Abstract
We used a recently developed method to produce mutant alleles of five endogenous Drosophila genes, including the homolog of the p53 tumor suppressor. Transgenic expression of the FLP site-specific recombinase and the I-SceI endonuclease generates extrachromosomal linear DNA molecules in vivo. These molecules undergo homologous recombination with the corresponding chromosomal locus to generate targeted alterations of the host genome. The results address several questions about the general utility of this technique. We show that genes not near telomeres can be efficiently targeted; that no knowledge of the mutant phenotype is needed for targeting; and that insertional mutations and allelic substitutions can be easily produced.
Keywords: Gene targeting, Drosophila, recombination, FLP, I-SceI
We recently described a method for targeted modification of the Drosophila genome through homologous recombination (HR). The ability to engineer specific changes into the genome is a highly useful adjunct to genetic investigation in any organism, but especially in a species with a completely determined genome sequence such as Drosophila melanogaster (Adams et al. 2000). This procedure had, until recently, been lacking in Drosophila. In our previous reports, we targeted two genes, rescuing a mutant allele of the first and generating a mutant allele of the second (Rong and Golic 2000, 2001). At this time there is a clear need for demonstrations of the generality of this technique. That is, can a variety of genes in different locations be modified by HR? There is also a need for the development of techniques that can produce mutant alleles of target genes. In this work, we address both issues by applying new methods for targeted mutagenesis of five autosomal genes.
A variety of schemes has been produced for targeted gene modification in organisms such as yeast and mice (Rothstein 1991; Muller 1999). However, these methods rely critically on the ability to culture single cells and carry out selections for rare events. Because the targeting technique we use occurs in whole animals, we devised variant approaches for introducing mutations into chromosomal genes. The methods are mechanistically similar to those developed for yeast and mouse, but procedurally quite different, as they do not rely on chemical selections. Instead, at each step, arbitrary genetic markers with simple visible phenotypes are used for genetic screening. In our previous experiments, the frequency of targeted gene modification through HR varied from ∼1 in 500 gametes to ∼1 in 30,000 gametes. These frequencies are easily within reach of the power provided by genetic screening.
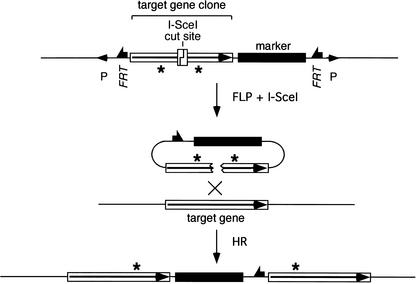
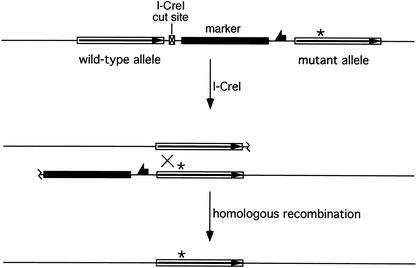
To perform gene targeting in flies we use transgenic expression of FLP site-specific recombinase and I-SceI endonuclease to generate a targeting donor molecule in vivo. This donor molecule is derived from a third transgenic element: a P element carrying DNA homologous to the target locus. Within the P element, FLP Recombinase Target sites (FRTs) flank a segment of DNA from the target locus, and an I-SceI recognition site is placed within the target-homologous sequence. The expression of FLP and I-SceI excises the donor sequence from its initially random chromosomal location and generates a double-strand break (DSB) within the sequence that is homologous to the target gene. The DSB stimulates HR of the donor and the corresponding chromosomal target locus (Fig. 1). Actual targeting events may be recovered by screening for movement of an eye-color marker gene that is carried into the target locus as part of the donor DNA. An alternative, more rapid, screening method has also been described (Rong and Golic 2001). The process of gene targeting in Drosophila is diagramed in Figure 2.
Figure 1.
General form of ends-in gene targeting. The recombinogenic donor molecule is generated by FLP and I-SceI action on a P-element donor construct, causing excision of the target-homologous sequence and the marker gene, and cutting at the I-SceI site. HR with the chromosomal target locus generates a tandem duplication of the sequence that was present in the donor with incorporation of the marker gene between duplicated target segments. Mutations engineered into the donor (indicated as asterisks) may be carried into both copies of the target gene.
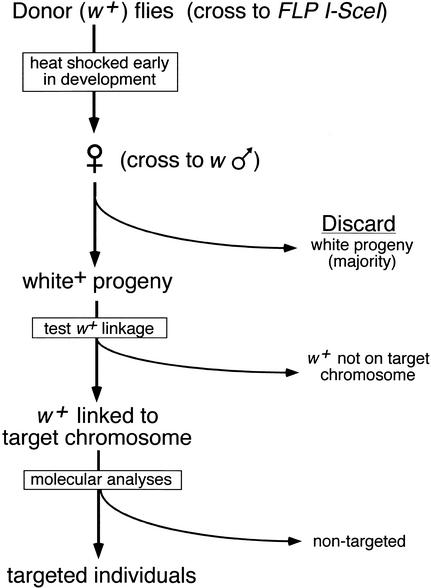
Figure 2.
