Abstract
Extraembryonic endoderm (ExE) is differentiated from the inner cell mass of the late blastocyst-stage embryo to form visceral and parietal endoderm, both of which have an important role in early embryogenesis. The essential roles of Gata-6 and Gata-4 on differentiation of visceral endoderm have been identified by analyses of knockout mice. Here we report that forced expression of either Gata-6 or Gata-4 in embryonic stem (ES) cells is sufficient to induce the proper differentiation program towards ExE. We believe that this is the first report of a physiological differentiation event induced by the ectopic expression of a transcription factor in ES cells.
Keywords: Gata-6, Gata-4, transcription factor, extraembryonic endoderm, embryonic stem cells
During early mouse development, the zinc finger-containing transcription factors, Gata-6 and Gata-4, are expressed in the extraembryonic endoderm (ExE) lineages, primitive endoderm (PrE), and the PrE derivatives: visceral endoderm (VE) and parietal endoderm (PE) (Morrisey et al. 1996). Gata-6 null mice die at embryonic day (E)5.5–E7.5 because of defects in VE formation and subsequent extraembryonic development (Morrisey et al. 1998). Mice null for Gata-4 die between E8 and E9 because of defects in heart morphogenesis (Kuo et al. 1997; Molkentin et al. 1997). Gata-4 null embryonic stem (ES) cells can generate cardiac myocytes but are partially defective in their ability to generate proper visceral endoderm and definitive endoderm of the foregut (Narita et al. 1997). This data suggested that such GATA factors might hold a pivotal role in the initiation and promotion of differentiation of ExE.
In our previous work, Gata-4 mRNA was increased during differentiation induced by elevation of Oct-3/4 or withdrawal of leukemia inhibitory factor (LIF) in ES cells (Niwa et al. 2000). To examine whether the induction of GATA factors is sufficient to drive the proper differentiation program toward ExE, we forced Gata-6 or Gata-4 overexpression in ES cells.
Results and Discussion
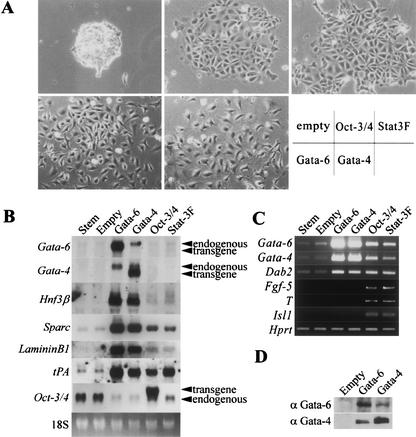
A large number of transcription factors, GATA factors, and hepatocyte nuclear factors (HNFs) are expressed in ExE. Gata-6 and Gata-4 are master regulator candidates because loss-of-function of either Gata-4 or Gata-6 resulted in loss of expression of all HNFs. In contrast, GATA factors were retained in mutant embryos lacking HNFs (Duncan et al. 1997, 1998; Barbacci et al. 1999; Coffinier et al. 1999). We introduced various endoderm-associated transcription factor genes into the supertransfection vector pCAG-IP (Niwa et al. 2002), and analyzed their gain-of-function phenotypes in MG 1.19 ES cells using episomal supertransfection (Gassmann et al. 1995; Niwa et al. 1998). This method allowed for efficient production of stable supertransfectants, reaching 50% of the treated cells. Thus, differentiation events induced by overexpression of transgenes can be easily confirmed by observing the morphology of primary transfectants. Among them, Hnf3α, Hnf4α, Hnf1α, Pdx1, Pax6, and Pax4 expression vectors cannot induce any morphological change (data not shown). The Ngn3 and Hnf3β vectors yielded no colonies, presumably due to toxic effects of overexpression. In contrast, we noticed altered morphologies of colonies that were transfected with Gata-6 and Gata-4 (Fig. 1A), which were dispersed refractive cells with stellate morphology. The morphology of these cells was reminiscent of PE cells (Jetten et al. 1979). The Gata-6 and Gata-4 transfectants were clearly distinguishable from Oct-3/4 (Niwa et al. 2000) and Stat3F (Niwa et al. 1998) expressing cells, which displayed pavements of closely apposed, large, flattened cells similar to ES cells cultured without LIF and were identified as mixed populations of different germ layer cells by differentiation marker gene expression patterns. Similar results were observed by conventional transfection of these vectors into germline-competent EB3 ES cells (data not shown).
Figure 1.
