sample) and subtracting the apparent IP efficiency of a control DNA segment (an internal fragment of the POL1 structural gene), which is arbitrarily defined as 1.0. For the experiment in Figure 3D, the apparent IP efficiencies of all genomic regions are presented directly without any background subtraction
Acknowledgments
We thank Tony Lee for designing the ORF controls present on our microarrays; Nancy Hannett, Chris Harbisson, and Itamar Simon for technical support; Marian Carlson and Brehon Laurent for the rsc8 and sth1 mutant strains; and Mark Longtine for plasmids. This work was supported by postdoctoral fellowships from the Cancer Research Fund of the Damon Runyon Walter Winchell Foundation (H.H.N.) and the National Cancer Institute of Canada (F.R.) and research grants from the National Institutes of Health to R.A.Y. and K.S. (GM30186 and GM53720).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kevin@hms.harvard.edu; FAX (617) 432-2104.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.978902.
References
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 2001;498:98–103. doi: 10.1016/s0014-5793(01)02503-0. [DOI] [PubMed] [Google Scholar]
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. doi: 10.1016/s1097-2765(01)00219-2. [DOI] [PubMed] [Google Scholar]
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Cao Y, Cairns BR, Kornberg RD, Laurent BC. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dimova D, Nackerdien Z, Furgeson S, Eguchi S, Osley MA. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- Du J, Nasir I, Benton BK, Kladde MP, Laurent BC. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Kooperberg C, Goldmark JP, Neal C, Basom R, Delrow J, Tsukiyama T. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felici F, Cesareni G, Hughes JM. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for cell growth. Mol Cell Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. Statistical methods for research workers. 12th ed. Edinburgh, Scotland: Oliver and Boyd; 1954. [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Vignali M, Reese JC, Workman JL. Promoter targeting of chromatin-modifying complexes. Front Biosci. 2001;6:1054–1064. doi: 10.2741/hassan. [DOI] [PubMed] [Google Scholar]
- Hofman-Bang J. Nitrogen-catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes & Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISW1 homologs Isw1p and Isw2p. Genes & Dev. 2001;15:619–629. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Fry CJ, Samuels ML, Peterson CL. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- Kuo M-H, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Evans CF, Engelke DR. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc Natl Acad Sci. 1991;88:6986–6990. doi: 10.1073/pnas.88.16.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu XL, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- Mai X, Chou S, Struhl K. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol Cell Biol. 2000;20:6668–6676. doi: 10.1128/mcb.20.18.6668-6676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Morgan BA, Mittman BA, Smith MM. Genetic analysis of histone H4: Essential role of lysines subject to reversible acetylation. Science. 1990;247:841–845. doi: 10.1126/science.2106160. [DOI] [PubMed] [Google Scholar]
- Moran L, Norris D, Osley MA. A yeast H2A-H2B promoter can be regulated by changes in histone gene copy number. Genes & Dev. 1990;4:752–763. doi: 10.1101/gad.4.5.752. [DOI] [PubMed] [Google Scholar]
- Moreira JM, Holmberg S. Transcriptional repression of the yeast CHA1 gene requires the chromatin-remodeling complex RSC. EMBO J. 1999;18:2836–2844. doi: 10.1093/emboj/18.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Mittman BA, Smith MM. The highly conserved N-terminal domains of histones H3 and H4 are required for normal cell cycle progression. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol Cell Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA, Gould J, Kim S-Y, Kayne M, Hereford L. Identification of sequences in a yeast histone promoter involved in periodic transcription. Cell. 1986;23:537–544. doi: 10.1016/0092-8674(86)90285-0. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev. 2000;10:187–192. doi: 10.1016/s0959-437x(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Sengupta SM, VanKanegan M, Persinger J, Logie C, Cairns BR, Peterson CL, Bartholomew B. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J Biol Chem. 2001;276:12636–12644. doi: 10.1074/jbc.m010470200. [DOI] [PubMed] [Google Scholar]
- Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Spector MS, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Winston F. The Swi/Snf family of nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I, Carlson M. Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:1768–1775. doi: 10.1128/mcb.17.4.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Hosotani T, Miyakawa T. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 1998;26:3286–3292. doi: 10.1093/nar/26.13.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes & Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8–Tup1 transcriptional corepressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Wolffe AP. Chromatin remodeling and transcriptional activation: The cast (in order of appearance) Oncogene. 2001;20:2991–3006. doi: 10.1038/sj.onc.1204323. [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- Warner JR. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Wu J, Grunstein M. 25 years after the nucleosome model: Chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: High resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bone JR, Edmondson DG, Turner BM, Roth SY. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]