Abstract
The prion protein (PrP) propensity to adopt different structures is a clue to its biological role. PrP oligomers have been previously reported to bear prion infectivity or toxicity and were also found along the pathway of in vitro amyloid formation. In the present report, kinetic and structural analysis of ovine PrP (OvPrP) oligomerization showed that three distinct oligomeric species were formed in parallel, independent kinetic pathways. Only the largest oligomer gave rise to fibrillar structures at high concentration. The refolding of OvPrP into these different oligomers was investigated by analysis of hydrogen/deuterium exchange and introduction of disulfide bonds. These experiments revealed that, before oligomerization, separation of contacts in the globular part (residues 127–234) occurred between the S1–H1–S2 domain (residues 132–167) and the H2–H3 bundle (residues 174–230), implying a conformational change of the S2–H2 loop (residues 168–173). The type of oligomer to be formed depended on the site where the expansion of the OvPrP monomer was initiated. Our data bring a detailed insight into the earlier conformational changes during PrP oligomerization and account for the diversity of oligomeric entities. The kinetic and structural mechanisms proposed here might constitute a physicochemical basis of prion strain genesis.
Keywords: folding, kinetics, oligomers, strain
Prion diseases or transmissible spongiform encephalopathies are deadly neurodegenerative pathologies affecting human and other mammalian species. According to the prion (or “protein-only”) hypothesis (1), the key event in the pathogenesis is the conversion of the α-helix-rich host-encoded prion protein (PrP) (PrPC) into a pathogenic conformer (PrPSc) characterized by a higher content in β-sheet and a polymeric state (2, 3). Emerging features of these pathologies are the appearance of so-called atypical cases (4) and the co-occurrence of various PrPSc species in one individual (5, 6), questioning the basis of the strain concept (7). Based on the protein-only hypothesis, the physical basis of strain multiplicity relies on PrPSc structural diversity (8).
Strong experimental evidence brought support to the prion hypothesis, relying on the synthetic production in acidic conditions of β-sheeted recombinant PrP-soluble oligomers endowed with pathogenic properties (9). The ability of PrP to fibrilize was widely studied previously (10–14), but only recently amyloidogenic oligomeric structures have been associated with the manifested biological effects. Indeed, recombinant PrP oligomers exhibited neurotoxic activities (15) in agreement with other studies on neurodegenerative diseases like Alzheimer's disease and other brain amyloidoses that clearly demonstrated the existence of toxic oligomers or protofibrils of amyloidogenic proteins in vivo and in vitro (16). One recent report showed that, in detergent-treated fractions of hamster brain PrPSc, the highest titers of transmissible spongiform encephalopathy infectivity were associated with soluble assemblies of 14–28 PrP molecules (17). Thus, in vitro and in vivo independent studies converge on the key role of PrP oligomers. Considering that any conformational change requires an unfolding event (18), the unraveling of PrP unfolding/folding pathways is a critical step in understanding the oligomerization mechanisms implied in the pathological evolution.
We previously showed that the recombinant full-length ovine PrP (OvPrP) formed discrete soluble oligomeric species upon thermal unfolding (12). From these former observations we analyzed the in vitro mechanisms of PrP oligomerization, addressing two main questions: (i) Can multiple pathways take place simultaneously, leading to different entities from the initial homogeneous protein and accounting for biochemical or structural diversity? (ii) What are the critical events involved in PrP oligomerization? These topics recall main unsolved biological questions in the prion field: the physicochemical basis of strain types, the critical steps contributing to initiate the pathogenic process, and their dependence on the PrP local concentration or on specific structural motifs enciphered in the PrP sequence.
Here we show that multiple assemblies could be formed via independent folding pathways, and we propose a kinetic scheme for in vitro PrP oligomerization. This scheme was substantiated by comparison of hydrogen/deuterium exchange (H/D exchange) experiments for OvPrP monomer and oligomers and by introduction of a covalent linkage between OvPrP secondary structure elements. Taken together, these experiments allowed for the identification of the sites causing PrP to unfold before the formation of each oligomer: (i) oligomerization required the unfolding of the H1 α-helix and the opening of the globular domain between the S1–H1–S2 and the H2–H3 moieties, and (ii) the type of oligomer formed is controlled by the conformation of the S2–H2 loop and the location of the site at which opening of the OvPrP globular core is initiated.
This refined mechanistic study brings clues to the genesis of the observed diversity in PrP oligomeric structures and enlightens the wide spectrum of possible PrP roles, in either a physiological or pathological context. The existence of multiple folding pathways could explain how different strains of prion are generated and how multiple biological properties could be associated to the single PrP.