Schematic representation of the targeting process. In the first step, donor-bearing flies are crossed to flies carrying the heat-inducible FLP and I-SceI genes. The progeny of this cross are heat-shocked during the first few days of their development, and the eclosing daughters are crossed to w males. Because FLP-mediated excision is very efficient (typically >99% with the 38°C, 1-h heat shock used here; Golic and Golic 1996), most progeny have white eyes. The w+ progeny are screened to look for movement of the w+ marker from the donor chromosome to the target chromosome using test crosses with marked chromosomes (Rong and Golic 2000). Balancer chromosomes may be incorporated in the initial crosses to facilitate the process. Alternatively, a second round of FLP induction can be used to rapidly identify nonexcised donors and exclude them from further analysis (Rong and Golic 2001). This method allows the use of donor insertions that lie on the same chromosome as the target locus. Potential targeting events are confirmed by molecular analysis of genomic DNA. This figure presumes that a w+ gene is used in the donor to track movement of the targeting molecule, and that the flies carry white null mutations on their X chromosomes.
In this paper we apply two new procedures to generate mutant alleles of genes that are identified only by DNA sequence. Knowledge of the mutant phenotype is not required or relied on in these procedures. Moreover, the end product of one protocol is a precise substitution of an engineered mutant allele for the wild-type allele: no exogenous DNA is left behind at the target locus other than the introduced mutation.
We used these procedures to specifically mutate several endogenous genes at a variety of chromosomal loci. We show that the process is sufficiently efficient and flexible to be generally useful for modification of the Drosophila genome.
Results
General parameters of gene targeting in Drosophila
We constructed a P-element vector, pTV2, to be used as a general transformation vector for gene targeting. This vector carries an FRT-flanked w+ gene (whs) that is used for transformation, and to track movement of the donor to detect gene targeting. We used transformants of donor constructs in TV2 to carry out targeted mutagenesis of five genes (Fig. 3). Only one of these genes was known by existing mutants prior to this work. Table 1 shows the efficiency of targeting obtained for each gene. Two general trends appear. First, targeting is substantially more efficient in the female germ line than in the male germ line. Looking at the data for pug (pugilist; Rong and Golic 1998) and p53 (encoding the Drosophila homolog of the p53 tumor suppressor; Brodsky et al. 2000; Ollmann et al. 2000), females produced targeting events in ∼3% of all vials (33 independent events in 1118 vials), whereas in males the frequency was approximately sixfold lower (5 independent events in 1012 vials; P = 0.001). A similar bias was apparent in our original work (Rong and Golic 2000), where a gene on the X chromosome was targeted. These results extend that observation to show that a significantly enhanced efficiency of targeting in females also applies to autosomal target genes. We conclude that relatively inefficient targeting in males is not related to lack of a homolog because this difference occurs for both autosomal and X-linked genes.
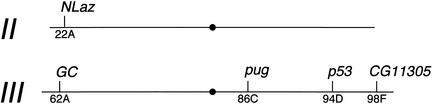
Figure 3.
Targeted genes. The names (above) and cytological positions (below) of the genes that we have targeted in this work are indicated.
Table 1.
Recovery of gene targeting events
| Gene
|
Female germ line
|
Male germ line
|
||||
|---|---|---|---|---|---|---|
| N
|
T
|
NT
|
N
|
T
|
NT
|
|
| NLaz (5.1 kb) | ||||||
| on X | 505 | 2 | 2 | |||
| GC (6.9 kb) | ||||||
| on X | 85 | 1 | 0 | |||
| on 2a | 131 | 0 | 0 | |||
| on 3a | 31 | 0 | 0 | |||
| on 3a | 93 | 0 | 0 | |||
| pug (8.9 kb) | ||||||
| on X | 173 | 7b | 0 | 304 | 3c | 0 |
| on 2 | 194 | 0 | 0 | 304 | 0 | 2 |
| on 3 | 221 | 3 | 4 | 121 | 2 | 2 |
| on 3 | 133 | 8d | 3 | 75 | 0 | 3 |
| p53 (8.6 kb) | ||||||
| on X | 158 | 9e | 1 | |||
| on 2a | 171 | 5 | 0 | 146 | 0 | 1 |
| on 3a | 68 | 1 | 0 | 62 | 0 | 1 |
| on 3a | 20 | 0 | 0 | |||
| CG11305 (5 kb) | ||||||
| on X | 50 | 3f | 1 | |||
| on 3a | 17 | 0 | 1 | |||
| on 3a | 37 | 1 | 0 | |||
| on 3a | 34 | 4 | 3 | |||
| Totals | 2121 | 44 | 15 | 1012 | 5 | 9 |
The target gene, the extent of donor–target homology, and the chromosome where the donor was inserted are given in the left column. N, number of vials screened; T, vials with targeting events; NT, vials with non-targeted events.
In these crosses only half of the tested females carried the donor element—we have corrected by multiplying the number of vials tested by 0.5.
Two were Class IV events.
One was a Class IV event.
Two were Class III events.
One was a Class IV event.
Two were Class III events.
Another significant difference between males and females is that the targeted (homologous) recombinants outnumber the nontargeted (illegitimate) events by 3:1 in females, but in males that relationship is reversed, with nontargeted events outnumbering targeted recombinants by almost 2:1 (P = 0.005).
Because of the increased efficiency of targeting in females, targeting of NLaz (Neural Lazarillo; Sanchez et al. 2000), GC (γ-glutamyl carboxylase; Li et al. 2000; Walker et al. 2001), and CG11305 (a homolog of the yeast Sir2 gene; Gasser and Cockell 2001) was done only through females. For the genes targeted in this work, independent homologous recombination events were recovered from females at an average (unweighted) rate of ∼1 per 40 vials. These crosses typically produce ∼100 progeny per vial. Targeting events were often recovered in clusters (but scored as single events), making the per gamete rate of targeting better than 1 in 4000. Because there are at least two and sometimes four (in G2) target chromosomes per cell, the cellular targeting rate is at least 1 in 2000.
