Abstract
Laonastes aenigmamus is an enigmatic rodent first described in 2005. Molecular and morphological data suggested that it is the sole representative of a new mammalian family, the Laonastidae, and a member of the Hystricognathi. However, the validity of this family is controversial because fossil-based phylogenetic analyses suggest that Laonastes is a surviving member of the Diatomyidae, a family considered to have been extinct for 11 million years. According to these data, Laonastes and Diatomyidae are the sister clade of extant Ctenodactylidae (i.e., gundies) and do not belong to the Hystricognathi. To solve the phylogenetic position of Laonastes, we conducted a large-scale molecular phylogeny of rodents. The analysis includes representatives of all major rodent taxonomic groups and was based on 5.5 kb of sequence data from four nuclear and two mitochondrial genes. To further validate the obtained results, a short interspersed element insertion analysis including 11 informative loci was also performed. Our molecular data based on sequence and short interspersed element analyses unambiguously placed Laonastes as a sister clade of gundies. All alternative hypotheses were significantly rejected based on Shimodaira–Hasegawa tests, supporting the idea that Laonastes does not belong to the Hystricognathi. Molecular dating analysis also supports an ancient divergence, ≈44 Mya ago, between Ctenodactylidae and Laonastes. These combined analyses support the hypothesis that Laonastes is indeed a living fossil. Protection of this surviving species would conserve an ancient mammalian family.
Keywords: Laonastes aenigmamus, molecular phylogeny, rodent, retroposons
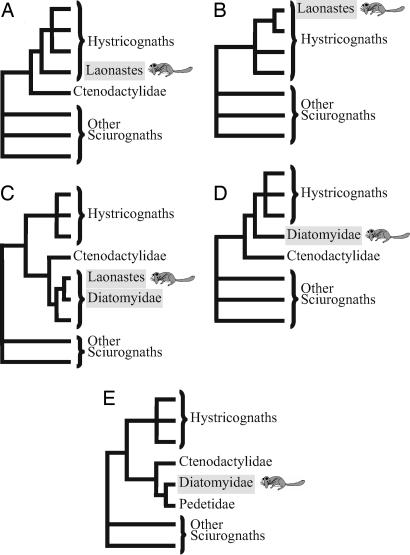
The discovery of a new mammalian family is a rare event; the last two were described in 1974 and 2005, with the discoveries of the bumblebee bat (1) and the Laotian rock rat or Kha-nyou (Laonastes aenigmamus) (2), respectively. L. aenigmamus is a rat-like rodent with an elongated head and squirrel-like tail. It was first found in a Laotian food market in 1996 and was introduced to the world in live video images in June 2006 (Fig. 1). Jenkins et al. (2) classified it as a new rodent family (Laonastidae) in the suborder Hystricognatha (currently classified as the infraorder Hystricognathi), which includes South American (e.g., guinea pigs, chinchillas), African, and Asian species (e.g., mole rats, porcupines). However, the position of Laonastidae among hystricognaths could not be conclusively determined, because morphological comparisons with extant rodent species suggested that the Laonastidae are the oldest hystricognath family (Fig. 2A), whereas analyses of molecular data indicated that they are closely related to the African family Bathyergidae (blesmoles and mole rats) or Petromuridae (dassie rats) (Fig. 2B) (2). Dawson et al. (3) then reanalyzed the morphological characters of Laonastes, integrating fossil rodent data in their analysis. They concluded that Laonastes is, rather, a member of the family Diatomyidae. According to the fossil record, Diatomyidae family members were thought to have lived from 33.9 Mya (Early Oligocene) to 11.6 Mya (Late Miocene), and, until now, to have been extinct for 11 million years. Thus, their classification as a diatomyid suggests that Laonastes is a living fossil and a “Lazarus taxon.”
Fig. 1.
A juvenile L. aenigmamus, captured and released after photography, provides evidence that this species is, indeed, very much alive. Known locally as Kha-nyou, the Laotian rock rat possesses a rat-like head with long whiskers and a furry squirrel-like tail. It lives in the limestone rock outcroppings of central Lao People's Democratic Republic. (Photo by Uthai Treesucon, David Redfield 2006 Lao expedition, and used with permission from Florida State University Research in Review Magazine).
