Abstract
Fibroblast growth factor (FGF) signaling is involved in skeletal development of the vertebrate. Gain-of-function mutations of FGF receptors (FGFR) cause craniosynostosis, premature fusion of the skull, and dwarfism syndromes. Disruption of Fgfr3 results in prolonged growth of long bones and vertebrae. However, the role that FGFs actually play in skeletal development in the embryo remains unclear. Here we show that Fgf18 is expressed in and required for osteogenesis and chondrogenesis in the mouse embryo. Fgf18 is expressed in both osteogenic mesenchymal cells and differentiating osteoblasts during calvarial bone development. In addition, Fgf18 is expressed in the perichondrium and joints of developing long bones. In calvarial bone development of Fgf18-deficient mice generated by gene targeting, the progress of suture closure is delayed. Furthermore, proliferation of calvarial osteogenic mesenchymal cells is decreased, and terminal differentiation to calvarial osteoblasts is specifically delayed. Delay of osteogenic differentiation is also observed in the developing long bones of this mutant. Conversely, chondrocyte proliferation and the number of differentiated chondrocytes are increased. Therefore, FGF18 appears to regulate cell proliferation and differentiation positively in osteogenesis and negatively in chondrogenesis.
Keywords: FGF, osteogenesis, chondrogenesis, craniosynostosis, achondroplasia, gene targeting
The vertebrate skeleton is generated by two distinct mechanisms: intramembranous ossification and endochondral ossification. In intramembranous ossification, mesenchymal precursor cells condense and convert directly to bone-forming osteoblasts that secrete bone matrix proteins. This process is responsible for generating the flat bones of the skull in vertebrates (Opperman 2000). Endochondral ossification accounts for the formation of the vertebrae and long bones, and in this process mesenchymal cells first differentiate into cartilage, which provides the template for bone formation by osteoblasts (Olsen et al. 2000).
In both mechanisms, the entire process is coordinated by the action of members of secreting signaling molecule families, including FGF, TGF-β, IGF, Wnt, and hedgehog (Hartmann and Tabin 2000; Olsen et al. 2000; Opperman 2000). Evidence from human and mouse genetics indicates contribution of FGF signaling in these two mechanisms. Gain-of-function mutations in genes encoding FGFR1, FGFR2, and FGFR3 cause craniosynostosis, which is characterized by premature fusion of the cranial sutures (Malcolm and Reardon 1996; Webster and Donoghue 1997; Wilkie 1997; Burke et al. 1998). In addition to genetic studies, implantation of beads soaked with FGF2 or FGF4 around sutures results in promotion of osteogenic differentiation and suture closure (Iseki et al. 1997, 1999; Sarkar et al. 2001). FGF signaling is therefore considered to be involved in intramembranous ossification. However, studies on null mutations in Fgfr1 and Fgfr2 have shown early embryonic lethality (Deng et al. 1994; Yamaguchi et al. 1994; Arman et al. 1998; Xu et al. 1998), which has so far precluded analysis of the function of these genes in osteogenesis, whereas no calvarial defects have been observed in Fgfr3 loss-of-function mutants (Colvin et al. 1996; Deng et al. 1996). Null mutants for Fgf2 show abnormalities in maintaining bone mass and bone formation in the adult mouse, but not in the embryo (Zhou et al. 1998; Montero et al. 2000). Thus, it has not yet been revealed that FGF signaling is actually required for calvarial development. On the other hand, gain-of-function mutations in Fgfr3 also result in dwarfism, of which the most common is achondroplasia (Rousseau et al. 1994; Shiang et al. 1994; Webster and Donoghue 1996; Naski et al. 1998; Chen et al. 1999; Wang et al. 1999; McLean and Olsen 2001). In addition, gene disruption studies of mouse Fgfr3 have revealed that FGFR3-mediated signaling inhibits proliferation and differentiation of chondrocytes in the formation of the vertebrae and long bones, indicating that FGF signaling is essential for chondrogenesis in the process of endochondral ossification (Colvin et al. 1996; Deng et al. 1996). However, uncertainty remains as to which members of the Fgf family are actually required for chondrogenesis.
During both osteogenesis and chondrogenesis, FGF signaling is considered to regulate cell proliferation and differentiation. In calvarial ossification, several lines of evidence show that FGF signaling inhibits proliferation and/or accelerates differentiation of osteogenic cells. For instance, Iseki et al. (1997, 1999) showed that suture closure induced by grafting FGF2-soaked beads is associated with a localized decrease in cell proliferation and increased expression of osteopontin (Opn), a marker for osteogenic differentiation. In contrast, evidence also indicates that FGF2 enhances cell proliferation in cultures of mesencephalic neural crest cells, which give rise to osteoblasts (Sarkar et al. 2001). This apparent inconsistency in the action of FGF signaling is probably owing to variations in the differentiation stage of osteogenic cells used for individual experiments. In fact, Mansukhani et al. (2000) showed that immature osteoblasts respond to FGF treatment with increased proliferation, whereas FGF in differentiating osteoblasts does not induce DNA synthesis. To clarify the real function of FGF signaling in osteogenesis, genetic analysis of members of the FGF family and components of this signaling pathway should be important. On the other hand, FGF signaling appears to negatively regulate both proliferation and differentiation of chondrocytes. Fgfr3-deficient mice show expansion of proliferating and hypertrophic chondrocytes, resulting in progressive outgrowth of the long bones (Colvin et al. 1996; Deng et al. 1996).
The FGF family of cell signaling molecules is composed of at least 22 members in the mouse (Ornitz and Itoh 2001). Although FGF signaling has been implicated in bone development, studies on null mutant mice have not yet revealed the role of this family in skeletal development (Ornitz and Itoh 2001). This suggests that another candidate Fgf gene that plays an essential role in bone development may exist. Interestingly, a member of this family, FGF18 (Hu et al. 1998; Ohbayashi et al. 1998), is known to show regulatory activity in bone development. Ectopic application of FGF18 protein in the chick limb bud inhibits bone growth in the limb (Ohuchi et al. 2000). As an attempt to identify another candidate Fgf gene essential for bone formation, we have therefore precisely examined the expression of mouse Fgf18 in comparison with those of Fgfrs using in situ hybridization and show that Fgf18 is coexpressed with some of the Fgfrs during calvarial and long bone development. Furthermore, to investigate the role of FGF18 in vivo, we generated a null allele of the Fgf18 locus by homologous recombination in ES cells. Homozygous mutant mice for the targeted Fgf18 allele showed striking defects in both osteogenesis and chondrogenesis.