Substantial variation in efficiency occurred at different target genes. At CG11305, targeting averaged 1 event per 17 vials, but at GC, it was as infrequent as 1 event per 340 vials (P = 0.0003). It is likely that some of this variation is attributable to the donor/target DNA sequences or their chromosomal context.
In most cases, targeting resulted in the predicted precise duplication of the target DNA (referred to as Class II events; Rong and Golic 2000). However, a small number of Class III events (having insertions or deletions within one or both of the duplicated segments) and Class IV events (triplications of the target locus) were also produced (Table 1). Molecular confirmation of targeting was usually accomplished by genomic Southern blotting (Fig. 4), except that PCR was used for NLaz targeting (see Materials and Methods).
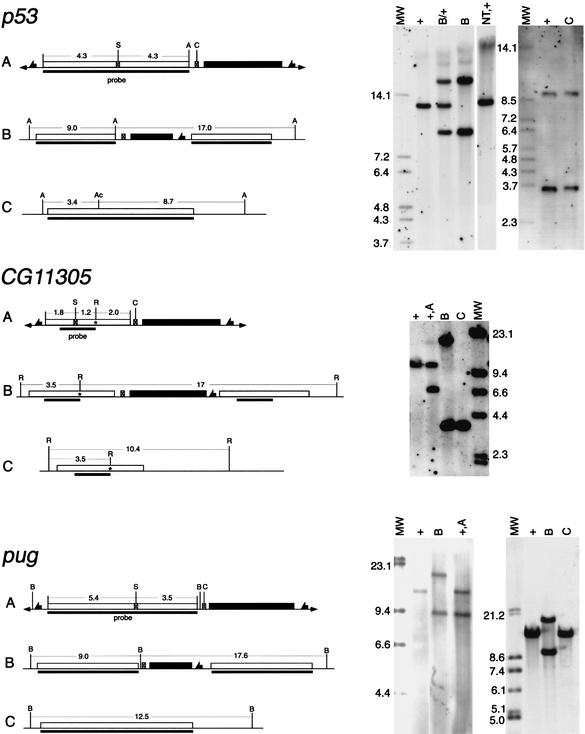
Figure 4.
Molecular verification of homologous recombination. A sample of the molecular data for targeting of three genes is shown here. On the left are shown diagrams of (A) the donor P element, (B) the Class II targeted allele, and (C) the single-copy reduction. The wild-type allele is identical to C except at CG11305, where it is indicated separately. The asterisk in CG11305 indicates the introduced mutation, which adds a new EcoRI site. In each diagram the target-homologous region is indicated by an open box; the whs marker gene by a filled box; fragment sizes are indicated in kilobases; the region used as a probe is indicated by a solid bar below each line. The endonuclease sites are: (A) Acc65I; (Ac) AccIII; (B) BamHI; (C) I-CreI; (R) EcoRI; (S) I-SceI. On the right side, the corresponding Southern blots are shown. Above each lane, the genotype of the flies from which DNA was extracted is indicated with reference to the diagrams to the left. Additional genotypes are: (+) wild-type (at the target locus); (NT) nontargeted event; (MW) molecular weight (mass) markers with sizes given (in kilobases) beside that lane. For p53, DNA in the left panel was digested with Acc65I, and in the right panel with Acc65I and AccIII. For CG11305, DNA was digested with EcoRI. For pug, DNA was digested with BamHI.
In these experiments, different donor insertions targeted with differing efficiency. In the pug-targeting experiments, one insertion of the donor gave targeting at a rate of 1 in 17 vials, but another gave no targeting events in almost 200 vials (P = 0.002 for pug targeting). Variation was also observed in p53 and CG11305 targeting, although it was not statistically significant. The basis for this difference may be related to the three-dimensional location of donor and target within nuclei, but because donor location was not determined in detail, the nature of such a relationship is unknown. There was no obvious advantage to using donor insertions located on the same chromosome as the target locus.
Another source of variation in targeting efficiency may be the extent of homology that a donor molecule carries to the target locus. To examine this issue we compared the frequency of pug targeting obtained in these experiments (with 8.9 kb of donor:target homology) with previous experiments that used only 2.5 kb of homology (Rong and Golic 2001). Targeting efficiency increased more than fivefold in the current experiments (18 independent events in 721 vials here vs. 2 in 455 vials previously), suggesting that the extent of donor:target homology plays a significant role in targeting efficiency. This is also the case for targeting in other systems (Deng and Capecchi 1992; Papadopoulou and Dumas 1997; Gray and Honigberg 2001).
Introducing point mutations
In these experiments two methods were used to produce mutant alleles of the target gene. Because the ends-in, or insertional, targeting approach that we used here generates a target site duplication, both copies of the target gene must be mutated to generate a mutant allele. In previous work we did this by using only a small portion of the target gene in the donor, so that each of the copies generated by targeting carried only a partial gene and they were nonfunctional (Rong and Golic 2001). In the present experiments, we took a different approach: We chose to engineer point mutations within the coding regions of the target genes. These mutations ranged from single-base changes to insertions of 10–20 nucleotides. Each copy of the target gene produced by ends-in targeting is composed of sequence derived from the target and from the donor. Thus, mutations engineered to both sides of the I-SceI site can be carried into the target gene to generate two mutant copies.
We used this method to generate a mutant allele of pug by introducing stop codons into both copies of a targeted pug allele. The mutants had the same recessive eye color defect that was previously observed for null alleles of this gene (Rong and Golic 1998). A mutant allele of GC was generated in the same fashion.
