Abstract
Previous studies of mitosis show that capture of single kinetochores by microtubules from both centrosomes (merotelic orientation) is a major cause of aneuploidy. We have characterized hcp-6, a temperature-sensitive chromosome segregation mutant in C. elegans that exhibits chromosomes attached to both poles via a single sister kinetochore. We demonstrate that the primary defect in this mutant is a failure to fully condense chromosomes during prophase. Although centromere formation and sister centromere resolution remain unaffected in hcp-6, the chromosomes lack the rigidity of wild-type chromosomes and twist around the long axis of the chromosome. As such, they are unable to establish a proper orientation at prometaphase, allowing individual kinetochores to be captured by microtubules from both poles. We therefore propose that chromosome rigidity plays an essential role in maintaining chromosome orientation to prevent merotelic capture.
Keywords: C. elegans, centromere, kinetochore, chromosome condensation, bipolar attachment, aneuploidy
The faithful segregation of chromosomes at mitosis is an essential aspect of eukaryotic cell division. Accurate segregation requires that the chromosomes achieve bipolar attachment, such that one kinetochore is bound exclusively by microtubules radiating from only one pole, while its sister kinetochore is attached only to the opposing pole (for review, see Nicklas 1997; Rieder and Salmon 1998). The capacity to attain this state is established prior to nuclear envelope breakdown. It is during this time that sister kinetochores form in a specific back-to-back orientation with the associated microtubule binding sites directed away from the chromosome (Roos 1973; Heneen 1975; Moore and Roth 2001). Owing to this orientation, which is maintained by the natural rigidity of the chromosome, capture of a kinetochore by one pole positions its sister kinetochore such that it faces toward, and is thus likely to be captured by, the opposing pole (Nicklas 1997).
Nevertheless, this system is not error-free, as seen by recent work that proposes that failures in bipolar attachment play a significant role in generating aneuploidy in mammalian cells (Cimini et al. 2001, 2002). The authors are able to detect, at various points following nuclear envelope breakdown, chromosomes attached to microtubules from both poles via single kinetochores. Defects of this nature are referred to as merotelically oriented chromosomes and lead to lagging anaphase chromosomes. The occurrence of these chromosomes can be enhanced by treatment with colcemid or nocodazole, which seem to induce morphological changes in the kinetochore, making them appear stretched or curled (Ladrach and LaFountain 1986; Cimini et al. 2001). This suggests that alterations in kinetochore structure and flexibility can prevent the establishment and maintenance of bipolar attachment. However, the mechanism by which these maloriented chromosomes naturally arise remains unclear.
In an effort to expand these observations, we have begun to investigate how chromosome orientation is regulated in the nematode Caenorhabditis elegans. Our decision to work with this organism is due in large part to its holocentric chromosomes, which provide an increased capacity for cytological examination (Albertson and Thomson 1982; Pidoux and Allshire 2000). Whereas the centromere of monocentric organisms is present at a discrete region of the chromosome, and directs the assembly of a localized kinetochore, the centromere of C. elegans assembles a holocentric kinetochore that extends the length of the chromosome. This magnified centromere provides a unique marker with which to examine the structure and orientation of not only the centromere, but of the mitotic chromosomes themselves. Components of the centromere and kinetochore have been conserved throughout evolution, suggesting that the centromeres of monocentric and holocentric organisms share functional similarity (Henikoff et al. 2000; Pidoux and Allshire 2000). Furthermore, detailed structural analysis of mammalian centromeres indicates that these structures are composed of discrete units brought together during mitosis, a situation analogous to what is observed in C. elegans (Zinkowski et al. 1991; Buchwitz et al. 1999).
Because we are interested in understanding how chromosome orientation is established in mitosis, we began by examining a collection of chromosome segregation mutants to identify mutants with defects in this process. We present here the characterization of holocentric protein 6 (hcp-6), a temperature-sensitive mutant whose chromosomes become attached to both poles via a single sister kinetochore, identical to previous descriptions of merotelic chromosomes. This defect results from a partial failure in chromosome condensation, which does not affect centromere formation or resolution of sister centromeres, but causes hcp-6 chromosomes to be less rigid than wild-type chromosomes. This prohibits hcp-6 from establishing and maintaining proper chromosome orientation, resulting in a failure to achieve bipolar attachment and a dramatic chromosome segregation defect.
Results
Isolation and cloning of hcp-6
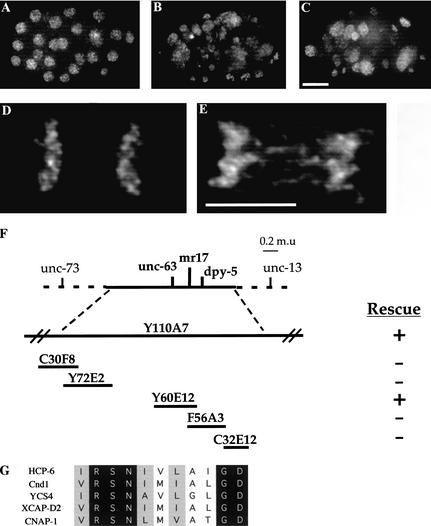
We began to examine the underlying cellular mechanisms that establish chromosome orientation by first isolating mutants with an impaired ability to segregate chromosomes. To do so, we assembled a collection of 250 temperature-sensitive embryo-lethal mutants in C. elegans. Using 4‘,6-diamidino-3-phenylindole dihydrochloride (DAPI) to visualize DNA, we identified 22 mutants in this collection that exhibited segregation defects following a shift to the nonpermissive temperature (26°C vs. 15°C). We chose one mutant, mr17, for further characterization, based on a drastic failure in chromosome segregation (Fig. 1, cf. A and B). Of particular interest were the anaphase phenotypes exhibited in this mutant, indicating a possible defect in chromosome orientation. These included extensive anaphase bridging and a high frequency of lagging chromosomes (Fig. 1, cf. D and E). Based on these results, we proceeded to clone the corresponding gene, which we eventually named holocentric protein 6 (hcp-6).
