Abstract
Small RNA molecules have been found to be specifically associated with posttranscriptional gene silencing (PTGS) in both plants and animals. Here, we find that small sense and antisense RNAs are also involved in PTGS in Neurospora crassa. The accumulation of these RNA molecules depends on the presence of functional qde-1 and qde-3 genes previously shown to be essential for gene silencing, but does not depend on a functional qde-2, indicating that this gene is involved in a downstream step of the gene silencing pathway. Supporting this idea, a purified QDE2 protein complex was found to contain small RNA molecules, suggesting that QDE2 could be part of a small RNA-directed ribonuclease complex involved in sequence-specific mRNA degradation.
Keywords: Posttranscriptional gene silencing, double-stranded RNA, short interfering RNA, quelling defective, small temporal RNA, RNA-induced silencing complex
Posttranscriptional gene silencing (PTGS) occurs in fungi, plants, and animals as a response to various types of foreign nucleic acids including transgenes, transposons, viral RNAs, and double-stranded RNA (dsRNA; Baulcombe 1999; Bosher and Labouesse 2000; Cogoni and Macino 2000; Waterhouse et al. 2001; Zamore 2001). In this process RNA molecules with sequence similarity to the introduced nucleic acid are degraded in a sequence-specific manner. Gene silencing is thought to represent an ancient natural defense system against viruses and transposons that has been conserved through evolution (Matzke et al. 2000). Recent studies have, indeed, indicated that the molecular basis of the PTGS mechanism is similar in different organisms. In particular, genetic screens carried out in Neurospora crassa, Arabidopsis thaliana, and Caenorhabditis elegans have identified homologous genes required for PTGS (Cogoni and Macino 1997; Ketting and Plasterk 2000; Mourrain et al. 2000). An emerging common model for PTGS is that transgenes or transposons are the source RNA molecules that are recognized by the silencing machinery and that subsequently activate a cascade of events leading to sequence-specific mRNA degradation. A key step in this process is likely to involve RNA-dependent RNA polymerases (RdRPs) that have been found to be required for PTGS in plants (Dalmay et al. 2000; Mourrain et al. 2000), fungi (Cogoni and Macino 1999a), and animals (Smardon et al. 2000). It has been proposed that the RdRP can recognize RNAs produced from transgenes or transposons and convert them into dsRNAs. Recent findings, obtained using cell-free extracts of Drosophila, indicate that dsRNA is processed into small RNA molecules of 21–23 nucleotides by an enzyme called Dicer, which contains both an RNase III and an RNA helicase domain (Zamore at al. 2000; Bernstein et al. 2001). Similar-sized small RNAs have also been associated with transgene-induced silencing in plants (Hamilton and Baulcombe 1999), suggesting that these RNAs play a central role in PTGS. Moreover, the introduction of these small RNAs (also called siRNA, short interfering RNA) in mammalian cells has been observed to specifically suppress the expression of endogenous genes, suggesting that siRNAs can induce sequence-specific RNA degradation (Elbashir et al. 2001). The hypothesis that siRNAs can work as guide molecules in inducing RNA degradation is further supported by the finding that siRNAs are associated with the Drosophila RNA-induced silencing complex (RISC), showing a sequence-specific RNAse activity that degrades mRNAs homologous to the silencing trigger (Hammond et al. 2000). Recently, one of the proteins belonging to the RISC multicomponent nuclease has been characterized and found to be a member of the Argonaute family of proteins (Hammond et al. 2001), which was previously found to be essential for PTGS in Neurospora (Catalanotto et al. 2000), C. elegans (Tabara et al. 1999), and Arabidopsis (Fagard et al. 2000). This observation suggests that similar siRNA-directed nuclease complexes might be conserved in eukaryotic organisms belonging to all three kingdoms. Moreover, it also indicates that different foreign nucleic acid molecules, either transgenic DNA or dsRNA, can converge to activate a similar RNA-degrading machinery.
In this work, to establish the universality of the PTGS mechanism, we looked for siRNAs in Neurospora crassa. We detected short sense and antisense RNAs 25-nt long that specifically accumulated in silenced transgenic strains of Neurospora. Moreover, we found that siRNAs derive from the vector portion of the transgenic construct, indicating that the transgenic transcripts are recognized by the silencing machinery, converted into dsRNA, and then diced into small RNA molecules. Additionally, we show that a functional qde-2 (quelling defective) gene (Catalanotto et al. 2000), although essential for PTGS, is not necessary for siRNA accumulation, suggesting that QDE2 could be required in an siRNA-directed RNA-degradation step. Supporting this role of QDE2 in the mRNA-degradation step, we found that the QDE2 protein copurifies with siRNAs, which suggests that QDE2 is a component of the siRNA-directed nuclease complex.