Results
Expression of Fgf18 during bone development in the mouse
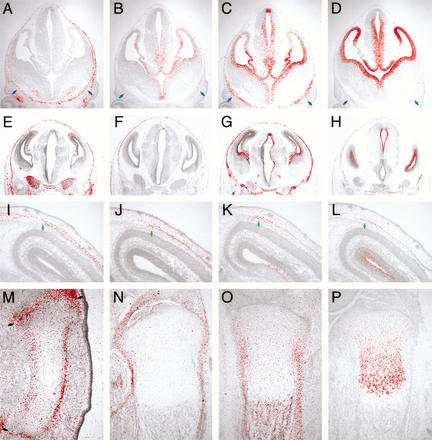
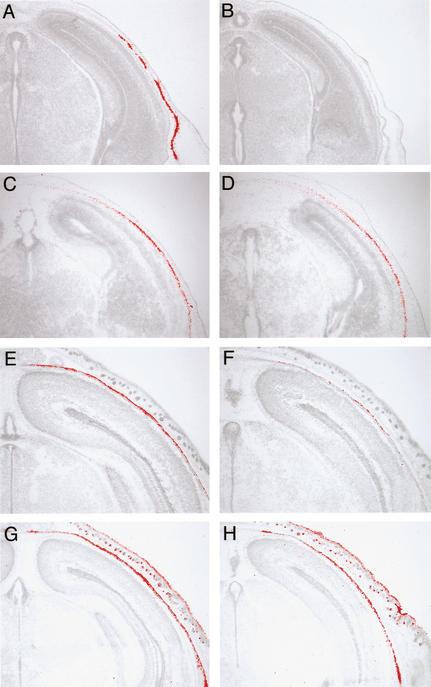
In calvarial development, the expression of Fgf18 was first detected in osteogenic mesenchymal cells surrounding the brain at 12.5 days postcoitum (dpc; Fig. 1A). At this stage, Fgfr1c, Fgfr2c, and Fgfr3c were expressed in ventral mesenchyme, where ossification is initiated (Fig. 1B–D, blue arrows). At 14.5 dpc, these three Fgfr genes were expressed in addition to Fgf18 in osteogenic mesenchyme (Fig. 1E–H). After 16.5 dpc, Fgf18 was expressed in differentiating osteoblasts and osteogenic mesenchyme between the osteogenic fronts (Fig. 1I). Fgfr1c was expressed in cells at the osteogenic front and in osteoblasts, whereas Fgfr2c and Fgfr3c were expressed intensely in cells at the osteogenic front (Fig. 1J–L; Kim et al. 1998; Johnson et al. 2000; Rice et al. 2000).
Figure 1.
Expression of Fgf18 and Fgf receptors in bone development. Localization of Fgf18 (A,E,I,M,N), Fgfr1c (B,F,J), Fgfr2c (C,G,K,O), and Fgfr3c (D,H,L,P) mRNA during calvarial bone development at 12.5 dpc (A–D), 14.5 dpc (E–H), and 16.5 dpc (I–L), and during tibial development at 14.5 dpc (M) and 18.5 dpc (N–P). 35S-labeled RNA probes of these genes were hybridized on cranial frontal sections and tibial longitudinal sections. (Blue arrows) Ventral osteogenic mesenchyme, (green arrows) osteogenic fronts, and (black arrows) developing joint.
In addition, Fgf18 expression was detected in developing long bones. Fgf18 was strongly expressed in the perichondrium and developing joint at 14.5 dpc (Fig. 1M). The similar expression pattern was observed at 18.5 dpc, along with expression in the perichondrium (Fig. 1N). At this stage, Fgfr2c was expressed in the perichondrium, periosteum, and endosteum (Fig. 1O; Orr-Urtreger et al. 1991; Delezoide et al. 1998; Britto et al. 2001), whereas Fgfr3c was expressed in proliferating and prehypertrophic chondrocytes (Fig. 1P; Peters et al. 1993; Wuechner et al. 1996; Delezoide et al. 1998).
Targeted disruption of Fgf18 gene
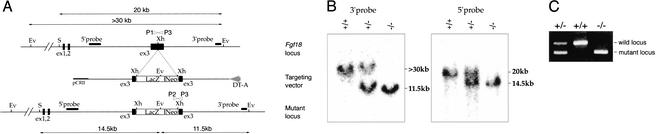
To investigate the functions of FGF18 in mouse development, we generated a null allele by inserting an IRES LacZ gene into the third exon of Fgf18 using the gene targeting method (Fig. 2A). In this construct, the IRES LacZ sequence was inserted at the position corresponding to the 16th amino acid downstream of the signal peptide cleavage site (Hu et al. 1998). Because all alleles of the FGF family members that have been truncated around this position are inactive, the successive targeting allele of Fgf18 was expected to be null (Ornitz and Itoh 2001). A genomic Southern blot with 5′ and 3′ flanking probes and PCR analysis confirmed that the predicted target allele was generated (Fig. 2B,C). Targeted ES cells were injected into C57BL/6 blastocysts, and the resulting male chimeric offspring were bred to C57BL/6 females. Mice carrying a heterozygous mutation in Fgf18 were viable, fertile, and morphologically normal except for slight distortions at the end of the tail (data not shown). LacZ staining in Fgf18+/− embryos revealed that expression of the LacZ reporter was detected as predicted by in situ hybridization analysis (data not shown). Interbreeding of Fgf18+/− mice to generate Fgf18−/− progeny resulted only in wild-type and heterozygote offspring in a 1:2 ratio, indicating that loss of Fgf18 results in embryonic lethality (Table 1). To determine when embryonic lethality occurred, genotyping of litters taken at 9.5–18.5 dpc was performed. Of the embryos recovered at each embryonic stage, ∼25% were homozygotes. Therefore, loss of FGF18 function leads to lethality just before or at birth.
