Most of the world relies on plants as their dietary source of iron (1, 2). Unfortunately, plants are not a good source of this essential nutrient for two reasons: iron deficiency often limits plant growth, and the iron that accumulates in plants is not readily available. The first problem stems not from the abundance of iron in soil but rather from its solubility, with pH and oxygen controlling the amount of free iron found in solution. When faced with an iron shortage, plants either use a reduction strategy (strategy I) and take up Fe2+ or use a chelation strategy (strategy II) and take up Fe3+ (Fig. 1). However, not all plants are proficient at mounting these iron-deficiency responses. Rice, one of the world's most important food crops, is much more susceptible to iron deficiency than other grasses, presumably because it releases low quantities of phytosiderophores that act as Fe3+ chelators (3). Although rice plants are also capable of taking up Fe2+ (4), rice does not have an inducible ferric chelate reductase activity, as do plants that use the reduction strategy to generate Fe2+ from the more abundant Fe3+ found in aerobic soils. In this issue of PNAS, Ishimaru et al. (5) ask whether introducing this activity into rice would improve its iron uptake ability. They report an impressive 7.9-fold increase in grain yield for transgenic rice plants engineered to express a ferric chelate reductase when these plants are grown in calcareous soil. Importantly, the ferric chelate reductase that they introduced had been selected for better performance at high pH because the native enzyme has an acidic pH optimum (6). Because one-third of the world's soils are alkaline, the results presented here could have a major impact on rice production, allowing rice to grow in soils now considered marginal and increasing crop biomass in soils now in cultivation. Furthermore, all plants, except the grasses, use the reduction strategy to mobilize iron from the rhizosphere, and ferric chelate reductase activity has been shown to be the rate-limiting step for iron uptake (7). Thus, all plants would benefit from increased expression of a ferric chelate reductase that functions better at alkaline pH.
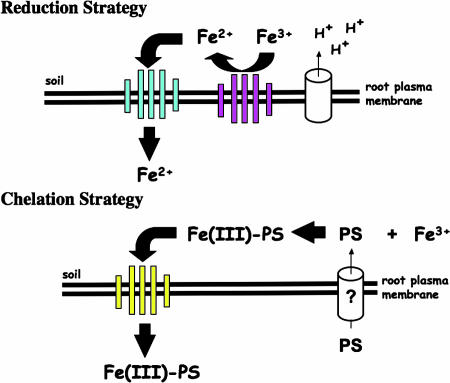
Fig. 1.
Iron deficiency responses. (Upper) The reduction strategy as typified by Arabidopsis is shown. An inducible ferric chelate reductase, FRO2, reduces Fe(III) chelates in the soil, releasing Fe2+ for transport across the plasma membrane by the Fe2+ transporter IRT1. Plasma membrane ATPase activity is also increased under iron deficiency, acidifying the rhizosphere and increasing Fe solubility. (Lower) The chelation strategy as typified by maize and rice is shown. Phytosiderophores (PS) are synthesized in the cytoplasm. PS are released into the soil, and Fe–PS complexes are subsequently transported across the membrane via the YS1 transporter. See Grotz and Guerinot (8) for a more detailed description of these responses.
For their experiments, Ishimaru et al. (5) used a yeast ferric chelate reductase gene that had been selected for improved activity at high pH (6). The allele, refre1/372, encodes three substitutions relative to the wild-type gene, including a replacement of methionine at position 312 with arginine. A substitution of methionine with arginine or lysine at this position was common to most of the high-activity variants recovered in the screen. Interestingly, this mutation is near one of four heme-coordinating histidine codons. The refre1/372 allele had been previously introduced into tobacco under control of a strong, ubiquitously expressed promoter and conferred enhanced tolerance to low Fe availability (6). Ishimaru et al. used the promoter from the Fe2+ transporter gene, OsIRT1, to drive expression of the refre1/372 gene to ensure that ferric chelate reductase expression was coupled to expression of the Fe2+ transporter. The OsIRT1 promoter is highly iron regulated and is expressed in the epidermis of the root (4), exactly when and where ferric chelate reductase activity would be needed. In addition to monitoring ferric chelate reductase activity, growth, and yield, Ishimaru et al. directly measured iron uptake by using a positron-emitting tracer imaging system. Transgenic plants had initial rates of iron uptake that were twice those of vector controls.
It was disappointing, but perhaps not surprising, that the increase in iron uptake did not lead to an increase in the iron content of the rice grain. We still know very little about how iron is distributed in plants (8). It seems that iron homeostasis is such that an increased ability to take up Fe leads to increased growth and seed yield but not to more Fe being stored in the seed. To drive more Fe into the seed, it will probably be necessary to create an enhanced “sink” for Fe in the seed. A number of studies have tried to increase seed Fe content by expressing the Fe storage protein, ferritin, under the control of promoters that are specifically expressed in rice seed (9–12). However, this technique has yielded modest, 2- to 3-fold increases in seed Fe content, apparently because of insufficient iron uptake (12, 13). Recently, it appeared that iron may be stored in the vacuoles of provascular cells in Arabidopsis seed (14), suggesting another possible storage sink for Fe in the seed.
There was a previous attempt to express the Arabidopsis ferric chelate reductase gene FRO2 from its endogenous promoter in rice, but no expression of the transgene was observed (15). Expression of the Arabidopsis ferric chelate reductase gene FRO2 from the 35S promoter in soybean did improve growth under alkaline conditions but did not result in more iron in the seed when plants were grown in the greenhouse (16). No information was given as to whether there was any change in seed yield. There did appear to be a penalty for constitutive expression of the FRO2 gene under iron-sufficient conditions (16), underscoring why the choice of the OsIRT1 promoter to drive expression of the ferric chelate reductase gene makes good biological sense.