Results and Discussion
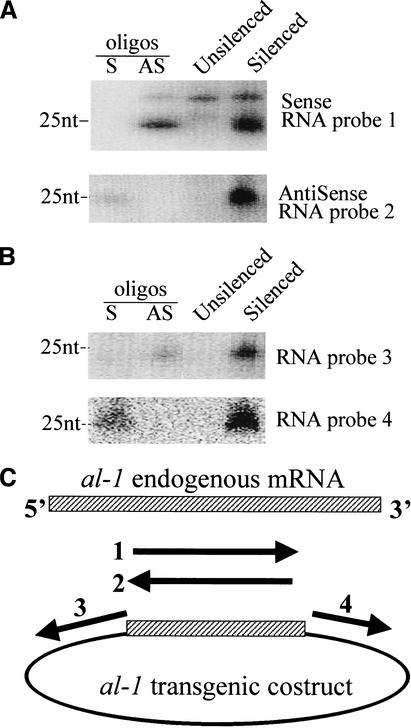
To investigate the PTGS mechanism in fungi, we looked for the presence of small RNAs in an N. crassa strain in which the endogenous gene albino 1 (al-1) is posttranscriptionally silenced as a consequence of the introduction of cognate transgenes. By using both sense and antisense RNA probes corresponding to the region of the al-1 gene present in the transgenic construct (Fig. 1A,C), we found that small RNA molecules in both sense and antisense polarity with a size of ∼25 nt were present in the al-1 silenced strain but not in a wild-type unsilenced strain. This result provides evidence that small RNA intermediates are a general feature of posttranscriptional gene silencing. Although the introduced transgenic construct does not contain a promoter, previous results (Cogoni et al. 1996) indicated that unexpected transcription of transgenic loci was associated with the occurrence of gene silencing. In particular, chimeric transgenic transcripts containing sequences corresponding both to the al-1 gene and to the bacterial vector were specifically found in silenced strains (Cogoni et al. 1996). These chimeric transcripts could be recognized by the cell as aberrant and therefore able to induce the cascade of events leading to RNA degradation. To analyze if chimeric transgenic transcripts are, indeed, templates for production of sense/antisense small RNA, we used vector sequences as probes (Fig. 1C). We found that small RNAs corresponding to the vector sequences are specifically accumulated in the silenced strains (Fig. 1B). Moreover, the small RNAs were found in both sense and antisense polarity, suggesting that, to produce siRNAs, transgenic sense RNA transcripts should first be converted to dsRNA. Then, in keeping with findings in other systems (Hammond et al. 2001), the dsRNA is likely to be processed into small sense/antisense RNA by an RNase III activity as in the case of Dicer in Drosophila. The fact that previous experiments in Neurospora failed to detect transgenic RNA in antisense polarity, only detecting sense transgenic RNA (Cogoni et al. 1996), might indicate that dsRNA is immediately processed on production, thus making its detection difficult.
Figure 1.
Small 25-nt sense and antisense RNAs derived from transgenic transcripts are associated with PTGS. RNA blot hybridization: 50 μg (each lane) of enriched low-molecular-weight RNAs prepared from either untransformed wild-type (unsilenced) or al-1 silenced strain (6xw). Identical RNA blots were hybridized with the following RNA probes: (A) Probes 1 and 2 corresponding to the duplicated al-1 region that are able to recognize small RNAs derived from both the transgenic and endogenous al-1 gene. The presence of the upper bands in wild-type and 6xw lanes were due to nonspecific hybridization of the RNA probe. (B) Probes 3 and 4 were, instead, specific for the transgenic chimeric RNA. 30 pmoles per lane of 25-nt DNA oligonucleotides in sense (S) and antisense (AS) orientation were used to verify the hybridization specificity and also as size markers in both A and B. (C) Schematic representation of plasmid pal-1.6 and RNA probes.
