Abstract
The frontal cortex (FC) is the seat of higher cognition. The genetic mechanisms that control formation of the functionally distinct subdivisions of the FC are unknown. Using a set of gene expression markers that distinguish subdivisions of the newborn mouse FC, we show that loss of Fgf17 selectively reduces the size of the dorsal FC whereas ventral/orbital FC appears normal. These changes are complemented by a rostral shift of sensory cortical areas. Thus, Fgf17 functions similar to Fgf8 in patterning the overall neocortical map but has a more selective role in regulating the properties of the dorsal but not ventral FC.
Keywords: arealization, regionalization, forebrain, protomap, neocortex
The frontal cortex (FC) consists of prefrontal, premotor, and motor areas that play a central role in cognition, movement, and behavior (1). The adult rodent prefrontal cortex (PFC) can be divided into medial and orbital regions that are thought to have homologues in primates (2). The medial PFC (mPFC) can be further subdivided into the dorsal mPFC, which includes frontal association, anterior cingulate, and dorsal prelimbic areas; and the ventral mPFC, which consists of ventral prelimbic, infralimbic, and medial orbital areas (3). The developmental mechanisms that generate FC subdivisions are unknown, in part, because of the lack of markers that distinguish these regions. In addition, most known mouse mutants that affect cortical patterning die at birth, precluding later analysis, when individual areas are distinguishable by classical histological methods.
Current evidence shows that neocortical areas are presaged by regionalized expression of transcription factors and other regulatory genes in the cortical neuroepithelium and cortical plate, supporting the protomap model (4–7). Members of the fibroblast growth factor (Fgf) family of genes have been implicated in controlling neocortical regionalization. Fgf8 and Fgf17 encode secreted signaling proteins and are expressed in a partially overlapping pattern in the rostral forebrain patterning center immediately adjacent to the developing FC [Fig. 1A and supporting information (SI) Fig. 7] (8–11). Fgf8 patterns the neocortex in part by regulating the expression of transcription factor gradients in the cortical neuroepithelium (7, 12–16). Although ectopic expression of Fgf17 has been reported to have effects similar to that of Fgf8 in mediating overall patterning of the neocortical map (13), the role of endogenous Fgf17 in forebrain development is unknown.
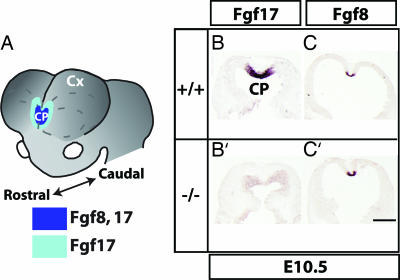
Fig. 1.
Fgf8 and Fgf17 expression overlap in the forebrain rostral patterning center, and Fgf8 expression is maintained in the Fgf17−/− mutant. (A) Fgf17 and Fgf8 RNA expression in the rostral forebrain patterning center. CP, commissural plate; Cx, cortex. (B, B′, C, and C′) Fgf17 and Fgf8 in situ hybridization (ISH) on horizontal sections from embryonic day 10.5 Fgf17+/+ (B and C) and Fgf17−/− (B′ and C′) forebrain. Rostral is at the top. (Scale bar: 0.5 mm.)
In this study we devised a panel of gene expression markers to examine the role of Fgf17 in the regionalization of the rodent FC using Fgf17-null mice (Fgf17−/−) (17). We report that the dorsal FC of Fgf17−/− mice was reduced in size, whereas ventral and orbital FC regions appeared normal. The reduction in the dorsal FC area was complemented by a rostromedial shift of caudal cortical areas. These changes in regionalization persisted into adulthood and were accompanied by a reduction in FC projections to subcortical targets. Thus, in addition to an overall effect on neocortical patterning, Fgf17 has an unexpectedly selective role in regulating dorsal FC development.
Results
We examined Fgf8 expression in the rostral patterning center of Fgf17−/− mutants given Fgf8's important function in telencephalic patterning (12, 14–16). At embryonic day 10.5, telencephalic expression of Fgf8 appeared the same in wild-type and Fgf17−/− littermate embryos (Fig. 1 C and C′), suggesting that Fgf17 does not affect cortical development by regulating Fgf8 expression.
The Fgf17−/− forebrain lacked overt morphological defects (SI Fig. 8 A, A′, B, and B′). Although we found no significant difference in cortical surface area in postnatal day 0 (P0) brains (SI Fig. 8 C), adult cortical surface area was slightly (≈7%) reduced (SI Fig. 8 D). In addition, the olfactory bulbs and basal ganglia, which are severely reduced in Fgf8neo/neo and Fgf8neo/null hypomorphic mutants, respectively (14, 16), are roughly normal in size and exhibited no differences in histology or gene expression in Fgf17−/− mutants (SI Fig. 9). This suggests that, compared with Fgf8, Fgf17 has only a minor role in regulating the overall growth of the telencephalon.
