Abstract
Dorsoventral asymmetry of the Antirrhinum corolla depends on expression of the CYC and DICH genes in dorsal petals. One role of these genes is to inhibit DIVARICATA (DIV), a determinant of ventral identity. Therefore, in cyc;dich double mutants ventral identity spreads all around the flower. We show that DIV encodes a protein belonging to the MYB family of transcription factors. Early on in corolla development, DIV affects specifically the growth of ventral and lateral petals but is transcribed in all petals. Analysis of a closely related gene suggests that the lack of effect on dorsal petals is not due to redundancy. More likely, therefore, DIV is regulated posttranscriptionally through a mechanism that depends on CYC and DICH. Later on, DIV affects growth and cell types and is transcribed mostly in a single layer of cells of ventral and lateral petals. This late pattern may itself depend on DIV activity because it fails to be established in a transcribed but inactive div mutant and, conversely, spreads all around the flower in cyc;dich double mutants.
Keywords: Flower development, dorsoventrality, Antirrhinum, DIVARICATA, MYB, transposons
Dorsoventral asymmetry of flowers is thought to have evolved several times from a radially symmetric condition. In Antirrhinum and Linaria, two closely related members of the Scrophulariacae, this asymmetry depends on expression of the CYCLOIDEA (CYC) gene in dorsal regions of the floral meristem (Luo et al. 1996; Cubas et al. 1999a). A similar pattern of expression has been found for a counterpart of CYC in Arabidopsis, a species with symmetric flowers that is distantly related to Antirrhinum (Cubas et al. 2001). This has suggested that a common ancestor with symmetric flowers already had an asymmetric pattern of gene expression that has been recruited several times to generate asymmetric flowers. Changes in gene interactions involving a CYC gene from this ancestor would account for the morphological manifestation of dorsoventral asymmetry. Gene interactions might in addition help explain how particular dorsoventral patterns evolved. To investigate such interactions we analyzed DIVARICATA (DIV), a gene that interacts with CYC to determine the dorsoventral pattern in Antirrhinum flowers (Almeida et al. 1997).
Antirrhinum flowers have four types of organs: sepals, petals, stamens, and carpels arranged in concentric whorls. Within each whorl there are several organs with different identities depending on their positions relative to a dorsoventral axis of the flower (Fig. 1). Dorsoventral asymmetry is particularly conspicuous in the corolla, which comprises five petals of three identities; two dorsal, two lateral, and one ventral. Each of the dorsal and lateral petals is itself dorsoventrally asymmetric, whereas the ventral petal is symmetric about the single plane of floral bilateral symmetry. Analysis of mutants in which this pattern is altered has led to the identification of four genes, CYC, RADIALIS (RAD), DICHOTOMA (DICH), and DIVARICATA, that control the overall asymmetry of the corolla or the asymmetries of individual petals in Antirrhinum (Stubbe 1966; Carpenter and Coen 1990; Luo et al. 1996, 1999; Almeida et al. 1997).
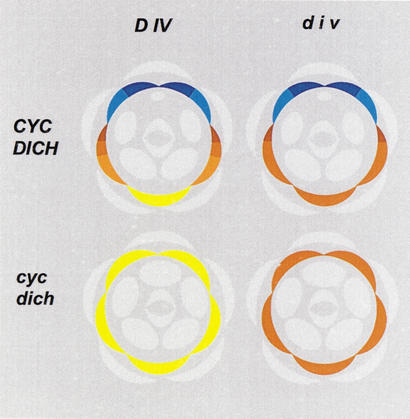
Figure 1.
Floral diagrams of wild type and dorsoventral mutants in Antirrhinum. Petals are in various colors, with all other organs in pale gray. Ventral identity is in yellow, lateral in brown, and dorsal in blue. The dorsal petal is divided in dorsal (dark blue) and lateral (pale blue) halves. In single dich mutants the dorsal half adopts a pale blue identity. The lateral petal is divided in three regions (shades of brown). div has very little or no effect on its most dorsal region (darkest brown) and affects its most ventral region (palest brown). In Div;div heterozygotes the lateral petal remains as in wild type while the ventral petal adopts the palest brown identity (see Almeida et al. 1997). Note that unlike cyc;dich, the div mutation has no effect on stamens.
CYC is required for lateral and dorsal identities as its inactivation results in partially ventralized phenotypes in which the lateral petals have symmetric ventral identity and the dorsal petals have laterodorsal, still asymmetric, identity. A similar ventralized phenotype is conferred by mutations in RAD. In dich mutants, the ventral and lateral petals remain as in wild type, but the asymmetry of the dorsal petals becomes reduced. The effects of cyc and dich on individual petals relate to a partially redundant role of the two genes in determining dorsoventral asymmetry. Thus, in double cyc;dich mutants, dorsoventral asymmetry is completely lost in the flower as a whole, which becomes radially symmetrical, and in individual petals, which adopt ventral identity (Fig. 1). In line with their redundant role, CYC and DICH are both specifically expressed in a dorsal domain of the floral meristem and later on in dorsal organs, and encode similar proteins belonging to the TCP family of transcription factors (Cubas et al. 1999b). However, within dorsal petals, the late domain of DICH expression is restricted to their most dorsal half, but that of CYC spreads through almost the entire petal.
Ventralization in the mutants has suggested that CYC and DICH have a role in inhibiting the spread of ventral identity toward dorsal positions. A candidate target for such inhibition is DIV, a determinant of ventral and lateral identities (Almeida et al. 1997). Similarly to cyc and dich, div mutants show a reduction in dorsoventral asymmetry. However, the polarity of the effect of div is opposite to that of cyc and dich. Thus, in div mutants a domain spanning the ventral petal and adjacent parts of the lateral petals adopts a more lateral identity, whereas the dorsal petals and the most dorsal part of the lateral petals remain as in wild type (Fig. 1). Consistent with a view that CYC and DICH inhibit DIV, the DIV domain spreads all around the flower in a background mutant for both cyc and dich as shown by the analysis of mutant combinations. Although double cyc;dich mutants are completely ventralized, triple cyc;dich;div mutants have lateral identity all around the flower.
We describe the isolation and analysis of DIV and of a series of div mutant alleles and show that DIV encodes a protein of the MYB family of transcription factors. At early stages of corolla development, DIV influences specifically the growth of ventral and lateral petals. However, DIV is transcribed in all primordia irrespective of their dorsoventral positions. It is unlikely that the lack of effect of DIV in dorsal petals is due to redundancy, because inactivation of a DIV-like gene (DVL) either on its own or in combination with a div null mutant has no effect on corolla morphology. Instead, the patterns of expression and activity suggest that DIV is inhibited posttranscriptionally in dorsal petals through a mechanism that depends on CYC and DICH. Late in corolla development, high levels of DIV mRNA accumulate in the inner epidermal layer of ventral and lateral petals, whereas low expression occurs across layers in dorsal regions. This ventral pattern of transcription appears to depend on DIV activity because it fails to be established in a transcribed but inactive div mutant. In contrast, in cyc;dich double mutants DIV is active in all petals, and therefore its ventral pattern of transcription spreads all around the flower.