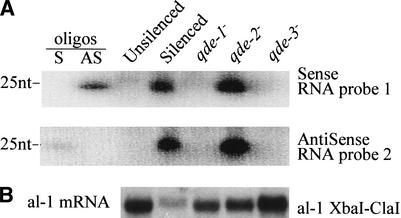
The silencing of the al-1 gene can be relieved by mutations occurring in three loci called qde, which encode cellular components of the silencing machinery (Cogoni and Macino 1997). To establish the role of the qde genes in siRNA accumulation, we looked at the presence of siRNA in the three qde mutants. We found that mutations in qde-1 and qde-3 genes but not in the qde-2 gene abolish the accumulation of siRNAs (Fig. 2), suggesting that both qde-1 and qde-3 gene products are involved either directly in generating the small RNAs or in steps required upstream in the silencing pathway. This finding is consistent with the proposed roles of these two genes. qde-1 encodes a putative RNA-dependent RNA polymerase enzyme (Cogoni and Macino 1999a) that has been hypothesized to be involved in the production of antisense RNA using transgenic RNAs as templates, leading to the formation of dsRNAs, which are subsequently processed into small RNAs by a RNase III-like activity (Bernstein et al. 2001). The role of qde-3, which encodes a DNA helicase (Cogoni and Macino 1999b), has been proposed to be involved in a nuclear step necessary for the activation of gene silencing, perhaps by modifying the chromatin state of the transgenic loci, allowing for the production of the silencing RNA signal.
Figure 2.
Analysis of small RNA accumulation in qde mutants. (A) RNA blot hybridization of low-molecular-weight RNAs prepared from the 107 strain (qde-1−), the 820 strain (qde-2−), the 627 strain (qde-3−), the untransformed wild-type strain (unsilenced), and an al-1 silenced strain (6xw). Sense and antisense probes are the same as in Figure 1. (B) Northern blot on total RNA extracts hybridized with a probe specific for al-1 mRNA.
The fact that in a qde-2 mutant background the al-1 mRNA is not degraded even in the presence of the same amount of small RNA found in a silenced strain (Fig. 2) indicates that the qde-2 gene product is not necessary for siRNA production but is essential for some steps required in the sequence-specific RNA-degradation process guided by siRNAs. Consistent with this hypothesis, AGO2, a homolog of QDE2, has recently been found to be a component of a multiprotein complex (RISC; Hammond et al. 2001) that is required in dsRNA-induced gene silencing (also called RNA interference, RNAi) in Drosophila. Purified RISC complexes were found to retain the ability to induce degradation of target RNAs and to contain small RNAs that are hypothesized to confer sequence specificity to the ribonuclease complex (Hammond et al. 2000).
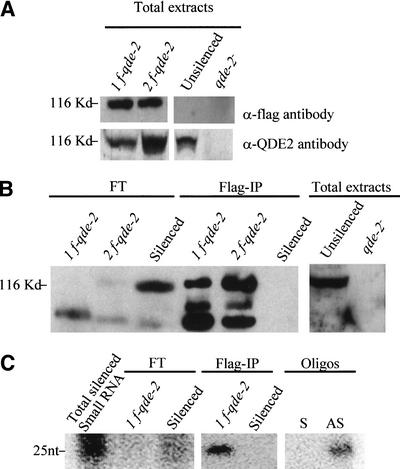
We tested whether QDE2, like its Drosophila homolog AGO2, is associated with small RNAs in the posttranscriptional mechanism in Neurospora. With this aim, we purified the QDE2 protein complex from a Neurospora strain expressing a Flag-epitope-tagged version of QDE2. We transformed an al-1 transgenic strain (820) in which the silencing of the al-1 gene was released by a deletion of the qde-2 gene, with a plasmid containing the qde-2 gene tagged at its N terminus with the Flag epitope and under the control of its own promoter. Several strains showing the recovery of al-1 silencing were isolated, indicating that Flag–QDE2 functions in gene silencing as the wild-type QDE2 does in vivo. The expression of the Flag–QDE2 was confirmed by Western analysis using an anti-Flag antibody on extracts from two independent transgenic lines (Fig. 3A). Moreover, the Flag–QDE2 was also recognized by antibodies against QDE2, and appears to be roughly the same size as the wild-type QDE2 (Fig. 3A). We purified Flag–QDE2 from Neurospora whole-cell extracts by immunopurification with anti-Flag affinity gel beads and elution by competition with Flag peptide. Protein extracts were prepared from an al-1 silenced strain expressing Flag–QDE2 and in parallel, as negative control, from two al-1 silenced strains (6xw) containing a native un-Flagged version of QDE2. SDS-PAGE and silver staining revealed several proteins specifically present in the immunopurification (IP) from the strains expressing Flag–QDE2 (data not shown). Furthermore, three of these bands were recognized by immunoblotting analysis using an anti-QDE2 antibody (Fig. 3B). Presumably, the species with the lowest mobility corresponds to the full-length Flag–QDE2, and the two faster-migrating species could be produced by proteolysis of the full-length form. The same three species were also detected by using a QDE2-specific antibody. In contrast, the native QDE2 protein of the 6xw strain was only detected in flowthrough of the IP but not in IP eluates, indicating that native QDE2 protein was not bound by Flag gel beads.