Figure 1.
hcp-6 embryos contain aneuploid nuclei, suggesting a defect in mitotic chromosome segregation. Embryos were isolated from adults and stained with DAPI to visualize DNA. (A) Note the uniform size and distribution of the nuclei in a wild-type embryo. (B) In hcp-6 embryos, the size and intensity of the DAPI staining vary greatly, indicating that DNA content is not consistent between individual cells. Furthermore, the nuclei are unevenly dispersed throughout the embryo, indicating multinucleate blastomeres. (C) RNAi of Y110A7A.1 generates a chromosome segregation phenotype identical to that of hcp-6. (D,E) Anaphase is disrupted in hcp-6 embryos. One-cell embryos undergoing anaphase were examined. (D) In the wild type, note the clean separation of sister chromatids. (E) In contrast, observe the strings of DNA suspended between the sister chromatids in hcp-6, indicative of anaphase bridging. (F) Cloning of HCP-6. Genetic and physical maps of the region surrounding hcp-6. Two YACs (Y110A7 and Y60E12) were able to rescue the embryo lethal phenotype associated with hcp-6. (G) Alignment of HCP-6 with XCAP-D2-like proteins from S. pombe (T43519), S. cerevisiae (S51408), X. laevis (AAC64359), and H. sapiens (NP055680) (GenBank accession numbers in parentheses). Bar, 5 μm.
Genetic analysis revealed that this locus was in the middle of chromosome I, in the 0.35 m.u. region between dpy-5 and unc-63 (see Materials and Methods). Transgenic rescue experiments (Fig. 1F), in conjunction with sequence data obtained from the C. elegans Sequencing Consortium, led us to focus our attention on four candidate open reading frames (ORFs). RNA-mediated interference (RNAi) (Fire et al. 1998) experiments revealed that only one of these, Y110A7A.1, generated a phenocopy of the hcp-6(mr17) phenotype. RNAi with this ORF not only produced approximately 100% embryo lethality, but DAPI staining of RNAi embryos revealed chromosome segregation defects comparable to hcp-6(mr17) (Fig. 1C). Previous RNAi analysis of this gene has yielded similar phenotypes, namely embryo lethality and multinucleate cells (Fraser et al. 2000; Piano et al. 2000).
To demonstrate ORF-specific rescue, we PCR-amplified the Y110A7A.1 ORF, along with flanking genomic sequences, and used this DNA to generate transgenic lines (Mello and Fire 1995). Introduction of this sequence into hcp-6(mr17) animals was sufficient to rescue the temperature-sensitive embryo lethality (see Materials and Methods), supporting the notion that the function of the Y110A7A.1 gene product was disrupted in hcp-6(mr17). To confirm this identification, we sequenced the Y110A7A.1 gene from hcp-6(mr17) genomic DNA and observed a single G to A transition at position 3073. This mutation lies within the predicted protein-coding region and converts glycine 1024 to glutamic acid. Based on these results we concluded that we had identified the hcp-6 gene.
Y110A7A.1 is predicted to encode a protein of 1724 amino acids. BLAST searches of protein sequence databases revealed that this gene product contains a short stretch of sequence that is conserved among XCAP-D2/CNAP1/Cnd1/YCS4 (Fig. 1G). These proteins are homologs of a conserved subunit of the condensin complex, which has been shown to play an important role in regulating mitotic chromosome condensation and chromosome segregation (for review, see Koshland and Strunnikov 1996; Hirano 2000). For example, mutations in the mix-1 gene, which encodes a core component of the condensin complex in C. elegans, generate a variety of mitotic defects, including lagging chromosomes and aneuploidy (Lieb et al. 1998). Similar phenotypes are observed in other organisms following disruption of condensin function (Saka et al. 1994; Strunnikov et al. 1995; Sutani et al. 1999; Freeman et al. 2000; Schmiesing et al. 2000; Bhalla et al. 2002). However, the exact mechanisms by which defects in chromosome condensation translate into chromosome missegregation remain unclear.
HCP-6 localizes to the centromere
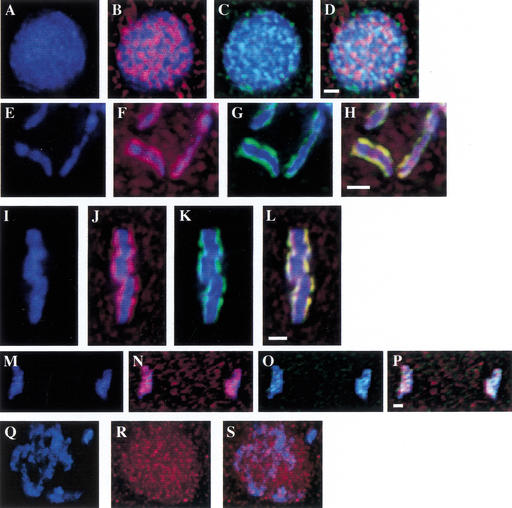
To determine the subcellular localization of this protein, we raised a polyclonal antibody against HCP-6 (see Materials and Methods). Immunofluorescence microscopy demonstrated that this antigen localizes to the poleward faces of the metaphase plate, a pattern previously shown to correspond with the holocentric centromere of C. elegans (Buchwitz et al. 1999). To confirm that this protein is present at the centromere, we performed a double staining experiment with HCP-3, a centromeric histone H3-like protein (Buchwitz et al. 1999). Although both of these proteins display a punctate staining pattern in interphase, there was no detectable overlap between the two (Fig. 2A–D). In contrast, examination of prophase, metaphase, and anaphase chromosomes revealed that there was nearly complete colocalization between these two proteins (Fig. 2E–P). These results confirm that HCP-6 localizes to the C. elegans centromere during mitosis. Due to this localization, we referred to this gene as holocentric protein 6 (hcp-6).