Fig. 2.
Phylogenetic hypotheses for L. aenigmamus and Diatomyidae. (A) Based on morphological characters of extant species, Laonastes constitutes a new rodent family at the base of the hystricognaths (2). (B) Based on partial mitochondrial gene sequences, Laonastes constitutes a new rodent family nested within the hystricognaths (2). (C) Based on dental, cranial, and postcranial characters of fossils and extant species, Laonastes is a living fossil of the Diatomyidae family (3). (D) Based on fossil dental evidence, the family Diatomyidae is inferred to be a sister clade of hystricognaths (5). (E) Based on incisor enamel microstructure and cranial characters, Diatomyidae are related to the Pedetidae and Ctenodactylidae (i.e., sciurognathy and hystricomorphy) (6, 7).
The two research teams also disagreed on the taxonomic position of Laonastes. According to Jenkins et al. (2), Laonastes is either the most basal group of the hystricognaths (Fig. 2A) or nested within the hystricognaths (Fig. 2B). According to Dawson et al. (3), Laonastes and the other Diatomyidae are the sister clade of the family Ctenodactylidae (i.e., gundies), a family that does not belong to the Hystricognathi, but to which it is considered closely related (4) (Fig. 2C). However, the phylogenetic position of Diatomyidae is also debated in the paleontological literature. A cladistic assessment of fossil dental evidence suggests that Diatomyidae are a sister clade of hystricognaths (5) (Fig. 2D), but other studies have suggested that the fossils Fallomus and Diatomys, now considered to be diatomyids, are the sister clade of the sciurognath family Pedetidae and that both families are related to Ctenodactylidae (6, 7) (Fig. 2E).
In this study, we performed two independent phylogenetic analyses aimed at resolving the debated phylogenetic position of Laonastes. First, >5 kb of sequence data from a large taxonomic sample of rodents were analyzed by using probabilistic evolutionary models. Second, short interspersed element (SINE) insertions from representatives of all major rodent lineages were examined.
Results and Discussion
We obtained 5.5 kb of sequences from two Laonastes individuals, including 4.5 kb from portions of four nuclear genes [the alpha 2B adrenergic receptor (ADRA2B), the growth hormone receptor (GHR), the interphotoreceptor retinoid binding protein (IRBP), and the von Willebrand factor (vWF)] and 1 kb from portions of two mitochondrial genes [the cytochrome b (cyt b) and the small ribosomal subunit RNA (12S rRNA)]. The second approach analyzed SINE insertions from 16 genomic loci. SINEs make very useful phylogenetic markers because the integration of a particular element at a location in the genome is, to all intents and purposes, irreversible and of known polarity. Although some similar morphological and molecular features may be homoplasious (i.e., similar characteristics shared by a set of species but not present in their common ancestor), retroposed genetic elements integrate randomly into genomes with negligible probabilities of the same element integrating independently into orthologous positions in different species. Thus, analyzing the presence and/or absence of these elements is an advantageous strategy for molecular systematics (8, 9).
Finally, to complement the phylogenetic reconstructions, sequence data were also used to evaluate the congruence between molecular and paleontological dating estimations. Molecular divergence dates of Laonastes and its closest outgroups were computed by using two different methods for relaxing molecular clock models (see Materials and Methods) and compared with paleontological estimates (3).