Results
Isolation of the DIV gene
Transposon tagging has been the strategy adopted for gene isolation based on mutant phenotypes in Antirrhinum (e.g., Martin et al. 1985; Coen et al. 1990). This strategy requires the availability of unstable mutants carrying transposon insertions. However, in the case of the classical div mutant allele (div-35), no instability has ever been detected, suggesting that div-35 carries either a mutation unrelated to transposons or a stable insertion. Therefore, to isolate DIV, we first sought to obtain novel div mutants caused by transposon insertions by screening populations derived from selfed, highly inbred lines carrying active transposons (Carpenter and Coen 1990; Luo et al. 1991).
The classical div mutant allele (div-35) is semidominant to wild type. Both ventral and lateral petals are affected in mutant homozygotes (strong phenotype). In heterozygotes the ventral petal is affected, giving a weak but clearly mutant phenotype (Fig. 2). This semidominance is due to haploinsufficiency of the wild-type allele rather than to modified activity of the mutant allele (Almeida et al. 1997). Therefore, we screened the transposon-mutagenized populations for insertions that either abolish or strongly reduce DIV activity (as in the strong phenotype) or that reduce weakly the activity of DIV (as in the weak phenotype of heterozygotes). With this screen, we identified plants with the weak phenotype in two independent families. Selfing these plants gave only progeny with the weak phenotype, which suggests that they were homozygous for weak div mutant alleles rather than heterozygous for wild type and a strong allele. Consistent with this, crossing the novel mutants to div-35 gave a moderately strong phenotype in the F1 generation (Fig. 2). In the F2 generation, no wild-type phenotypes occurred. Instead, a 1:2:1 segregation of strong, moderately strong, and weak phenotypes was observed, showing that the new mutants were allelic to div-35. Thus, as expected, crossing the new mutants to wild type gave only wild-type or nearly wild-type progeny in the F1 generation, which in the F2 generation segregated 3 wild-type or nearly wild-type to 1 weak. We therefore concluded that in both cases the weak phenotypes were due to mutations in the DIV locus that caused partial reductions in DIV activity. These novel div alleles were named div-1 and div-2.
Figure 2.
Phenotypes of wild type and div mutants. Flowers are oriented to show the ventral side of the corolla. (A) Wild type. (B) Weak mutant phenotype of heterozygotes for wild type and div null alleles or of homozygotes for weak alleles. (C) Moderately strong phenotype of heterozygotes for weak and null alleles. (D) Strong phenotype of null alleles. The petals are fused at the base, forming a tube that ends in the lobes. The arrows indicate the position where the tube ends in the ventral petal. In wild type this position is hidden by the petal lobe. At the boundary between lateral and dorsal petals, tube length is similar for all phenotypes (for a detailed description of phenotypes, see Almeida et al. 1997).
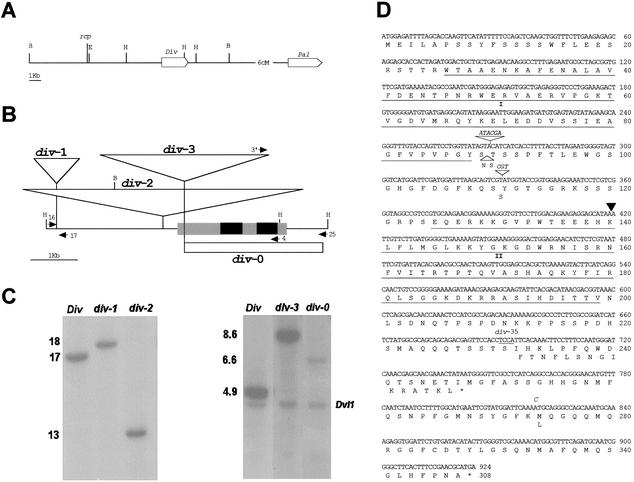
Neither of the two weak alleles gave wild-type revertant progeny, and no linkage between div phenotypes and various Antirrhinum transposons was detected by Southern analysis of segregating families. Therefore, we adopted an alternative, map-based, strategy for gene isolation. DIV is located 6 cM away from PALLIDA (PAL), a gene controlling flower color (Martin et al. 1985). Null pal mutants have ivory flowers unlike the red wild-type flowers. Several pal alleles conferring altered color patterns have been described (Coen et al. 1986; Almeida et al. 1989). Most of these alleles carry small promoter deletions that, relative to wild type, change the color pattern along the length of the flower (tube/lobe). However, an exceptional allele, pal-41, carries a large-scale inversion with a breakpoint in the pal promoter and the other at a locus, reciprocal (rcp), which is tightly linked to div (Fig. 3A; Robbins et al. 1989; Almeida et al. 1997). This allele confers an altered dorsoventral pattern of color, with red pigment accumulating mostly in ventral and lateral petals. One explanation might be that the inversion brought the PAL gene under the control of sequences normally regulating DIV. This would imply that DIV was located between RCP and PAL and only a few kilobases away from RCP, because the inversion has no effect on flower shape.
Figure 3.
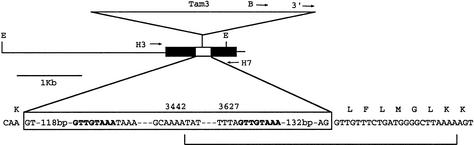
The div locus and mutants. (A) Chromosomal positions of rcp, div, and pal. The boxes indicate genes and their orientations. Restriction sites are EcoRI (E), BamHI (B), and HindIII (H). (B) Maps of DIV and div alleles. Black boxes are coding segments and gray boxes transcribed nontranslated regions. Transcription is from left to right. Triangles represent transposons. The white box shows the deletion in div-0. Arrows represent oligonucleotides (see Materials and Methods). (C) Southern blots of genomic DNA from the alleles indicated above the lanes. BamHI digests were probed with the BamHI–EcoRI segment containing the rcp locus (left panel). HindIII digests were probed with a cDNA clone (right panel). The cDNA initiates 166 bp to the left of the Tam3 insertion site, thus detecting a faint 6.6-kb band in div-0. A segment of the DVL1 gene is detected in the right panel. (D) Sequences of wild type and div-35. The main sequence is that of DIV in line JI2. The black triangle shows the position of the intron. Insertions or substitutions in DIV-25 and div-35 (shown in italic) result in the amino acid changes indicated below the main sequence. The underlined TCCA sequence is deleted in div-35 with the resulting frame-shift shown below the main sequence. Underlined amino acid sequences contain the MYB domains (I and II).