Figure 3.
QDE2 copurifies with small RNAs. (A) Characterization of antibodies. α-Flag monoclonal and α-QDE2 antibodies were used in Western analysis of total proteins extracted from two deletion qde-2 mutants that express Flag–QDE2 protein (lines 1f-qde-2 and 2f-qde-2), a wild-type (unsilenced) strain, and a qde-2 deletion mutant strain 820 (qde-2−). The estimated protein size is shown on the left. (B) QDE2 protein was immunopurified with anti-FLAG affinity gel beads from protein extracts of two Flag–QDE2 strains and of the al-1 silenced strain 6xw (silenced). Western analysis, using as probe α-QDE2 antibody, was performed on the flowthrough fraction (FT) and on the immunopurified fraction (Flag-IP). Three bands were specifically detected only in immunopurified fractions of Flag–QDE2-expressing strains (lines 1f-qde-2 and 2f-qde-2). The slower-migrating band comigrates with the wild-type version of QDE2 (unsilenced), total extract, indicating that the two additional faster-migrating bands could be proteolyzed forms produced in the IP elution step. A band corresponding to QDE2 is present in the flowthrough (FT) fraction of the silenced (6xw) strain only, indicating that the wild-type QDE2 is not bound by the anti-Flag affinity gel beads. (C) Small RNA molecules were extracted from the Flag-IP protein fractions and analyzed by Northern blotting using a riboprobe specific for the al-1 sequence. Small RNAs of ∼25 nt were detected in the RNA preparations from the immunopurified fraction of the Flag–QDE2 strain (line Flag-IP/1f-qde-2) but not in the 6xw strain (Flag-IP/6xw). In contrast, small RNAs are detected in the flowthrough of 6xw (FT/Silenced) but not in the Flag–QDE2 strain (FT/1f-qde-2). The weakness of the signal in the flowthrough fraction recovered from the 6xw strain is explained by the high dilution of this fraction. A small-RNA-enriched preparation from the total RNA extracted from the 6xw strain is used as a positive control. Sense (S) and antisense (AS) 25-nt oligonucleotides are used as size markers.
To verify whether small RNA molecules were associated with the QDE2 protein, immunopurified protein fractions from both the Flag–QDE2-expressing strain and the silenced (6xw) strain were used to extract RNA, which was then analyzed by Northern blotting with an al-1 riboprobe. We found that 25-nt RNAs corresponding to the al-1 gene were present in the eluate of the Flag–QDE2-expressing strain; in contrast, these RNAs were not detected in the eluate of the silenced strain (Fig. 3C). Moreover, small RNAs were detected in the flowthrough fraction of the 6xw strain, indicating that siRNAs copurify with QDE2.