Figure 2.
Targeted disruption of the Fgf18 gene. (A) Diagram of the Fgf18 locus (top), the targeting vector (middle), and the mutant allele (bottom). The location of the fragments used as probes in Southern blotting are shown, in addition to the size of restriction fragments detected by the probe. Open arrowheads (P1, P2, and P3) represent primers used in PCR screening. (Ev) EcoRV; (S) SacII; (Xh) XhoI. (B) Southern blot analysis showing accurate 3′ and 5′ targeting of the Fgf18 locus. The >30-kb and 11.5-kb fragments detected with the 3′ probe correspond to the wild-type and mutant alleles, respectively. The 20-kb and 14.5-kb fragments detected with the 5′ probe correspond to the wild-type and mutant alleles, respectively. (C) PCR genotyping of embryos showing fragments from the wild-type (489 bp; P1/P3) and mutant (335 bp; P2/P3) alleles.
Table 1.
Embryonic and postnatal survival of progeny obtained from Fgf18+/− intercrosses
| Stage
|
Genotype
|
Total
|
||
|---|---|---|---|---|
| +/+
|
+/−
|
−/−
|
||
| E9.5 | 39 (21.3%) | 99 (54.1%) | 45 (24.6%) | 183 |
| E12.5 | 43 (32.6%) | 56 (42.4%) | 33 (25.0%) | 132 |
| E14.5 | 32 (26.7%) | 61 (50.8%) | 27 (22.5%) | 120 |
| E16.5 | 23 (21.9%) | 49 (46.7%) | 33 (31.4%) | 105 |
| E18.5 | 51 (24.3%) | 105 (50.0%) | 54 (25.7%) | 210 |
| P0 | 16 (38.1%) | 26 (61.9%) | 0 (0.0%) | 42 |
Skeletal abnormalities in Fgf18-deficient embryos
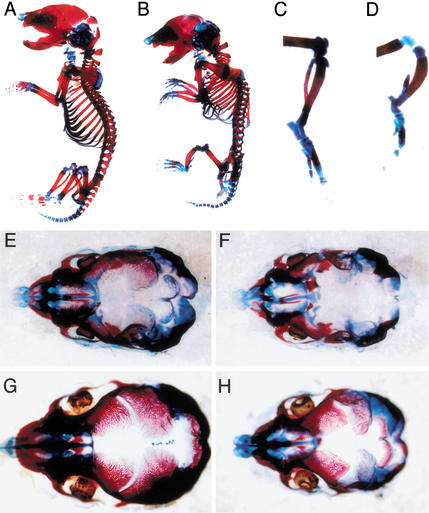
To assess whether the loss of Fgf18 causes defects in bone development, embryos at 16.5 and 18.5 dpc were stained with Alcian blue and Alizarin red (Fig. 3A–H). Ossification of frontal and parietal bones is initiated at the ventrolateral sides of plates of osteogenic mesenchyme overlying the brain, and progresses dorsomedially along each plate. In Fgf18−/− embryos, unossified areas appeared wider than in wild-type embryos in the metopic and saggital sutures, which separate the frontal and parietal bones, respectively, at the midline (Fig. 3E–H). Ossification appeared to be delayed by 1–2 d in Fgf18−/− embryos at 18.5 dpc. Unossified areas were also wider in the coronal sutures, located between the frontal and parietal bones (data not shown). A cleft palate was also observed in Fgf18−/− embryos at 18.5 dpc (data not shown). The progress of calvarial ossification was therefore delayed in Fgf18−/− mice.
Figure 3.
Skeletal analysis of Fgf18−/− embryos. Skeletal preparations of whole structures (A,B), hindlimbs (C,D), and crania (E–H) at 18.5 dpc (A–D,G,H) and 16.5 dpc (E,F) stained with Alizarin red and Alcian blue in wild-type (A,C,E,G) and Fgf18−/− (B,D,F,H) embryos.
In addition to the calvarial defect, skeletal abnormalities were observed in the vertebrae and long bones. In the vertebrae, the prominent phenotype was kyphosis in the cervical and upper thoracic spine observed at 18.5 dpc (Fig. 3A,B). Histological analysis revealed that bend of the spinal column was first observed at 13.5 dpc (data not shown). The shape of the ribs was also abnormal, resulting in reduction of the volume of the thoracic cavity (Fig. 3B). The long bones in both the fore- and hindlimbs appeared thick and short. In particular, the lengths of the ulna, radius, and tibia were decreased, and ossification of the fibula appeared incomplete (Fig. 3C,D).
Calvarial bone defects in Fgf18 mutants
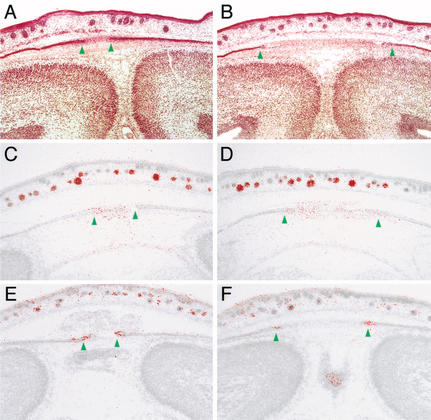
To examine delayed calvarial ossification in Fgf18−/− embryos more precisely, we next performed histological analysis and in situ hybridization, focusing on expression of Msx2 and Fgfr2c, which are normally expressed in the osteogenic mesenchyme and osteogenic front, respectively (Kim et al. 1998; Rice et al. 2000). The results clearly showed that the distance between osteogenic fronts in the metopic and saggital sutures appeared wider in Fgf18−/− embryos than in wild-type embryos at 18.5 and 16.5 dpc, but not in the presumptive sutures at 14.5 dpc (Fig. 4A–F; data not shown). Suture closure was therefore actually delayed in Fgf18−/− embryos after 16.5 dpc. On the contrary, no obvious abnormalities were observed in tissues outside of the skeletal system, suggesting that delay of the suture closure is likely to be a direct effect of Fgf18 on the skeletal cells.