Supertransfection of Gata-4 and Gata-6 expression vector into MG 1.19 embryonic stem (ES) cells. (A) Photographs of colonies 7 d after transfection with each expression vector and selected in the presence of leukemia inhibitory factor (LIF). Gata-6 and Gata-4 transfectants show similar PE like morphology. (B) Northern blot analysis of endoderm markers in total RNAs from undifferentiated (stem), empty, Gata-6, Gata-4, Oct-3/4, and Stat3F supertransfected MG 1.19 ES cells. Reciprocal transactivation between Gata-6 and Gata-4 results in endoderm marker gene expression. (C) RT–PCR analysis of germ layer markers. (D) Western blots of nuclear extracts from empty, Gata-6, and Gata-4 vector transfected MG 1.19 cells. Induced expression of Gata-6 protein results in Gata-4 protein expression, and vice versa.
ExE gene marker expression was tested by Northern blot and RT–PCR in several supertransfectants (Fig. 1B,C). Induction of endogenous Gata-6 and Gata-4 was detected in both Gata-6 and Gata-4 transfectants. This is indicative of cross- and self-transactivation between these two GATA factors. Western blotting confirmed the proper production of GATA-4 and GATA-6 proteins in these transfectants (Fig. 1D). Neither Gata-6 nor Gata-4 was detectable in parental ES cells, empty vector expressing clones, Oct-3/4 transfectants, or Stat3F transfectants by Northern blot (Fig. 1B). However, a faint band for Gata-6 and Gata-4 was detected in undifferentiated ES cells and empty vector expressing clones by RT–PCR (Fig. 1C), which was compatible to the previous report for Gata-6 expression in ES cells as well as in blastocysts (Koutsourakis et al. 1999). Gata-6 and Gata-4 were also weakly induced in Oct-3/4 or Stat3F clones (Fig. 1C). The promotion of Gata-4 expression in Oct-3/4 transfectants was in agreement with our previous report (Niwa et al. 2000). The level of induced expression of the endogenous Gata-4 was much lower than the level of Gata4 transgene and endogenous Gata-4 expression induced by ectopic Gata-6 expression. Hnf3β, an early marker for endoderm differentiation (Farrington et al. 1997), and the parietal endoderm marker genes Sparc (osteonectin; Mason et al. 1986), LamininB1 (Senior et al. 1988), tissue plasminogen activator (tPA; Strickland et al. 1976), and disabled homolog 2 (Dab2; Morrisey et al. 2000) were induced strongly in Gata-4 and Gata-6 transfectants in parallel to the increased expression levels of GATA factors (Fig. 1B). This suggested a quantitative effect of these GATA factors on ExE differentiation.
To examine whether GATA overexpression in ES cells restricts their cell fate to the PrE lineage, we studied the expression of other differentiation marker genes. Exogenous GATA expression resulted in more obvious down-regulation of Oct-3/4 than in Oct-3/4 or Stat3F transfectants (Fig. 1B). These results suggested that the differentiation process induced by GATA factors may include active repression of Oct-3/4. Fibroblast growth factor (Fgf)-5, a marker of primitive ectoderm in early development (Hebert et al. 1991), was detected in Oct-3/4 and Stat3F transfectants, but not in GATA transfectants by our RT–PCR analysis (Fig. 1C). The mesoderm marker Brachyury (T; Wilkinson et al. 1990) and the neurectodermal motor neuron marker islet-1 (Isl1 Ericson et al. 1992) were also activated in Oct-3/4 and Stat3F, but not in GATA transfectants (Fig. 1C). This data clearly indicated that GATA factors restrict cell fate to ExE whereas Oct-3/4 and Stat3F interfere with ES self-renewal without tight restriction of cell fates.