On the whole, our results are in accordance with other findings obtained in Drosophila in vitro systems, which indicate that AGO2, a homolog of QDE2, is one constituent of a multicomponent nuclease complex involved in RNAi and also contains small RNA as cofactors (Hammond et al. 2001). Furthermore, members of this protein family occur in almost all the eukaryotes, and several of them are implicated in gene silencing. In addition to qde-2 of Neurospora and ago-2 of Drosophila, rde-1 is required for RNAi in C. elegans (Tabara et al. 1999), and argonaute 1 (ago-1) is involved in PTGS in Arabidopsis (Fagard et al. 2000). Although the role of the QDE2/AGO2/RDE1/AGO1 family in gene silencing is well established, their biochemical function is still unknown, as is also still unclear whether they participate in the same step of the silencing pathway. These proteins have in common two motifs: a domain called PAZ and a domain called PIWI. The function of both these domains has yet to be defined; however, it has been suggested that the PAZ domain (Cerutti et al. 2000), which is also contained in Dicer, could be involved in mediating protein–protein interactions occurring between Dicer and AGO2 (Hammond et al. 2001). Using an approach combining genetic and biochemical analysis, we found that QDE2 is dispensable for siRNA accumulation but required for steps in posttranscriptional gene silencing in which siRNA are used as cofactors to direct sequence-specific degradation. However, further studies will be required to understand if small RNAs interact directly with QDE2 or if they are bound to another protein belonging to the QDE2 protein complex and that copurifies with QDE2. Of particular interest is that the components and biochemical mechanism underlining the targeting and degradation of mRNA appear to be very similar in gene silencing phenomena induced either by trangenes as in Neurospora or by dsRNA as in Drosophila. Therefore, the emerging view is that different gene silencing pathways might converge in a universally conserved biochemical process that involves enzymes able to process dsRNA into small RNAs and specialized proteins like QDE2/AGO2 that use these small RNAs as guides to direct the RNA degradation. However, although in RNAi phenomena the injected dsRNA is the direct source of siRNA, in transgene-induced gene silencing additional steps are required to generate dsRNA molecules and eventually siRNA. These might involve the recognition and modification of transgenes at the DNA level. For instance, PTGS is not efficiently maintained in Arabidopsis mutants that are impaired in a SWI2/SNF2 chromatin component (ddm1; Jeddeloh et al. 1999) or in the major DNA methyltransferase (met1; Finnegan and Dannis 1993), suggesting that DNA methylation and chromatin structure play a key role in transgene-induced gene silencing (Morel et al. 2000). A similar role in modifications of chromatin of transgenic loci has been also hypothesized for the RecQ DNA-helicase QDE3 in N. crassa.
An important part of the silencing mechanism is also played by the RNA-dependent RNA polymerases (RdRP). Although the role of RdRP is still not completely clear, evidence exists that in transgene-induced gene silencing phenomena, RdRPs are required to convert transgenic RNA to dsRNA (Dalmay et al. 2000). However, a putative RdRP encoded by ego-1 was also found to be partially required for RNAi in C. elegans, in which the silencing inducer is a dsRNA, suggesting that the function of RdRPs is not merely the production of an initial dsRNA from a single-stranded RNA template. Indeed, recent findings indicate that siRNAs can work as primers for RdRP activity both on dsRNA and sense-targeted mRNA templates (Lipardi et al. 2001). Thus, RdRP could work in a silencing amplification step by copying dsRNA, increasing their concentration, or converting part of the target mRNA into dsRNA. This amplification step could be important to increase the effectiveness of PTGS (Lipardi et al. 2001; Sijen et al. 2001). However, our results show that a mutation in qde-1 completely abolishes both target mRNA degradation and siRNA accumulation, suggesting that the RdRP codified by qde-1 function is not limited to the ancillary function of increasing silencing strength but is likely to be required for the initial step of dsRNA production using single-stranded transgenic RNAs as templates.
In addition to the role played in posttranscriptional gene silencing, the QDE2 family of proteins has been shown to have important functions in development. For example, in Drosophila, the piwi gene is necessary for the maintenance of the germ-line stem cell population (Cox at al. 1998), and ago1 is required for embryogenesis (Kataoka et al. 2001). In C. elegans, two other members of this family, alg-1 and alg-2, are also involved in development (Grishok et al. 2001). It is interesting to note that endogenously produced small RNAs, called small temporal RNAs (stRNAs), similar in size with siRNA are also involved in specific developmental gene regulation events in C. elegans. However, whereas in gene silencing siRNAs are used as guides to induce degradation of homologous mRNA, in developmental gene regulation stRNAs work as guides to block translation of target mRNA containing the 3′ untranslated region sites complementary to the stRNA. Thus, it appears that small RNAs in association with proteins belonging to the QDE2 family may be part of a general conserved mechanism to control gene expression either at the posttranscriptional or translational level. The recent identification of a number of genes coding for small RNAs in vertebrates and invertebrates (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001) further supports the argument for the widespread nature of such a regulatory mechanism. Therefore, the characterization of the function of small RNAs and the QDE2 protein family has implications not only in gene silencing but also in gene regulation in general.