Figure 2.
Localization of HCP-6 throughout the cell cycle. (A–P) Wild-type embryos showing DNA (blue), anti-HCP-6 (red), or anti-HCP-3 (green) staining, or merged images. Colocalization between HCP-6 and HCP-3 is shown as yellow. (A–D) At interphase, this antigen is visible as punctate dots distributed throughout the nucleus. (E–H) In prophase nuclei, HCP-6 forms two parallel lines on individual prophase chromosomes, a pattern suggesting centromere localization. (I–L) At metaphase, the HCP-6 protein is present on the poleward faces of the metaphase plate. (M–P) During anaphase, HCP-6 remains associated with the separating groups of sister chromatids. (Q–S) hcp-3(RNAi) embryos stained with anti-HCP-6 (red). In the absence of HCP-3, HCP-6 does not assemble onto mitotic chromosomes properly. Bars, 1 μm.
Having demonstrated that HCP-6 is present at the centromeres of mitotic chromosomes, we were interested in examining whether this localization would be maintained in the absence of a functional centromere. To perform this experiment, we reduced HCP-3 expression with RNAi, and stained the resulting embryos with anti-HCP-6 antibody. Although HCP-6 was present in the nucleus, it was not possible to detect parallel lines of HCP-6 staining on condensed prophase chromosomes (Fig. 2Q–S). Similar results were observed when expression of HCP-4, the C. elegans homolog of the mammalian CenP-C protein (Moore and Roth 2001), was disrupted via RNAi (data not shown). Therefore, organization of HCP-6 on chromosomes is dependent on proper centromere assembly.
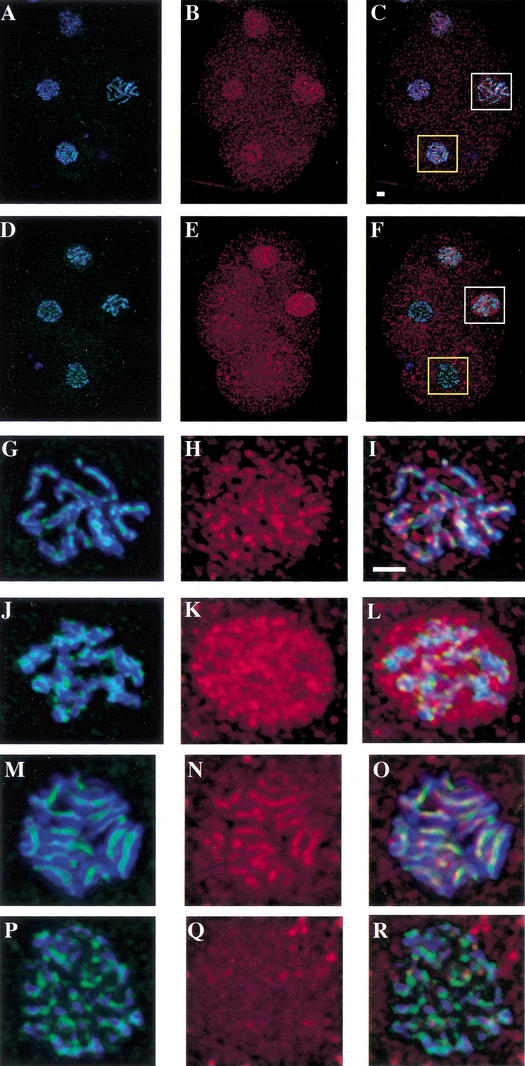
After establishing the localization of HCP-6 in wild-type embryos, we were interested in determining whether this pattern was altered in hcp-6(mr17) embryos following a shift to the nonpermissive temperature (Fig. 3A–F). No discernible differences were detected during interphase (data not shown). However, as the chromosomes began to individualize and condense, in wild-type nuclei HCP-6 was concentrated on the chromosomes (Fig. 3G–I), whereas prophase staining in hcp-6(mr17) cells remained punctate and no association with chromosomes was detected (Fig. 3J–L). Furthermore, while HCP-6 was clearly present at the centromeres of metaphase chromosomes in wild-type cells (Fig. 3M–O), the protein was completely absent from metaphase plates in mutant embryos (Fig. 3P–R). The altered localization of HCP-6 in hcp-6(mr17) embryos at the nonpermissive temperature is consistent with this allele being a temperature-sensitive, loss-of-function mutation.
Figure 3.
HCP-6 fails to localize to mitotic chromosomes in hcp-6 mutants. Following a 1-h shift to 26°C, embryos were stained with anti-HCP-6 (red) and anti-HCP-3 (green). DNA was visualized with DAPI (blue). Four-cell embryos at the same stage of development were examined. The AB blastomeres (bottom and left) are in metaphase. The remaining two blastomeres are in prophase, with the EMS blastomere (right) at a slightly later point in mitosis. (A–C) In wild-type cells, as was described above, HCP-6 can be seen associating with both prophase and metaphase chromosomes. (D–F) In contrast, although the HCP-6 antigen is present in prophase nuclei, it is completely absent from hcp-6 metaphase chromosomes. (G–R) Magnified views of the EMS prophase nuclei (white boxes, G–L) and the AB metaphase plates (yellow boxes, M–R). In wild-type cells (G–I), the HCP-6 protein can be seen associating with prophase chromosomes. However, in hcp-6 (J–L), this antigen is distributed throughout the nucleus, and shows no enrichment at the chromosomes. Inspection of wild-type metaphase plates (M–O) demonstrates that HCP-6 is easily detectable at the centromere of metaphase chromosomes. In contrast, although HCP-3 localizes to the centromere of mutant chromosomes (P–R), no HCP-6 staining is observed. Bars, 1 μm.