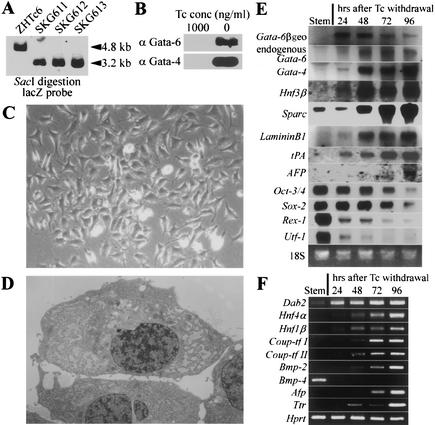
Analyses of knockout mice indicated that Gata-6 could be regarded as an upstream regulator of Gata-4 because loss of Gata-6 expression resulted in the absence of Gata-4, whereas loss of Gata-4 lead to the up-regulation of Gata-6 (Kuo et al. 1997; Morrisey et al. 1998). Because overproduction of both GATA factors caused similar differentiation events, as shown above and forced expression of one GATA factor resulted in activation of both endogenous genes, we focused on the function of Gata-6 in further analysis. To monitor the differentiation process triggered by Gata-6 more precisely, we generated an inducible Gata-6 transgene integration by supertargeting in ZHTc6 ES cells (Niwa et al. 1998). This strategy allowed for the establishment of ES cell lines with an ideally regulatable transgene expression controlled by tetracycline (Tc). Correctly targeted clones were selected by Southern blot analysis (Fig. 2A), and withdrawal of Tc from these clones, to induce Gata-6 transgene expression, resulted in the induction of uniform differentiation as observed in supertransfection (Fig. 2C). Out of these supertargeted clones, we used SKG612 as a representative clone for detailed analysis. Gata-6 transgene expression was evident from 24 h after withdrawal of Tc by Northern blot, and then endogenous Gata-6, Gata-4, Hnf3β, Sparc, LamininB1, tPA, and the VE marker α-fetoprotein (Afp; Dziadek and Adamson 1978) were gradually up-regulated (Fig. 2E). We could detect Gata-6 protein on Western blots after 24 h of induction by Tc withdrawal. Gata-4 protein up-regulated by Gata-6 was also detected after 24 h of Gata-6 induction (Fig. 2B). Morphological changes in SKG612 cells were apparent from 72 h after Tc withdrawal. From then the cells proliferated slowly and migrated while still maintaining their stellate shapes and multiple branching filopodia (Fig. 2C). The electron micrograph (Fig. 2D) showed a dilated rough endoplasmic reticulum, characteristic of PE (Enders et al. 1978). Stem cell marker genes Oct-3/4 and Sox-2 (Yuan et al. 1995) were gradually down-regulated. In contrast, the zinc finger protein Rex-1/Zfp42 and the transcriptional co-factor Utf-1, which were identified as targets of Oct-3/4 (Ben-Shushan et al. 1998; Nishimoto et al. 1999), were down-regulated more rapidly than Oct-3/4 (Fig. 2E). RT–PCR analysis (Fig. 2F) revealed that chicken ovalbumin upstream promoter transcription factor (Coup-tf)-I and -II, which were known to be up-regulated in PrE in blastocysts (Murray and Edgar 2001), were up-regulated in addition to Hnf4α and Hnf1β. It was reported previously that bone morphogenetic protein (Bmp)-2 expressed in VE, whereas Bmp-4 is found in the inner cell mass (ICM) and embryonic ectoderm of the embryo (Coucouvanis and Martin 1999). In our Gata-6 inducing cells, Bmp-2 is gradually up-regulated while Bmp-4 is extinguished immediately. In addition to the PE markers, the early VE markers Afp and transthyretin (Ttr; Abe et al. 1996) were also gradually up-regulated. This data confirmed that Gata-6 triggered the proper differentiation program toward ExE, especially towards PE.
Figure 2.
Gata-6 overexpression in stem cells leads to endoderm differentiation and blocks stem cell states. (A) An inducible Gata-6 transgene integration by supertargeting. For diagnosis of the supertargeting event, the selected clones were analyzed by Southern blot hybridization as reported previously (Niwa et al. 1998). A 3.2-kb SacI fragment was detected with a probe from the 5′ end of lacZ, indicative of the replacement of the Oct-3/4 cDNA with Gata-6. Original ZHTc6 cells retained the 4.8-kb fragment of the preexisting Oct-3/4 transgene. Supertargeting events had taken place in all clones tested, and SKG612 is representing one of such clones. (B) Western blot analysis of SKG612 embyonic stem (ES) cells using anti-Gata-6 or anti-Gata-4 antibody after incubation for 24 h at the indicated concentrations of tetracycline (Tc). Induced expression of Gata-6 and Gata-4 protein was detected in SKG612 cells. (C) Colony morphology of SKG612 cells cultured for 4 d in the presence of leukemia inhibitory factor (LIF) and absence of Tc. (D) Electron micrograph of differentiated SKG612 cells. Magnification, ×4000. (E) Time course for the activation of endoderm associated genes and the repression of stem associated genes during the differentiation of SKG612 cells, analyzed by Northern blot. (F) Time course for the activation of endoderm associated genes and the repression of Bmp-4, analyzed by RT–PCR.