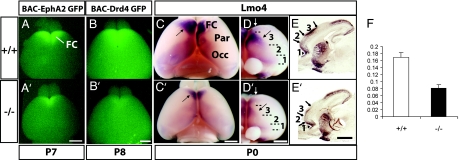
To assess whether the Fgf17−/− mutation altered rostral parts of the telencephalon, we focused on the FC. To this end, we introduced BAC-EphA2 and BAC-Drd4 alleles, which express GFP in specific FC domains (18). Fgf17−/− mice at P0, P7, and P8 had a smaller domain of FC GFP fluorescence (Fig. 2A, A′, B, and B′ and data not shown), suggesting a decrease in FC size. We similarly observed reduced dorsal FC Lmo4 RNA expression in Fgf17−/− whole-mount brains at P0 (Fig. 2 C, C′, D, and D′), providing evidence that the small FC is not due to the BAC transgene. Lmo4+ dorsal FC area was reduced by 52% after correcting for overall cortex size (Fig. 2F). Interestingly, Lmo4 expression in the medial and orbital FC was not overtly affected (Fig. 2 D and D′), suggesting that Fgf17 has a selective role in patterning FC subdivisions.
Fig. 2.
Reduced FC size in Fgf17−/− mice. Arrows signify shifted boundaries, and arrowheads signify maintained boundaries. (A and A′) Dorsal views of P7 Fgf17+/+ and Fgf17−/− brains positive for the BAC-EphA2 GFP transgene. The GFP+ domain that marks the FC was reduced in Fgf17−/− mutants. (B and B′) Dorsal views of P8 Fgf17+/+ and Fgf17−/− brains positive for the BAC-Drd4 GFP transgene. (C and C′) Dorsal views of Lmo4 whole-mount ISH on P0 Fgf17+/+ and Fgf17−/− brains. Par, parietal cortex; Occ, occipital cortex. (D and D′) Frontal views of the same brains in C and C′ reveal gene expression boundaries that distinguish three early FC subdivisions that we have labeled 1–3. (E and E′) Sagittal sections processed for Lmo4 ISH on P0 Fgf17+/+ and Fgf17−/− brains reveal sharp gene expression boundaries within the FC. White arrows in D and D′ indicate the approximate plane of section in E and E′. (F) Ratio of Lmo4+ dorsal FC area to total cortex area in Fgf17+/+ (n = 4) and Fgf17−/− (n = 4) P0 hemispheres (Student's t test; t = 5.21 and P < 0.01). (Scale bars: 1 mm.)
To distinguish between a reduction in expression levels and a shift in area properties, we examined Lmo4 expression in sagittal sections. We observed no change in the level of Lmo4 RNA or in the layer-specific pattern, but rather a rostral shift of the sharp borders that approximate neocortical areal subdivisions (Fig. 2 E and E′). In the sagittal view the dorsal FC domain was rostrally shifted (3 in Fig. 2 E and E′), whereas the ventral domain was less affected (1 in Fig. 2 E and E′). Other brain structures such as the striatum, olfactory tubercle, and hippocampus displayed normal Lmo4 expression.
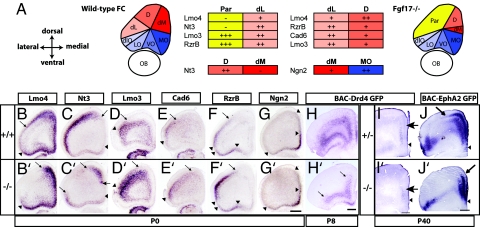
We explored the possibility that Fgf17 has a selective role in dorsal FC patterning using a panel of gene expression markers on a series of coronal sections that span the FC at P0 (Fig. 3 and SI Figs. 10 and 11). Based on the expression domains and complementary borders of BAC-Drd4 GFP, Lmo4, Cad8, Nt3, Steel, Ngn2, Rzr-β, Cad6, Lmo3, EphrinA5, and Id2, we distinguished subdivisions in the rostral cortex that correlate with presumptive anatomical cortical areas (Fig. 3A, SI Fig. 10, Table 1, and SI Table 2). In the dorsal cortex we defined three FC subdivisions, dorsomedial, dorsal, and dorsolateral, and a single parietal cortex subdivision, whereas the ventral FC consisted of orbital subdivisions (medial orbital cortex, ventral orbital cortex, lateral orbital cortex, and dorsolateral orbital cortex), and more caudally the agranular insular and infralimbic areas (Fig. 3A, SI Fig. 10, Table 1, and SI Table 2).