On the above assumption, we compared the restriction patterns of div-1 and div-2 with their wild-type isogenic sibs by Southern analysis using the BamHI–EcoRI fragment containing rcp as probe (Fig. 3A). Both mutants had altered restriction patterns relative to wild type. For example, with BamHI, wild type gave a 17-kb segment whereas div-1 and div-2 gave 18-kb and 13-kb segments, respectively (Fig. 3B,C). Together with the tight linkage between div and rcp (further extended by analysis of families segregating for the novel alleles), this result indicated that the region of the 17-kb BamHI segment contained the DIV gene. This was confirmed by molecular cloning and characterization of that region in wild-type and div mutant alleles, as described below (see also Materials and Methods).
DIV encodes a MYB protein
Using a genomic segment of the DIV locus as probe, we isolated a clone from an inflorescence cDNA library (Simon et al. 1994). The DIV cDNA had an open reading frame (ORF) encoding a protein of 307 amino acids. Mapping the transcription initiation site for the corresponding mRNA showed that the ORF was preceded by a long leader of 0.9 kb. As determined by aligning cDNA and genomic sequences, DIV had a single intron of 260 nucleotides. RFLP analysis using the cDNA as probe revealed that there was an unlinked closely related gene in the Antirrhinum genome. Isolation and analysis of this gene (DIVARICATA-like, DVL1) showed that it encoded a protein of 291 amino acids with 72% identity to DIV. DVL1 also had a single intron at the same position and with the same size as the intron in DIV.
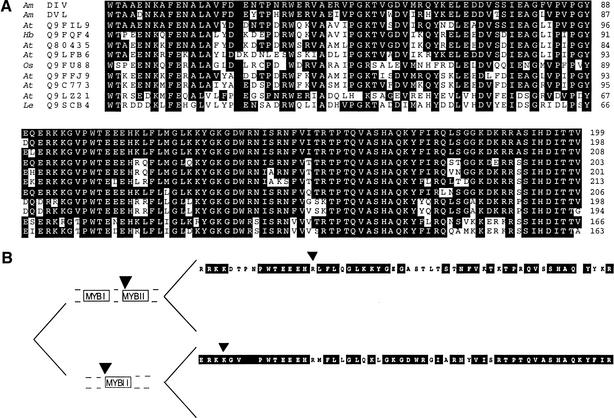
Searching databases showed that DIV had significant similarity to proteins of the MYB family of transcription factors. As first found in animals, these factors contain three characteristic imperfect repeats of a domain of ∼50 amino acids that is involved in DNA binding (Tanikawa et al. 1993). In plants, numerous variants of MYB proteins containing one to three of the characteristic repeats have been described (Jin and Martin 1999). DIV has two regions with MYB-like domains (Fig. 3D). The proteins with similarity to DIV could be divided in three classes according to similarities in the MYB domains.
Proteins in class I had identity to DIV in both MYB domains and a similar overall structure with the MYB I domain near the N terminus, separated from the MYB II domain by a variable spacer and with a variable C-terminal segment. In addition, all class I genes had a single intron at the same position different from other MYB genes. One of the class I proteins, LeMYBI (Q9SCB4; Fig. 4) of tomato, has been shown to bind DNA and activate transcription (Rose et al. 1999). It is unlikely that LeMYBI is a DIV ortholog because several proteins with higher identity to DIV than LeMYBI were found in Arabidopsis, Hevea, and rice, all species less closely related to Antirrhinum than tomato. The identity between DIV and DVL1 was higher than between any of these two proteins and a number of class I proteins from Arabidopsis. This suggests that DIV and DVL1 arose following a duplication that occurred after the split between the lineages leading to Antirrhinum and Arabidopsis.
Figure 4.
(A) Alignment of regions containing the MYB I (top) and II (bottom) domains of class I proteins in order of their relatedness to DIV. (Am) Antirrhinum majus, (At) Arabidopsis thaliana, (Os) Oryza sativa, (Hb) Hevea brasiliensis, (Le) Lycopersicum esculentum. Identities and alignments were according to Altschul et al. (1990) and Thompson et al. (1994). (B) Evolution of MYB II domains. The sequence above is that of SQY9, an Arabidopsis class I protein not shown in A. The sequence below is that of MYBSt1, a class II protein (one-MYB repeat). Residues in the black boxes are identical to those in DIV. Triangles represent introns. In having two MYB domains and from the position of its intron, DIV belongs to class I (above). However, DIV is more similar to MYBSt1 than to SQY9.
Class II proteins had only the MYB II domain and included MYBSt1, the first protein found in plants with a single MYB domain (Baranowskij et al. 1994). Both class I and class II had the characteristic motif SHAQKY in the MYB II domain (Rose et al. 1999). The MYB II domain of DIV had higher identity to that of MYBSt1 than to MYB II domains of some class I proteins. This suggests that diversity of MYB II domains reflects functional constraints under which they have evolved, because all class I genes have a similar overall structure different from that of class II genes (Fig. 4B).
Proteins of class III had only the MYB I domain and included a number of proteins from plants for which no biochemical or other role is known and proteins from animals (e.g., MIDA1) that function in the control of aspects of cell division (Shoji et al. 1995). Unlike in the case of the MYB II domain, which has been shown to bind DNA, the MYB I domain of class I proteins appears to be dispensable for DNA binding (Rose et al. 1999). This has led to the suggestion that the two domains may function in different ways, consistent with the observation that within class I proteins, the MYB I domains were more variable than the MYB II domains (Fig. 4A).