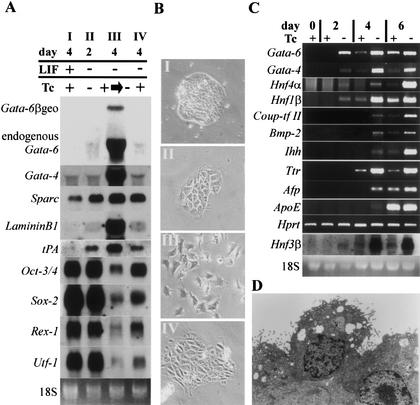
As shown above, overexpression of GATAs induced terminal differentiation of PE, although weak expression of GATAs were induced in PrE. To examine if strong Gata-6 expression acts to promote terminal differentiation, we induced Gata-6 expression in cells differentiated by withdrawal of LIF, which would become a mixture of PrE and mesoderm. Significant levels of endogenous Gata-6 transcripts were induced after 48 h of culture in a LIF-depleted medium. As shown in Figure 3A, endogenous Gata-6 and Gata-4 appeared at day 2 and day 4 in the LIF depleted culture, and both were strongly up-regulated when Gata-6 transgene expression was induced at day 4. Endoderm structural genes Sparc, LamininB1, and tPA were only weakly induced under LIF-depleted culture conditions, but the differentiating cells, in which Gata-6 was induced after LIF withdrawal, strongly expressed these transcripts. Stem-associated genes were gradually down-regulated under LIF depleted culture conditions, but when Gata-6 is induced after LIF withdrawal, the expression of such genes was tightly down-regulated, as found at 96 h after Tc withdrawal in SKG612 cells (Fig. 2E). As shown in Figure 3B, significant morphological change was led by induced Gata-6 expression, suggesting that strong Gata-6 expression might be involved in promoting the terminal differentiation of PrE to PE.
Figure 3.
Gata-6 promotes terminal differentiation of extraembryonic endoderm (ExE) after withdrawal of leukemia inhibitory factor (LIF) and promotes visceral endoderm (VE) differentiation under aggregated cell culture. (A) Northern blot analysis of SKG612 cells after (I) 4 d culture in the presence of LIF and tetracycline (Tc), (II) 2 d culture in the absence of LIF and presence of Tc, (III) further 2 d (total 4 d) following the withdrawal of Tc from a culture that was grown for 2 d in the absence of LIF and presence of Tc, (IV) 4 d culture in the absence of LIF and the presence of Tc. Gata-6 works not only as initiator but also as promoter of endoderm differentiation. (B) Photographs of SKG612 cells cultured under conditions described above. (C) SKG612 cells were cultured in suspension for 0, 2, 4, or 6 d in the presence of LIF and in the presence or absence of Tc. RNA was isolated at each time point and analyzed by RT–PCR. Note the correlation between the expression of endoderm-associated genes and concurrent or prior expression of Gata-6. (D) Electron micrograph of differentiated SKG612 cells in aggregated culture. Magnification, ×6000.
In Gata-6 knockout embryos, cellular defects were evident in VE (Morrisey et al. 1998), but overexpression of Gata-6 in ES cells mainly induced the PE phenotype. Because the VE phenotype can only be maintained in proper culture conditions, and VE cells can transdifferentiate into PE cells in two-dimensional culture (Hogan and Tilly 1981), it could be possible that we missed the induction of VE in our normal culture system. Because VE differentiation is maintained by embryoid body formation (Abe et al. 1996), we examined the effect of Gata-6 expression on VE differentiation by culturing SKG612 ES cells in the presence or absence of Tc in suspension, and compared the cells' ability to express VE marker genes. In this case, as shown in Figure 3C, the precocious expression of ExE transcription factors Hnf3β, Hnf4α, Hnf1β, and Coup-tf II were observed on Gata-6 induction. VE markers Bmp-2, Indian hedgehog (Ihh; Maye et al. 2000), Ttr, Afp, and apolipoprotein E (ApoE; Basheeruddin et al. 1987) were also enhanced by induction of Gata-6. Electron microscopic analysis confirmed that the cells that covered the surface of the embryoid body formed in the absence of Tc have a morphological future as VE (Fig. 3D). These cells displayed many microvilli and micropinocytotic vesicles and were joined together by small apical junctions. This data indicated that Gata-6 can enhance VE differentiation in the proper culture conditions, without enforcing differentiation to PE.