Figure 4.
Delay of suture closure in Fgf18−/− embryos. Frontal sections around the saggital sutures of wild-type (A,C,E) and Fgf18−/− (B,D,F) embryos at 18.5 dpc stained with hematoxilin-eosin (A,B), and hybridized with Msx2 (C,D) and Fgfr2c (E,F) probes. Green arrowheads indicate the positions of the osteogenic fronts.
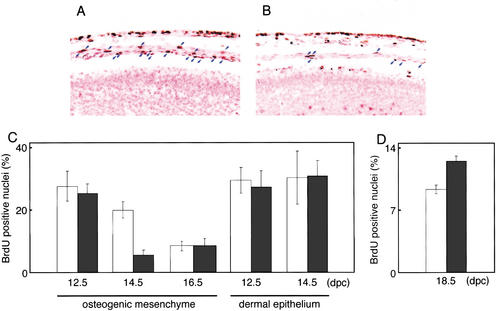
Some evidence has suggested that proliferation of osteogenic cells may be involved in the process of suture closure. For instance, implantation of FGF4-soaked beads results in increased proliferation of osteogenic mesenchymal cells at the suture in addition to accelerating suture closure (Kim et al. 1998). The P250R mutation in Fgfr1 produces premature suture fusion in mice, and increased proliferation of osteogenic cells at the suture is again observed (Zhou et al. 2000). Therefore, we next investigated proliferation of osteogenic cells in Fgf18−/− embryos using BrdU immunohistochemistry. At 12.5 dpc, no obvious difference in the proportion of BrdU-positive cells was observed in the cranial region (Fig. 5C). At 14.5 dpc, the proportion of BrdU-positive cells was specifically reduced in osteogenic mesenchymal cells overlying the brain (Fig. 5A–C). The proportion of BrdU-positive osteogenic cells was decreased even in the wild type at 16.5 dpc, but a difference was no longer observed between wild-type and Fgf18−/− embryos (Fig. 5C). These results indicate that FGF18 is required for maintenance of the highly proliferative ability of the osteogenic mesenchymal cells at 14.5 dpc.
Figure 5.
Reduced proliferation of osteogenic cells and increased proliferation of chondrocytes in Fgf18−/− embryos. (A,B) BrdU labeling in the mediodorsal regions of the crania of wild-type (A) and Fgf18−/− (B) embryos in the same litter at 14.5 dpc. Blue arrows indicate osteogenic mesenchymal cells labeled with BrdU. Counterstaining was performed with nuclear fast red. Artifactual separation of the layers of osteogenic mesenchyme from the dermal epithelium and neuroepithelium occurred during the process of fixation. (C,D) The proportion of BrdU-positive cells in osteogenic mesenchyme and in dermal epithelium in the mediodorsal regions of the crania at 12.5, 14.5, and 16.5 dpc (C) and in the proliferating chondrocytes at 18.5 dpc (D) of wild-type (open bars) and Fgf18−/− (solid bars) mice.
We next examined whether osteoblast differentiation is defective in Fgf18−/− embryos. Opn and osteocalcin (Oc) are normally expressed in differentiated osteoblasts (Nakase et al. 1994; Iseki et al. 1997, 1999). However, expression of Opn was severely decreased in calvarial osteoblasts at 14.5 dpc (Fig. 6A,B), whereas its expression appeared at the same level as the wild type at 16.5 dpc (data not shown). Expression of Oc was also severely decreased at 18.5 dpc (Fig. 6E,F). In addition, Alizarin red staining showed decreased calvarial bone mineralization (data not shown). In contrast, expression of cbfa1 normally begins at an earlier stage of osteogenic differentiation than Opn and Oc (Komori et al. 1997; Otto et al. 1997), but was clearly detected in calvarial osteoblasts of Fgf18−/− embryos from 12.5 dpc to 18.5 dpc (Fig. 6C,D,G,H; data not shown). These results indicated that FGF18 is not required for an earlier differentiation to cbfa1-expressing preosteoblasts, but is required for the normal progression of terminal differentiation to osteoblasts, which express Opn and Oc in addition to cbfa1. Therefore, both defective proliferation of osteogenic mesenchyme and delay of terminal differentiation to osteoblasts are likely to contribute to delay of ossification in Fgf18−/− embryos.
Figure 6.
Delay in maturation of osteoblasts in Fgf18−/− calvarial bone. Expression of Opn (A,B), Oc (E,F), and Cbfa1 (C,D,G,H) in wild-type (A,C,E,G) and Fgf18−/− (B,D,F,H) embryos at E14.5 (A–D) and E18.5 (E–H). 35S-labeled RNA probes of these genes are hybridized on cranial frontal sections. Note that the expression of Opn and Oc in osteoblasts is obviously reduced, whereas expression of Cbfa1 is unchanged.