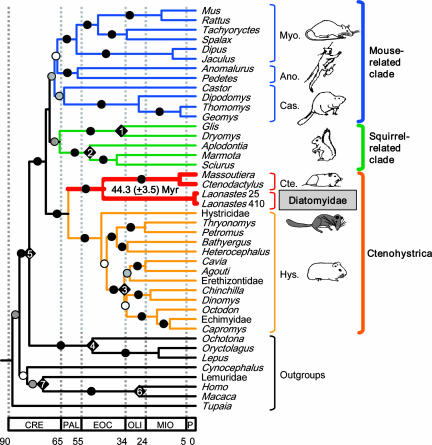
Phylogenetic analyses based on the combined data sets resulted in a well resolved phylogeny [Fig. 3 and supporting information (SI) Fig. 5], in agreement with the division of rodents into five suborders [bootstrap percentage (BP) ≥ 99% and posterior probability (PP) = 1.0, Sciuromorpha, Myodonta (or Myomorpha), Anomaluromorpha, Castorimorpha, and Ctenohystrica (or Hystricomorpha: family Ctenodactylidae + infraorder Hystricognathi)] as suggested by Carleton and Musser (10) and supported by molecular studies (11–13). These five suborders can be further grouped into three rodent lineages (BP > 95% and PP = 1.0): a mouse-related clade (Myodonta, Anomaluromorpha, Castorimorpha), a squirrel-related clade (Sciuromorpha), and Ctenohystrica (Hystricomorpha) (12). It is worth mentioning that previous molecular studies have failed to show significant support for the division of rodents into three lineages. However, alternatives to the squirrel- and mouse-related clades could not be statistically rejected [Shimodaira–Hasegawa (SH) test; Table 1]. Additional data are thus needed to solve the base of the rodent tree.
Fig. 3.
Molecular time scale for the order Rodentia. The chronogram was obtained by using the topology of the best ML tree and a Bayesian relaxed clock method with different substitution models for each gene partition. Fossil constraints are indicated by diamonds on the corresponding nodes: 1, 28.5–50 Mya; 2, >37 Mya; 3, 28.5–37 Mya; 4, 37–90 Mya; 5, 55.4–90 Mya; 6, 25–35 Mya; and 7, 63–90 Mya (detailed references are provided in SI Appendix). The divergence date and the confidence interval of the Laonastes/Ctenodactylidae split are indicated. Circles indicate the phylogenetic support of the corresponding branches. Solid circles indicate branches with maximal support value (ML BP = 100 and Bayesian PP = 1.0); gray circles indicate branches with high support value (100 > BP > 90; PP = 1.0); and white circles indicate nodes with moderate support values (90 > BP > 50; 1.0 > PP > 0.75). Ano., Anomaluromorpha; Cas., Castorimorpha; Cte., Ctenodactylidae; Dia., Diatomyidae; Hys., Hystricognathi; Myo., Myodonta. The terms hystricomorph, myomorph, and sciuromorph indicate both character states and suborders. However, the character states and their corresponding taxonomic divisions disagree. For example, the Gliridae have myomorph characteristics but belong to the sciuromorphs. To avoid confusion, we thus prefer to use the terms, Ctenohystrica, Myodonta, and squirrel-related clade rather than Hystricomorpha, Myomorpha, and Sciuromorpha. CRE, Cretaceous; PAL, Paleocene; EOC, Eocene; OLI, Oligocene; MIO, Miocene; P, PlioPleistocene. Laonastes 25 and 410, specimen vouchers BMNH1998.25 and BMNH1998.410, respectively.
Table 1.
Comparison of competing hypotheses using the SH test
| Phylogenetic hypothesis | − Ln L | Diff − ln L | P value |
|---|---|---|---|
| Best ML tree | 72,989.92 | — | — |
| [Laonastes+ Ctenodactylidae] paraphyly | 73,117.72 | 127.81 | 0.0002* |
| [Laonastes+ Ctenodactylidae + Hystricognathi] paraphyly | 73,079.93 | 90.02 | 0.0253* |
| [Laonastes + Phiomorpha s.s.] monophyly | 73,298.38 | 308.46 | < 0.0001* |
| [(Laonastes + Pedetidae)+Ctenodactylidae] monophyly | 73,326.78 | 336.87 | < 0.0001* |
| Hystricognathi paraphyly | 73,166.14 | 176.22 | 0.0001* |
| Caviomorpha paraphyly | 73,107.07 | 117.16 | 0.0045* |
| [Sciuroidea + Gliridae] paraphyly | 73,014.30 | 24.38 | 0.6169 |
| [Muroidea + Dipodidae + Anomaluromorpha + Geomyoidea + Castoridae] paraphyly | 73,022.30 | 32.39 | 0.4718 |
*Indicates significant P values.