Results
Multiple Pathways Generate Distinct Oligomers.
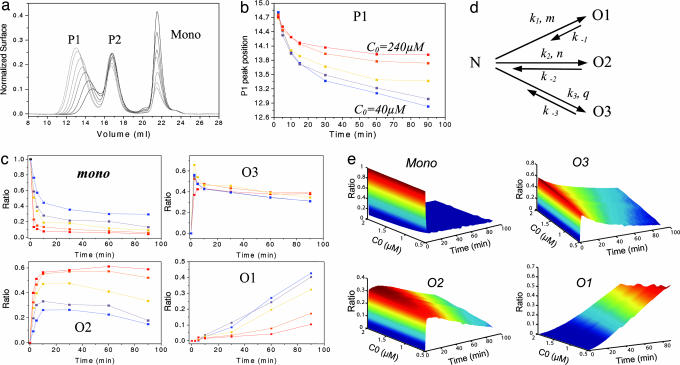
We previously showed that thermal unfolding of monomeric OvPrP led to formation of discrete oligomers eluting as two peaks named P1 and P2 according to their order of appearance on a size-exclusion column (Fig. 1a) and estimated to be 36mers and 12mers, respectively, based on SAXS measurements (12). The elution volume and peak profile of the larger oligomeric species (P1) varied as a function of heating time (2.5–90 min) (Fig. 1a) or with the initial PrP concentration (C0) (Fig. 1b), whereas the P2 elution volume remained constant.
Fig. 1.
Effect of heating time and monomer concentration on PrP oligomerization and proposed kinetic scheme for oligomerization. (a) Size-exclusion chromatogram of OvPrP oligomers formed by heating the monomer (C0 = 70 μM) at 48°C for 2.5–90 min. Two major peaks, P1 and P2, are observed. (b) Dependency of the P1 elution volume on C0 and heating time. Lower concentration favors larger oligomers. (c) After P1 and P2 deconvolution, the ratios of monomer (mono) and O3, O2, and O1 oligomers were plotted versus heating time and C0. Lower C0 favors O1 oligomer. (d) Proposed kinetic scheme describing the oligomerization process. k constants are the kinetic constants for each pathway, and m, n, and q are the reaction kinetic orders. (e) Numerical simulation of the proposed kinetic scheme relating the effect of C0 on the evolution of monomer, O3, O2, and O1 ratios. The simulated kinetics of oligomerization (e) fit well to the experimental data (c). In all experiments, C0 varies from 40 μM (blue) to 240 μM (red).
These observations suggested that the P1 peak contained more than one oligomer. Should OvPrP oligomerization proceed as a sequence of two irreversible steps, the formation of the largest oligomer would be favored at higher C0, even if fragmentation/nucleation occurs subsequently [see supporting information (SI) Fig. 5]. Intriguingly, the elution volume of the P1 peak decreased (P1 shifting to higher molecular weight) when C0 decreased (Fig. 1b), indicating that (i) the P1 peak contains at least two oligomers, (ii) these oligomers are not kinetically related and originate from at least two distinct reaction pathways with different kinetic constants, and (iii) the kinetic order for the formation of the largest oligomers is lower than for the smaller oligomers constituting the P1 peak.
To accurately predict the diversity of species generated during the OvPrP oligomerization, we deconvoluted the chromatograms using a home-made procedure. Three distinct oligomers were predicted with this method, referred to as O1, O2, and O3, with two of these species (O1 and O2) underlying the P1 peak (see SI Fig. 6). The ratios of deconvoluted populations of O1, O2, and O3 were plotted as a function of heating time and C0 (Fig. 1c). Consistent with an oligomerization reaction, the rate of OvPrP monomer disappearance increased as a function of C0 (Fig. 1c Upper Left). The O3 oligomer formation was slightly favored at higher initial protein concentrations (Fig. 1c Upper Right). The largest oligomer (O1) was more readily formed at lower monomer concentrations (Fig. 1c Lower Right), whereas the smaller oligomer (O2) observed in the P1 peak was favored when C0 was higher (Fig. 1c Lower Left).
Several reaction schemes were numerically simulated to test their ability to account for (i) the rate of O1, O2, and O3 formation, and (ii) the dependency of oligomer amounts on C0. Only the scheme shown in Fig. 1d satisfied all conditions and allowed suitable numerical simulations (Fig. 1e). According to this scheme, OvPrP monomers “N” partially unfold and oligomerize to form O1, O2, and O3 independently. The predicted kinetic rate orders of polymerization were lower for O1 than for O2 and O3 (m < n, q), thereby explaining the intriguing effect of C0 on the formation rate of O1 (Fig. 1e) and thus the P1 peak shift. Furthermore, this scheme showed that depolymerization of the OvPrP oligomers should occur as a first-order disintegration rather than a sequential, piece-by-piece disassembly. In addition, it predicts a depolymerization rate constant lower for O1 than for O2 and O3 (k−1 ≪ k−2, k−3). This implies that the O1 oligomer is the most thermodynamically stable of the three OvPrP oligomeric species and that most of the initial monomeric species will ultimately be converted into O1, which defines it as an attractor in the kinetic scheme.