Long bone defects in Fgf18 mutants
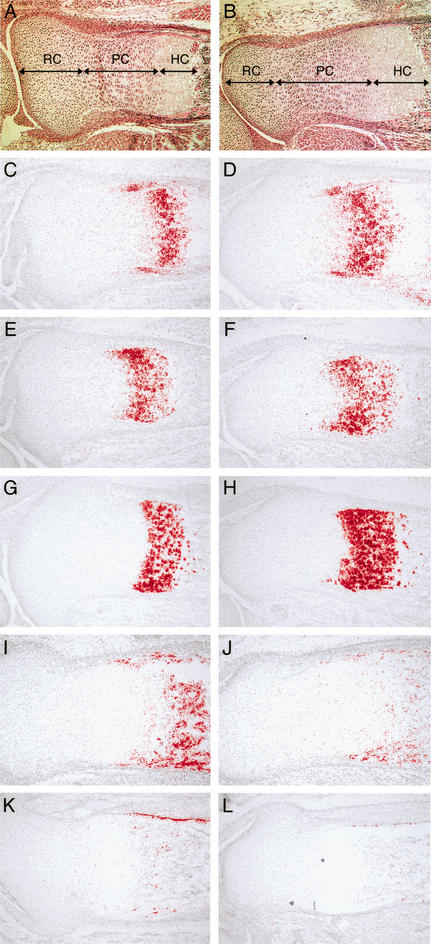
To examine defects in the long bones of Fgf18−/− embryos more closely, we first performed histological analysis. In endochondral ossification, chondrocytes sequentially transit through resting, proliferating, prehypertrophic, and hypertrophic stages. Histology of the growth plate indicated increased numbers of prehypertrophic and hypertrophic chondrocytes in Fgf18−/− embryos at 18.5 dpc (Fig. 7A,B). To confirm this histological observation, in situ hybridization analysis was performed using several markers expressed during chondrogenesis. PTHrP receptor (PTHrP-R) and Indian hedgehog (Ihh) are normally expressed in prehypertrophic chondrocytes and prehypertrophic and early hypertrophic chondrocytes, respectively (Lanske et al. 1996; Vortkamp et al. 1996; St-Jacques et al. 1999). The number of cells expressing PTHrP-R and Ihh was increased in Fgf18−/− embryos (Fig. 7C–F). Furthermore, the number of cells expressing type X collagen (ColX), a marker for the hypertrophic chondrocyte (Iyama et al. 1991), was also increased (Fig. 7G,H). These results indicated that FGF18 is required for negative regulation of differentiation into prehypertrophic and hypertrophic chondrocytes. In addition, histological analysis indicated that the number of proliferating chondrocytes was increased in Fgf18−/− embryos (Fig. 7A,B). To examine proliferation of chondrogenic cells in Fgf18−/− embryos, BrdU labeling analysis of the growth plate was performed. At 18.5 dpc, the proportion of BrdU-positive cells was specifically increased in proliferating chondrocytes in Fgf18−/− embryos (Fig. 5D). This result indicates that the absence of FGF18 leads to increased proliferation of chondrocytes.
Figure 7.
Enlarged proliferative and hypertrophic zone of chondrocytes and inhibited differentiation to osteoblasts in Fgf18−/− embryos. (A–L) Longitudinal sections of the proximal tibia of wild-type (A,C,E,G,I,K) and Fgf18−/− (B,D,F,H,J,L) embryos at 18.5 dpc stained with hematoxilin-eosin (A,B), and hybridized with PTHrP-R (C,D), Ihh (E,F), ColX (G,H), Opn (I,J), and Oc (K,L) probes. (RC) Resting chondrocytes; (PC) proliferating chondrocytes; (HC) hypertrophic chondrocytes.
To examine whether osteogenic differentiation is defective in the developing long bones of Fgf18−/− embryos, expression of Opn and Oc was analyzed by in situ hybridization. Expression of Opn and Oc was decreased in the tibia and metatarsal bones (Fig. 7I–L; data not shown). Therefore, FGF18 is required for the differentiation to osteoblasts in the development of the long bones as well as in that of the calvarial bones.
Discussion
FGF18 is required for both osteogenesis and chondrogenesis in bone development
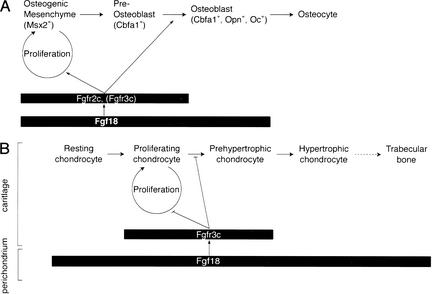
The characterization of Fgf18−/− embryos showed that this gene is required for both osteogenesis and chondrogenesis in the skeletal development of the mouse (Fig. 8). In calvarial development, Fgf18−/− embryos showed decreases in both proliferation of osteogenic mesenchymal cells (Fig. 5) and progression to the terminally differentiated osteoblasts (Fig. 6). Furthermore, in developing long bones of Fgf18−/− embryos, decreased terminal differentiation to Opn- and Oc-positive osteoblasts was also observed (Fig. 7I– L). Based on these results, we consider that FGF18 is required for the promotion of both proliferation of osteogenic mesenchymal cells and terminal differentiation to mature osteoblasts.
Figure 8.
Schematic representation of the expression and the roles of FGF18 in osteogenesis (A) and chondrogenesis (B). The differentiation stages and domains at which Fgf18 and Fgfrs are expressed are indicated by the rectangles. (A) In calvarial osteogenesis, osteogenic mesenchymal cells, which are characterized by expression of Msx2, differentiate to osteocytes through preosteoblasts, which express Cbfa1, but not Opn and Oc, and osteoblasts, which express Cbfa1, Opn, and Oc. Fgf18 is expressed in osteogenic mesenchyme, preosteoblasts, and osteoblasts. FGF18 positively regulates the proliferation of osteogenic mesenchyme and the differentiation to osteoblasts. FGFR2c, which can bind with high affinity to FGF18 and is transiently coexpressed with FGF18, may act as a receptor for FGF18 in this process. FGFR3c may function redundantly with FGFR2c (see Discussion). (B) In chondrogenesis of the long bone, the maturation of chondrocytes takes place through resting, proliferating, prehypertrophic, and hypertrophic stages. The hypertrophic chondrocyte is finally replaced by osteoblasts, which give rise to trabecular bone. FGF18 signaling negatively regulates the proliferation and differentiation of chondrocytes. In this case, FGF18 is expressed in the perichondrium and is likely to act on chondrocytes through FGFR3c.
On the other hand, Fgf18−/− embryos showed increased numbers of proliferating chondrocytes in addition to prehypertrophic and hypertrophic chondrocytes in the developing long bone (Fig. 7A–H). Furthermore, an increased proportion of BrdU-positive proliferating chondrocytes was observed in Fgf18−/− embryos, indicating that the posited negative regulation by FGF18 on chondrogenic proliferation. FGF18 therefore appears to play opposing roles during cell proliferation and differentiation in chondrogenesis to those it plays in osteogenesis.