In this maximum-likelihood (ML) tree, Laonastes is the sister clade of Ctenodactylidae. This grouping received a maximal support value (Fig. 3; BP > 100% and PP = 1.0). In addition, when analyzed separately, all of the genes studied, except the partial 12S gene, also support the clustering of Laonastes with Ctenodactylidae (SI Fig. 6). The phylogenetic tree obtained is, thus, in agreement with the hypothesis of Dawson et al. (3) as depicted in Fig. 2C. All other hypotheses (Fig. 2 A, B, D, and E) do not support the grouping of Laonastes with Ctenodactylidae. Indeed, these various alternatives, when compared with the best tree, were significantly rejected (P < 0.0002) based on the SH likelihood-based test (Table 1) (14).
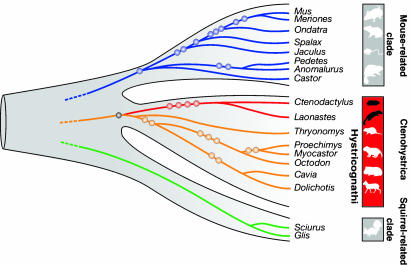
In the analysis of retroposed elements, we found four SINEs common to L. aenigmamus and Ctenodactylus gundi that were clearly absent in all of the other investigated rodent species (Fig. 4 and SI Fig. 7). The shared presence of four perfect orthologous insertions of retroposed elements in Laonastes and Ctenodactylus implies their acquisition via a common ancestry, whereas the orthologous absence of these elements in more distant taxa (i.e., Hystricognathi) indicates the ancestral condition before integration (15). These results provide significant confirmation for the evolutionary relationship of Laonastes and gundies and clearly reject the grouping of Laonastes and hystricognaths.
Fig. 4.
Phylogenetic affiliations of Laonastes based on presence/absence patterns of retroposed SINEs. The phylogenetic tree indicates the three major rodent clades: the mouse-related clade, the Ctenohystrica, and the squirrel-related clade. Eleven retroposed elements, present in certain mouse-related species (blue circles), were clearly absent in Laonastes. Nine other SINEs, present in certain Hystricognathi species (orange circles), were also absent in Laonastes. The monophyly marker of Ctenohystrica was present in all representatives including Laonastes (black circle) and was absent in members of both the mouse- and squirrel-related clades. Four diagnostic markers were present in Laonastes and Ctenodactylus (red circles) but absent in all other investigated rodents. Detailed information on the 11 investigated loci and diagnostic SINE markers are given in SI Appendix.
The Laonastes and Ctenodactylidae clade was found to be the sister clade of the Hystricognathi (i.e., Ctenohystrica monophyly). This clustering also received maximal support (BP = 100% and PP = 1.0). The SH test significantly rejected the alternative hypothesis, in which Laonastes and Ctenodactylidae do not group with the Hystricognathi (P = 0.0253; Table 1). In addition, one SINE was found to support the grouping of Laonastes, Ctenodactylidae, and Hystricognathi (Fig. 4).
Dawson et al. (3) grouped Laonastes within the extinct family Diatomyidae. The Diatomyidae were suggested to be a sister clade of Ctenodactylidae and the clade containing Diatomyidae and Ctenodactylidae clustered with Hystricognathi. Our molecular trees (Figs. 3 and 4) are thus in full agreement with Dawson et al.'s morphological tree. Because fossil Diatomyidae are too old to still contain DNA, molecular data alone cannot determine whether Laonastes is a member of the family Diatomyidae. The complete agreement between the morphological, sequence-based, and SINE trees, and the ability to use likelihood-based tests to reject alternative hypotheses, do, however, clearly suggest that Laonastes is a diatomyid.
A time scale for the evolution of the order Rodentia based on Bayesian dating analysis is depicted in Fig. 3. It is worth noting that similar dating results were obtained with different methods and tree topologies (SI Appendix). Molecular estimations indicate that Laonastes and extant Ctenodactylidae diverged 44.3 (±3.5) Mya. All extant rodent families diverged no more than 38 (±2.5) Mya (Fig. 3). Thus, the molecular dating analysis suggests that Laonastes and Ctenodactylidae belong to different rodent families. Our molecular dating results are in agreement with the divergence times estimated by Dawson et al. (3). In their estimation based on the paleontological record, Diatomyidae and Ctenodactylidae diverged ≈41 Mya, which is not significantly different from the 44.3 Mya estimate obtained here, given the standard error on molecular dating. The agreement in divergence times further strengthens the hypothesis that Laonastes is a “Lazarus” diatomyid.