It is interesting to speculate as to why rice does not have an inducible ferric chelate reductase. The rice genome encodes two ferric chelate reductase genes that have high similarity to the Arabidopsis FRO genes (17). Ishimaru et al. (5) posit that the progenitors of modern-day rice came from swampy, waterlogged soils, where Fe2+ would be found in abundance, thus precluding a need for ferric chelate reductase activity. And why do the grasses have a chelation strategy, whereas other plants do not? This question is harder to answer. The chelation strategy is clearly less sensitive to pH than the reduction strategy, but there is a cost to synthesizing and releasing phytosiderophores because they may or may not be recovered after release into the rhizosphere.
So, what about enhancing the chelation strategy? For strategy II plants, there is a strong, positive correlation between the amounts of phytosiderophores released and the resistance of plants to iron deficiency (3). The biosynthetic pathway that leads to the formation of the rice phytosiderophore, deoxymugineic acid, provides several targets for increasing the amount of phytosiderophore produced. Takahashi et al. (18) introduced two barley genes encoding nicotianamine aminotransferase (NAAT), the enzyme that catalyzes the second step in the biosynthesis of deoxymugineic acid, into rice. Under iron deficiency, these transgenic rice plants released more phytosiderophores that correlated with improved growth in alkaline soils. Similar to what was seen with the plants engineered to express the ferric chelate reductase, the NAAT plants yielded four times as much grain as control plants. Engineering other steps in the phytosiderophore biosynthetic pathway could further increase phytosiderophore release. For example, NAAT may be limited for its substrate, nicotianamine, that is synthesized from S-adenosyl methionine. Nicotianamine itself is thought to play a role in iron homeostasis, serving as an iron chelator (8). Thus, another question to be answered is whether diverting more nicotianamine into the production of phytosiderophores will have any effect on iron distribution in the plant itself. And then there is the question of diverting more S-adenosyl methionine, a primary donor of methyl groups, into the production of nicotianamine.
Given the concern with introduction of genes from other species into crop plants, it would seem that the next step should be selection for improved expression of rice's own ferric chelate gene(s). The “to do” list also includes field tests, because the impressive yield increases reported here are for plants grown in the greenhouse. And most importantly, the transgenic plants are now in hand to test whether plants with increased phytosiderophore production do better than plants expressing a high-pH-adapted Fe(III) chelate reductase or whether plants with both enhancements do best of all.
Footnotes
The author declares no conflict of interest.
See companion article on page 7373.
References
- 1.McGuire J. SCN News. 1993;9:1–10. [Google Scholar]
- 2.Graham RD, Welch RM, Saunders DA, Ortiz-Monasterio I, Bouis HE, Bonierbale M, de Haan S, Burgos G, Thiele G, Hobbs PR, et al. Adv Agron. 2007;92:1–74. [Google Scholar]
- 3.Marschner H. Mineral Nutrition of Higher Plants. Boston: Academic; 1995. [Google Scholar]
- 4.Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. Plant J. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- 5.Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Proc Natl Acad Sci USA. 2007;104:7373–7378. doi: 10.1073/pnas.0610555104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oki H, Kim S, Nakanishi H, Takahashi M, Yamaguchi H, Mori S, Nishizawa NK. Soil Sci Plant Nutr. 2004;50:1159–1165. [Google Scholar]
- 7.Connolly EL, Campbell N, Grotz N, Prichard CL, Guerinot ML. Plant Physiol. 2003;133:1102–1110. doi: 10.1104/pp.103.025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grotz N, Guerinot ML. Biochim Biophys Acta. 2006;1763:595–608. doi: 10.1016/j.bbamcr.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F. Nat Biotechnol. 1999;17:282–286. doi: 10.1038/7029. [DOI] [PubMed] [Google Scholar]
- 10.Lucca P, Hurrell R, Potrykus I. J Sci Food Agric. 2001;81:828–834. [Google Scholar]
- 11.Vasconcelos M, Datta K, Oliva N, Khalekuzzaman M, Torrizo L, Krishnan S, Oliveira M, Goto F, Datta SK. Plant Sci. 2003;164:371–378. [Google Scholar]
- 12.Qu L-Q, Yoshihara T, Ooyama A, Goto F, Takaiwa I. Planta. 2005;222:225–233. doi: 10.1007/s00425-005-1530-8. [DOI] [PubMed] [Google Scholar]
- 13.Van Wuytswinkel O, Vansuyt G, Grignon N, Fourcroy P, Briat J-F. Plant J. 1999;17:93–97. doi: 10.1046/j.1365-313x.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- 14.Kim SA, Punshon T, Lanzirotti A, Li L, Alonzo JM, Ecker JR, Kaplan J, Guerinot ML. Science. 2006;314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- 15.Vasconcelos M, Musetti V, Li C-M, Datta SK, Grusak MA. Soil Sci Plant Nutr. 2004;50:1151–1157. [Google Scholar]
- 16.Vasconcelos M, Eckert H, Arahana V, Graef G, Grusak MA, Clemente T. Planta. 2006;224:1116–1128. doi: 10.1007/s00425-006-0293-1. [DOI] [PubMed] [Google Scholar]
- 17.Gross J, Stein RJ, Fett-Neto AG, Fett JP. Genet Mol Biol. 2003;26:477–497. [Google Scholar]
- 18.Takahashi MT, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S. Nat Biotechnol. 2001;19:466–469. doi: 10.1038/88143. [DOI] [PubMed] [Google Scholar]