Mitotic chromosomes fail to align properly in hcp-6
To examine whether the anaphase defects present in hcp-6 result from a failure in chromosome orientation, we set out to establish a detailed description of the hcp-6 phenotype. In particular, we wanted to investigate whether the defects seen in hcp-6 are analogous to previous descriptions of merotelically oriented chromosomes (Yu and Dawe 2000; Cimini et al. 2001, 2002). These reports demonstrate that lagging anaphase chromosomes often consist of a sister chromatid with a single kinetochore that is connected to microtubules from both poles. To test whether the lagging chromosomes present in hcp-6 represent single chromatids, we stained embryos with anti-HCP-3.
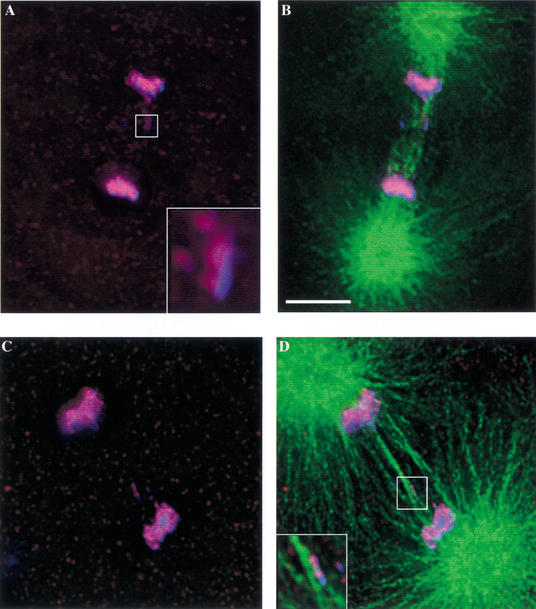
Because a shift to the restrictive temperature (26°C) generates such a severe anaphase phenotype, we performed a shift to a semipermissive temperature (23°C) to optimize our ability to observe single lagging chromosomes. Under these conditions, we were able to test the hypothesis that merotelically oriented chromosomes are generated in hcp-6. An examination of wild-type one-cell embryos demonstrated that lagging chromosomes were detectable in less than 1% of anaphases (n = 112). In contrast, 96% of anaphases (n = 92) in hcp-6 one-cell embryos displayed one or more lagging chromosome fragments, many of which appeared to interact with microtubules from both poles (Fig. 4A–D, inset in D). Furthermore, none of these chromosomal bodies displayed double lines of HCP-3 staining. Instead, the majority possessed a single line of reactivity (Fig. 4A, inset), consistent with the idea that they represent individual sister chromatids. The high frequency with which merotelic chromosomes are observed, even at a semipermissive temperature, suggests that they play a major role in generating the extreme aneuploidy seen in hcp-6.
Figure 4.
hcp-6 chromosomes display merotelic orientation. hcp-6 embryos were stained with anti-HCP-3 (red), anti-tubulin (green), and DAPI (blue) following a 2-h shift to 23°C. Images of one-cell embryos undergoing anaphase were then collected. (B, D, and D, inset) The tubulin staining demonstrates that microtubules from both poles are associating with the lagging chromosomes in hcp-6 embryos. A higher-resolution image of the lagging chromosome (A, inset) reveals that only one line of HCP-3 is present, suggesting that this DNA body represents a single chromatid. Taken together, these observations indicate that merotelic chromosomes are generated in the hcp-6 background. Bar, 5 μm.
At this point, having shown that maloriented chromosomes are present in hcp-6, we sought to expand upon previous studies and investigate the mechanism(s) by which orientation defects can arise. We began to address this question by defining earlier points in mitosis where orientation defects could still be observed. We focused initially on metaphase, because defects in the maintenance of bipolar attachment would be particularly obvious at this stage of the cell cycle (Fig. 5G). In wild-type embryos, as has been previously described, staining with anti-HCP-3 revealed that this antigen is concentrated on the poleward faces of the metaphase plate (Fig. 5A–C). Although linear aggregates of HCP-3 staining are visible in hcp-6 metaphase plates, this reactivity is not distributed exclusively onto the poleward faces (Fig. 5D–F). Instead, individual centromeres can be seen running through the metaphase plate in an extremely disorganized fashion. Strikingly, it is possible to detect single centromeres traversing the entire width of the metaphase plate, potentially associating with both poles (Fig. 5E, see arrow). One model to explain this observed misalignment is that hcp-6 chromosomes lack the structural integrity of wild-type chromosomes, inhibiting their ability to orient properly.
Figure 5.
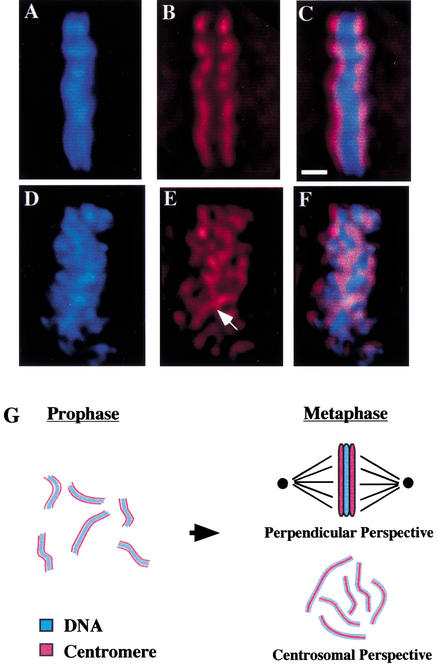
Centromere organization is disrupted in hcp-6. All data were collected from one-cell embryos following a 1-h shift to 26°C and show DNA (blue), anti-HCP-3 (red), or a merged image of both. (A–F) HCP-6 chromosomes are not organized properly within the metaphase plate (perpendicular perspective). (A–C) In wild-type cells, HCP-3 localizes to the poleward faces of the metaphase plate, as described above. (D–F) In hcp-6 embryos, although linear aggregates of HCP-3 staining, corresponding to the centromeres of individual chromosomes are visible at metaphase, their localization is not restricted to the poleward faces of the metaphase plate. Instead, they run throughout the width of the metaphase plate and associate with both poles simultaneously (see arrow). (G) At metaphase, all of the chromosomes have aligned themselves onto the metaphase plate, a disc-like structure which lies in the middle of the cell. When viewed from the “perpendicular perspective,” (top right) such that the poles lie to the left and right of the image, the chromosomes in the metaphase plates merge together and appear as a solid bar of DNA. The centromeres, which have maintained their 180° of opposition, are oriented towards the poles, and lie on either side of the DNA. It is also possible to view a metaphase plate from the “centrosomal perspective” (bottom right) such that the metaphase plate lies flat on the plane of the page, and the centrosomes lie above and below the page. In this case, one is able to look down at the metaphase plate and individual sister centromeres, each lying on top of its corresponding metaphase chromosome. If one were able to view further down through the page, the opposing set of sister centromeres would then become visible. Bar, 1 μm.