Molecular findings are often in disagreement with traditional morphologically based trees. In some cases, the molecular analysis has shaken the morphological tree, leading paleontologists to reconsider fossils and missing links. One classical example is the evolutionary origin of Cetacea (e.g., dolphins, whales). The unorthodox clustering of whales within artiodactyls was first suggested based on molecular data and is now supported by strong fossil evidence (16). In the case of Laonastes we have the opposite situation. The first tree was based on both molecular and morphological data. The paleontological analysis then suggested an entirely different phylogeny, and, surprisingly, our molecular data analysis is in agreement with the paleontological tree rather than with the previous molecular tree.
The phylogenetic position of Laonastes highlights its importance with respect to mammalian biodiversity. Numerous theoretical and empirical studies have stressed that phylogenetic relationships among taxa may be a more inclusive measure than species numbers for conservation biology (17, 18). The importance of a given taxon in conservation biology is thus inversely proportional to the relative number and closeness of its phylogenetic relatives (18). Laonastes, the sole known representative of an extinct mammal family, and distantly related to the Ctenodactylidae, which include several fossil taxa but only five extant species, would appear to be a key taxon to protect.
Materials and Methods
Sequencing of Nuclear and Mitochondrial Genes.
The taxon sampling includes representatives of the three major lineages of rodents (i.e., mouse-related clade, Ctenohystrica and squirrel-related clade). Primate, tree shrew, and flying lemur sequences served as outgroups. In total, 34 rodent and eight outgroup taxa were considered. Origin of samples, PCR primers, and protocols are described in detail SI Appendix. Detailed information on the genes and species amplified is given in SI Table 2.
Sequence Alignments.
To respect codon boundaries in the DNA alignments, protein sequences of the five coding genes were aligned by using PROBCONS (19), and DNA alignments were performed by hand based on the protein alignments. All codon positions were considered except for the cytochrome b gene. We excluded the third codon position of this gene because preliminary analysis has shown that this position was saturated. The alignment of 12S rRNA was performed with ClustalX (20) with default parameter settings and refined by hand to minimize the number of indels (insertions–deletions) in stems (21, 22). Finally, gaps present in >25% of the taxa were removed from the analyses for each data set. The characteristics of each data set are indicated in SI Table 3.
Sequence-Based Phylogenetic Reconstructions.
Phylogenetic tree reconstructions were performed on each individual gene and on the concatenated data set. For each data set, two tree reconstructions were conducted: a ML and a Bayesian analysis. The program MODELTEST 3.07 (23) was used to determine the best probabilistic model of sequence evolution by using the Akaike information criterion. The models selected are indicated for each data set in SI Table 3.
ML searches for the best trees were performed with the program PAUP* (24). The parameters of the model and the best ML tree were then determined in an iterative way. The initial parameter values were those estimated by MODELTEST 3.07; those values were used for a first round of heuristic search starting with a neighbor-joining (NJ) tree and using tree-bisection-reconnection (TBR) branch-swapping. Parameters were then estimated on the resulting tree and used for another round of heuristic search. The process was repeated until all parameters were stable. BPs were estimated after 500 replicates by using the best estimated parameters, a NJ starting tree, and TBR branch-swapping.
Bayesian inferences used the program MrBayes 3.1.2 (25). The analyses of individual genes were performed under a single model of sequence evolution (no partition), whereas the analysis of the combined data set was performed on partitioned data with each of the six genes evolving with independent model parameters. MrBayes 3.1 does not implement all of the models available in MODELTEST and PAUP*. The general time-reversible model of evolution was thus chosen for all genes because it was the closest to the model used in the ML analyses. For each analysis, two simultaneous independent runs were performed. For each run, four chains were sampled every 100 generations for 5,000,000 generations after the burn-in cycles. Length of burn-in cycles varied depending on the ability of the data set to converge. For the four nuclear genes and the combined data set, chains were run for 7,500,000 generations (burn-in 25,000 trees). For the two mitochondrial genes, chains were run for 20,000,000 generations (burn-in 150,000 trees). In all cases, the average SD of split frequencies remained <0.005 after the burn-in threshold; additionally, the potential scale reduction factors of the parameters were close to or equal to 1, which indicates that the runs had most probably converged.