Fig. 3.
Selective changes in dorsal FC molecular properties revealed by a panel of gene expression markers. Arrows signify shifted boundaries, and arrowheads signify maintained boundaries. (A) Schema of wild-type and Fgf17−/− mutant FC subdivisions based on a panel of gene expression markers at P0. Dorsal and ventral FC subdivisions are shaded in red and blue, respectively. The parietal cortex is shaded in yellow. The table focuses on key wild-type subdivision distinctions with levels of expression for each gene: +++, strong expression; ++, moderate expression; +, weak expression; −, no detectable expression. See Table 1 for corresponding anatomical areas and SI Tables 2 and 3 for a more detailed analysis. (B–G and B′–G′) ISH for Lmo4, Nt3, Lmo3, Cad6, Rzr-β, and Ngn2 on Fgf17+/+ and Fgf17−/− littermate P0 coronal sections. Note the shift in dorsal expression borders (arrows) but maintenance of ventral borders (arrowheads). (H, H′, I, and I′) Anti-GFP immunohistochemistry on coronal sections from P8 (H and H′) and P40 (I and I′) mice containing the BAC-Drd4 GFP transgene. Note that the expression is much broader in the FC at P8 but is restricted in the mPFC at P40. (J and J′) Anti-GFP immunohistochemistry on coronal sections from P40 mice containing the BAC-EphA2 GFP transgene. D, dorsal FC; dlO, dorsolateral orbital cortex; dM, dorsomedial FC; LO, lateral orbital cortex; MO, medial orbital cortex; Par, parietal cortex; VO, ventral orbital cortex. (Scale bars: 0.5 mm.)
Table 1.
Frontal cortex subdivision definitions
| Gene-defined region | Anatomical areas |
|
|---|---|---|
| Zilles and Wree (40) | Krettek and Price (41) | |
| Dorsolateral | Fr1, Fr3 | PrCl |
| Dorsal | Fr1, Fr2 | PrCl, PrCm |
| Dorsomedial | Cg1, Cg2, Cg3 | ACd, ACv, PL |
| Infralimbic | IL | IL |
| Medial orbital | MO | MO |
| Ventral orbital | VO | VO |
| Lateral orbital | LO | LO |
| Dorsolateral orbital | DLO | DLO |
| Agranular insular | AID/AIV | AId/AIv |
| Parietal | Par1 | S1 |
We used this gene expression panel to determine how individual regional FC subdivisions were altered in P0 Fgf17−/− mutants. In this analysis we compared matched coronal sections (Fig. 3) and a whole series of coronal sections (SI Figs. 10 and 11). We focused on some key subdivision distinctions that showed the most obvious changes in gene expression (Fig. 3A and SI Table 3). Fgf17−/− mice had medially shifted dorsal FC expression borders of Lmo4, Nt3, BAC-Drd4 GFP, and Cad8 (Fig. 3 B, B′, C, and C′ and SI Figs. 10 and 11), whereas parietal cortex markers Lmo3, Cad6, Rzr-β, and EphrinA5 showed a complementary expansion from more caudal cortex into this region (Fig. 3 D, D′, E, E′, F, and F′ and SI Figs. 10 and 11). By contrast, the ventral FC showed no changes in gene expression (Fig. 3 and SI Figs. 10 and 11). Together, the pattern of changes suggests that subdivisions of the dorsal FC (dorsal, dorsolateral, and dorsomedial regions) are reduced, the parietal cortex expands into the FC, and ventral FC subdivisions are not affected (Fig. 3A and SI Fig. 11). This provides strong evidence that Fgf17 has a selective role in regulating the regional properties of the dorsal but not ventral FC.
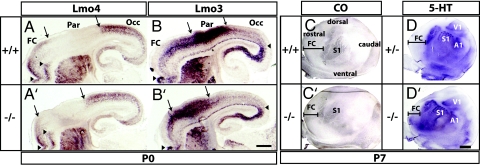
To explore further whether there is a rostral shift of caudal cortical regions, we examined gene expression in P0 sagittal sections. This revealed a rostral shift of parietal and occipital domains delimited by Lmo4 and Lmo3 expression (Fig. 4A, A′, B, and B′ and SI Fig. 12). These observations were confirmed at P7 based on a rostral shift of the somatosensory and visual cortex in flat-mount preparations stained for cytochrome oxidase and 5-hydroxytryptamine (5-HT, serotonin) (Fig. 4 C, C′, D, and D′) (19). Therefore, in addition to regulating the size of dorsal FC areas, Fgf17 controls the position of sensory neocortical areas along the rostral–caudal axis.
Fig. 4.