Isolation and Characterization of the Various Oligomers.
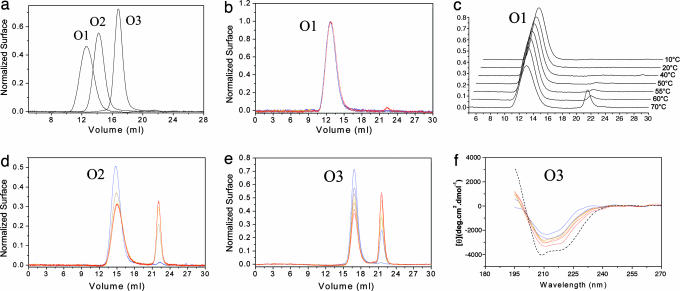
As predicted by chromatogram deconvolution and kinetic scheme (see SI Figs. 6 and 7), we were able to separate up to three different species (Fig. 2a). The kinetics of depolymerization of each purified oligomer were analyzed by size-exclusion chromatography at low protein concentration (8 μM monomer equivalents) to avoid reoligomerization of the newly formed monomer. We observed that (i) for each purified oligomer the depolymerization process occurred in one step without observation of any intermediate oligomers (Fig. 2 b–d), even at low temperature (10°C incubation for 15 days; data not shown); (ii) in our conditions, a given OvPrP oligomer was neither a precursor nor a product of another one; (iii) O1 was the most stable oligomer (Fig. 2 b and c), thus constituting the attractor as predicted; and (iv) depolymerization involved a transition from a β-sheet-enriched conformation to an α-helical conformation identical to that of the initial OvPrP monomer, as shown by CD (Fig. 2f).
Fig. 2.
Purification and depolymerization of PrP oligomers. (a) Overlay of size-exclusion chromatograms of the three purified OvPrP oligomers. (b, d, and e) O1, O2, and O3 were depolymerized by heating at 55°C from 0 min (blue) to 60 min (red). Size-exclusion analysis revealed a one-step depolymerization without intermediates for O2 and O3, whereas O1 remained stable. (c) Thermal depolymerization of O1 (6 μM) after 15 min of incubation at different temperatures (from 20°C to 75°C). O1 depolymerization only occurred above 55°C. (f) The variation of the secondary structure of O3 (10 μM) incubated at 55°C from 0 min (blue) to 60 min (red) showing an increase in α-helix content. The dash curve indicates the monomer CD spectrum at pH 3.4.
Taken together, the experimental data and the predictions from the kinetic scheme (Fig. 1d) converge toward an oligomerization process in which the OvPrP monomer partially unfolds into three intermediate states, all of which oligomerize reversibly and independently from each other to form O3, O2, and O1.
Generation of Larger Molecular Species from OvPrP Oligomers.
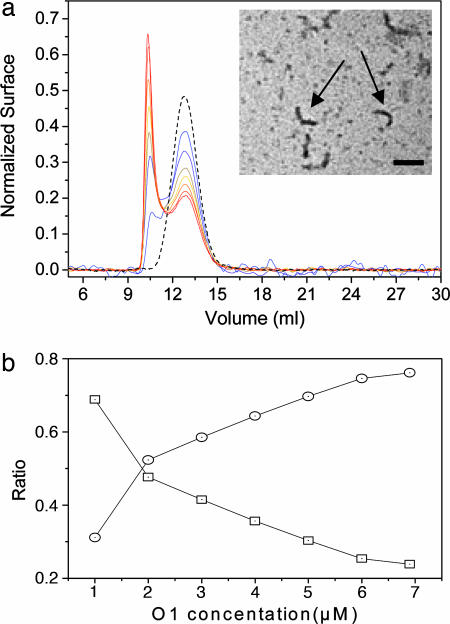
As for other amyloidogenic proteins, the PrP fibrilization process can take place only when the concentration of the amyloidogenic precursor overcomes a critical concentration. To determine whether our oligomers might constitute such precursors, a high concentration was induced by ultracentrifugation of oligomer solutions in conditions where no pellet was formed (Fig. 3a). For O3 and O2, no change was observed in the size-exclusion chromatograms after centrifugation, independent of the initial oligomer concentration or centrifugation time (up to 4 h; data not shown). For O1 a new peak (P0) appeared in the column void volume (Fig. 3a). Whereas a time-dependent seeding oligomerization presents a sigmoid curve, the hyperbolic shape of the P0 formation rate as a function of initial O1 concentration was consistent with a helical or tubular seeding oligomerization process (19, 20) (Fig. 3b). The electron microscopic analysis of purified P0 revealed small curved fibrils (Fig. 3a Inset). Therefore, this experiment suggested that only the O1 oligomer was able to fibrilize in the conditions used.