We have clearly shown that expression of these GATA factors is sufficient for triggering differentiation toward ExE. There have been several trials to induce differentiation in ES cells by overexpression of tissue-specific transcription factors; however, we believe that this is the first report of the induction of a physiological differentiation event by ectopic expression of a transcription factor in ES cells. ES cells might have a limited choice to undergo differentiation directly into PrE, trophectoderm (TE), and epiblast/primitive ectoderm. They can also differentiate by formation of embryoid bodies (EBs) via transition to primitive ectoderm, which might be essential to acquire competency for differentiation into embryonic germ layers. If so, forced expression of transcription factors specifically expressed in embryonic tissues cannot induce differentiation because of the lack of competency. Gata-4 overexpression causes a different response in ES and P19 embryonal carcinoma (EC) cells, inducing ExE and cardiomyocyte (Grepin et al. 1997), respectively. This difference might be based on the differential competency between these pluripotent cells because the P19 EC cells bear a characteristic resemblance to the primitive ectoderm.
What is the trigger of GATA expression in the natural course of ExE differentiation? Gata-6 and Gata-4 mRNAs were detectable in undifferentiated ES cells by RT–PCR, but not by Northern blot analysis, indicating very weak expression of these genes. Gata-4 transcripts were increased during differentiation induced by Oct-3/4 overexpression, Stat3F expression, withdrawal of LIF, or EB formation. We found that forced expression of Gata-6 or Gata-4 was sufficient to induce differentiation in the presence of LIF. The induction of endogenous Gata-4 and Gata-6 indicated the presence of cross- and auto-regulation pathways whose function could override the mechanism to maintain ES self-renewal. This data suggested that the repression of this GATA network is an important part of the molecular mechanism to maintain the ES cells in an undifferentiated state, a state that was released by withdrawal of LIF or up-regulation of Oct-3/4. However, down-regulation of Oct-3/4 induced TE transcription factors such as Hand-1 and Cdx-2 but not Gata-4 and Gata-6 (Niwa et al. 2000), which indicated that Oct-3/4 did not repress the GATA network directly. Next, we considered the endoderm differentiation model shown in Figure 4C. We hypothesized that unidentified transcription factor(s) activated by the cooperation of Oct-3/4 and its cofactor(s) acts as a repressor of Gata-6. During ES self-renewal, expression of Gata-6 is repressed by the hypothetical repressor X, at a point below the threshold level necessary to trigger differentiation; however, once expression of the repressor X is down-regulated, Gata-6 expression is up-regulated initially, then Gata-6 and Gata-4 trans- and self-activate themselves. Once the GATA protein expression level is increased beyond the threshold, the ExE differentiation process can progress.
Figure 4.
Forced expression of Coup-tf I represses stem cell marker genes. (A) Northern blot analysis of stem cell and differentiation marker genes in total RNAs from MG1.19 embryonic stem (ES) cells supertransfected with empty or Coup-tf I expression vectors. Overexpression of Coup-tf I results in down-regulation of stem cell marker genes Oct-3/4 and Utf-1. (B) Colony morphology of Coup-tf I supertransfectants. These cells show epithelial-like morphology distinct from extraembryonic endoderm (ExE). (C) Model for early endoderm differentiation of ES cells. See the text for details.
Because it was reported that COUP-TF could act as a repressor on the Oct-3/4 promoter (Ben-Shushan et al. 1995), COUP-TF induced by GATA expression might play some role to block stem cell self-renewal. Indeed, overexpression of Coup-tf I in ES cells using supertransfection repressed endogenous Oct-3/4 expression. This resulted in differentiation to epithelial cells that expressed the TE marker gene Hand-1 (Fig. 4A), a marker gene that was never induced in GATA transfectants (data not shown). However, the morphology of these epithelial cells was different from that of the TE-like cells induced by the down-regulation of Oct-3/4 (Fig. 4B). This data also suggested that COUP-TF alone is not sufficient to mimic the effect of GATA factors, and that the cooperative function of the GATA factors and their target genes, including Coup-tf, might be important to trigger the proper differentiation program toward ExE, and the down-regulation of stem-cell-specific genes.
Why is weak induction of GATA factors by overexpression of Oct-3/4 or inactivation of STAT-3 not sufficient to induce the terminal differentiation of the ExE? Our data clearly indicates that there is a threshold level for the promotion of a terminal differentiation event. Because strong expression of GATA factors can trigger the differentiation of ES cells, it might be possible that their weak induction is sufficient for initiation of differentiation but not for promotion. Alternatively, weak GATA induction could be just a result of PrE differentiation. It will be necessary for us to clarify this point by both in vitro and in vivo studies, in addition to identifying the repressor of Gata-6.