Rostral shift of the neocortical map in Fgf17−/− mutants. Rostral is to the left. Arrows signify shifted boundaries, and arrowheads signify maintained boundaries. (A, A′, B, and B′) Lmo4 and Lmo3 ISH on sagittal sections from Fgf17+/+ and Fgf17−/− P0 brains mark complementary cortical domains: frontal/occipital (FC/Occ) and parietal (Par) cortex, respectively. (Scale bar: 0.5 mm.) (C, C′, D, and D′) Cytochrome oxidase (CO) and anti-serotonin (5-HT) immunohistochemistry on tangential sections of flattened P7 cortices reveal a rostrodorsal shift of primary sensory areas. S1, somatosensory; V1, visual; A1, auditory. (Scale bar: 1 mm.)
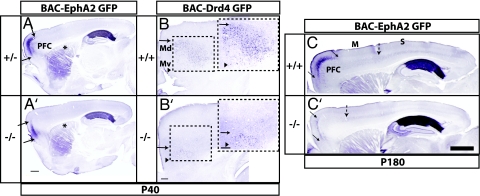
Next we assessed whether these early postnatal alterations in cortical areas were observed in the mature brain by examining expression of BAC-Drd4 GFP and BAC-EphA2 GFP in P40 and 6-month-old mice. At P40, BAC-Drd4 GFP labeled a discrete domain of cells in the medial FC that correlates with the prelimbic area (Figs. 3I and 5B) (20). Although the position of the ventral border and layer-specificity of GFP+ cells was maintained, the extent of this domain was reduced in the Fgf17−/− brain (Figs. 3I′ and 5B′), suggesting that the prelimbic cortical area (ventral part of dorsomedial region) is reduced in size. At P40 and in 6-month-old mice, BAC-EphA2 GFP labels the PFC (including frontal association, anterior cingulate, prelimbic, and infralimbic areas) (Figs. 3J and 5 A and C and SI Figs. 13 and 14). In the Fgf17−/− mutant, the dorsal expression border was shifted medially, whereas the ventral border was maintained (Figs. 3J′ and 5A′ and C′ and SI Figs. 13 and 14), confirming that the PFC is smaller.
Fig. 5.
Persistence of changes in FC molecular regionalization in mature Fgf17−/− mice. Arrows signify shifted boundaries, and arrowheads signify maintained boundaries. (A, A′, B, and B′) Sagittal sections of brains from P40 mice carrying either the BAC-EphA2 GFP or BAC-Drd4 GFP transgene processed for anti-GFP immunohistochemistry. Fiber staining in the dorsal striatum in BAC-EphA2 GFP+ mice corresponds to projections from the FC (asterisks in A and A′). Boxed areas, consisting of dorsomedial (Md) and ventromedial (Mv) subdivisions of the PFC, are shown in a magnified view (Right Insets in B and B′). (Scale bars: 0.5 mm.) (C and C′) Sagittal sections of cortex from adult (P180) BAC-EphA2+, Fgf17+/+, and Fgf17−/− mice processed for anti-GFP immunohistochemistry. The broken arrow approximates the sensory (S)–motor (M) boundary. (Scale bar: 1 mm.)
The dorsal FC sends projections through the dorsal striatum, which can be visualized in mice expressing BAC-EphA2 GFP (3, 18). In Fgf17+/− BAC-EphA2 GFP+ mice, GFP+ fiber staining is apparent throughout most of the dorsal striatum (Fig. 5A and SI Fig. 13). However, in Fgf17−/− mice, GFP+ fiber labeling in the dorsolateral striatum appeared reduced, consistent with the smaller domain of BAC-EphA2 GFP+ cells in the dorsal FC (Fig. 5A′ and SI Fig. 13).
BAC-EphA2 GFP is expressed at higher levels in the somatosensory cortex and at lower levels in the motor cortex at P180 (Fig. 5C). The sensory–motor boundary was shifted to a more rostral position in the Fgf17−/− cortex (dashed arrows in Fig. 5 C and C′). Consistent with this gene expression shift, cytochrome oxidase staining of adjacent sections revealed that the somatosensory barrels were shifted rostrally in the Fgf17−/− mutant (SI Fig. 14). Together, these findings suggest that the early pattern of changes in regional molecular properties (Figs. 2–4) results in a permanent change in the distribution of adult cortical areas.