The div mutant alleles
Weak alleles carry transposon insertions upstream of the transcribed region
The weak alleles div-1 and div-2 carried transposon insertions upstream of the DIV transcription unit (Fig. 3). div-2 had an insertion of Ram1, a transposon previously trapped at the INCOLORATA locus (S. Doyle, R. Carpenter, and E. Coen, pers. comm.). div-1 had an insertion of a novel transposon of ∼1 kb (Tam11). Unlike most Antirrhinum transposons so far characterized, which have short terminal inverted repeats and cause target duplications of 3 or 8 bp, Tam11 had long terminal inverted repeats and caused the duplication of a 9-bp target. Southern analysis indicated that the Antirrhinum genome carried several copies of Tam11 (<10; data not shown). No long ORF was found within Tam11, suggesting that the copy at div may be a nonautonomous one. Although Tam11 resembled Mu elements of maize in having long terminal inverted repeats (Barker et al. 1984), database searches revealed no homologs of Tam11 in other species. The observation that these insertions confer weak phenotypes indicates that they partially reduce the expression of DIV. Presumably, the insertions could disrupt upstream DIV sequences that are important for normal expression. Alternatively, the transposons carry sequences that partially inhibit DIV expression.
div-35 encodes a truncated protein
Northern and RT–PCR analysis showed that the strong div-35 allele was transcribed (data not shown), suggesting that it might carry a mutation in the coding region. The sequence of div-35 cDNA showed several changes in the coding region relative to our wild-type reference sequence (stock JI2). All but one of these changes were in-frame insertions or silent or conservative nucleotide substitutions (Fig. 3D). The one exceptional change that could likely explain the div-35 strong phenotype was a 4-bp deletion that would result in a C-terminal truncated protein. This was tested by sequencing a wild-type DIV allele contained in stock JI25 (DIV-25). As shown previously, although JI25 is wild type with respect to DIV, its restriction pattern in the region of rcp is identical to that in div-35 and different from that in JI2, suggesting that div-35 could have arisen from DIV-25 (Almeida et al. 1997). The sequence of DIV-25 was fully consistent with our expectations because, except for the 4-bp deletion, DIV-25 was identical to div-35.
Deletion of the DIV gene (div-0) confers a strong phenotype
The finding that div-35 was transcribed and encoded a truncated protein retaining the MYB domains raised the question of whether it might have some activity. To address this problem, we generated a deletion mutant of DIV by following a two-step procedure. The first step involved selecting for an insertion of the transposon Tam3 in DIV, by PCR-based screening of a library of transposon-mutagenized plants (Davies et al. 1999). Tam3 has the advantages that its activity is favored in plants grown at 15°C and that it can give rise to deletions of flanking sequences (Ingram et al. 1997). Therefore, in a second step, these properties should enable us to select for a transposon-induced deletion of DIV.
The first step yielded a weak allele, div-3, carrying Tam3 in the transcribed, nontranslated leader of the DIV gene (Fig. 3C). div-3 was unstable, or leaky, producing flowers with phenotypes in the weak to wild-type range. As expected, wild-type revertants carrying typical Tam 3 excision footprints (Coen et al. 1989) occurred in progenies of div-3 homozygotes (data not shown). For the second step, we carried out a PCR-based selection for deletions affecting the DIV coding region. This screen yielded a deletion, div-0, with a break-point at the 3′ end of Tam3 and the other 744 bp downstream of the DIV transcription termination site, hence removing the entire DIV coding sequence. div-0 conferred a strong phenotype identical to that of div-35. We therefore concluded that div-35 is inactive and that the DIV gene is specifically required for determining the identities of ventral and lateral petals.
Phenotypic effects of DIV during flower development
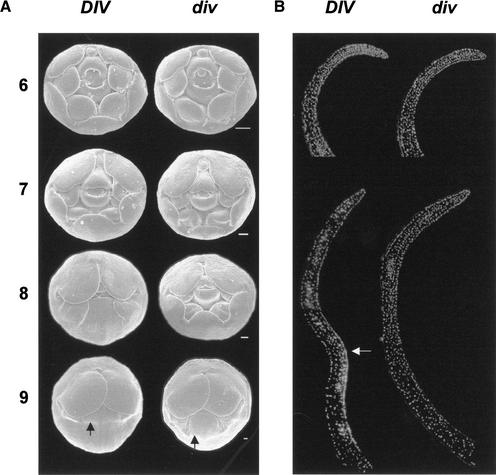
The phenotypic consequences of DIV activity were analyzed by comparing wild-type and div-0 mutant flowers at different developmental stages by scanning electron microscopy and histological analysis. Wild-type flower development has been divided into 15 stages, each comprising 3–5 nodes, that is, about 2 d (10 h separate each node at 25°C; Carpenter et al. 1995; Luo et al. 1996, 1999; C. Vincent, R. Carpenter, and E. Coen, pers. comm.). Wild-type and div-0 phenotypes were similar from stages 0 to 5 (data not shown). During these stages, bracts arose in the periphery of the apical inflorescence meristem (stage 0), floral meristems were generated and grew in the axils of bracts (stages 1–3), and floral organs arose sequentially in floral meristems (sepals at stage 4 and petals and stamens at stage 5).
For both wild-type and div-0, dorsoventral asymmetry of the flower, which is established before stage 5 (Luo et al. 1996), was already evident in the corolla at stage 6 (a stage defined by the emergence of two carpels in the center of the flower). At this stage the dorsal petals differed in shape from the ventral and lateral ones, and the most dorsal stamen showed retarded growth relative to the remaining stamens (Fig. 5A). However, individual organs showed little if any dorsoventral asymmetry. It is possible that at stage 6 the ventral and lateral petals had increased in length to a slightly greater extent in wild-type than in mutant flowers. This effect on growth became more obvious through stages 7 and 8, when in wild-type plants the margins of the ventral and lateral petals came closer to the carpel covering the stamens completely, but in the mutant plants the stamens and the ventral carpel remained exposed. Therefore, a clear early effect of DIV was an increase in the length of ventral and lateral petals. This enhanced growth was accompanied by an effect on petal shape. Up to stage 7, ventral and lateral petals had a similar round shape in both wild type and div-0. During stages 7 and 8, the wild-type ventral petal progressively adopted a more rectangular shape in contrast to the roundness of the lateral petals. In the mutant, however, the ventral petal retained the round shape at stage 8.
Figure 5.
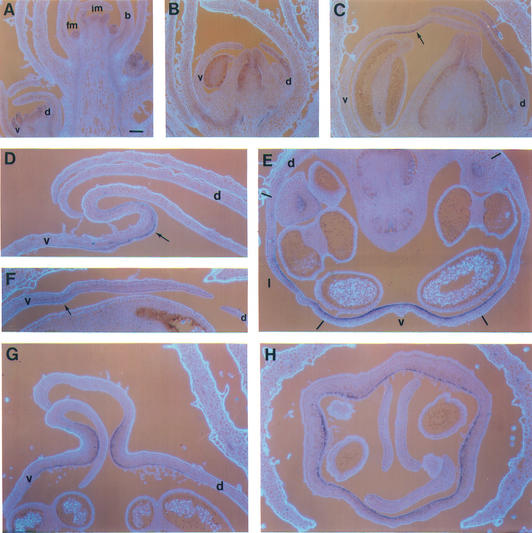
Development of wild type and div-0. (A) Scanning electron micrographs of buds from stages 6 to 9 (dorsal is to the top). Sepals were removed to show the corolla on the outside. The arrows show the ventral furrow. Scale bar, 100 μm. (B) Medial longitudinal sections of ventral petals stained with DAPI to show nuclei. The inner side (carpel side) of the petals is to the right. (Top left) Wild type at stage 7, (top right) div-0 at stage 8. (Bottom) Stage 9 sections. The arrow indicates the epidermal layer around the furrow with high density of cells. Note that the base of the wild-type petal at stage 9 is not shown.