Materials and methods
Plasmid construction
Mouse Hnf3α, mouse Hnf3β, rat Hnf4α, rat Hnf1α, mouse Pdx1, mouse Ngn3, mouse Pax6, mouse Pax4, mouse Gata-6, mouse Gata-4, mouse Oct-3/4, human Coup-tf I, or Stat3F cDNA were introduced between EcoRI or BstXI sites of pCAG-IP (Niwa et al. 2002) for episomal expression. For supertargeting, mouse Gata-6 cDNA was introduced between the SfiI sites of the pSuperKO vector (Niwa et al. 1998).
ES cell culture and transfection
ZHTc6, MG 1.19, and their derivatives were cultured without feeders in LIF-supplemented medium as described previously (Niwa et al. 1998). Transfection of episomal expression vectors into MG 1.19 ES cells (supertransfection) was done as described previously using Lipofectamine 2000 (Invitrogen) (Niwa et al. 2002). For supertargeting, 1 × 107 ZHTc6 cells were electroporated with 100 μg of linearized SuperKO–Gata-6 plasmid DNA at 800 V and 3 μF using a Bio-Rad Gene Pulser, and then selected in the presence of 300 μg/mL G418 and 1 μg/mL Tc. Targeted clones were maintained in the continuous presence of Tc (1 μg/mL), and the supertargeting events were confirmed by Southern blot analysis as described previously (Niwa et al. 1998).
RNA isolation, Northern blot, and RT–PCR analyses
Total RNA was isolated from ES cells by TRIZOL Reagent (Invitrogen). Next, 5 μg of total RNA was separated on a denaturing formaldehyde gel of 1% agarose, blotted on to a Hybond-N nylon membrane (Amersham), and then analyzed by nonradioactive filter hybridization (Gene Image, Amersham). For RT–PCR analyses, cDNA was synthesized from 1 μg of total RNA, with an oligo-dT primer and Maloney murine leukemia virus RT (ReverTra Ace, Toyobo). One twentieth of the single strand cDNA products were used for each PCR amplification. Primer sets are listed in Table 1. By applying the indicated numbers of PCR cycles in Table 1, all cDNAs were amplified in quantitative ranges, which were confirmed by testing various cycles for the samples giving maximum levels of signals in each primer sets.
Table 1.
List of gene specific primers
| Gene
|
Forward primer
|
Reverse primer
|
Size of product (bp)
|
No. of cycle
|
GenBank accession no.
|
|---|---|---|---|---|---|
| Gata-6 | gcaatgcatgcggtctctac | ctcttggtagcaccagctca | 571 | 35 | AF179425 |
| Gata-4 | gcctgtatgtaatgcctgcg | ccgagcaggaatttgaagagg | 469 | 30 | U85046 |
| Dab2 | ggcaacaggctgaaccattagt | ttggtgtcgatttcagagtttagat | 283 | 35 | U18869 |
| Fgf-5 | aaagtcaatggctcccacgaa | cttcagtctgtacttcactgg | 465 | 30 | M30643 |
| T | atgccaaagaaagaaacgac | agaggctgtagaacatgatt | 835 | 30 | NM009309 |
| Is11 | agcaagaacgacttcgtgatg | gactgagagggtctccagctc | 187 | 35 | AJ132765 |
| Hnf4α | tgccctctcacctcagcaatg | cccctcagcacacggttttg | 357 | 49 | NM008261 |
| Hnf1β | gaaagcaacgggagatcctccgac | cctccactaaggcctccctctcttcc | 279 | 40 | AB008174 |
| Coup-tfI | agccatcgtgctattcacg | ttctcaccagacacgaggtc | 570 | 40 | MMU07625 |
| Coup-tfII | gcaagagcttcttcaagcg | gcttctccacttgctcttgg | 621 | 35 | U07635 |
| Bmp-2 | gtttgtgtttggcttgacgc | agacgtcctcagcgaatttg | 720 | 35 | NM007553 |
| Bmp-4 | tgtgaggagtttccatcacg | ttattcttcttcctggaccg | 566 | 35 | X56848 |
| Afp | tcgtattccaacaggagg | aggcttttgcttcaccag | 174 | 35 | V00743 |
| Ttr | agtcctggatgctgtccgag | ttcctgagctgctaacacgg | 440 | 30 | NM013697 |
| Ihh | acgtgcattgctctgtcaagt | ctggaaagctctcagccggtt | 221 | 35 | X76291 |
| ApoE | aggatgcctagccgagggagagc | tagatcctccatgtcggctccgagt | 322 | 35 | M12414 |
| Hprt | ctcgaagtgttggatacagg | tggcctataggctcatagtg | 350 | 25 | J00423 |
Nuclear extracts and Western blot analyses
ES cells were washed with PBS, scraped off the plates, and collected by centrifugation. Nuclear extracts were prepared as described by Gobert et al. (1996). Aliquots (7 μg) of nuclear extracts were fractionated on a 10% SDS–polyacrylamide gel and electroblotted onto PVDF membrane (Immobiron, Millipore). After treatment in blocking buffer (1× TTBS plus 3% skimmed milk), membranes were sequentially probed with the anti-Gata-6 (sc-7244, Santa Cruz) or anti-Gata-4 (sc-9053, Santa Cruz) HRP-conjugated anti-goat or rabbit IgG, then further developed using ECL reagents (Amersham)