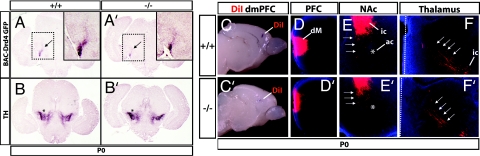
A subset of FC projections extends to the substantia nigra pars compacta and adjacent ventral tegmental area (3). These projections can be visualized in mice expressing the BAC-Drd4 GFP transgene (Fig. 6A and SI Fig. 15) (18). Staining of these projections was reduced in the Fgf17−/− mutant (Fig. 6A′), consistent with the reduction of BAC-Drd4 GFP+ cells in the FC (Fig. 3H′ and SI Fig. 11). This suggests that the Fgf17−/− mutation has a quantitative effect on dorsal FC cell number but does not have an overt qualitative effect on the pathfinding properties of the remaining BAC-Drd4 GFP+ axons. Furthermore, despite the reduction in BAC-Drd4 GFP+ FC axons, staining for tyrosine hydroxylase, a marker of midbrain dopamine neurons, did not show a discernable change (Fig. 6 B and B′), suggesting that the Fgf17−/− mutation does not overtly affect midbrain dopamine cell number.
Fig. 6.
FC connectivity in Fgf17−/− mice. (A and A′) Reduction in FC projections to the ventral midbrain revealed by anti-GFP immunohistochemistry on P0 coronal sections from Fgf17+/+ and Fgf17−/− mice containing the BAC-Drd4 GFP transgene. Note the reduced staining of fibers emanating from the cerebral peduncle (arrows). Insets show magnified views of the boxed areas. (B and B′) Anti-tyrosine hydroxylase (TH) immunohistochemistry on sections adjacent to those shown in A and A′. Staining of the substantia nigra pars compacta and ventral tegmental area (asterisks) was similar between genotypes. (C and C′) DiI crystal placements in the dorsomedial FC of P3 Fgf17+/+ and Fgf17−/− brains viewed from the medial side. (D and D′) Restricted DiI-labeled field in the dorsomedial (dM) PFC in coronal section rostral to the crystal placement. Medial is to the left. (E and E′) Projections to the nucleus accumbens (NAc) (arrows). Anterograde-labeled fibers in the internal capsule (ic) restricted to the ventromedial striatum are also present. The anterior commisure provides a landmark (asterisk). Medial is to the left. (F and F′) Corticothalamic fibers (arrows) emanate from the internal capsule (ic) and are present in similar locations in both genotypes. A dotted line designates the thalamic midline.
Finally, we examined immature FC connectivity in P0 and P3 Fgf17+/+ and Fgf17−/− mutant brains using the lipophilic dye (1,1′-dioctadecyl 3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) (Fig. 6 and SI Fig. 16). Placement of DiI crystals in the mPFC (Fig. 6C and SI Fig. 16) (dorsomedial and ventromedial regions) labeled cells and fibers in restricted domains within the mPFC and did not back-label cells or fibers in other parts of the cortex (Fig. 6D and SI Fig. 16). We observed a subset of projections oriented toward the nucleus accumbens (Fig. 6E and SI Fig. 16), other fibers tightly localized within the internal capsule in the medial striatum (Fig. 6E and SI Fig. 16), and labeling in the medial thalamus (Fig. 6F and SI Fig. 16). We did not observe major differences between genotypes in these labeling patterns (Fig. 6 C′, D′, E′, and F′ and SI Fig. 16).
Discussion
We provide evidence that Fgf17 has a selective function in the regionalization of FC subdivisions. Our analysis required identifying a panel of gene expression markers that distinguish dorsal and ventral subdivisions of the newborn rodent FC (Fig. 3, SI Fig. 10, Table 1, and SI Tables 2 and 3). In addition, we have characterized two lines of BAC transgenic mice that provide specific GFP labeling of the FC, and we show that they can be used as readouts of cortical regionalization and projections patterns from the FC (Figs. 2, 3, and 5 and SI Figs. 10, 13, and 14). The expression pattern of many of these genes revealed distinct borders within the FC at birth, suggesting a genetic partitioning of this cortical region before overt cytoarchitectonic differentiation. Although expression of none of the genes was limited to a single cortical region, combinations of genes were useful in defining regional subdivisions that correlated with histologically defined cortical areas (Table 1).
The lack of overt forebrain morphologic defects in Fgf17−/− mutants and the mildly reduced cortical surface area in the adult (SI Fig. 8) show that Fgf17 does not have a major effect on forebrain growth. Rather, the results suggest that Fgf17 has a more specific function in regulating regional specification, particularly within the FC. By contrast, severe reductions in Fgf8 levels result in gross forebrain morphologic defects that are likely due to a combination of abnormal regional specification, decreased proliferation, and increased apoptosis (15, 16). In addition to spatiotemporal differences in Fgf8 and Fgf17 expression (17) (Fig. 1), differences in ligand affinity for FGF receptors and ability to regulate gene expression may explain the differences in phenotypic effects (21, 22).