By stage 9, a furrow that corresponds to the boundary between corolla tube and lobes developed across ventral and lateral petals in both wild-type and mutant plants. From stage 9 onward this furrow became a pronounced fold in the wild type but not in the mutant, as can be seen in longitudinal sections at stage 10 (Fig. 6D,E). In the wild type, this was accompanied by a much greater increase of tube length in the ventral petal and adjacent regions of the lateral petals than in the dorsal region of the lateral petal. As a result, the wild-type lateral petal became highly asymmetric (see stage 15 in Fig. 2; Almeida et al. 1997). In the mutant, tube length of the ventral petal and adjacent parts of the lateral petals did not increase to the same extent as in wild type, hence the reduction in lateral asymmetry.
Figure 6.
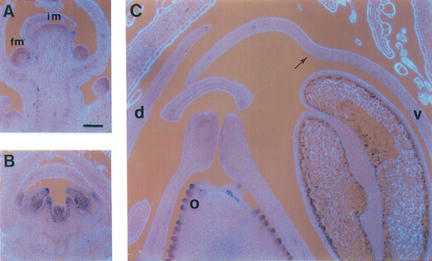
RNA in situ hybridization of wild-type and mutant sections probed with DIV. (A) Longitudinal section through a wild-type inflorescence showing the apical inflorescence meristem (im), bracts (b), floral meristem (fm), and a stage 6 bud with ventral (v) and dorsal (d) petals. (B,C) Medial longitudinal sections through wild-type buds at stages 7 (B) and 9 (C). The arrow in C points to the inner layer of the furrow. (D,E) Medial longitudinal sections of ventral petals of wild type (D) and div-35 (E) at stage 10 showing the fold in wild type (arrow). (F) Tranverse section of wild type at stage 10. Dashes show the positions of petal boundaries (which are in the medial planes of stamens). Lateral petals are marked (l). (G,H) Medial longitudinal (G) and transverse (H) sections of cyc;dich double-mutant buds at stage 10. Scale bar, 100 μm.
To investigate further the effect of DIV , we compared medial longitudinal sections of wild-type and mutant ventral petals at several developmental stages. Sections stained with DAPI to reveal nuclei showed that the ventral petals in wild type at stage 7 and mutant at stage 8 were similar with rather evenly spread cells (Fig. 5B). At stage 9, cells became unevenly spread in wild type, with the highest density of nuclei in a domain of the inner epidermis around the furrow. In contrast, no obvious asymmetry in the density of nuclei was detected in the mutant at stage 9.
In addition to affecting growth and shape, DIV had effects on cell types. For example, stripes of trichomes occur on the internal surface of the tube around the boundaries between ventral and lateral petals of wild type but not in the mutant (Almeida et al. 1997). These trichomes were clearly visible by stage 9 in wild type and did not arise in div-0 buds at any stage (data not shown).
Patterns of DIV transcription
Mutant analysis indicated that DIV activity had consequences specifically in ventral and lateral petals. To determine whether this reflected a specific pattern of DIV transcription, we carried out RNA in situ hybridization on sections of wild-type inflorescences (Fig. 6). In longitudinal sections, DIV mRNA was detected in the apical inflorescence meristem, in bract primordia arising in its periphery and in floral meristems produced in the axils of bracts (stages 0–3; Fig. 6A,B). From stage 3 to stage 8, transcripts were detected in all floral organs irrespective of their dorsoventral positions (Fig. 6B). As a control, the div-0 mutant did not give detectable signal (data not shown). Therefore, DIV transcription largely preceded any obvious DIV-dependent morphological effect and incorporated but was not restricted to the region affected in mutants.
From stage 9 onward DIV mRNA became barely detectable in bracts, sepals, and stamens. In the corolla, however, DIV expression was maintained and enhanced in some regions. Within ventral and lateral petals at stage 9, DIV adopted an asymmetric pattern of expression with high levels of transcripts in the inner epidermis of the furrow and very reduced levels in the remaining cell layers (Fig. 6C). Thus, DIV was expressed at stage 9 mostly in the domain of high density of cells revealed in DAPI-stained sections, a pattern that was maintained through later stages (Fig. 6D). In the dorsal petals, from stage 9 onward, DIV transcripts were still detected but with a more even distribution across cell layers than in the ventral petal. To determine the extent to which the inner–outer asymmetry of DIV transcription spread along the dorsoventral axis, we analyzed transverse sections through wild-type buds at stage 10. The domain expressing high levels of DIV mRNA in the inner epidermis ended or started fading within the lateral petals (Fig. 6E). Thus, the dorsoventral domain of high DIV expression coincided with the region affected in mutants.
To investigate whether the late ventral pattern of DIV transcription itself depended on DIV activity, we analyzed the expression pattern of div-35, an allele that, as described above, is transcribed but inactive. From stage 9 onward, div-35 mRNA was detected across cell layers in the ventral region without any obvious inner–outer asymmetry (Fig. 6F). Therefore, establishment of the late ventral pattern of DIV transcription depends, directly or indirectly, on DIV activity. This predicted that in cyc; dich double mutants the late ventral pattern of DIV transcription should spread all around the flower because in such mutant background DIV is active in all petals. Analysis of the pattern of DIV transcription in cyc;dich mutant flowers confirmed this prediction. In these flowers, DIV mRNA accumulated mostly in the inner epidermis of the corolla furrow, as seen in longitudinal sections (Fig. 6G), and all around the flower, as seen in transverse sections (Fig. 6H).
Analysis of redundancy in DIV function
Loss of DIV function had no effect in dorsal petals, although these petals expressed DIV mRNA. This lack of effect might be caused by redundancy, a possibility raised by the finding that DIV had a closely related gene, DVL1. Analysis of the pattern of DVL1 expression showed that it was transcribed in meristems and in all floral organs at early stages, similarly to DIV (Fig. 7). At later stages, however, DVL1 was transcribed mostly in ovules and showed very little expression in the region of the corolla furrow (Fig. 7C). Such a pattern would be consistent with DVL1 and DIV having an overlapping early role, but divergent functions at later stages.
Figure 7.
RNA in situ hybridization of wild-type sections probed with DVL1. (A) Inflorescence (im) and floral (fm) meristems. (B) Stage 6 bud. (C) Stage 9 bud showing low expression in the ventral furrow (arrow) and high expression in ovules (o). Scale bar, 100 μm.