Cellular exonucleases are expected to enlarge the DSB made by I-SceI into a gap. In the course of HR, this gap is filled by copying the corresponding information from the chromosomal target (Szostak et al. 1983). The consequence of gap repair is that some proportion of targeted genes will not carry the introduced mutation because exonucleolytic digestion removed it prior to HR. A good example is provided by the NLaz targeting, where the two recovered targeting events each carried only one of the two engineered mutations present in the donor.
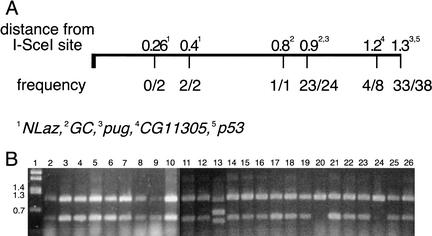
The practical implication of gap enlargement is that there is more surety of introducing a mutation to the target if the mutation is farther from the cut site. The results we obtained in targeting five genes are summarized in Figure 5. We successfully introduced mutations to the target gene at distances ranging from 400 bp to 1300 bp from the I-SceI cut site. In most cases, the majority of the targeted alleles carried the mutation in question. We did not succeed in introducing a mutation only 260 bp away from the cut site, but only two events were examined. A more extensive screen might recover targeting events that incorporated the closer mutation.
Figure 5.
Retention of engineered mutations. (A) The fractions of targeting events that incorporated mutations placed in the donor are given as a function of distance from the I-SceI site. The data were summed in cases where mutations were located similar distances from the I-SceI site. (B) A sample of PCR results used to determine the presence of the upstream point mutation (at the NruI site, 1.3 kb from I-SceI: see Materials and Methods) in targeted pug alleles. The smaller (490-bp) band indicates the presence of the mutant allele. Lane 1 is a molecular weight marker with band sizes indicated to the left. Lanes 2–8 and 11–26 use genomic DNA from flies with targeting events as template. The template DNA for lane 9 comes from flies with an I-CreI-stimulated reduction event (which retained the mutation; see Fig. 7). Lane 10 is from pug+/+ flies that carried the donor P element. The mutation was present in all targeting events except those represented by lanes 20 and 24. Lane 13 represents a Class III targeting event.
Two-step allelic substitution
There are many cases in which the method of introducing two point mutations might be difficult to apply. For instance, in the case of genes that are very small, the constraint of placing the I-SceI site and the two mutations within the gene may cause difficulties because the donor mutations will frequently be lost to gap enlargement. In addition, one of the two mutations may be forced rather far downstream in the gene, allowing the possibility that a hypomorphic or neomorphic peptide may still be encoded.
To deal with these situations we designed a method for carrying out allelic substitutions in two steps (Fig. 6). The goal of this scheme is to make a simple substitution of an engineered mutant allele for the wild-type allele in the chromosome, and to do so with no limitations on the location of the mutation within the gene and no knowledge of the phenotype produced by the mutant allele. In the first step, an ends-in targeting event carries a single mutation into the chromosome at the target locus. In the second step, the target site duplication is collapsed to a single copy of the target that carries the introduced mutation; all other DNA extrinsic to the locus is eliminated.
Figure 6.
Allelic substitution. In the first step, a standard ends-in targeting is used to carry a single point mutation into the target locus (see Fig. 1). In the second step (shown here), the target locus duplication is reduced to a single copy by HR between the repeated sequence elements. This event is stimulated by an I-CreI-generated DSB between the repeats.
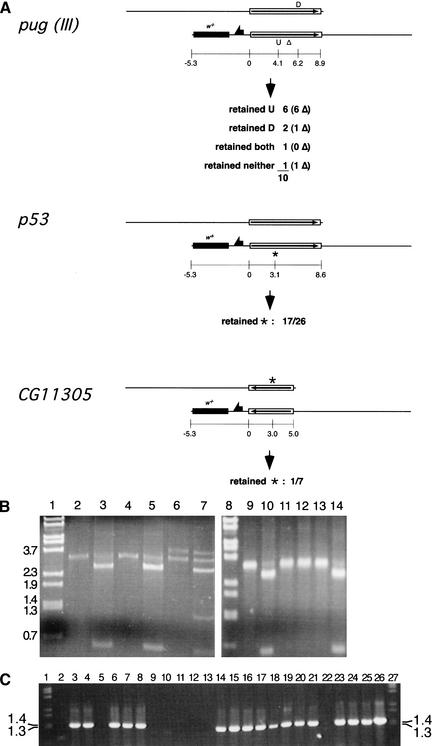
The first step is achieved by ends-in targeting with TV2. This vector also carries one additional element between the marker gene and the NotI and KpnI cloning sites: the 22-bp recognition site for the rare-cutting endonuclease I-CreI (Thompson et al. 1992). When I-CreI is expressed in flies with a targeted allele, the DSB produced by I-CreI is most often repaired by recombination between the homologous regions that flank the DSB. This eliminates the intervening marker gene, providing an easy screen for the event, and produces a chromosome carrying a single copy of the target locus. Some fraction of these reduction events will carry the mutation from the donor. We generated mutant alleles of four out of the five target genes using this method. (Reductions at GC were not tested for the point mutations.)