The Fgf17−/− mutant displays a rostral shift of caudal cortical areas (Fig. 4 and SI Fig. 12) that is less severe than in Fgf8neo/neo mildly hypomorphic mutants (14), suggesting that Fgf17 has a more subtle role than Fgf8 in patterning the neocortical areal map. Our postnatal analyses do not clarify whether the rostral shift in sensory cortices is only due to hypoplasia of the FC and/or due to respecification of the rostral progenitor domain to develop as more caudal cortex.
Analysis of FC subdivisions unexpectedly identified that Fgf17 selectively regulates the sizes and positions of dorsal, but not ventral, FC subdivisions (Fig. 3 and SI Figs. 10 and 11). In Fgf8neo/neo mutants both the dorsal and ventral FC are reduced (ref. 14 and J.A.C. and J.L.R.R., unpublished data). These phenotypic differences may be explained in part by the observations that (i) Fgf17 was expressed in a broader domain in the rostral patterning center than Fgf8 (particularly in its dorsal extent), and (ii) Fgf8 expression was normal in the Fgf17−/− embryonic brain (Fig. 1). We propose that reduced FGF signaling specifically in the dorsal part of the rostral patterning center could selectively affect dorsal FC regionalization while preserving the ventral FC. Current evidence, which is consistent with the protomap model, suggests that FGFs produced by the rostral patterning center regulate the regional expression of transcription factors in the neuroepithelium to specify cortical areal identity (4–7).
The neocortex of Fgf17−/− mutants shows correct area-specific thalamic innervation, as exemplified by the presence of somatosensory barrels, detected by both cytochrome oxidase staining and serotonin immunohistochemistry at P7 (Fig. 4). However, the position of the somatosensory barrel fields is shifted rostrally, showing that thalamocortical innervation shifts in concert with the shift in areal molecular markers. In both the Fgf17−/− and Fgf8neo/neo mutants, there was no detectable difference in thalamocortical innervation at P0 (14) (SI Fig. 16 and data not shown). Ectopic Fgf8 expression experiments, which result in viable animals, suggest that rerouting of thalamocortical axons to area-specific targets occurs postnatally within the cortex (23). This could account for the rostral shift of the innervation of somatosensory barrel fields in the Fgf17−/− mutant.
Recent evidence suggests that Fgf8 plays a role in regulating patterns of intracortical connectivity (24). Unlike in the Fgf8neo/neo mutant, we found no evidence for ectopic rostral projections of caudally located cortical neurons in the Fgf17−/− brain (data not shown), consistent with the subtler phenotype of the Fgf17−/− mutants.
Although Fgf17−/− mutants did not show an overt qualitative defect in dorsomedial FC connectivity/projections (Fig. 6 and SI Fig. 16), there was evidence for a quantitative reduction in its subcortical projections based on BAC-EphA2 and BAC-Drd4 GFP expression in the striatum and ventral midbrain, respectively (Figs. 5 and 6 and SI Figs. 13 and 15). We propose that this is secondary to a reduction in the number of FC-specified neurons. Reduced PFC output to striatal or midbrain dopaminergic neurons may have important physiologic ramifications for the regulation of neural pathways involved in reward, cognition, and social behavior (25–29).
Dorsal and ventral FC subdivisions have distinct roles in regulating cognition and behavior in rodents (3, 26) and primates including humans (25, 30, 31). For example, subdivisions of the dorsal PFC are implicated in working memory, attention, response selection, temporal processing of information, effort-related decision making, and social valuation, whereas ventromedial and orbital subdivisions are implicated in behavioral flexibility, emotional regulation, delay-related decision making, evaluation of rewards, and autonomic control (1, 3, 25, 26, 29, 30, 32–34). Therefore, the Fgf17−/− mutant provides a unique opportunity to examine the behavioral and neurophysiologic consequences of an early developmental genetic lesion that selectively affects the dorsal FC. We have identified circumscribed behavioral deficits in Fgf17−/− mutants that affect social interactions (K. Scearce-Levie, E. Roberson, J.A.C., J.L.R.R., and L. Mucke, unpublished data). We propose that elucidating the signaling pathways downstream of Fgf17 will provide important insights into the genetic pathways that regulate FC development and that may be disrupted in disorders that affect cognition, emotion, and social interactions.
Methods
Detailed Methods.
SI Methods contains a more detailed description of methods used.
Animals and Tissue Preparation.
All mice were housed and handled in accordance with the Institutional Animal Care and Use Committee of the University of California, San Francisco. Fgf17−/− mice and embryos were generated by mating male and female heterozygotes (Fgf17+/−) (17). BAC transgenic lines BAC-Drd4 GFP and BAC-EphA2 GFP (18) were mated to Fgf17−/− mice to generate double heterozygotes, which were then crossed to Fgf17+/− mice to generate Fgf17+/+ and Fgf17−/− BAC transgene-positive littermates. All tissue was harvested, fixed, and cryopreserved according to standard methods. Sections were cut on either a cryostat or a freezing microtome.