To investigate this further, we carried out a PCR-based screen for transposon insertions in DVL1. The screen yielded an allele, dvl1-3, carrying Tam3 in the intron (Fig. 8). There was no obvious phenotype, possibly because the insertion had not inactivated the gene. We therefore carried out a second PCR-based selection for deletions adjacent to Tam3 in progenies of plants carrying dvl1-3. This screen gave an allele, dvl1-0, carrying a deletion with a breakpoint in Tam3 and the other in the second exon (Fig. 8). Thus, the 3′-splice acceptor site together with a coding segment of the conserved MYB II domain were removed. dvl1-0 plants had the same morphology as wild type. To test whether this was because DIV could completely substitute for a function of DVL1, we constructed div-0;dvl1-0 double mutants. These mutants had the div-0 phenotype without any additional obvious morphological change. Observations of ovules, which expressed high levels of DVL1 mRNA, showed no difference between dvl1 mutants and wild type, and the mutants were fully fertile and produced normal seedlings.
Figure 8.
Structure of dvl1 alleles. Black boxes are coding segments, and the white box is the intron. Transcription is from left to right. Arrows are oligonucleotides (see Materials and Methods). (E) EcoRI. The boxed sequence is from the intron and Tam3. Numbers above the box are positions in Tam3 (Hehl et al. 1991). The sequence in bold is the target duplication produced by the insertion of Tam3. The line at the bottom shows the deletion in dvl1-0.
Discussion
We have isolated the DIV gene using a map-based strategy combined with transposon mutagenesis. DIV encodes a protein belonging to the MYB family of transcription factors. In plants, this is a family that comprises numerous members with diverse functions. For example, in Antirrhinum, the MIXTA gene controls cell shape, and PHANTASTICA is involved in leaf development (Noda et al. 1994; Waites et al. 1998). MYB genes can be divided in various classes according to structural similarities. Members of the particular class to which DIV belongs have two MYB domains separated by a variable spacer and a variable C-terminal segment. An additional diagnostic property is that genes in the DIV class have a single intron at the same position. One of these genes encodes the I-box binding factor of tomato, which has been shown to bind DNA and activate transcription, although its function is unknown by mutational analysis (Rose et al. 1999). In the case of DIV, comparison of flower development in wild type and null mutants revealed specific effects on the identities of ventral and lateral petals that can be divided into early effects on growth and late effects on growth and cell types.
The earliest detectable consequence of DIV activity is an increase in the length of ventral and lateral petals at stage 6, corresponding to ∼2 d after petal primordia arise in the floral meristem (stage 5). This effect becomes more obvious in subsequent stages, so that by stage 8 the ventral and lateral petals are considerably longer in wild type than in div mutants. DIV could promote growth through an effect on cell proliferation, as suggested by the observation that the wild-type ventral petal at stage 7 resembles that of the div mutant at stage 8 in size and density of cells. The effect on growth is restricted to the ventral petal and its adjacent regions of the lateral petals. Thus, DIV activity results in the establishment of distinctions between ventral and lateral petals together with an increase in the asymmetry of lateral petals.
Although the DIV-dependent increase in length is restricted to ventral and lateral petals, DIV is transcribed in all petals. Therefore, early on in corolla development, the regional specificity of DIV activity is not attributable to transcriptional regulation. Instead, the lack of effect of DIV in dorsal petals could be caused by posttranscriptional regulation or redundancy. To test for redundancy, we have analyzed a gene, DVL1, encoding a protein with 72% identity to DIV and a similar early pattern of expression. A dvl1 null mutant, on its own or in combination with a div null mutant, had no further consequences on corolla morphology, suggesting that the lack of effect of DIV in dorsal regions is not due to redundancy. It is therefore possible that DIV is inhibited posttranscriptionally in dorsal petals. Such inhibition depends on the action of the dorsal genes CYC and DICH. However, these genes encode transcription factors and are expressed dorsally through the same early stages as DIV (Luo et al. 1996, 1999). Therefore, the negative effect of CYC and DICH on DIV is likely to be indirect.
By later stages of corolla development (stage 9), effects of DIV on cell types become manifest. The most obvious effect is the formation of stripes of trichomes on the inner surface of the corolla to either side of the boundary between ventral and lateral petals. In addition, DIV has further late effects on growth. At stage 9, a furrow that corresponds to the boundary between corolla tube and lobes develops across the ventral and lateral petals in wild type and mutant. However, a domain of closely spaced cells is formed in the inner epidermis around this furrow in wild type but not in the div mutant. These cells express high levels of DIV transcripts, whereas cells in other layers of the furrow or in dorsal petals express low levels. From stage 9 onward, this pattern is maintained while the furrow grows to form a pronounced fold in wild type but not in the mutant. This growth might again reflect an effect of DIV on cell proliferation in line with the finding that cells in the inner layer around the furrow also show preferential expression of cell cycle related genes such as CYCLIN D3b and HISTONE H4 (Gaudin et al. 2000).
The late ventral pattern of DIV transcription becomes established at stage 9, whereas effects of DIV on growth are already manifest at stage 6. Furthermore, this pattern fails to be established in div-35, a transcribed but inactive mutant. This suggests that DIV activity is required for generating the ventral pattern of DIV transcription. One consequence is that this pattern spreads all around the flower in cyc;dich double mutants because DIV activity is no longer inhibited in dorsal regions. A requirement for DIV in determining its own expression pattern could reflect its involvement in an autoregulatory loop. An example of one such loop involving a MYB gene required for cell proliferation is that of B-MYB in animals. The activity of B-MYB is enhanced through phosphorylation by CYCLIN A1/CDK2 complexes, whereas transcription of the CYCLIN A1 and B-MYB genes themselves is regulated by B-MYB (Muller-Tidow et al. 2001). No such cases have been documented in plants, although some cell cycle related genes are thought to be regulated by MYB factors (e.g., Ito et al. 2001).
The results above might help explain how the dorsoventral pattern of Antirrhinum flowers evolved. It is unlikely that the generation of dorsoventral asymmetry in the Antirrhinum lineage resulted directly from coupling the activities of DIV and CYC ancestral genes, because div mutant flowers are still asymmetrical. More likely, once dorsoventral asymmetry became established, DIV was recruited for further elaboration of the dorsoventral pattern. This could have occurred by cis-regulatory changes in DIV that enhanced and prolonged its expression. Such changes would have consequences restricted to ventral and lateral petals because DIV is inhibited posttranscriptionally in dorsal petals through a mechanism that depends on CYC. Therefore, the establishment of dorsoventral asymmetry would have been a prerequisite for the role of DIV to evolve. An outcome of the enhanced DIV-dependent growth was that the corolla became closed. Closed corollas also occur in asymmetric flowers of some members of diverse families (e.g., Utricularia in the Lentibulariaceae), suggesting that this configuration may have evolved several times independently, perhaps as an adaptation to the protection of reproductive organs (Olmstead et al. 1992; Endress 1996). Analysis of DIV counterparts in these families might therefore provide further insight into how this trait may have evolved.