The second step of this screen, reduction of the tandem duplication to a single copy, is very efficient. In three cases (NLaz, GC, and pug) we scored the frequency of w+ marker loss. After a 36°C, 1-h heat shock applied in the first 2–3 d of development to induce expression of the 70I-CreI transgene, w+ loss averaged 36% in the male germ line (Table 2). We typically carry out this reduction step in males, simply as a matter of convenience. As judged by Southern blotting (Fig. 4; Table 2), nearly all (94%) of these w+ loss events occurred by simple HR between the flanking repeats, leaving a single copy of the target locus. Three of the w+ loss events at pug were not straightforward reductions to single copy, and probably resulted from nonhomologous end-joining (NHEJ) (Critchlow and Jackson 1998), which deleted all, or part of, the w gene. Two of these three were also associated with linked recessive lethal mutations.
Table 2.
Reduction of duplicated target loci to single copy
| Gene
|
I-CreI-stimulated loss of whitea
|
Reduction to single copyb
|
||
|---|---|---|---|---|
| N
|
Frequency
|
N
|
Frequency
|
|
| NLaz | 125 | 0.32 | ND | |
| GC | 256 | 0.57 | 6 | 1.00 |
| pug(II)c | 1008 | 0.28 | ND | |
| pug(III)c | 1229 | 0.28 | 13 | 0.77 |
| p53 | ND | 18 | 1.00 | |
| CG11305 | ND | 7 | 1.00 | |
The targeted chromosome or its homolog was dominantly marked. Heat-shocked 70I-CreI-bearing males were crossed to w1118 females with the appropriate balancer chromosome for recovery of the reduction allele. Flies that received the targeted chromosome were scored as white+ or white. The frequency of white-eyed flies is reported here.
Chromosomes in which the target locus had lost the w+ marker were examined by Southern blotting. The frequency of reduction to single copy by homologous recombination (as diagrammed in Fig. 6) is reported here.
Numerals in parentheses refer to a Class II targeting event (from canonical HR), and a Class III targeting event (with a small deletion in the right hand copy of the pug gene). All other targeting events were Class II.
ND, Not done.
With four of the targeted genes, we recovered single-copy reduction events that carried an engineered mutation (those shown in Fig. 7 and NLaz). In the case of pug, we used this procedure on a targeted allele that had incorporated mutations into both copies of the gene. In addition, this was a Class III pug allele, with the right-hand copy carrying a small deletion (∼550 bp). We obtained reduction events with one, both, or neither of the two introduced mutations, including alleles with and without the deletion.
Figure 7.
Single-copy reduction results. (A) Three targeted genes were reduced to single copy and assayed for the retention of point mutations located as shown (coordinates given in kilobases from the left end of the duplicated target segment). For pug, a Class III allele having a small deletion in the right-hand (lower) copy of the gene was used. The number of each type that also have the deletion is given in parentheses. Class II alleles were used in the other cases. (B) A sample of the PCR assay data for retention of mutations in pug reduction alleles. Lanes 1 and 8 are molecular weight markers with sizes indicated to the left. Genomic DNA from homozygous flies was used as template for PCR with the primers ProxH5′ and MTH4826u, which amplify a 3.7-kb fragment from a wild-type pug allele and a 3.2-kb band from the deletion copy. Lanes 2, 4, 6, 9, 11, and 13 represent the amplification products from potential reduction events. All except lane 6 represent actual single-copy reduction that retained the deletion; lane 6 is an example of white+ loss without reduction to single copy, and both the left-hand pug gene copy (full length) and right-hand deletion-bearing copy are present. Lanes 3, 5, 7, 10, 12, and 14 are the corresponding PCR products digested with SpeI to indicate the presence or absence of the engineered mutations. All the single-copy reduction alleles shown here retained only the mutation represented as U in A, except the event represented by lane 12, that retained neither. (C) A sample of the allele-specific PCR data used to determine whether the p53 single-copy reduction alleles carried the engineered mutation. The presence of the 1.3-kb band indicates the presence of the mutant allele. These results were confirmed by the complementary PCR that amplified only the wild-type allele (data not shown). Lanes 1 and 27 are molecular weight markers; lanes 2–25 each represent PCR using genomic DNA from independently isolated reduction homozygotes as template. Lane 26 is a PCR amplification from the p53 donor construct.
Most of these events can be formally explained as simple crossovers between homologous regions to each side of the break. However, some events appear to require multiple exchanges. A more likely explanation is the formation of heteroduplex DNA by single-strand annealing (SSA; Rudin and Haber 1988; Lin et al. 1990; Maryon and Carroll 1991). SSA relies on the action of a 5′–3′ endonuclease to generate regions of single-stranded DNA that can anneal by base complementarity. Then, degradation of protruding single-strand tails, polymerization to fill any remaining single-stranded regions, and ligation finish the process. If heteroduplex DNA is generated by strand annealing, it may be repaired so that a single reduction event appears to have undergone multiple exchanges. SSA is a highly efficient mechanism to repair a DSB between repeated sequences (Carroll 1996; Paques and Haber 1999).
Further examination of the results of Figure 7 provides added support for the SSA mechanism. If SSA is a very efficient mechanism (as the reduction step is), then it is easy to imagine that the entire top copy (as drawn) of the target locus, or nearly so, will become single-stranded by the time the left end of the bottom copy becomes single-stranded. Rapid annealing of the single-stranded complementary portions predicts that retention of the mutations that are furthest from the site of the DSB (those located in the bottom copy) will be favored, and this was observed. SSA in other organisms shows similar characteristics (Carroll 1996). The location of a mutation in the top or bottom copy of the gene depends simply on the orientation of the target-homologous DNA within pTV2. It follows that, for maximal efficiency, the engineered segment should be oriented with the mutation on the side of the I-SceI site that is farther away from the I-CreI site, giving the targeted arrangement diagramed in Figure 6.