ISH and Immunohistochemistry.
Digoxigenin-labeled riboprobes were generated for the following genes: Cadherin-6, Cadherin-8 (35), Fgf8 (36), Fgf17 (10), Id-2 (35), Lmo3, and Lmo4 (37), Neurogenin-2 (38), Neurotrophin-3 (gift from Luis Parada, UT Southwestern, Dallas, TX), RZR-β (35), and Steel (gift from E. Grove, University of Chicago, Chicago, IL). Section and whole-mount ISH were performed as described previously in refs. 35 and 19, respectively.
Immunohistochemistry was performed by using standard protocols (35) with rabbit anti-GFP (1:1,000; Invitrogen, Carlsbad, CA), rabbit anti-tyrosine hydroxylase (1:500; Chemicon, Temecula, CA), rabbit anti-serotonin (1:50,000; Immunostar, Hudson, WI) antibodies and detected with goat anti-rabbit biotinylated secondary antibody (1:200–1:400; Vector Laboratories, Burlingame, CA) and the ABC kit (Vector Laboratories).
Axon Tracing.
P0–P3 brains were stored in 4% PFA in PBS at 4°C. Single crystals of the fluorescent carbocyanide dye DiI (Invitrogen) were placed in various cortical locations (39). After diffusion, sections were cut on a vibratome and immediately mounted on slides using Vectashield mounting medium with DAPI (Vector Laboratories).
Digital Imaging and Quantification of Cortical Areas.
Whole brains and sections were photographed by using SPOT (Diagnostic Instruments, Sterling Heights, MI) and Olympus digital cameras and imaging software. Areas were determined by using photos of dorsally viewed whole-mount brains in ImageJ (National Institutes of Health, Bethesda, MD). Excel (Microsoft, Redmond, WA) was used for calculations and statistical analysis.
Supplementary Material
Acknowledgments
We thank David Ornitz for providing the Fgf17 mice; Kimberly Scearce-Levie, Erik Roberson, Lennart Mucke, and members of the J.L.R.R. laboratory for insightful discussion and valuable comments regarding the manuscript; and Dianna Kahn for technical assistance. This work was supported by the Medical Scientist Training Program of the University of California, San Francisco (J.A.C.); Nina Ireland (J.L.R.R.); the Larry L. Hillblom Foundation (J.L.R.R.); and National Institutes of Health Grants NS34661-01A1 and K05 MH065670 (both to J.L.R.R.).
Abbreviations
- FC
frontal cortex
- PFC
prefrontal cortex
- mPFC
medial PFC
- Pn
postnatal day n
- DiI
1,1′-dioctadecyl 3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- ISH
in situ hybridization.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702225104/DC1.
References
- 1.Fuster JM. Neuron. 2001;30:319–333. doi: 10.1016/s0896-6273(01)00285-9. [DOI] [PubMed] [Google Scholar]
- 2.Uylings HB, Groenewegen HJ, Kolb B. Behav Brain Res. 2003;146:3–17. doi: 10.1016/j.bbr.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 3.Heidbreder CA, Groenewegen HJ. Neurosci Biobehav Rev. 2003;27:555–579. doi: 10.1016/j.neubiorev.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Rakic P. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 5.O'Leary DD, Nakagawa Y. Curr Opin Neurobiol. 2002;12:14–25. doi: 10.1016/s0959-4388(02)00285-4. [DOI] [PubMed] [Google Scholar]
- 6.Grove EA, Fukuchi-Shimogori T. Annu Rev Neurosci. 2003;26:355–380. doi: 10.1146/annurev.neuro.26.041002.131137. [DOI] [PubMed] [Google Scholar]
- 7.Sur M, Rubenstein JL. Science. 2005;310:805–810. doi: 10.1126/science.1112070. [DOI] [PubMed] [Google Scholar]
- 8.Maruoka Y, Ohbayashi N, Hoshikawa M, Itoh N, Hogan BL, Furuta Y. Mech Dev. 1998;74:175–177. doi: 10.1016/s0925-4773(98)00061-6. [DOI] [PubMed] [Google Scholar]
- 9.Hoshikawa M, Ohbayashi N, Yonamine A, Konishi M, Ozaki K, Fukui S, Itoh N. Biochem Biophys Res Commun. 1998;244:187–191. doi: 10.1006/bbrc.1998.8239. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Lawshe A, MacArthur CA, Ornitz DM. Mech Dev. 1999;83:165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 11.Bachler M, Neubuser A. Mech Dev. 2001;100:313–316. doi: 10.1016/s0925-4773(00)00518-9. [DOI] [PubMed] [Google Scholar]