Materials and methods
Plants
Alleles div-1 and div-2 were selected for in M2 families of a transposon mutagenesis experiment described elsewhere (Carpenter and Coen 1990; Luo et al. 1991) and kindly provided by R. Carpenter and E. Coen. The families in which these novel mutants arose were derived from JI2. The div-3 and div-0 alleles were also derived from selfed JI2 plants as described below. The classical div-35 allele, originally contained in line JI35 (Almeida et al. 1989), and the cyc;dich double mutant have been described elsewhere (Luo et al. 1996; Stubbe 1966; Almeida et al. 1997). The dvl1 allele carrying Tam3 was derived from selfed plants of line JI98. For double-mutant construction, single div-0 was crossed to single dvl1-0. In the resulting F2 families all possible genotypic combinations were then identified by Southern analysis.
Isolation and analysis of DIV and DVL1
Wild-type genomic DNA was digested with BamHI and fractionated by agarose gel electrophoresis. A fraction of ∼17 kb was purified and used to prepare a partial library in λ DASH, which was probed with a 5-kb BamHI–EcoRI segment of the rcp locus (Fig. 3). Various segments of a wild-type λ clone obtained in this way were subcloned into pBluescript (Stratagene) and used as probes against Northern blots of inflorescence mRNA. A 2-kb-long mRNA was detected with a 4-kb PstI–XbaI segment located to the right side of the insertion in div-2 (Fig. 3). No transcripts were detected using probes to the left of the insertion in div-2. The PstI–XbaI fragment was therefore used to isolate a clone from an inflorescence cDNA library. The sequence of the cDNA was matched against that of a 6720-bp genomic region from 958 bp to the left side of Tam11 to the HindIII site on the right side of DIV (Fig. 3). The DIV transcription start site was determined by 5′-RACE (GIBCO BRL). A complete cDNA was then obtained by RT–PCR.
Probing a family derived by selfing a DIV;div-35 heterozygote with a subclone of the DIV cDNA that comprised both MYB domains revealed an additional locus giving 3.5- or 12-kb EcoRI bands that segregated independently of div. The 3.5-kb segment was isolated from a partial library in λ ZAP (Stratagene) and shown to contain the 5′ part of the DVL1 gene. The 3′ end was obtained by 3′-RACE using standard primers for the 3′ end and primer H3 (CCTCCCAAATCTCTGCATTCTTCAC, 84 bp upstream of ATG toward 3′; Fig. 8).
Analysis of mutant alleles and PCR-based selection for mutants
A clone of div-2 was isolated from a partial library of the 13-kb BamHI genomic fraction, constructed in λ DASH as for wild type. The structure of the div-2 allele was determined by sequencing a subclone containing the left end of the insertion (Ram 1) and the flanking sequence of DIV. Ram 1 was inserted 288 bp upstream of the DIV transcription initiation site. The insertion in div-1 was mapped by Southern analysis and then characterized by sequencing a PCR product obtained with primers D16 (GAGAGTTGAGAACCGTG) and D17 (CTATAGTG CACTGGTGC) for DIV flanking sequences (Fig. 3). Tam 11 was inserted 2599 bp upstream of the DIV transcription initiation site, duplicating the target sequence GTTTGGCGG. div-35 was characterized by sequencing both a cDNA spanning the entire coding region obtained by RT–PCR and a genomic segment amplified by PCR. The sequence of DIV-25 was likewise obtained from PCR-amplified genomic DNA.
The libraries used in the selection for transposon insertions were described elsewhere (Davies et al. 1999) and kindly provided by E. Keck, R. Carpenter, and E. Coen (John Innes Institute, Norwich, UK). The libraries were screened with various primers for DIV or DVL1 in pairs with primers for either the 3′ or 5′ end of Tam3. div-3 was detected using primer D4 (CAT GCGTTCGGAAAGTGAAG, 3′ end of coding region toward 5′; Fig. 3) and a primer for the 3′ end of Tam3 (Ingram et al. 1997) in a library derived from line JI2. The insertion in DVL1 was detected using primer H7 (CTGAACCCAAATTGACAGGC ATC; Fig. 8) and the 3′ end of Tam3 in a library from line JI98. To select for Tam3-induced deletions, plants carrying the div-3 or dvl1-3 alleles were grown at 15°C. Pooled cotyledons from progeny seedlings of those plants were then used to prepare DNA, which was screened for deletions by PCR, following essentially the strategy described by Ingram et al. (1997). However, no permanent libraries were in this case established because, for economy of space, only the selected plants were potted. The div-0 allele was detected using primer D25 (CACTC GTCCCTGCCAATTTCTGTC; Fig. 3) and the 3′ end of Tam3. The dvl1-0 allele was detected using primer H7 and an internal primer for Tam3, primer B (GAAACGGTCTTGGCAATGGA TGGA; Fig. 8). The structures of all alleles were determined by sequencing the PCR products and confirmed by Southern analysis of genomic DNA in segregating families.
Microscopy and in situ hybridization
Preparation of samples for scanning electron microscopy was as described by Carpenter et al. (1995). For cell nuclei detection, tissue was prepared and stained as described by Fobert et al. (1994). In situ hybridization was essentially as described by Bradley et al. (1993). cDNA segments from DIV or DVL1 were subcloned in pBluescript and used for producing digoxigenin-labeled RNA probes. Segments corresponding to the entire DIV cDNA or to its 5′ or 3′ nontranslated regions all gave the same result. The specificity of the DIV in situ signal was further confirmed by Northern and RT–PCR analysis of RNA from dissected corollas of wild-type or div-35 buds at stages 9–11. Northerns showed that DIV or div-35 mRNA accumulated in all petals of wild-type or div-35 buds, respectively, whereas, as a control, CYC mRNA was detected only in dorsal petals. RT–PCR using primers between positions 247 and 456 (Fig. 3D), which detect a div-35 segment of 219 bp and a wild type segment of 210 bp, showed that wild-type buds only produced a wild-type transcript in all petals, and div-35 buds only produced a div-35 transcript in all petals.
Acknowledgments
We thank E. Coen, R. Carpenter, E. Keck, L. Copsey, and C. Vincent for providing mutants and mutant libraries and for advice on SEM; and E. Coen for discussions and constructive criticism of the manuscript. We thank T. Nunes and T.G. Antunes for the use of the SEM facility at the FCUL. This work was funded by the Portuguese Government (FCT-Sapiens 36221) and by the EU.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL almeida@itqb.unl.pt; FAX 351-214411277.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.221002.