Materials and methods
Plasmids and Neurospora strains
The 6xw stable silenced strain of Neurospora was obtained by transforming spheroplasts of strain FGSC no. 3958 a (qa-2; aro-9) with pX16 containing the qa-2 gene as a selectable marker and a 1500-bp fragment of the coding sequence of al-1. The qde mutants used for small RNA detection were obtained by UV mutagenesis of the 6xw strain (Cogoni and Macino 1997). The 820 strain was obtained by transforming 6xw with pMXY2, which contains a benomyl-resistance β-tubulin gene that functions as a dominant selectable marker in N. crassa. Benomyl-resistant transformants were selected, and the orange phenotype was rescued. Southern analysis revealed the deletion of the qde-2 locus probably as a consequence of recombination of integrated plasmids (PMYX2; data not shown).
The pal-1.6 plasmid was derived from a pBluescript II SK+ vector in which a 1500-bp XbaI/ClaI fragment of al-1 was cloned. The pal-1.6 plasmid was digested with ClaI and transcribed with a T3 RNA polymerase to obtain a sense transcript (probe 1); it was then digested by XbaI and transcribed with a T7 RNA polymerase to obtain an antisense al-1 fragment (probe 2). A pBluescript II SK+ was digested with PvuII, transcribed with T3 RNA polymerase for the RNA molecules, and used as probe 3. A pBluescript II SK+ was digested with SspI, transcribed with T7 RNA polymerase for the RNA molecules, and used as probe 4. To produce the plasmid for Flag–QDE2 expression, a sequence containing the entire qde-2 gene with its own promoter was cloned into the BamHI site of a pBluescript II SK+ without its HindIII site in the polylinker. To introduce the Flag epitope at the N terminus of the protein, an 80-bp fragment was removed and was substituted with a PCR homologous fragment amplified with 5′-oligo: 5′-CCCAAGCTTGACTACAAGGAC GACGATGACAAATCGCTCAGCGAGAAGGAG-3′ (containing a HindIII restriction site, a 24-bp sequence coding for Flag epitope, and a region of homology to qde-2 sequence) and with 3′-oligo: 5′-CCCAAGCT TCACCTTCTCGCCCATGGT-3′.
QDE2 purification and Western analysis
Frozen mycelia were homogenized in 10% glycerol, 50 mM HEPES, and 135 mM KCl. Extracts were incubated 5 min on ice. After microcentrifugation at 4°C for 10 min, SDS-loading buffer was added to supernatants, and proteins were denatured at 94°C for 5 min. All protein buffers contained leupeptin (1 μM), pepstatin (1 μM), and phenylmethanesulfonyl fluoride (50 μM). To purify the Flag–QDE2 protein, mycelia of strains expressing the Flag–QDE2 and of the 6xw strain as a control were filtered through cheesecloth, resuspended in 140 mM KCl, 20 mM Tris-HCl (pH 8), and 5 mM MgCl2, and homogenized. Total extracts were centrifuged at 4°C for 20 min, and supernatants were incubated with α-flag M2 affinity gel. After 3 h, beads were harvested and washed with resuspending buffer. Elution was done in the same buffer containing 100 μg/mL Flag peptide.
Western blot analyses were performed as follows. Neurospora crassa protein extracts were separated by electrophoresis on 7% SDS-polyacrylamide gel and electrotransferred to nitrocellulose membrane. Blots were probed with α-flag monoclonal (Sigma) or α-QDE2 polyclonal antibody. Polyclonal antibodies were raised against two QDE2 peptides: amino acids 115–129 (WTEPSSNQNLPSKPQ) and amino acids 316–330 (GDERGKQKDGKEVRY). Two rabbits were immunized with a mix of the coupled peptides, and the antiserum was used for Western analysis in a dilution of 1:1000. All blots were blocked and washed in TBST with 5% nonfat dry milk, followed by horseradish peroxidase-labeled anti-mouse or anti-rabbit IgG (Sigma, BioRad) secondary antibody incubation. The ECL Western blot chemiluminescence detection kit was applied for immunodetection (Amersham).