Fig. 3.
Generation of protofibrils by the O1 oligomer of OvPrP. (a) The experiment was performed with initial O1 concentrations (expressed in monomer equivalents) varying from 1 μM (blue line) to 7 μM (red line) with 1-μM steps. The dashed line represents the O1 profile at 7 μM before ultracentrifugation. The larger oligomer (P0) generated by ultracentrifugation had a fibrillar morphology, as seen by transmission EM. (Inset Scale bar: 0.2 μm.) (b) After chromatogram deconvolution variation of O1 (square) and P0 (circle) amounts were plotted as a function of O1 initial concentration. The hyperbolic variations of O1 and P0 as a function of the initial concentration of O1 suggested a seeding oligomerization process.
H/D Exchange Patterns of OvPrP Monomer and Oligomers.
To probe the PrP regions undergoing structural changes upon oligomerization, we resorted to the H/D exchange technique. Only O1 and O3 oligomers were stable under the experimental conditions, whereas O2 exhibited significant depolymerization under these conditions and thus could not be studied. After hydrogen exchange and pepsin digestion of the monomer and oligomers, several peptides corresponding to amino acids 135–175 of the polypeptide chain were identified (see SI Fig. 8). No peptide stemmed from the helices H2 and H3, probably as a consequence of the higher stability of this segment of OvPrP (where the native disulfide bond is located). Conversely, the high accessibility of the N-terminal region probably caused its extensive digestion by pepsin, preventing further MS analysis.
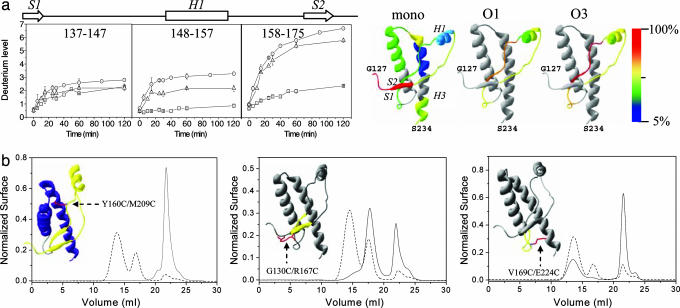
Whereas a reduced hydrogen exchange is expected at the monomer–monomer interface in the oligomers, we did not measure any decrease in the deuterium incorporation in the analyzed OvPrP peptides upon oligomerization. This observation suggested that the 135–175 region of OvPrP does not constitute any monomer–monomer interface in O1 and O3, which is consistent with the fact that this region was accessible to pepsin in these oligomers. Most surprisingly, some of the analyzed peptides exhibited a strong exchange increase when the protein oligomerized (Fig. 4a). Peptides corresponding to the region 148–157 (helix H1) showed up to 2.5 and 3.3 exchanged hydrogens in O1 and O3, respectively, compared with 0.9 hydrogens for the monomer. Similarly, the peptides in the region 158–175 exchanged up to 5.8 hydrogens in O1 and 6.7 in O3, compared with 2.4 hydrogens in the monomer. This significant increase in deuterium incorporation reflects major structural changes in these parts of the protein accompanying oligomerization. The increase in hydrogen exchange in the region 148–157 may reflect a partial or complete unfolding of the H1 α-helix. For the region 158–175, the strong increase in hydrogen exchange is believed to reflect an unfolding of the H1–S2 structural elements of the protein. Furthermore, the small difference between O1 and O3 H/D exchange profiles might reflect a difference in the subunit conformation between these two oligomers.
Fig. 4.