References
- Almeida J, Carpenter R, Robbins TP, Martin C, Coen ES. Genetic interactions underlying flower color patterns in Antirrhinum majus. Genes & Dev. 1989;3:1758–1767. doi: 10.1101/gad.3.11.1758. [DOI] [PubMed] [Google Scholar]
- Almeida J, Rocheta M, Galego L. Genetic control of flower shape in Antirrhinum majus. Development. 1997;124:1387–1392. doi: 10.1242/dev.124.7.1387. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L. A novel DNA binding protein with homology to myb containing only one repeat can function as a transcription activator. EMBO J. 1994;13:5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker RF, Thompson DV, Talbot DR, Swanson J, Bennetzen J. Nucleotide sequence of the maize transposable element Mu1. Nucleic Acids Res. 1984;12:5955–5967. doi: 10.1093/nar/12.15.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes results from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell. 1993;72:85–95. doi: 10.1016/0092-8674(93)90052-r. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Coen ES. Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes & Dev. 1990;4:1483–1493. doi: 10.1101/gad.4.9.1483. [DOI] [PubMed] [Google Scholar]
- Carpenter R, Copsey L, Vincent C, Doyle S, Magrath R, Coen E. Control of flower development and phyllotaxy by meristem identity genes in Antirrhinum. Plant Cell. 1995;7:2001–2011. doi: 10.1105/tpc.7.12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E, Carpenter R, Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986;47:285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- Coen ES, Robbins TP, Almeida J, Hudson A, Carpenter R. Consequences and mechanisms of transposition in Antirrhinum majus. In: Berg D, Howe M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 411–434. [Google Scholar]
- Coen E, Romero JM, Doyle S, Elliot R, Murphy G, Carpenter R. Floricaula: A homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999a;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: Motif found in proteins regulating plant growth and development. Plant J. 1999b;18:215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- Cubas P, Coen E, Zapater JM. Ancient asymmetries in the evolution of flowers. Curr Biol. 2001;10:1050–1052. doi: 10.1016/s0960-9822(01)00295-0. [DOI] [PubMed] [Google Scholar]
- Davies B, Motte P, Keck E, Saedler H, Sommer H, Schwarz-Sommer Z. PLENA and FARINELLI: Redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 1999;18:4023–4034. doi: 10.1093/emboj/18.14.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. Diversity and evolutionary biology of tropical flowers. Cambridge Tropical Biology Series. Cambridge: Cambridge University Press; 1996. [Google Scholar]
- Fobert PR, Coen E, Murphy GJP, Doonan J. Patterns of cell division revealed by transcriptional regulation of genes during the cell cycle in plants. EMBO J. 1994;13:616–624. doi: 10.1002/j.1460-2075.1994.tb06299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamilichi C, Murray JAH, Coen E, Doonan J. The expression of D-Cyclin genes defines distinct developmental zones in Snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 2000;122:1137–1148. doi: 10.1104/pp.122.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl R, Nacken WK, Krause A, Saedler H, Sommer H. Structural analysis of Tam3, a transposon from Antirrhinum majus, reveals homologies to the Ac element of maize. Plant Mol Biol. 1991;16:369–371. doi: 10.1007/BF00020572. [DOI] [PubMed] [Google Scholar]
- Ingram GC, Doyle S, Carpenter R, Schultz EA, Simon R, Coen E. Dual role for fimbriata in regulating floral homeotic genes and cell division in Antirrhinum. EMBO J. 1997;16:6521–6534. doi: 10.1093/emboj/16.21.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y, Doonan J, Watanabe A. G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell. 2001;13:1891–1905. doi: 10.1105/TPC.010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB gene family. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Doyle S, Coen E. Pigmentation mutants produced by transposon mutagenesis in Antirrhinum majus. Plant J. 1991;1:59–69. [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen ES. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–799. doi: 10.1038/383794a0. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen ES. Control of organ asymmetry in flowers of Antirrhinum. Cell. 1999;99:367–376. doi: 10.1016/s0092-8674(00)81523-8. [DOI] [PubMed] [Google Scholar]
- Martin C, Carpenter R, Sommer H, Saedler H, Coen E. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO J. 1985;4:1625–1630. doi: 10.1002/j.1460-2075.1985.tb03829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Tidow C, Wang W, Idos GE, Diederichs S, Yang R, Readhead C, Berdel WE, Serve H, Saville M, Watson R, et al. Cyclin A1 directly interacts with B-myb and cyclinA1/cdk2 phosphorylate B-myb at functionally important serine and threonine residues: Tissue-specific regulation of B-myb function. Blood. 2001;97:2091–2097. doi: 10.1182/blood.v97.7.2091. [DOI] [PubMed] [Google Scholar]
- Noda K, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369:661–664. doi: 10.1038/369661a0. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Michaels HJ, Scott KM, Palmer JD. Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Ann Missouri Botanical Garden. 1992;79:249–265. [Google Scholar]
- Robbins TP, Carpenter R, Coen E. A chromosome rearrangement suggests that donor and recipient sites are associated during Tam 3 transposition in Antirrhinum majus. EMBO J. 1989;8:5–13. doi: 10.1002/j.1460-2075.1989.tb03342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A, Meier I, Wienand U. The tomato I-box binding factor LeMYBI is a member of a novel class of Myb-like proteins. Plant J. 1999;20:641–652. doi: 10.1046/j.1365-313x.1999.00638.x. [DOI] [PubMed] [Google Scholar]
- Shoji W, Inoue T, Yamamoto T, Obinata M. MIDA1, a protein associated with Id, regulates cell growth. J Biol Chem. 1995;270:24818–24825. doi: 10.1074/jbc.270.42.24818. [DOI] [PubMed] [Google Scholar]
- Simon R, Carpenter R, Doyle S, Coen E. Fimbriata controls flower development by mediating between meristem and organ identity genes. Cell. 1994;78:99–107. doi: 10.1016/0092-8674(94)90576-2. [DOI] [PubMed] [Google Scholar]
- Stubbe H. Genetik und Zytologie von Antirrhinum L. sect. Antirrhinum. Jena: VEB Gustav Fischer Verlag; 1966. [Google Scholar]
- Tanikawa J, Yasukawa T, Enarmi M, Ogata K, Nishimura Y, Ishii S, Sarai A. Recognition of specific DNA sequences by the c-myb protooncogene product: Role of three repeat units in the DNA binding domain. Proc Natl Acad Sci. 1993;90:9320–9324. doi: 10.1073/pnas.90.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Selvadurai HRN, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]