hcp-6 exhibits a defect in mitotic chromosome condensation
To evaluate the structure of mutant and wild-type chromosomes, we examined metaphase cells, when chromosomes are fully compacted (Fig. 6A–F). An inspection of hcp-6 metaphase plates (Fig. 6D–F) reveals that distinct chromosomal bodies are evident, indicating that a certain degree of condensation and individualization has been achieved (Fig. 6D). Nevertheless, the mutant chromosomes appear structurally different than the wild type. Most noticeably, they look much stringier and do not seem to have reached the level of condensation observed in the wild type. In another deviation from wild-type, viewing serial sections of metaphase plates shows that hcp-6 chromosomes are twisted such that sister centromeres no longer lie in a single plane (see Supplementary movie at http://www.genesdev.org). In the two-dimensional projections displayed in the figure, these twists are visible as gaps in centromeric staining (Fig. 6F), in contrast to the completely contiguous lines detectable in the wild type (Fig. 6C).
Figure 6.
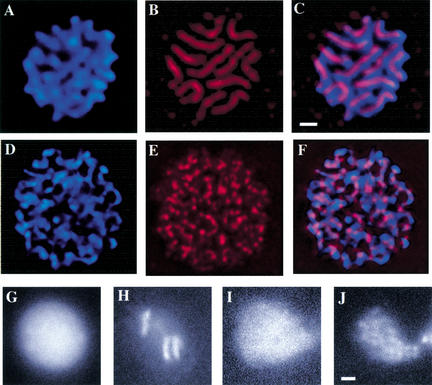
hcp-6 possesses a chromosome condensation defect. (A–F) Metaphase plates in AB blastomeres (4-cell embryo) stained with anti-HCP-3 (red) and DAPI (blue) and viewed from the centrosomal perspective. (A–C) In wild-type cells, 12 fully compact chromosomal masses are evident, each with a corresponding centromere that runs the length of the chromosome. (D–F) The chromosomes in an hcp-6 metaphase plate are longer and thinner, indicating a failure to fully condense. Although aggregates of HCP-3 are visible along the chromosomes, uninterrupted stretches of centromeric staining cannot be detected. (G–J) hcp-6 chromosomes display a prophase condensation defect. The images shown here represent GFP∷ histone-labeled nuclei at interphase (M,O) and prophase (N,P). (N) Note the fully condensed chromosomes that are detectable in the wild type. (P) In hcp-6, although some degree of condensation is evident, the chromosomes have not achieved a wild-type level of compaction. Bars, 1 μm.
These results indicate that the failure of hcp-6 chromosomes to achieve bipolar attachment at metaphase is due to a partial defect in chromosome condensation. Because this process represents one of the earliest steps in mitosis, it raised the possibility that events occurring prior to nuclear envelope breakdown play a critical role in establishing and maintaining bipolar attachment. To investigate whether hcp-6 displays defects in mitotic chromosome condensation during prophase, we obtained a transgenic strain containing an integrated copy of a GFP∷histone H2B fusion construct (Praitis et al. 2001). In this strain, DNA can be detected by a fluorescence microscope within living embryos. Beginning at interphase (Fig. 6G,I), we collected images every 45 sec until sister chromatid separation at anaphase was achieved (data not shown). Once this sequence of images was obtained, it was possible to go back and define the point of maximal chromosome condensation, which occurs shortly before nuclear envelope breakdown. In wild-type blastomeres, fully condensed chromosomes are clearly evident (Fig. 6H), whereas mutant chromosomes fail to achieve this level of compaction (Fig. 6J). One explanation for how this prophase phenotype translates into the observed orientation defects is that the partially decondensed hcp-6 chromosomes lack the rigidity of wild-type chromosomes, allowing them to twist such that a single centromere could become attached to both poles following nuclear envelope breakdown.
hcp-6 chromosomes are more susceptible to twisting
To determine whether hcp-6 chromosomes exhibit an increased frequency of twisting in prophase, we used HCP-3 staining as a marker for chromosome orientation. It was also important to define a brief period in prophase where we could better compare mutant and wild-type chromosomes. To accomplish this, we costained with anti-HCP-1, a component of the C. elegans kinetochore that assembles onto mitotic chromosomes late in prophase (Moore et al. 1999; data not shown). In both wild-type and hcp-6, the majority of chromosomes displayed parallel lines of centromeric staining running the length of chromosome (Fig. 7A,F). A small percentage of prophase chromosomes contained a single 180° twist (Fig. 7B,C,G,H). This configuration was observed in 7% of the wild-type chromosomes examined. An increased frequency of twisted chromosomes (28%) was seen in hcp-6 (Fig. 7K). Furthermore, 4% of hcp-6 chromosomes (n = 47) display two or more twists, patterns which are not detected in wild-type (Fig. 7D,E,I,J). These results support the proposed model that hcp-6 chromosomes lack the rigidity and structural integrity of wild-type chromosomes at prophase. In turn, this allows single kinetochores to be captured by both poles simultaneously, generating merotelically oriented chromosomes and profound defects in mitotic chromosome segregation.
Figure 7.