Conformational dynamics during PrP oligomerization. (a) H/D exchanges in the OvPrP monomer and oligomers. (Left) Number of deuteriums incorporated for the exchange kinetics, as analyzed by MS, in peptides generated from pepsin digestion of the monomer (squares), O3 (circles), and O1 (triangles) after incubation in a deuterated buffer (from 0 to 120 min). The error bars result from three independent experiments. (Right) Deuterium incorporation after 2 h, normalized to peptide length and averaged between peptides spanning over a common region (see SI Fig. 8 for each individual peptide), is visualized on the OvPrP structure, revealing an increase of accessibility and structural changes near the H1–S2 region. (b) Effect of intramolecular covalent bonds on OvPrP oligomerization. (Left) After 90 min at 50°C, the wild-type OvPrP monomer (80 μM) oligomerizes to yield the O1, O2, and O3 oligomers, which elute as two peaks (dotted line). In contrast, the Y160C/M209C mutant (80 μM) does not generate any oligomeric species in the same conditions (solid line). (Center) In the same conditions, the G130C/R167C mutant (80 μM) oligomerizes to generate major amounts of O3, low amounts of O2 (eluting at 15 ml), and no O1. (Right) The V169C/E224C mutant (100 μM) forms a mixture of O2 and O1 and no O3 oligomer, whereas in the same conditions wild type generates O3 oligomers. Insets show the positions of the additional disulfide bonds.
Structural Dynamics During Oligomerization.
If, as suggested by the above experiments, OvPrP has to locally unfold in different regions to form different oligomers, then locking the structural mobility of regions evidenced by H/D exchange should alter the population of generated oligomers. Double cysteine mutants were designed to covalently link different PrP subdomains by disulfide bonds. G130C/R167C links the two β-strands of OvPrP (Fig. 4b Center Inset), V169C/E224C links the S2–H2 loop to the H3 helix (Fig. 4b Right Inset), and Y160C/M209C links the H1 and H3 helices (Fig. 4b Left Inset). When exposed to oligomerization conditions, all mutants exhibited strongly modified kinetics of oligomer accumulation. The Y160C/M209C mutant displayed complete inhibition of oligomerization (Fig. 4b Left), suggesting that oligomer formation required the dissociation of the contacts between H1–S2 and H3. The G130C/R167C mutant led to the accumulation of O3, whereas O2 and O1 formation was considerably slowed down, suggesting that the flexibility of the β-strand region plays an important role in the formation of O1 and O2 (Fig. 4b Center). The V169C/E224C mutant formed the O1 oligomer, whereas the O3 oligomer was not observed at all (Fig. 4b Right). Formation of O3 therefore involves the opening of the S2–H2 hinge-loop. The different behavior of these three mutants provides structural support for the parallel oligomerization scheme proposed here (Fig. 1d). These results indicate that OvPrP oligomerization requires a physical separation of the S1–H1–S2 segment from the H2–H3 α-helical bundle and that the type of oligomer produced after unfolding is strongly dependent on the site at which this dissociation occurs.
Discussion
The high propensity of the PrP to form various structures is likely to be the clue to its role in transmissible spongiform encephalopathy pathogenesis as well as to its putative physiological functions. Based on thermodynamic considerations, the prerequisite events triggering these conformational changes led to an unfolding process generating a partially unfolded state prone to evolve to new conformations. From this point of view, the effect of the infectious agent can be reduced to a simple physicochemical perturbation promoting the unfolding of PrPC and favoring its structural conversion. The present work shows that OvPrP undergoes parallel and reversible unfolding/refolding processes upon heating/cooling leading to distinct quaternary structure assemblies: O3, O2, and O1. By analyzing H/D exchange patterns of OvPrP in its different forms and the effects of the introduction of disulfide bonds to restrain the dynamics of protein unfolding, we showed that, before oligomerization, the S1–H1–S2 domain must unfold and physically separate from the H2–H3 domain. Moreover, the type of oligomer to be formed was precisely controlled by the site where this expansion was initiated. Our data (i) provide insight to the mechanisms of unfolding/folding of the PrP, (ii) account for the diversity of PrP assemblies, and (iii) enlighten the physical origin of prion strains.
Parallel PrP Oligomerization Pathways.
The intriguing dependence of the ratio of oligomeric species on the initial monomer concentration (a lower C0 enhanced the formation rate of the larger oligomer O1) led us to propose a kinetic scheme in which each oligomer directly originates from the native state. Assuming that a sequential oligomerization process is similar to a phase condensation process (19, 21), a sequential process could not account for the oligomerization behavior of the double cysteine mutants. As a whole, a set of parallel oligomerization pathways remains the best model to describe OvPrP oligomerization. The fact that O1 was experimentally confirmed to be the most stable of the three oligomers implies that, according to the oligomerization scheme proposed here (Fig. 1d), the formation of O3 and O2 would never reach an equilibrium, and thus that these two oligomers constitute transient species (22). Noticeably, depolymerization of all types of oligomers never generated assemblies of intermediate size but led to formation of α-helix-rich monomers.