Intrinsic I-CreI sites
The reduction to single copy stimulated by I-CreI cutting is very efficient. However, the 36% reduction seen here is much less than we observed previously, where I-SceI was used to generate the DSB between repeated sequences (Rong and Golic 2000). In that case we observed almost 90% reduction to single copy. Such a high level could not be achieved with I-CreI because, unlike I-SceI, a high level of I-CreI expression strongly reduces viability. The I-CreI recognition site lies in a highly conserved region of the Chlamydomonas gene that encodes the 23S ribosomal RNA subunit—17 of 22 bp of this site are identical in the Drosophila 28S gene. The differences that do exist have all been observed in substrate sequences that can be cut by this enzyme in vitro (Argast et al. 1998). When I-CreI is expressed in flies it does cut within the rDNA clusters located in the heterochromatin of the X and Y chromosomes (data not shown). This, presumably, is the basis for the lethality produced by I-CreI expression. Consequently, a moderate heat shock is used to induce I-CreI expression so that the flies will survive to reproduce.
A concern with using this procedure might be that DSBs in rDNA could result in mutations that would interfere with subsequent analysis. This seems unlikely to be a problem for analysis of targeted autosomal genes. Fresh X and Y chromosomes can be easily substituted into the mutant flies after the reduction event, and a straightforward molecular analysis of the target locus should reveal any unexpected rearrangements at that site. In the case of X-linked target genes, a lesser heat shock could be used to reduce the chance of coincident damage to rDNA arrays of the X. The screen is very easy and would work well even with very low rates of reduction to single copy.
None of the target genes reported here are vital genes, and mutant alleles of the four genes produced in this fashion (expected to be null alleles) were viable in the homozygous condition. In the vast majority of cases, neither lethal nor visible mutations arose elsewhere on the target chromosomes coincident to the reduction events. In only three instances (at pug and NLaz) did a new lethal occur on a chromosome with an apparent (judged by loss of w+) reduction allele. One such pug allele was obviously not the expected reduction event, as judged by Southern blotting, and on this basis could easily be excluded from use in further experiments. The nature of the second pug allele was not definitively determined, but was consistent with a large deletion. The recessive lethal chromosome generated during NLaz reduction did not carry the mutant allele, and was not examined further.
The occasional occurrence of lethal mutations on the target chromosome, unrelated to the target locus, does point out the necessity, as with any mutational technique, of verifying that observed phenotypes are attributable to the mutation of interest. Techniques for establishing this are well known and include mapping, complementation with a wild-type transgene, and the use of multiple independently derived alleles.
Discussion
The results presented here address the most important questions about the usefulness of this gene-targeting technique for Drosophila. First (Engels 2000), can genes that are not close to telomeres be targeted? Second (Anonymous 2000), can general methods be developed for targeted mutagenesis? And third (Gloor 2001), can the procedure be made efficient enough to serve as a general method for gene disruption? Our results show that the answer to all these questions is “yes.”
In the case of the first question, Engels proposed that the mechanism of gene targeting involved break-induced replication (BIR), wherein targeting would occur by one-sided recombination events that required invasion of the homolog and unscheduled DNA synthesis to the telomere to finish. Part of the evidence that supported his thesis was that targeting of the X-linked yellow gene was more efficient in females (with two Xs) than in males (with one X). However, we found that targeting of autosomal genes was also more efficient in females. It seems, instead, that it is the physiology of the female germ line that makes the difference, and not the presence or absence of a homolog. It is also clear that targeting can work efficiently for genes that are very far from telomeres. The pug gene is ∼20 Mb away from the nearest telomere and was targeted with reasonable efficiency (>1/4000 gametes) in this work.
We previously showed one method for targeted gene disruption. Two additional techniques for producing mutant alleles of targeted genes are shown in this work. Although the ends-in targeting that we used generates a duplication of the donor DNA at the target locus, this has not been an impediment to the generation of mutant alleles. In two cases the duplicated gene segments each carried a point mutation, producing a mutant allele despite the duplication of target sequence. This procedure has the advantage of generating a mutant allele as the direct result of targeting, providing the most rapid route to mutant flies for phenotypic analyses. Another benefit of this approach is that the mutant allele is tagged with the w+ marker, making it easy to follow in crosses.
Alternatively, a single mutation may be introduced into one of the two copies of the gene, and in a second step, the targeted locus is reduced to a single copy by an I-CreI-mediated DSB. There are also a number of advantages to this approach. Because a mutation is introduced to only one side of the I-SceI site, much more flexibility is allowed when building the donor construct. The I-SceI site need not be placed within the gene, and the mutation may be located essentially anywhere in the gene. This provides the ability to carry out a very fine analysis of gene function. Alleles with specific single-amino-acid changes can be produced and analyzed in homozygous condition without the potential confusion of other alleles being present. Furthermore, a series of specifically altered alleles can be generated and tested in isolation or combinatorially. Similar methods have been proven highly useful in yeast and mice (Rothstein 1991; Muller 1999). However, their usefulness depends on the use of selective methods to efficiently recover the reduction allele. The use of I-CreI to stimulate the reduction event provides a reasonable substitute for application to Drosophila.
This method may prove especially useful if there is concern that the targeted gene may be haploinsufficient. In such cases the donor construct may be designed so that the targeted allele still has one good copy of the target gene and the mutant allele is generated only after reduction to single copy. The recovery of only wild-type and no mutant alleles in the reduction step may indicate haploinsufficiency.
Another significant feature of this method is that no foreign DNA is left behind at the target locus (except the mutation) after reduction to single copy. This can be particularly useful to fully ensure that a gene's regulation is not altered by the introduction of a marker gene. In mammalian systems, Cre-mediated site-specific recombination has been used to remove marker genes after transformation or targeting (Torres and Kuhn 1997), but a lox site is left behind in such instances.