- 12.Fukuchi-Shimogori T, Grove EA. Science. 2001;294:1071–1074. doi: 10.1126/science.1064252. [DOI] [PubMed] [Google Scholar]
- 13.Fukuchi-Shimogori T, Grove EA. Nat Neurosci. 2003;6:825–831. doi: 10.1038/nn1093. [DOI] [PubMed] [Google Scholar]
- 14.Garel S, Huffman KJ, Rubenstein JL. Development (Cambridge, UK) 2003;130:1903–1914. doi: 10.1242/dev.00416. [DOI] [PubMed] [Google Scholar]
- 15.Storm EE, Rubenstein JL, Martin GR. Proc Natl Acad Sci USA. 2003;100:1757–1762. doi: 10.1073/pnas.0337736100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storm EE, Garel S, Borello U, Hebert JM, Martinez S, McConnell SK, Martin GR, Rubenstein JL. Development (Cambridge, UK) 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Liu Z, Ornitz DM. Development (Cambridge, UK) 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- 18.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 19.Hamasaki T, Leingartner A, Ringstedt T, O'Leary DD. Neuron. 2004;43:359–372. doi: 10.1016/j.neuron.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 20.Noain D, Avale ME, Wedemeyer C, Calvo D, Peper M, Rubinstein M. Eur J Neurosci. 2006;24:2429–2438. doi: 10.1111/j.1460-9568.2006.05148.x. [DOI] [PubMed] [Google Scholar]
- 21.Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu A, Li JY, Bromleigh C, Lao Z, Niswander LA, Joyner AL. Development (Cambridge, UK) 2003;130:6175–6185. doi: 10.1242/dev.00845. [DOI] [PubMed] [Google Scholar]
- 23.Shimogori T, Grove EA. J Neurosci. 2005;25:6550–6560. doi: 10.1523/JNEUROSCI.0453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huffman KJ, Garel S, Rubenstein JL. J Neurosci. 2004;24:8917–8923. doi: 10.1523/JNEUROSCI.2086-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman-Rakic PS. Proc Natl Acad Sci USA. 1996;93:13473–13480. doi: 10.1073/pnas.93.24.13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalley JW, Cardinal RN, Robbins TW. Neurosci Biobehav Rev. 2004;28:771–784. doi: 10.1016/j.neubiorev.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Young LJ, Wang Z. Nat Neurosci. 2004;7:1048–1054. doi: 10.1038/nn1327. [DOI] [PubMed] [Google Scholar]
- 28.Meyer-Lindenberg A, Kohn PD, Kolachana B, Kippenhan S, McInerney-Leo A, Nussbaum R, Weinberger DR, Berman KF. Nat Neurosci. 2005;8:594–596. doi: 10.1038/nn1438. [DOI] [PubMed] [Google Scholar]
- 29.Kellendonk C, Simpson EH, Polan HJ, Malleret G, Vronskaya S, Winiger V, Moore H, Kandel ER. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Price JL. Prefrontal Cortex. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- 31.Amodio DM, Frith CD. Nat Rev Neurosci. 2006;7:268–277. doi: 10.1038/nrn1884. [DOI] [PubMed] [Google Scholar]
- 32.Rudebeck PH, Buckley MJ, Walton ME, Rushworth MF. Science. 2006;313:1310–1312. doi: 10.1126/science.1128197. [DOI] [PubMed] [Google Scholar]
- 33.Rudebeck PH, Walton ME, Smyth AN, Bannerman DM, Rushworth MF. Nat Neurosci. 2006;9:1161–1168. doi: 10.1038/nn1756. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell JP, Macrae CN, Banaji MR. Neuron. 2006;50:655–663. doi: 10.1016/j.neuron.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 35.Rubenstein JL, Anderson S, Shi L, Miyashita-Lin E, Bulfone A, Hevner R. Cereb Cortex. 1999;9:524–532. doi: 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- 36.Crossley PH, Martin GR. Development (Cambridge, UK) 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- 37.Bulchand S, Subramanian L, Tole S. Dev Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- 38.Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 39.Godement P, Vanselow J, Thanos S, Bonhoeffer F. Development (Cambridge, UK) 1987;101:697–713. doi: 10.1242/dev.101.4.697. [DOI] [PubMed] [Google Scholar]
- 40.Zilles K, Wree A. In: The Rat Nervous System. Paxinos G, editor. San Diego: Academic Press; 1995. pp. 649–685. [Google Scholar]
- 41.Krettek JE, Price JL. J Comp Neurol. 1977;171:157–191. doi: 10.1002/cne.901710204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.