The diversity of the heat-induced OvPrP oligomers is reminiscent of the numerous in vitro-generated PrP assemblies previously reported on the way to neuropathogenesis, amyloidogenesis, and fibril formation (11–14). Because only O1 constitutes the precursor of the P0 fibrillar assembly, a low monomer concentration enhancing O1 oligomerization might increase the P0 formation rate, which questions the link between PrPC expression level and fibril deposition. In addition, oligomerization of OvPrP into different species or the inverse depolymerization of PrP assemblies into the normal monomeric isoform could be modulated as a function of the cell type in which these processes occur.
Structural Dynamics During Oligomerization and PrPC/PrPSc Conversion.
The differences in H/D exchange patterns between monomer and oligomers revealed regions undergoing structural changes during oligomerization. Additionally, by locking subdomains of OvPrP with disulfide bonds we show that, before oligomerization, the S1–H1–S2 domain must physically separate from the H2–H3 domain, which implies a conformational change in the S2–H2 hinge-loop corresponding to residues 168–173.
Altogether our kinetic and structural data are consistent with observations related to the PrPC/PrPSc conversion in vivo. The conformation of the S2–H2 loop seems to play an important role in PrPC/PrPSc conversion. (i) Factor X was putatively proposed to target the S2–H2 loop and promote opening of the molecule like a lever (23). (ii) In sheep, mutation Q171R located in the S2–H2 loop confers a resistance phenotype to sheep scrapie (24). (iii) The conformation of this loop was proposed as the basis of transmissible spongiform encephalopathy susceptibility differences in domestic animals (25). Furthermore, because the separation of H1–S2 from the rest of the protein is a prerequisite unfolding event for oligomerization, all mutations affecting the H1–S2 expansion from the H2–H3 bundle may be involved in the PrPC/PrPSc conversion, as is the case for the human Q217R mutation. According to H/D experiments, H1 undergoes structural changes during oligomerization. Strikingly, several antibodies that inhibit prion replication in vivo (26–28) bind H1 and might act by preventing H1 unfolding. Taken together our results not only pinpoint the structural changes required before oligomerization but also enlighten the earlier events of the conversion process.
A Track to Understand Prion Strain Diversity.
Multiple PrP folding pathways offer a rationale to correlate structural dynamics and physical hypothesis about the origin of prion strains. So far the strain phenomenon is only attributed to biochemical and anatomopathology diversity. Assuming that PrPSc is the infectious agent, strain variety can be explained if one accepts that different PrPSc conformers do exist, each with a given biological activity, not excluding that some may be benign. The primary sequence variability among the different mammalian species as well as different tissue and cellular environments may affect the oligomerization pathways and the type of oligomers contributing to strain diversity. Several studies (8, 29–32) tend to prove the conformational diversity of PrPSc.
Overall our data present a kinetic overview of the early events on the pathway of PrP pathological conversion. Finally, the fact that PrP follows independent folding pathways leading to transient oligomers or fibrillar precursors addresses the controversial debate around linkage between neurodegeneration and amyloid deposits. The challenge is now to unravel these types of oligomers in vivo.
Materials and Methods
Analysis of Size-Exclusion Chromatograms and Kinetic Simulation of the OvPrP Oligomerization.
The generation of OvPrP oligomers and their analysis by size-exclusion chromatography were performed as described (12) by using the full-length recombinant A136R154Q171 variant. Briefly, OvPrP samples in 20 mM sodium citrate (pH 3.4) were incubated for 5–90 min at concentrations ranging between 50 and 230 μM or at temperatures ranging from 20°C to 70°C, then cooled down to 15°C, and analyzed on a 90-cm TSK4000SW column. The chromatograms were deconvoluted by using a home-made deconvolution method based on determination of the anisotropy of the chromatograms second derivative.
Based on the kinetic scheme presented in Fig. 1d, the differential equations describing the evolution of the amount of each species (N for the native monomer; O1, O2, and O3 for the heat-induced oligomers) as a function of time were established: dN/dt = k−1O1 + k−2O2 + k−3O3 − (mk1Nm + nk2Nn + qk3Nq); dO1/dt = mk1Nm − k−1O1; dO2/dt = nk2Nn − k−2O2; and dO3/dt = qk3Nq − k−3O3. These differential equations were numerically solved by using a Runge-Kutta solver (Fig. 1e). The kinetic orders m, n, and q were preset at 2, 3, and 3, respectively. According to depolymerization data, the reverse-kinetic constants (k−) were empirically preset as k−1 = 10−3 s−1, k−2 = 0.3 s−1, and k−3 = 0.1 s−1. The forward-kinetic constants were k1 = 5 × 103 M−1·s−1, k2 = 5 × 108 M−2·s−1, and k3 = 8 × 108 M−2·s−1. The simulation time scale was 90 min, and the initial value of N was fixed to unit. No singularity was observed for all C0, k−, k+, m, n, and q ≥ 0.