Testing Alternative Hypotheses.
The best ML tree was compared with several constrained topologies by using the SH test (14) as implemented in PAUP*. The tests were conducted on the combined data set with RELL optimization, 10,000,000 bootstrap replicates, and the parameters of the best ML tree. Eight alternative topologies were considered. (i) The best alternative that does not cluster Laonastes and Ctenodactylidae (in this tree Laonastes is the sister clade of the Hystricognathi). This tree corresponds to the hypotheses of Fig. 2 A and D (2, 5). (ii) The best alternative that does not cluster Laonastes, Ctenodactylidae, and Hystricognathi (in this tree the clade Laonastes + Ctenodactylidae is at the base of the rodents). (iii) The best alternative that groups Laonastes with Phiomorpha sensus stricto (i.e., Bathyergidae + Petromuridae + Thryonomyidae). This tree corresponds to the hypothesis of Fig. 2B (2). (iv) The best alternative that groups Laonastes and Pedetidae as sister clade of Ctenodactylidae. This tree corresponds to the hypothesis of Fig. 2E (6, 7). (v) The best alternative that does not support the monophyly of Hystricognathi. (vi) The best alternative that does not support the monophyly of Caviomorpha. (vii) The best alternative that does not support the monophyly of Sciuroidae + Gliridae. (viii) The best alternative that does not support the monophyly of Anomaluromorpha + Muroidea + Dipodoidea + Geomyoidea + Castoridae. The eight topologies were built by using constrained ML heuristic searches. Each search was conducted starting with a neighbor-joining tree, the tree-bisection-reconnection branch-swapping option, and the parameters of the best ML tree.
Molecular Dating.
Molecular dating estimations were computed by using two different approaches: a Bayesian method as implemented in the software Multidivtime (26) and a penalized likelihood method as implemented in the program r8s (27). To verify that the position of the root did not have a significant impact on the molecular dating results, we compared dating results obtained with two different positions of the root. The first topology, the ML tree, placed the tree shrew at the base of the tree (Fig. 3), whereas the second topology placed the tree shrew as a sister clade of Primates and Dermoptera [monophyly of Euarchonta (28)]. Molecular dating analyses are detailed in SI Appendix.
SINE Insertion Analysis.
We recently conducted an exhaustive, automated search for phylogenetic informative SINE markers in rodents, revealing 31 diagnostic SINEs from 16 genomic loci in representatives of all major rodent lineages (29). Using these as templates for PCR, in the present study we amplified the orthologous loci in samples of Laonastes DNA. For the SINE insertion analysis one specimen of Laonastes was purchased in 2006 in the market at Tha-Kek (Lao People's Democratic Republic). SINE amplification protocols and sequence alignments are given in SI Data Set. Phylogenetic relationships based on SINE data were reconstructed by using the maximum parsimony method. SINE presence/absence data resemble virtually homoplasy-free multistate characters with an extremely large possible number of unique character states. It is worth noting that Steel and Penny (30) suggested that for such data maximum parsimony converges to a ML estimator.
Supplementary Material
Acknowledgments
We thank F. Catzeflis (curator of the tissue collection of the Institut des Sciences de l'Evolution de Montpellier); C. Faulkes, M. Mensink, L. Contreras, M. Robinson, R. Timmins, P. Gouat, S. Honigs, C. Matthee, J.-C. Gautun, T. Arrizabalaga, R. S. Hoffmann, D. S. Semple, D. L. Dittmann, J. L. Patton, D. Eilam, J.-M. Duplantier, P. Gambarian, J. Terkel, E. Pelé, L. I. Grassman, and V. Volobouev for donating or collecting tissue; E. Douzery for advice and help with sequencing; V. Arnal and S. Kinamon for technical assistance; M. Bundman for revising the English text; and L. Marivaux and two anonymous reviewers for comments on the manuscript. This work was supported by United States–Israel Binational Science Foundation Grant 2004-407 (to D.H.), Deutsche Forschungsgemeinschaft Grant SCHM1469 (to J.S. and J.B.), and the Förderkreis Universität Münster (J.S.). This publication is contribution number ISEM2007-021 of the Institut des Sciences de l'Evolution de Montpellier (Unité Mixte de Recherche 5554, Centre National de la Recherche Scientifique).