Mechanism of calvarial development regulated by FGF18
The role of FGF18 in the proliferation of osteogenic mesenchyme may explain the considerable delay of suture closure in Fgf18−/− embryos. Gain-of-function mutations in Fgfr1, Fgfr2, and Fgfr3 are known to result in defects opposite in nature to those observed in Fgf18−/− embryos, namely, premature closure of the suture (Malcolm and Reardon 1996; Webster and Donoghue 1997; Wilkie 1997; Burke et al. 1998). Therefore, FGF signaling positively regulates the progression of suture closure. Interestingly, we found that the above three Fgfrs were transiently coexpressed with Fgf18 in the osteogenic mesenchyme at 12.5 dpc and 14.5 dpc and in the osteogenic front at 16.5 dpc (Fig. 1A–L). Therefore, delay of suture closure, a calvarial phenotype observed after 16.5 dpc in Fgf18−/− embryos, might be a consequence of earlier cellular events occurring in osteogenic mesenchymal cells and/or in preosteoblasts along the osteogenic front. In fact, decrease of cell proliferation in the osteogenic mesenchymal cells began at 14.5 dpc in Fgf18−/− embryos, whereas it began at 16.5 dpc in the wild type (Fig. 5A–C). Therefore, maintenance of proliferation of these cells may affect the progress of suture formation. If this hypothesis is true, the delay of suture closure in Fgf18−/− embryos would be attributable to decreased proliferation of osteogenic mesenchymal cells, whereas the premature suture closure observed in gain-of-function mutants of Fgfr1, Fgfr2, and Fgfr3 would be caused by increased proliferation of these cells. In addition, since another member of the FGF family, Fgf9, is still expressed in the osteogenic mesenchymal cells in Fgf18−/− embryos (data not shown), persistence of incomplete proliferative activity of the osteogenic mesenchymal cells caused by remaining FGF activity might result in delay, but not complete loss, of ossification in Fgf18−/− embryos.
FGF18 is also required for a process of the osteogenic differentiation specifically from cbfa1+, Opn−, and Oc− preosteoblasts to cbfa1+, Opn+, and Oc+ osteoblasts (Figs. 6 and 8). This event occurs in preosteoblasts that were left ventrolaterally to the osteogenic fronts in the dorsomedial progress of suture closure. Therefore, a defect in this event may not be a cause for delay of the progress of the osteogenic fronts in Fgf18−/− embryos. Rather, FGF18 appears to play a role in maturation of the osteoblasts, another process in the ossification. In vitro experiments show that FGF2 up-regulates Oc mRNA accumulation and Oc promoter activity in phenotypically immature MC3T3-E1 calvarial osteoblastic cells in the presence of cyclic AMP. FGF signaling may therefore directly activate the transcription of Opn and Oc (Boudreaux and Towler 1996; Newberry et al. 1997) in osteogenic cells in vivo.
Receptor for FGF18
As we have already mentioned, Fgfr1c, Fgfr2c, and Fgfr3c are transiently coexpressed with Fgf18 during calvarial development, suggesting that one or more of these proteins may act as a receptor for FGF18 during calvarial development (Fig. 1A–L).
Mitogenic activity of FGF18 for FGFRs-expressing cells showed that FGF18 can interact strongly with FGFR3c and modestly with FGFR2c, but not with FGFR1c or any b-type receptors (Xu et al. 2000). In addition, in vitro binding studies of FGF18 with a number of different FGF receptors using BIAcore have indicated that FGF18 can bind with high affinity to FGFR2c and FGFR3c, but not to FGFR1c (N. Itoh, unpubl.). FGFR2c and FGFR3c are, therefore, strong candidates to be bona fide receptors for FGF18, at least during calvarial development. However, gene disruption studies show that loss of FGFR2 results in early embryonic lethality (Arman et al. 1998; Xu et al. 1998), precluding an analysis of the role of FGFR2 in skeletal development. Loss of FGFR3 results in defective chondrogenesis, but no obvious defects in calvarial development (Colvin et al. 1996; Deng et al. 1996). FGFR2c may act as a receptor for FGF18 at a later stage than the development of lethality in null mutants, or these two receptors may be functionally redundant in calvarial development (Fig. 8A).
During chondrogenesis of the long bone, the phenotype of Fgf18−/− embryos resembles that of Fgfr3−/− embryos, indicating that FGFR3c acts as a receptor to FGF18 in this process. Because Fgfr3c is expressed in chondrocytes but not in the perichondrium (Fig. 1P; Peters et al. 1993; Wuechner et al. 1996; Delezoide et al. 1998), whereas Fgf18 is expressed in the perichondrium (Fig. 1M,N), FGF18 is therefore likely to act on chondrocytes through FGFR3c (Fig. 8B). However, the length of the long bone of Fgfr3−/− mice is considerably greater than the wild type, whereas that of Fgf18−/− mice appeared to be reduced. As mentioned above, FGF18 negatively regulates chondrogenesis, whereas it positively regulates osteogenesis. Therefore, reduced long bone length in Fgf18−/− embryos would represent dominance of the osteogenic defects to the chondrogenic ones in outgrowth of long bones (Fig. 3C,D). In contrast, greater long bone length in Fgfr3−/− mice would indicate that osteogenesis in these mice proceeds differently from that in Fgf18−/− embryos. Therefore, some other FGFR member is probably involved in osteogenesis during long bone development. Because Fgfr2c is expressed in the developing perichondrium, periosteum, and endosteum (Fig. 1O; Orr-Urtreger et al. 1991; Delezoide et al. 1998; Britto et al. 2001), it might represent a candidate for an FGF18 receptor during osteogenesis of developing long bones. However, we cannot exclude another possibility, that action of FGF18 on chondrocytes via another FGF receptor may also affect osteogenesis indirectly. To identify receptors actually receiving the FGF18 signal in vivo, examination of the genetic interactions between these FGFR candidates and FGF18 would be effective.
Materials and methods
Targeting vector
Recombinant phages containing the mouse Fgf18 locus were isolated from a 129/Sv genomic library (Stratagene) using Fgf18 cDNA as a probe. A 12.7-kb fragment including exons 3 and 4 was subcloned into pCRII. The neomycin phosphotransferase gene, followed by the lacZ gene placed between an independent ribosomal entry sequence (IRES) and an SV40 polyadenylation signal, was introduced at the XhoI site in the genomic DNA. A diphtheria toxin A (DTA) expression cassette was inserted at the 3′ end of the genomic DNA.