Small RNA purification and Northern analysis
Small RNA purification was performed as described by Hamilton and Baulcombe (1999) with minor modifications. Frozen mycelia were homogenized with a potter in 50 mM Tris-HCl (pH 9.0), 10 mM EDTA, 100 mM NaCl, and 2% SDS. The homogenates were extracted with an equal volume of phenol-chloroform, and the nucleic acids were precipitated by adding 3 volumes of absolute ethanol and 1/10 volume of 3 M sodium acetate (pH 5), over night at −20°C. After centrifugation the pellets were washed in 70% ethanol, dried, and resuspended in double distilled water. Incubating this solution for 30 min on ice with polyethylene glycol (MW 8000) at a final concentration of 5% and 500 mM NaCl, we precipitated nucleic acids with high molecular weight whereas the small RNA molecules remained in the solution. The supernatants were precipitated with ethanol as described above. The concentration of the RNA preparation was quantified by spectrophotometric analysis. Low-molecular-weight RNAs were separated by electrophoresis in 0.5× TBE through 15% polyacrylamide 7 M urea. Ethidium bromide staining was used to verify the correct loading. Then RNA was electrotransferred in 1× TBE onto Gene Screen Plus filters (New England Nuclear), and fixed by ultraviolet cross-linking. To control the size and polarity of low-molecular-weight RNAs, 25-mer oligonucleotides were used as molecular size markers. Oligonucleotides 5′-GCAAGAACGGAAAGAAGCCAAAGGC-3′ and 5′-CA TGAGCAACGTGACGACAAGGCGG-3′ were used as sense and antisense controls, respectively, in hybridizations performed with RNA probes 1 and 2; oligonucleotides 5′-CCTCGAGGGGGGGCCCGGTAC CCAA-3′ and 5′-AAAACGACGGCCAGTGAGCGCGCGT-3′ in those with RNA probes 3 and 4. Prehybridization and hybridization were at 35°C in 50% deionized formamide, 7% SDS, 250 mM NaCl, 125 mM sodium phosphate (pH 7.2), and sheared, denatured, salmon sperm DNA (100 mg/mL). After overnight hybridization, membranes were washed twice in 2× SSC and 0.2% SDS at 35°C for 30 min and once in 20 mM Tris-HCl (pH 7.5), 5 mM EDTA, 60 mM sodium chloride, and 10 μg/mL RNase A at 37°C for 1 h to remove unspecific background.
Single-stranded RNA probes were transcribed in both sense and antisense orientation by pal16 and pal17 plasmids with 32P-labeled uridine triphosphate (50 μCi per 20 μL reaction volume; specific activity 3000 Ci/mmole; New England Nuclear), using T3 RNA polymerase (Roche). To remove plasmid template, the reaction was incubated at 37°C for 15 min with RNase-free DNase I (Roche). To break labeled transcripts to an average size of 50 nt, 300 μL of 80 mM sodium bicarbonate and 120 mM sodium carbonate were added to the transcriptional reaction and incubated at 60°C for 3 h. To stop the hydrolysis reaction of the transcript, 20 μL of 3 M sodium acetate (pH 5.0) was added (Hamilton and Baulcombe 1999).
Acknowledgments
We thank Emma Forrest for revising the manuscript. This work was supported in part by grants from the Istituto Pasteur Fondazione Cenci Bolognetti, from the Ministero dell'Università e della Ricerca Scientifica e Tecnologica.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL carlo@bce.med.uniroma1.it; FAX 39-06-4462891.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.222402.
References
- Baulcombe DC. Gene silencing: RNA makes RNA makes no protein. Curr Biol. 1999;9:R599–R601. doi: 10.1016/s0960-9822(99)80383-2. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bosher JM, Labouesse M. RNA interference: Genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:31–36. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- Catalanotto C, Azzalin G, Macino G, Cogoni C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;10:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci. 1997;94:10233–10238. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999a;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- ————— Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999b;286:2342–2345. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- ————— Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev. 2000;10:638–643. doi: 10.1016/s0959-437x(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Irelan JT, Schumacher M, Schmidhauser T, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA–DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptionalgene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A novel species of small antisense RNA in post-transcriptional gene silencing. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond S, Boettcher S, Caudy A, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR. siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/s0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Mette MF, Matzke AJ. Transgene silencing by the host genome defense: Implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol. 2000;43:401–415. doi: 10.1023/a:1006484806925. [DOI] [PubMed] [Google Scholar]
- Morel JB, Mourrain P, Beclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RHA, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- Zamore PD. RNA interference: listening to the sound of silence. Nat Struct Biol. 2001;8:746–750. doi: 10.1038/nsb0901-746. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]