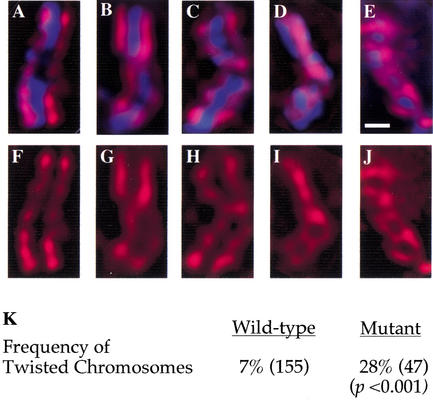
hcp-6 chromosomes are less rigid than wild-type. (A–J) Prophase nuclei from one-cell embryos were examined in both wild-type and hcp-6 backgrounds. Embryos were stained with DAPI to visualize DNA (blue) and anti-HCP-3 (red). Images of prophase chromosomes stained with both DAPI and anti-HCP-3 (A–E) or anti-HCP-3 alone (F–J). In both wild-type and hcp-6 embryos, the majority of prophase chromosomes displayed two parallel lines of HCP-3 staining which ran the length of the chromosome (A,F). More often in hcp-6, but also in the wild type, chromosomes displaying single 180° twists were observed (B,C,G,H). In hcp-6 embryos, chromosomes displaying two (D,I) or more (E,J) twists were also observed, patterns which were not seen in the wild type. The chromosomes displayed in (A–C,F–H) were obtained from wild-type embryos; (D,E,I,J) were collected from hcp-6 embryos. (K) Quantitative analysis of the incidence of twisted chromosomes. The number of chromosomes examined is in parentheses. The frequency of twisted chromosomes was increased fourfold in hcp-6 embryos. Bars, 1 μm.
Discussion
HCP-6 regulates chromosome condensation
This work centers on hcp-6, a mutant that was originally identified because it failed to attain bipolar attachment. A more detailed characterization of the hcp-6 phenotype has demonstrated that this gene plays an important role earlier in mitosis, mediating chromosome condensation and establishing the rigidity of mitotic chromosomes. However, it is essential to note that not all aspects of chromosome condensation are abrogated in this mutant. Although hcp-6 chromosomes are certainly less compact than wild-type, they do individualize into distinct units. Similar chromosome condensation phenotypes were described when components of the C. elegans condensin complex were disrupted via RNAi (Hagstrom et al. 2002). Furthermore, centromere condensation and sister centromere resolution appear normal in hcp-6 embryos. Finally, several molecular markers for chromosome condensation, such as the phosphorylation of Ser 10 on histone H3, remain unchanged in an hcp-6 background (data not shown). These results indicate that there are many factors involved in chromosome condensation, and that disrupting the function of a gene like hcp-6 only affects specific aspects of this process.
The centromeric localization of HCP-6 invites speculation about a possible role for the centromere in mediating chromosome condensation. Reports from other systems have demonstrated that several important condensation factors seem to localize to the centromere (Rattner et al. 1996; Torok et al. 1997), and recent studies of C. elegans have demonstrated that the two core components of the condensin complex are present at the centromere (Hagstrom et al. 2002). It will be interesting to explore this question further and attempt to incorporate these results into a more general model of chromosome condensation. Finally, previous work on chromosome condensation has demonstrated that disrupting this process in vivo leads to severe mitotic defects, including anaphase bridging and chromosome missegregation (Uemura et al. 1987; Strunnikov et al. 1995; Lieb et al. 1998; Schmiesing et al. 1998, 2000; Sutani et al. 1999; Freeman et al. 2000; Bhalla et al. 2002). Building on these observations, we present here the first high-resolution cell biological description of a chromosome condensation phenotype. These results provide a model to explain how defects in condensation can lead to chromosome missegregation and aneuploidy.
HCP-6 is required to maintain chromosome rigidity and ensure accurate segregation
The earliest detectable defect in hcp-6 embryos is a failure to properly condense chromosomes during prophase, resulting in less rigid chromosomes which are more susceptible to twisting. Following nuclear envelope breakdown, it is thus possible for hcp-6 chromosomes to become oriented such that a single centromere is connected to both poles simultaneously. Despite this failure to achieve bipolar attachment, these chromosomes are able to congress towards the center of the cell, assemble onto the metaphase plate, and begin to separate at anaphase (Khodjakov et al. 1997). However, these chromosomes fail to efficiently segregate into the daughter cells. Instead, they remain suspended in the center of the cell, or become stretched into chromatin bridges due to the strain of being pulled in two directions at once. Regardless, the end result is a marked failure in chromosome segregation.
An essential aspect of this model is that the rigidity of condensed wild-type chromosomes, although established earlier in mitosis, plays an important role in setting up bipolar attachment at prometaphase. It maintains the 180° opposition between sister centromeres, ensuring that once a sister centromere becomes captured by a pole, the opposing sister centromere will face the opposite pole (Roos 1973; Heneen 1975; Moore and Roth 2001). In addition, rigidity establishes the torsional integrity of wild-type chromosomes and prevents them from twisting. In wild-type cells, once a sister centromere is attached to a microtubule via the kinetochore, the chromosome is oriented such that the entire sister centromere is facing that pole (Rieder and Salmon 1998). The rigidity of wild-type chromosomes prevents them from twisting such that a single centromere is facing both poles simultaneously (Nicklas 1997). The less condensed chromosomes present in hcp-6 do not display any defects in centromere resolution, as seen by the back-to-back alignment maintained between their sister centromeres. However, their reduced rigidity permits them to become torqued or twisted along the long axis of the chromosome, such that a single centromere is oriented towards both poles, allowing microtubules from both centrosomes to capture a single sister chromatid.