Electron microscopy
Differentiated SKG612 ES cells were fixed with a fixative (2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M cacodylate buffer at pH 7.4) for 2 h at room temperature and stored at 4°C for several weeks. After washing with 0.1 M cacodylate buffer (pH 7.4) three times (5 min each), cells were postfixed with ice-cold 1% OsO4 in the same buffer for 2 h. The samples were rinsed with distilled water, stained with 0.5% aqueous uranyl acetate for 2 h at room temperature, dehydrated with ethanol, and embedded in Poly/Bed 812. Ultra-thin sections of these cells were cut, doubly stained with uranyl acetate and lead citrate, and viewed with a JEM 1010 transmission electron microscope (JEOL).
Acknowledgments
We thank Dr. M. Yamamoto (Tsukuba University) for providing Gata-6 cDNA, Dr. T. Morisaki (National Cardiovascular Center Research Institute) for providing Gata-4 cDNA, Dr. M. Takiguchi (Chiba University) for providing hCoup-tf I cDNA, Naoko Inoue (RIKEN) for technical assistance on electron microscopic analysis, and Dr. A. Smith (University of Edinburgh) and Neal M. Rao (RIKEN) for critical reading of the manuscript. This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Science, and Culture of Japan award to H.N.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL niwa@rtc.riken.go.jp; FAX 81-78-306-0101.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.968802.
References
- Abe K, Niwa H, Iwase K, Takiguchi M, Mori M, Abe S-I, Abe K, Yamamura K-I. Endoderm-specific gene-expression in embryonic stem cells differentiated to embryoid bodies. Exp Cell Res. 1996;229:27–34. doi: 10.1006/excr.1996.0340. [DOI] [PubMed] [Google Scholar]
- Barbacci E, Reber M, Ott MO, Breillat C, Huertz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- Basheeruddin K, Stein P, Strickland S, Williams DL. Expression of the murine apolipoprotein E gene is coupled to the differentiated state of F9 embryonal carcinoma cells. Proc Natl Acad Sci. 1987;84:709–713. doi: 10.1073/pnas.84.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shushan E, Sharir H, Pikarsky E, Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol Cell Biol. 1995;15:1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shushan E, Thompson JR, Gudas LJ, Bergman Y. Rex-1, a gene encoding a transcription factor expressed in the early embryo, is regulated via Oct-3/4 and Oct-6 binding to an octamer site and a novel protein, Rox-1, binding to an adjacent site. Mol Cell Biol. 1998;18:1866–1878. doi: 10.1128/mcb.18.4.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Thepot D, Babinet C, Yaniv M, Barra J. Essential role for the homeoprotein vHNF1/HNF1beta in visceral endoderm differentiation. Development. 1999;126:4785–4794. doi: 10.1242/dev.126.21.4785. [DOI] [PubMed] [Google Scholar]
- Coucouvanis E, Martin GR. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development. 1999;126:535–546. doi: 10.1242/dev.126.3.535. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: Tetraploid rescue of Hnf-4−/−. embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Navas MA, Dufort D, Rossant J, Stoffel M. Regulation of a transcription factor network required for differentiation and metabolism. Science. 1998;281:692–695. doi: 10.1126/science.281.5377.692. [DOI] [PubMed] [Google Scholar]
- Dziadek M, Adamson E. Localization and synthesis of alphafoetoprotein in post-implantation mouse embryos. J Embryol Exp Morphol. 1978;43:289–313. [PubMed] [Google Scholar]