The efficiency of targeting in these experiments was as high as 1 targeting event in ∼1500 gametes (CG11305), and ranged as low as 1 in 34,000 gametes (GC). It is possible to achieve mutagenesis rates in this range with chemical mutagens, and Gloor suggested that this targeting procedure is, therefore, no easier to use than classical mutagenesis methods. This criticism, of course, overlooks the fact that gene targeting requires no knowledge of the mutant phenotype, whereas traditional mutagenesis protocols are based on phenotypic screening, and often require more arduous crosses to look for recessive phenotypes. Moreover, the molecular identification of which gene has been mutated to produce a phenotype most often requires a large investment of time and effort. Phenotypic screening of random mutations is certainly very powerful, especially for implicating genes in a process where their involvement was not suspected. But, when a gene of interest has already been identified, perhaps through biochemical or informatic methods, gene targeting is a more direct and efficient route to obtaining mutants if they do not already exist.
We were surprised to find that the frequency of nontargeted insertion varied for different donors. We initially thought that nontargeted insertions would occur at a relatively constant background rate, having no particular relation to the frequency of homologous recombination. One explanation for the difference in nontargeted insertion frequency might be the rapidity of degradation of the cut donor. Because the recognition and recovery of insertions of any type require an intact white gene, it could be that larger donors buffer the white gene for a longer period, allowing it more opportunity to insert at nonhomologous sites. However (from females) we recovered a much higher rate of nontargeted events in the CG11305-targeting crosses (∼1 in 28 vials) than in the other targeting crosses (with pug being closest at ∼1 in 100 vials), and yet the CG11305 “buffer” sequence was the smallest used in this work. Instead, there appears to be a correlation (r = 0.86, P = 0.03), between the frequencies of targeted and nontargeted insertions. An explanation that accounts for this is to suppose that the DSB generated by I-SceI has a half-life that is at least partially controlled by the sequences that flank the cut site. Some sequences, for example those flanking the I-SceI site in the GC donor, may promote rapid intramolecular repair of the DSB, reducing the opportunity for both targeted and nontargeted integration. Another possibility to explain the low frequency of GC targeting may be that many Class II targeting events occurred but went undetected, because they were frequently reduced to Class I (allelic substitution) events during the targeting process. The observation that reduction to single copy at GC was nearly twice as efficient as at the other tested genes offers some support for this.
In these experiments we have not been concerned with generating donor DNAs that are iso-sequential with the target locus. Many of the constructs were built using PCR to produce segments of the donor. In other cases, subclones from genomic libraries were used. In no instance was particular care used to derive the donor from the actual chromosome that would be targeted. It is possible that targeting could be made more efficient by ensuring that donor and target are iso-sequential (as for mouse targeting; te Riele et al. 1992), but we conclude that it is not generally necessary for efficient targeting in flies.
Our findings contrast strikingly with the results of targeting in mouse ES cells with respect to the ratio of targeted to nontargeted insertions. We observed a much higher proportion of targeted insertions than is typical in ES cells. When positive–negative selection is not used in ES cell transformation, the majority of positively selected clones contain nontargeted insertions. These often outnumber targeted insertions by orders of magnitude (Mansour et al. 1988; Bollag et al. 1989). In Drosophila females, the majority of donor mobilization events were targeted, and even in males they constituted one-third of the total. In Drosophila, positive–negative selection is not needed to select the targeting events because most are targeted.
The difference between males and females in our results suggests that the arrays of DNA repair enzymes expressed by different cell types at least partly control targeting success, and that Drosophila may provide relatively advantageous proportions. Improving the efficiency of targeting by manipulating the levels of repair activities is not likely to be easy because the balance of activities that provides efficient gene targeting is probably complex (Pierce et al. 2001; Yanez and Porter 2002).
Another influence on the efficiency of gene targeting in the Drosophila system may be the fact that the donor DNA is derived from a preexisting chromosomal site, with the donor already packaged as chromatin. This may contribute to efficient targeting and might partly explain some of the variation seen when different insertions of the same donor element are used. They may be bound by different proteins and consequently target with different efficiencies.
General guidelines for efficient targeting may be deduced from our results. First, targeting is more efficient in females: enough so that it is probably not worth screening for targeting through the male germ line. Second, several different insertions of the donor element should be used because targeting efficiency can vary considerably for different sites of insertion. Finally, to increase the probability of efficient targeting it seems prudent to use as large an extent of homology as is feasible. Our results suggest that the donor should carry 5 kb or more of target site homology for reasonably efficient targeting.
Practical considerations of cloning will typically constrain the length of homology, but other factors may also enter into the decision. For instance, the use of a long target-homologous segment may result in an undesirable duplication of a neighboring gene. If the mutant is generated by the reduction to single copy, this need not be a concern because phenotypic analysis will be performed in a genotype without the duplication.
A target site duplication might be used to advantage by constructing a donor that generates experimentally useful alleles at the duplication stage, and after reduction to single copy. A particularly advantageous combination would be the generation of a gene that encodes a fusion of the wild-type protein and Green Fluorescent Protein (GFP; Chalfie et al. 1994) as the first step, and a null allele in the second step. The fusion protein would be used to report the expression pattern of the target gene and the subcellular location of the protein. After reduction to single copy, a mutant allele is produced for phenotypic analysis. Because the 5′ regulatory sequences of the target gene remain intact in their proper chromosomal context, expression of the GFP fusion protein is likely to reflect the wild-type expression pattern. Additionally, because the second copy of the target gene will be a mutant, the fusion protein can be tested in homozygous condition, or heterozygous with the single-copy mutant, for complementation of any phenotypes that are observed in the mutant. The results can confirm that localization of the GFP fusion protein reflects function (Wang and Hazelrigg 1994). An additional refinement may be the inclusion of a significant stretch of 3′ target sequence following GFP, so that some reduction events will produce the mutant allele and others the GFP fusion with no additional extrinsic DNA.