Targeted disruption and generation of Fgf18−/− embryos
CJ7 ES cells were electroporated with a NotI-linearized targeting vector and selected in G418 as described (Takada et al. 1994). Targeted clones were confirmed by Southern blot analysis of EcoRV-digested genomic DNA probed with the 3′ probe, and by Southern blot analysis of EcoRV/SacII-digested genomic DNA probed with the 5′ probe. Heterozygous ES cells were injected into blastocysts of C57BL/6 strain mice to generate germ-line chimeras. Chimeric males were mated with C57BL/6 females, and subsequent experimental procedures were performed on a mixed background. Genotypes were determined by Southern blotting as described above, or by PCR using the following three primers: P1, 5′- CCCAGATGTCATTGGGATAG-3′; P2, 5′- CCCGTGATATTGCTGAAGAG-3′; P3, 5′- TGAAT GGGAGGTCTCTAAGG-3′.
In situ hybridization
In situ hybridization was carried out on 4% paraformaldehyde fixed frozen sections until 12.5 dpc, and on unfixed frozen sections thereafter using 35S-labeled RNA probes as described (Yamasaki et al. 1996).
Skeletal preparation and histology
For skeletal preparations, embryos were eviscerated and fixed in 100% ethanol overnight and processed as described (Ikeya and Takada 2001). For histological analyses, unfixed frozen sections from crania and paraffin sections from hindlimbs of 18.5-dpc embryos were prepared and stained with hematoxylin-eosin according to standard procedures.
Analysis of BrdU incorporation
Pregnant mice were intraperitoneally injected with BrdU (100 mg/kg body weight) 2 h before being killed. Crania and hindlimbs of embryos were fixed overnight in Bouin's fixative at 4°C, dehydrated through an ethanol series, cleared in xylene, embedded in paraffin, and sectioned at 5 μm. BrdU was detected immunohistochemically with a Cell proliferation kit (Amersham) according to the manufacturer's directions, with the following modifications: sections were treated with 2% H2O2 in PBS at room temperature for 1 h and with 2 N HCl at 37°C for 1 h. Sections were then incubated with anti-BrdU antibody (Amersham) followed by incubation with a 1:200 dilution of biotinylated anti-mouse IgG antibody (Vector) in blocking buffer, and a 1:200 dilution of Vectastain ABC (Vector) in PBS. After labeling with anti-BrdU antibody, sections were stained with nuclear fast red. At least two litters were examined at each embryonic day.
Acknowledgments
We thank Dr. Komori for his donation of the mouse cbfa1 probe. We are grateful to Drs. Masatoshi Takeichi and Yo-ichi Nabeshima for helpful support. This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Culture and Sports of Japan; and grants from the Japan Science and Technology Corporation, Japan Society for the Promotion of Science, Kato Memorial Science Foundation, and Takeda Science Foundation to S.T.; and a grant-in-aid for scientific research from the Ministry of Education, Science, Culture and Sports of Japan and the Human Frontier Science Program, France to N.I.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL stakada@nibb.ac.jp; FAX 81-75-753-4265.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.965702.
References
- Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreaux JM, Towler DA. Synergistic induction of osteocalcin gene expression: Identification of a bipartite element conferring fibroblast growth factor 2 and cyclic AMP responsiveness in the rat osteocalcin promoter. J Biol Chem. 1996;271:7508–7515. doi: 10.1074/jbc.271.13.7508. [DOI] [PubMed] [Google Scholar]
- Britto JA, Chan JC, Evans RD, Hayward RD, Jones BM. Differential expression of fibroblast growth factor receptors in human digital development suggests common pathogenesis in complex acrosyndactyly and craniosynostosis. Plast Reconstr Surg. 2001;107:1331–1345. doi: 10.1097/00006534-200105000-00001. [DOI] [PubMed] [Google Scholar]
- Burke D, Wilkes D, Blundell TL, Malcolm S. Fibroblast growth factor receptors: Lessons from the genes. Trends Biochem Sci. 1998;23:59–62. doi: 10.1016/s0968-0004(97)01170-5. [DOI] [PubMed] [Google Scholar]
- Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- Delezoide AL, Benoist-Lasselin C, Legeai-Mallet L, Le Merrer M, Munnich A, Vekemans M, Bonaventure J. Spatio-temporal expression of FGFR 1, 2 and 3 genes during human embryo–fetal ossification. Mech Dev. 1998;77:19–30. doi: 10.1016/s0925-4773(98)00133-6. [DOI] [PubMed] [Google Scholar]
- Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes & Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development. 2000;127:3141–3159. doi: 10.1242/dev.127.14.3141. [DOI] [PubMed] [Google Scholar]
- Hu MC, Qiu WR, Wang YP, Hill D, Ring BD, Scully S, Bolon B, DeRose M, Luethy R, Simonet WS, et al. FGF-18, a novel member of the fibroblast growth factor family, stimulates hepatic and intestinal proliferation. Mol Cell Biol. 1998;18:6063–6074. doi: 10.1128/mcb.18.10.6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeya M, Takada S. Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech Dev. 2001;103:27–33. doi: 10.1016/s0925-4773(01)00338-0. [DOI] [PubMed] [Google Scholar]
- Iseki S, Wilkie AO, Heath JK, Ishimaru T, Eto K, Morriss-Kay GM. Fgfr2 and osteopontin domains in the developing skull vault are mutually exclusive and can be altered by locally applied FGF2. Development. 1997;124:3375–3384. doi: 10.1242/dev.124.17.3375. [DOI] [PubMed] [Google Scholar]
- Iseki S, Wilkie AO, Morriss-Kay GM. Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development. 1999;126:5611–5620. doi: 10.1242/dev.126.24.5611. [DOI] [PubMed] [Google Scholar]
- Iyama K, Ninomiya Y, Olsen BR, Linsenmayer TF, Trelstad RL, Hayashi M. Spatiotemporal pattern of type X collagen gene expression and collagen deposition in embryonic chick vertebrae undergoing endochondral ossification. Anat Rec. 1991;229:462–472. doi: 10.1002/ar.1092290405. [DOI] [PubMed] [Google Scholar]