- Enders AC, Given RL, Schlafke S. Differentiation and migration of endoderm in the rat and mouse at implantation. Anat Rec. 1978;190:65–78. doi: 10.1002/ar.1091900107. [DOI] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science. 1992;256:1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- Farrington SM, Belaoussoff M, Baron MH. Winged-helix, Hedgehog and Bmp genes are differentially expressed in distinct cell layers of the murine yolk sac. Mech Dev. 1997;62:197–211. doi: 10.1016/s0925-4773(97)00664-3. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Donoho G, Berg P. Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc Natl Acad Sci. 1995;92:1292–1296. doi: 10.1073/pnas.92.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert S, Chretien S, Gouilleux F, Muller O, Pallard C, Dusanter-Fourt I, Groner B, Lacombe C, Gisselbrecht S, Mayeux P. Identification of tyrosine residues within the intracellular domain of the erythropoietin receptor crucial for STAT5 activation. EMBO J. 1996;15:2434–2441. [PMC free article] [PubMed] [Google Scholar]
- Grepin C, Nemer G, Nemer M. Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development. 1997;124:2387–2395. doi: 10.1242/dev.124.12.2387. [DOI] [PubMed] [Google Scholar]
- Hebert M, Boyle M, Martin GR. mRNA localization studies suggest that murine FGF-5 plays a role in gastrulation. Development. 1991;112:407–415. doi: 10.1242/dev.112.2.407. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Tilly R. Cell interactions and endoderm differentiation in cultured mouse embryos. J Embryol Exp Morphol. 1981;62:379–394. [PubMed] [Google Scholar]
- Jetten AM, Jetten ME, Sherman MI. Analyses of cell surface and secreted proteins of primary cultures of mouse extraembryonic membranes. Dev Biol. 1979;70:89–104. doi: 10.1016/0012-1606(79)90009-5. [DOI] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor Gata6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes & Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Mason IJ, Taylor A, Williams JG, Sage H, Hogan BL. Evidence from molecular cloning that SPARC, a major product of mouse embryo parietal endoderm, is related to an endothelial cell 'culture shock' glycoprotein of Mr 43,000. EMBO J. 1986;5:1465–1472. doi: 10.1002/j.1460-2075.1986.tb04383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maye P, Becker S, Kasameyer E, Byrd N, Grabel L. Indian hedgehog signaling in extraembryonic endoderm and ectoderm differentiation in ES embryoid bodies. Mech Dev. 2000;94:117–132. doi: 10.1016/s0925-4773(00)00304-x. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes & Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. GATA-6: A zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol. 1996;177:309–322. doi: 10.1006/dbio.1996.0165. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes & Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey EE, Musco S, Chen MY, Lu MM, Leiden JM, Parmacek MS. The gene encoding the mitogen-responsive phosphoprotein Dab2 is differentially regulated by GATA-6 and GATA-4 in the visceral endoderm. J Biol Chem. 2000;275:19949–19954. doi: 10.1074/jbc.M001331200. [DOI] [PubMed] [Google Scholar]
- Murray P, Edgar D. Regulation of laminin and COUP-TF expression in extraembryonic endodermal cells. Mech Dev. 2001;101:213–215. doi: 10.1016/s0925-4773(00)00554-2. [DOI] [PubMed] [Google Scholar]
- Narita N, Bielinska M, Wilson DB. Wild-type endoderm abrogates the ventral developmental defects associated with GATA-4 deficiency in the mouse. Dev Biol. 1997;189:270–274. doi: 10.1006/dbio.1997.8684. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes & Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J-I, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J-I. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior PV, Critchley DR, Beck F, Walker RA, Varley JM. The localization of laminin mRNA and protein in the postimplantation embryo and placenta of the mouse: An in situ hybridization and immunocytochemical study. Development. 1988;104:431–446. doi: 10.1242/dev.104.3.431. [DOI] [PubMed] [Google Scholar]
- Strickland S, Reich E, Sherman MI. Plasminogen activator in early embryogenesis: Enzyme production by trophoblast and parietal endoderm. Cell. 1976;9:231–240. doi: 10.1016/0092-8674(76)90114-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Bhatt S, Herrmann BG. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature. 1990;343:657–659. doi: 10.1038/343657a0. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox-2 and Oct-3. Genes & Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]