The observation of merotelically oriented chromosomes in this mutant is particularly interesting, because they were recently proposed to play a major role in generating the aneuploidy seen in mammalian cells (Cimini et al. 2001, 2002). However, one question that remains unaddressed in these experiments is the mechanism by which merotelic chromosomes naturally arise. They are observed with an increased frequency following treatment with colcemid or nocadazole (Ladrach and LaFountain 1986; Cimini et al. 2001), which seem to bring about structural changes in the kinetochores, causing them to adopt a curled, crescent-like morphology and promoting merotelic attachment (Cimini et al. 2001). This observation is analogous to our results describing how the decondensed chromosomes of hcp-6 are more flexible, which leads to a failure in bipolar attachment. Based on this work, it seems possible that disrupting aspects of chromosome condensation in mammalian cells could promote the generation of merotelic chromosomes, and perhaps initiate tumor progression. This suggestion is consistent with the recent idea that defects in chromosome condensation may contribute to the genomic instability associated with cancer (Sen 2000). Our hope is that further examinations of the mechanisms of chromosome missegregation in model systems such as C. elegans will also provide valuable insights into important aspects of human biology.
Materials and methods
C. elegans strains and culture conditions
The N2 Bristol strain was used as the wild-type background. The following marker mutations were used, and are listed by chromosome: LGI dpy-5(e61), unc-13(e450), unc-73(e936), unc-38(x20), unc-63(x18); LGII rol-6(e187); LGIII unc-32(e189); LGIV unc-5(e53); LGV dpy-11(e224); LGX lon-2(e678). The integrated GFP∷histone strain AZ212 unc-119(ed3) ruIs32[pAZ132: pie-1∷GFP∷histoneH2B] III was generated by J. Austin as described by Praitis et al. (2001). The collection of temperature-sensitive embryo lethal mutants was generated using methods similar to those described by O’Connell et al. (1998). All hcp-6(mr17) stocks were maintained at a permissive temperature of 15°C and shifted to a restrictive temperature of 26°C for most experiments. Strains were cultured on NGM plates with E. coli OP50 as described by Brenner (1974).
Mapping and cloning of hcp-6
Initial mapping of hcp-6(mr17) demonstrated that it lies in the gene cluster of LGI. Fine-scale genetic mapping to the interval between dpy-5 and unc-63 was performed as follows: Progeny from hermaphrodites of genotype hcp-6/unc-63 dpy-5 were examined to identify Unc non-Dpy and Dpy non-Unc recombinants. Five out of 73 Unc non-Dpy recombinants carried the hcp-6(mr17) mutation, as did 68 of 77 Dpy non-Unc recombinants. Transgenic rescue experiments were performed in one of two ways. First, transgenic strains containing the YAC Y110A7 and a GFP marker plasmid (pPD118.20, which was constructed by A. Fire, Carnegie Institute, Baltimore, MD) were provided by Steve L'Hernault (Emory University, Atlanta, GA). The hcp-6(mr17) mutation was introduced into these strains using standard crosses. Second, exogenous DNA was injected into the gonads of hcp-6(mr17) adults (Mello and Fire 1995). YAC and cosmid (Fig. 2) DNA was prepared and injected at a concentration of 100ng/μL. A genomic clone of the entire hcp-6 gene was generated by PCR amplification of a 7697 bp sequence using the Elongase long-range PCR system (Gibco BRL) with the primers 5′-TCAACCTCGATTGCTGGCTG-3′ and 5′-CCTTCACAGC TCCTCCCATATC-3′. This construct represents the 6048 bp of hcp-6 plus approximately 1500bp of flanking genomic sequence. This construct was injected at a concentration of 5 ng/μL, along with yeast genomic DNA at a concentration of 80 ng/μL. In all cases, the dominant marker rol-6 was coinjected with the test DNA at a concentration of 100 ng/μL. To assay for rescue, individual F2 Rol adults from stable lines were shifted to 26°C and examined for their ability to generationally amplify. When the single Y110A7A.1 ORF was injected, four independent lines were obtained. On average, 10% of the adults from these lines were able to found a viable population of worms (2/32, 3/32, 4/24, 2/22).
Sequencing of the Y110A7A.1 gene from the hcp-6(mr17) genome was accomplished by purifying genomic DNA from a homozygous population of hcp-6(mr17) worms. Both strands of the gene were sequenced using a number of hcp-6 primers. The mutation site was confirmed in three independent reactions.
RNA interference
We generated a template for the production of Y110A7A.1 double stranded RNA as described by Moore et al. (1999). A 1000 bp sequence representing a portion of exon 8 was amplified from wild-type genomic DNA using the following primers: Sense 5′-gttggtcgaatggatgcaatg-3′; T7 sense 5′-GCGTAATAC GACTCACTATAGGGgttggtcgaatggatgcaatg-3′; Antisense 5′- ggattgcatctcgtaacataag-3′; T7 antisense 5′-GCGTAATACGA CTCACTATAGGGggattgcatctcgtaacataag-3′. dsRNA was synthesized using T7 RNA polymerase and brought up in water to a final concentration of 5mg/mL. This solution was injected into the syncytial gonad of adult N2 worms. Injected worms were allowed to recover for 18 h before embryo viability was assayed or embryo slides were prepared. hcp-3(RNAi) embryos were generated as described (Moore and Roth 2001).
Antibody preparation
A PCR fragment encoding amino acids 893–1247 of HCP-6 was cloned into the pET28a vector (Novagen). The resulting 6X His tagged recombinant protein was purified over a nickel column according to the manufacturer's instructions (QIAGEN). The purified protein was mixed with Freund's adjuvant and used to immunize mice. Boosts were performed every 2 wk, and serum was obtained after 6 wk. The sera was affinity-purified against membrane-bound antigen (Harlow and Lane 1988). The specificity of the antibody was demonstrated by three different criteria: a complete inhibition of staining following preincubation with the HCP-6 antigen, a significant decrease in the staining of hcp-6(RNAi) embryos, and an alteration of the staining pattern in hcp-6(mr17) embryos.