Purification and Analysis of the OvPrP Oligomers.
For oligomer purification, 500 μl of full-length recombinant ovine PrP was heated and cooled down in specified conditions (monomer concentration and temperature) depending on the type of oligomer to be purified (see SI Fig. 7). Fractions corresponding to each peak were collected and reinjected on the same size-exclusion column for analysis. The purified oligomers were depolymerized by heating at 55°C for various times between 0 and 60 min and then cooled down to 15°C. For all depolymerization experiments the oligomers concentration was below 12 μM (monomer equivalents) to prevent oligomerization back-pathway. For amyloidogenesis studies 400 μl of purified oligomers at a 6 μM (monomer equivalents) concentration was submitted to ultracentrifugation for 1 h at 84,000 × g and 15°C, leading to local concentration increases along the sedimentation profile. Mixing of the centrifuged samples disrupted the concentration gradient (Fig. 3a). Size-exclusion chromatography analysis was performed as described, and the newly formed species were collected after their elution at the void volume of the column. EM analysis of the P0 species was performed immediately after collection. The sample was loaded on a carbon grid and stained with 2% uranyl acetate. CD analysis of the OvPrP oligomers was performed with a thermostated Jasco (Tokyo, Japan) 810 spectropolarimeter by using a 500-μm path-length cuvette.
Design of the OvPrP Mutants.
Three double cysteine mutants were designed by modeling disulfide bonds on the OvPrP structure (Protein Data Bank ID code 1TPX) (33) according to the following criteria: (i) mutated residues had to be close enough for disulfide bond formation, (ii) the mutations had to ensure a good disulfide stereochemistry, and (iii) the bond had to be solvent-exposed so as to keep the integrity of the hydrophobic core of OvPrP. We therefore selected the following mutations: G130C/R167C, which links the two β-strands of OvPrP; V169C/E224C, which links the S2–H2 loop to the H3 helix; and Y160C/M209C, which links the H1 and H3 α-helices. The mutations were introduced into the ARQ gene cloned in a pET 22bC vector by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The wild-type PrP and mutants were purified as previously described (12). The expected S–S bonds were checked by combining proteolysis and MS.
H/D Exchange.
Purified OvPrP monomer (13 μM) and O1 and O3 oligomers (in the range of 5–10 μM) in 20 mM sodium citrate (pH 3.4) were used for H/D exchange experiments. Each step was carried out on ice (0°C) unless otherwise stated. H/D exchange was initiated by transferring 100 μl of each sample into deuterated 20 mM sodium citrate (pH 3.00) by using MicroSpin G-25 columns (Amersham Biosciences, Piscataway, NJ), equilibrated six times with 400 μl of the deuterated buffer. Samples were incubated at 15°C, and 20-μl aliquots were taken at several times. The H/D exchange was quenched by addition of 20 μl of 4% formic acid (pH 2.3). Each sample was then incubated for 5 min with 2 μl of 40 μM pepsin in 20 mM sodium citrate buffer (pH 3.4). The resulting peptides were desalted and concentrated by using C18 microcolumn tips (Agilent Technologies, Palo Alto, CA). Elution from the tips was performed with a H2O/MeOH/formic acid 49.9:49.9:0.2 (vol/vol/vol) solution at room temperature, and the peptides were immediately analyzed by using nano-ESI-FTMS. Spectra were recorded by an APEX III instrument (Bruker Daltonics, Bremen, Germany) equipped with a 7 T actively shielded magnet and an Apollo ESI source. Fifty scans were accumulated for each sample. Desalting and recording MS were performed within 2 min after digestion, with 5–10% back exchange. The H/D exchange results reported here are the averages of three independent experiments for each PrP assembly and are relative to the highest number of exchanged hydrogen for each peptide.
Supplementary Material
Acknowledgments
We thank G. Ohanessian for access to MS facilities and interest in the work, and F. Fraternali (King's College, London, U.K.) and P. Simister (Laboratoire de Virologie Moléculaire et Structurale, Gif-sur-Yvette, France) for critical reading of the manuscript and helpful discussions. T.D. received a grant from the EGIDE program. F.E. was supported by the Fondation pour la Recherche Médicale.
Abbreviations
- H/D exchange
hydrogen/deuterium exchange
- PrP
prion protein
- OvPrP
ovine recombinant full-length PrP.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607745104/DC1.