Abbreviations
- BP
bootstrap percentage
- ML
maximum likelihood
- PP
posterior probability
- SH
Shimodaira–Hasegawa
- SINE
short interspersed element.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AM407897–AM407933, DQ139933, and EF052254–EF052265).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701289104/DC1.
References
- 1.Hill JE. Bull Br Mus Nat Hist (Zool) 1974;27:301–336. [Google Scholar]
- 2.Jenkins PD, Kilpatrick CW, Robinson MF, Timmins RJ. Syst Biodivers. 2005;2:419–454. [Google Scholar]
- 3.Dawson MR, Marivaux L, Li CK, Beard KC, Metais G. Science. 2006;311:1456–1458. doi: 10.1126/science.1124187. [DOI] [PubMed] [Google Scholar]
- 4.Huchon D, Catzeflis FM, Douzery EJP. Proc R Soc London Ser B. 2000;267:393–402. doi: 10.1098/rspb.2000.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marivaux L, Vianey-Liaud M, Jaeger JJ. Zool J Linn Soc. 2004;142:105–134. [Google Scholar]
- 6.Martin T. Berliner Geowiss Abh E. 1995;16:693–707. [Google Scholar]
- 7.Flynn LJ, Jacobs LL, Cheema IU. Am Mus Novitates. 1986;2841:1–58. [Google Scholar]
- 8.Shedlock AM, Okada N. BioEssays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 9.Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J. PLoS Biol. 2006;4:537–544. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carleton MD, Musser GG. In: Mammal Species of the World: A Taxonomic and Geographic Reference. Wilson DE, Reeder DM, editors. Baltimore: Johns Hopkins Univ Press; 2005. pp. 745–752. [Google Scholar]
- 11.DeBry RW. Syst Biol. 2003;52:604–617. doi: 10.1080/10635150390235403. [DOI] [PubMed] [Google Scholar]
- 12.Huchon D, Madsen O, Sibbald MJJB, Ament K, Stanhope M, Catzeflis FM, de Jong WW, Douzery EJP. Mol Biol Evol. 2002;19:1053–1065. doi: 10.1093/oxfordjournals.molbev.a004164. [DOI] [PubMed] [Google Scholar]
- 13.Adkins RM, Walton AH, Honeycutt RL. Mol Phylogenet Evol. 2003;26:409–420. doi: 10.1016/s1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- 14.Shimodaira H, Hasegawa M. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 15.Schmitz J, Ohme M, Zischler H. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boisserie JR, Lihoreau F, Brunet M. Proc Natl Acad Sci USA. 2005;102:1537–1541. doi: 10.1073/pnas.0409518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith DP. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 18.Vane-Wright RI, Humphries CJ, Williams PH. Biol Conserv. 1991;55:235–254. [Google Scholar]
- 19.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Springer MS, Douzery E. J Mol Evol. 1996;43:357–373. doi: 10.1007/BF02339010. [DOI] [PubMed] [Google Scholar]
- 22.Douzery E, Catzeflis FM. J Mol Evol. 1995;41:622–636. doi: 10.1007/BF00175821. [DOI] [PubMed] [Google Scholar]
- 23.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 24.Swofford DL. PAUP* Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 25.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 26.Thorne JL, Kishino H. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 27.Sanderson MJ. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 28.Waddell PJ, Norihiro O, Hasegawa M. Syst Biol. 1999;48:1–5. [PubMed] [Google Scholar]
- 29.Farwick A, Jordan U, Fuellen G, Huchon D, Catzeflis FM, Brosius J, Schmitz J. Syst Biol. 2006;55:936–946. doi: 10.1080/10635150601064806. [DOI] [PubMed] [Google Scholar]
- 30.Steel M, Penny D. Mol Biol Evol. 2000;17:839–850. doi: 10.1093/oxfordjournals.molbev.a026364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.