Antibody staining and microscopy
The N,N-dimethylformamide/methanol fixation and staining protocol described by Moore et al. (1999) was used for this work. The following primary antibodies were used: anti-HCP-3 (Buchwitz et al. 1999), anti-HCP-1 (Moore et al. 1999), and YL1/2, an anti-tubulin antibody (Amersham Pharmacia Biotech). Staining was detected with AlexaFluor 594 and 488-conjugated secondary antibodies (Molecular Probes).
All images were gathered using a Deltavision multiple wavelength fluorescent microscope (Applied Precision). Stacks of 0.2 μm optical sections were collected for the indicated wavelengths and then deconvolved using Softworx software (Applied Precision). The figures displayed here are two-dimensional projections of multiple optical sections.
GFP microscopy
Wild-type and hcp-6(mr17) adults homozygous for the GFP∷histone H2B transgene (see above) were shifted to 26°C for 1 h. They were then sliced open with a syringe needle, and one-cell embryos were collected using a mouth pipette. These embryos were placed on an agarose pad (1% agarose in M9 buffer) and examined under a Deltavision microscope. The microscope stage was heated to 26°C to maintain the proper temperature conditions. The one-cell embryos were examined until one began to undergo its first cleavage division, which generates the AB and P1 blastomeres. Immediately following this division, the progression of the AB nucleus through the cell cycle began to be filmed, with time points collected every 45 sec. For every time point, 20 0.2 μm optical sections were taken, to account for nuclear migration through the blastomere. In most cases, a complete round of the cycle (i.e., interphase to telophase) could be observed.
Acknowledgments
We thank Sue Biggins, Brian Buchwitz, Jesse Goldmark, Landon Moore, and Todd Nystul for critical reading and help in preparing this manuscript. The mutant collection described here was generated by Mark Roth, Mike Morrisson, and Kevin Harris. Brian Buchwitz, Debbie Frank, Mike Morrisson, Landon Moore, and the authors all contributed to the screen itself. We are especially grateful to Barbara Page, Steve L'Hernault, and A.M. Rose for genetics advice and worm strains. Some of the strains used here were provided by the Caenorhabditis Genetics Center. J.S. was supported by NIH training grants T32GM07270 and T32CA09657. This work was supported by NIH grant R01GM48435-06 to M.B.R.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mroth@fred.fhcrc.org; FAX (206) 667-6877.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.989102.
References
- Albertson DG, Thomson JN. The kinetochores of Caenorhabditis elegans. Chromosoma. 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Cimini D, Fioravanti D, Salmon ED, Degrassi F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci. 2002;115:507–515. doi: 10.1242/jcs.115.3.507. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes & Dev. 2002;16:729–742. doi: 10.1101/gad.968302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1988. [Google Scholar]
- Heneen WK. Ultrastructure of the prophase kinetochore in cultured cells of rat-kangaroo (Potorous tridactylis) Hereditas. 1975;79:209–220. [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, McEwen BF, Buttle KF, Rieder CL. Chromosome fragments possessing only one kinetochore can congress to the spindle equator. J Cell Biol. 1997;136:229–240. doi: 10.1083/jcb.136.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Ladrach KS, LaFountain JR., Jr Malorientation and abnormal segregation of chromosomes during recovery from colcemid and nocodazole. Cell Motil Cytoskeleton. 1986;6:419–427. doi: 10.1002/cm.970060407. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Albrecht MR, Chuang PT, Meyer BJ. MIX-1: An essential component of the C. elegans mitotic machinery executes X chromosome dosage compensation. Cell. 1998;92:265–277. doi: 10.1016/s0092-8674(00)80920-4. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Moore LL, Morrison M, Roth MB. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans. J Cell Biol. 1999;147:471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore LL, Roth MB. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- O'Connell KF, Leys CM, White JG. A genetic screen for temperature-sensitive cell-division mutants of Caenorhabditis elegans. Genetics. 1998;149:1303–1321. doi: 10.1093/genetics/149.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F, Schetter AJ, Mangone M, Stein L, Kemphues KJ. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr Biol. 2000;10:1619–1622. doi: 10.1016/s0960-9822(00)00869-1. [DOI] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC. Centromeres: Getting a grip of chromosomes. Curr Opin Cell Biol. 2000;12:308–319. doi: 10.1016/s0955-0674(00)00094-6. [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Hendzel MJ, Furbee CS, Muller MT, Bazett-Jones DP. Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J Cell Biol. 1996;134:1097–1107. doi: 10.1083/jcb.134.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos UP. Light and electron microscopy of rat kangaroo cells in mitosis. II. Kinetochore structure and function. Chromosoma. 1973;41:195–220. doi: 10.1007/BF00319696. [DOI] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing JA, Ball AR, Jr, Gregson HC, Alderton JM, Zhou S, Yokomori K. Identification of two distinct human SMC protein complexes involved in mitotic chromosome dynamics. Proc Natl Acad Sci. 1998;95:12906–12911. doi: 10.1073/pnas.95.22.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing JA, Gregson HC, Zhou S, Yokomori K. A human condensin complex containing hCAP-C-hCAP-E and CNAP1, a homolog of Xenopus XCAP-D2, colocalizes with phosphorylated histone H3 during the early stage of mitotic chromosome condensation. Mol Cell Biol. 2000;20:6996–7006. doi: 10.1128/mcb.20.18.6996-7006.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S. Aneuploidy and cancer. Curr Opin Oncol. 2000;12:82–88. doi: 10.1097/00001622-200001000-00014. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok T, Harvie PD, Buratovich M, Bryant PJ. The product of proliferation disrupter is concentrated at centromeres and required for mitotic chromosome condensation and cell proliferation in Drosophila. Genes & Dev. 1997;11:213–225. doi: 10.1101/gad.11.2.213. [DOI] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Yu HG, Dawe RK. Functional redundancy in the maize meiotic kinetochore. J Cell Biol. 2000;151:131–142. doi: 10.1083/jcb.151.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere–kinetochore complex: A repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]