References
- 1.Prusiner SB. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 3.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benestad SL, Sarradin P, Thu B, Schonheit J, Tranulis MA, Bratberg B. Vet Rec. 2003;153:202–208. doi: 10.1136/vr.153.7.202. [DOI] [PubMed] [Google Scholar]
- 5.Puoti G, Giaccone G, Rossi G, Canciani B, Bugiani O, Tagliavini F. Neurology. 1999;53:2173–2176. doi: 10.1212/wnl.53.9.2173. [DOI] [PubMed] [Google Scholar]
- 6.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Lancet Neurol. 2005;4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 7.Bruce ME. Br Med Bull. 2003;66:99–108. doi: 10.1093/bmb/66.1.99. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka M, Collins SR, Toyama BH, Weissman JS. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 9.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 10.Swietnicki W, Morillas M, Chen SG, Gambetti P, Surewicz WK. Biochemistry. 2000;39:424–431. doi: 10.1021/bi991967m. [DOI] [PubMed] [Google Scholar]
- 11.Sokolowski F, Modler AJ, Masuch R, Zirwer D, Baier M, Lutsch G, Moss DA, Gast K, Naumann D. J Biol Chem. 2003;278:40481–40492. doi: 10.1074/jbc.M304391200. [DOI] [PubMed] [Google Scholar]
- 12.Rezaei H, Eghiaian F, Perez J, Doublet B, Choiset Y, Haertle T, Grosclaude J. J Mol Biol. 2005;347:665–679. doi: 10.1016/j.jmb.2005.01.043. [DOI] [PubMed] [Google Scholar]
- 13.Lu BY, Chang JY. Biochem J. 2002;364:81–87. doi: 10.1042/bj3640081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskakov IV, Legname G, Baldwin MA, Prusiner SB, Cohen FE. J Biol Chem. 2002;277:21140–21148. doi: 10.1074/jbc.M111402200. [DOI] [PubMed] [Google Scholar]
- 15.Kazlauskaite J, Young A, Gardner CE, Macpherson JV, Venien-Bryan C, Pinheiro TJ. Biochem Biophys Res Commun. 2005;328:292–305. doi: 10.1016/j.bbrc.2004.12.172. [DOI] [PubMed] [Google Scholar]
- 16.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 17.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ptitsyn OB. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 19.Oosawa F, Asakura S. Thermodynamics of the Polymerization of Protein. New York: Academic; 1971. [Google Scholar]
- 20.Powers ET, Powers DL. Biophys J. 2006;91:122–132. doi: 10.1529/biophysj.105.073767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop MF, Ferrone FA. Biophys J. 1984;46:631–644. doi: 10.1016/S0006-3495(84)84062-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson HP, Pantaloni D. Biophys J. 1981;34:293–309. doi: 10.1016/S0006-3495(81)84850-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko K, Zulianello L, Scott M, Cooper CM, Wallace AC, James TL, Cohen FE, Prusiner SB. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter N, Goldmann W, Benson G, Foster JD, Hope J. J Gen Virol. 1993;74:1025–1031. doi: 10.1099/0022-1317-74-6-1025. [DOI] [PubMed] [Google Scholar]
- 25.Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wuthrich K. Proc Natl Acad Sci USA. 2005;102:646–650. doi: 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enari M, Flechsig E, Weissmann C. Proc Natl Acad Sci USA. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peretz D, Williamson RA, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn IR, Legname G, Wormald MR, Rudd PM, et al. Nature. 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 28.White AR, Enever P, Tayebi M, Mushens R, Linehan J, Brandner S, Anstee D, Collinge J, Hawke S. Nature. 2003;422:80–83. doi: 10.1038/nature01457. [DOI] [PubMed] [Google Scholar]
- 29.Caughey B, Raymond GJ, Bessen RA. J Biol Chem. 1998;273:32230–32235. doi: 10.1074/jbc.273.48.32230. [DOI] [PubMed] [Google Scholar]
- 30.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 31.Mulcahy ER, Bessen RA. J Biol Chem. 2004;279:1643–1649. doi: 10.1074/jbc.M307844200. [DOI] [PubMed] [Google Scholar]
- 32.Peretz D, Scott MR, Groth D, Williamson RA, Burton DR, Cohen FE, Prusiner SB. Protein Sci. 2001;10:854–863. doi: 10.1110/ps.39201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eghiaian F, Grosclaude J, Lesceu S, Debey P, Doublet B, Treguer E, Rezaei H, Knossow M. Proc Natl Acad Sci USA. 2004;101:10254–10259. doi: 10.1073/pnas.0400014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.