- Johnson D, Iseki S, Wilkie AO, Morriss-Kay GM. Expression patterns of Twist and Fgfr1, -2 and -3 in the developing mouse coronal suture suggest a key role for twist in suture initiation and biogenesis. Mech Dev. 2000;91:341–345. doi: 10.1016/s0925-4773(99)00278-6. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Rice DP, Kettunen PJ, Thesleff I. FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lanske B, Karaplis AC, Lee K, Luz A, Vortkamp A, Pirro A, Karperien M, Defize LH, Ho C, Mulligan RC, et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- Malcolm S, Reardon W. Fibroblast growth factor receptor-2 mutations in craniosynostosis. Ann NY Acad Sci. 1996;785:164–170. doi: 10.1111/j.1749-6632.1996.tb56255.x. [DOI] [PubMed] [Google Scholar]
- Mansukhani A, Bellosta P, Sahni M, Basilico C. Signaling by fibroblast growth factors (FGF) and fibroblast growth factor receptor 2 (FGFR2)-activating mutations blocks mineralization and induces apoptosis in osteoblasts. J Cell Biol. 2000;149:1297–1308. doi: 10.1083/jcb.149.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean W, Olsen BR. Mouse models of abnormal skeletal development and homeostasis. Trends Genet. 2001;17:S38–S43. doi: 10.1016/s0168-9525(01)02458-1. [DOI] [PubMed] [Google Scholar]
- Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase T, Takaoka K, Hirakawa K, Hirota S, Takemura T, Onoue H, Takebayashi K, Kitamura Y, Nomura S. Alterations in the expression of osteonectin, osteopontin and osteocalcin mRNAs during the development of skeletal tissues in vivo. Bone Miner. 1994;26:109–122. doi: 10.1016/s0169-6009(08)80056-6. [DOI] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- Newberry EP, Boudreaux JM, Towler DA. Stimulus-selective inhibition of rat osteocalcin promoter induction and protein–DNA interactions by the homeodomain repressor Msx2. J Biol Chem. 1997;272:29607–29613. doi: 10.1074/jbc.272.47.29607. [DOI] [PubMed] [Google Scholar]
- Ohbayashi N, Hoshikawa M, Kimura S, Yamasaki M, Fukui S, Itoh N. Structure and expression of the mRNA encoding a novel fibroblast growth factor, FGF-18. J Biol Chem. 1998;273:18161–18164. doi: 10.1074/jbc.273.29.18161. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Kimura S, Watamoto M, Itoh N. Involvement of fibroblast growth factor (FGF)18-FGF8 signaling in specification of left–right asymmetry and brain and limb development of the chick embryo. Mech Dev. 2000;95:55–66. doi: 10.1016/s0925-4773(00)00331-2. [DOI] [PubMed] [Google Scholar]
- Olsen BR, Reginato AM, Wang W. Bone development. Ann Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- Opperman LA. Cranial sutures as intramembranous bone growth sites. Dev Dyn. 2000;219:472–485. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1073>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:3005.1–3005.12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Urtreger A, Givol D, Yayon A, Yarden Y, Lonai P. Developmental expression of two murine fibroblast growth factor receptors, flg and bek. Development. 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Peters K, Ornitz D, Werner S, Williams L. Unique expression pattern of the FGF receptor 3 gene during mouse organogenesis. Dev Biol. 1993;155:423–430. doi: 10.1006/dbio.1993.1040. [DOI] [PubMed] [Google Scholar]
- Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Petiot A, Copp A, Ferretti P, Thorogood P. FGF2 promotes skeletogenic differentiation of cranial neural crest cells. Development. 2001;128:2143–2152. doi: 10.1242/dev.128.11.2143. [DOI] [PubMed] [Google Scholar]
- Shiang R, Thompson LM, Zhu YZ, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Stark KL, Shea MJ, Vassileva G, McMahon JA, McMahon AP. Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes & Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- Wang Y, Spatz MK, Kannan K, Hayk H, Avivi A, Gorivodsky M, Pines M, Yayon A, Lonai P, Givol D. A mouse model for achondroplasia produced by targeting fibroblast growth factor receptor 3. Proc Natl Acad Sci. 1999;96:4455–4460. doi: 10.1073/pnas.96.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MK, Donoghue DJ. Constitutive activation of fibroblast growth factor receptor 3 by the transmembrane domain point mutation found in achondroplasia. EMBO J. 1996;15:520–527. [PMC free article] [PubMed] [Google Scholar]
- ————— FGFR activation in skeletal disorders: Too much of a good thing. Trends Genet. 1997;13:178–182. doi: 10.1016/s0168-9525(97)01131-1. [DOI] [PubMed] [Google Scholar]
- Wilkie AO. Craniosynostosis: Genes and mechanisms. Hum Mol Genet. 1997;6:1647–1656. doi: 10.1093/hmg/6.10.1647. [DOI] [PubMed] [Google Scholar]
- Wuechner C, Nordqvist AC, Winterpacht A, Zabel B, Schalling M. Developmental expression of splicing variants of fibroblast growth factor receptor 3 (FGFR3) in mouse. Int J Dev Biol. 1996;40:1185–1188. [PubMed] [Google Scholar]
- Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- Yamasaki M, Miyake A, Tagashira S, Itoh N. Structure and expression of the rat mRNA encoding a novel member of the fibroblast growth factor family. J Biol Chem. 1996;271:15918–15921. doi: 10.1074/jbc.271.27.15918. [DOI] [PubMed] [Google Scholar]
- Zhou M, Sutliff RL, Paul RJ, Lorenz JN, Hoying JB, Haudenschild CC, Yin M, Coffin JD, Kong L, Kranias EG, et al. Fibroblast growth factor 2 control of vascular tone. Nat Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YX, Xu X, Chen L, Li C, Brodie SG, Deng CX. A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 and premature fusion of calvarial sutures. Hum Mol Genet. 2000;9:2001–2008. doi: 10.1093/hmg/9.13.2001. [DOI] [PubMed] [Google Scholar]