In any method that involves the introduction of mutations, it is important to place them a sufficient distance from the I-SceI site to prevent inevitable loss to gap enlargement. Our results suggest that 400 bp is sufficient, but even greater distances may be advisable if a small screen is planned, so that even if only a few targeting events are recovered, the chance of incorporating the mutation will be high. Gloor et al. (1991) characterized the extent of gap enlargement during P-element-mediated gene conversion. Their results suggest that a mutation should lie ∼450 bp from the DSB for a 50% chance of being converted, which is not dissimilar to our findings for gene targeting.
We previously speculated that Class III events could be used as a potential route to generate mutants in a target gene (Rong and Golic 2000). One of the reduction events from the Class III pug targeting event carried the deletion that was generated during targeting, but neither of the engineered point mutations. This small deletion within the coding region of pug generated a null allele that gave the recessive pug phenotype (data not shown), confirming that this can be a useful method for generating mutants in a target gene.
It is interesting that the mutant alleles of all five genes that we targeted are homozygous viable, and lack obvious phenotypes. pug, p53, and CG11305 mutants do have subtle phenotypes (in eye pigmentation, cell death, and position effect variegation, respectively), but they are not immediately revealed to casual observation. This may be a reflection of the state of genetic research in Drosophila. Genes uncovered in standard screens most often have mutant phenotypes that can be recognized with reasonable facility. Genes that do not mutate to give an easily visible phenotype await analysis through the use of mutants recovered in other types of screens. These may include random transposon insertion collections (Spradling et al. 1999), screens of chemically treated chromosomes for randomly induced changes within a specific region (Bentley at al. 2000), or gene targeting as we have described here. These methods provide the keys to connect the identification of genes by genome sequencing projects with studies of their function.
Materials and methods
Plasmid construction
The targeting vector pTV2
Two oligonucleotides (5′-GTAC CAAAACGTCGTGAGACAGTTTGCCATG-3′ and 5′-GCAAA CTGTCTCACGACGTTTTGGTACCATG-3′), providing the recognition sites for I-CreI and KpnI, were annealed and ligated into SphI-cut pP[>whs•N>] (from J. Feder, University of Chicago, Illinois), which is a modified form of pP[>whs>] (Golic and Lindquist 1989). A clone was selected that had incorporated the oligonucleotide to provide the following orientation of sites: FRT, NotI, KpnI, I-CreI, whs, FRT. The NotI and KpnI sites are unique and useful for insertion of engineered donor constructs.
The NLaz donor element
The P1 clone DS08613 spanning the NLaz genomic sequence was used as template. The first two PCR reactions were with the Not1 forward primer (5′-TCT TATAAGCGGCCGCACATAAATCGAATGGC-3′) and the HindIII I–SceI reverse primer (5′-TATAATAAGCTTTAGG GATAACAGGGTAATGTCTAAACGATCAGTTGCAAGCG-3′) to give a 2125-bp product, and the HindIII forward (5′-TTATAAAAGCTTTCCTATTCTAAATCTATCAGAATG-3′) and Not1 reverse primer (5′-TCTTATAAGCGGCCGCATCT TGGCCATATCTCGGTTTAA-3′) to give a 3035-bp product. Herculase polymerase (Stratagene) and reaction conditions suggested by Stratagene were used. The reaction products were gel-purified, cut with HindIII, and ligated together. A dilute fraction of the ligation mix was used as template for a PCR reaction using the NotI forward and reverse primers. From this, a 5.1-kb band was gel-purified, cut with NotI, and ligated into the NotI-cut vector pHSS6 (Seifert et al. 1986), and transformed into bacteria. Plasmid DNA was digested with I-SceI to verify the integrity of the cut site in the targeting sequence.
Next, two premature stops were engineered into the targeting sequence of NLaz. The first stop in exon 2 was generated by cutting pHSS6+NLaz with XhoI, blunting with Klenow, and then religating to generate a new PvuI site and a stop. The second stop in exon 3 was generated by designing two primers that incorporated SpeI sites. Forward SpeI (5′-ATACGGAC TAGTCCATATACTGAAGTATTCGA-3′) and Reverse SpeI (5′-ATACGGACTAGTGCGTCTGGTACGAGTACGCA-3′) were used to amplify from the pHSS6+NLaz template a linear sequence with SpeI ends that included the vector and NLaz. DNA was run on an agarose gel, excised, purified, and then digested with SpeI. The DNA was purified from the digest, ligated, and transformed into bacteria. Plasmid DNA was cut with SpeI, NotI, and PvuI to verify the engineered stops. The pHSS6+NLaz with both verified stops was cut with I-SceI to reverify the integrity of that site. Finally, the engineered NLaz fragment was removed from pHSS6 by NotI digestion and ligated into NotI-cut pTV2. This donor P element was transformed into Drosophila by standard techniques (Rubin and Spradling 1982).
The GC donor element
For the GC donor element, 6.9 kb of DNA encoding the Drosophila γ-glutamyl carboxylase and flanking DNA (from −2261 to +4661, with nucleotide +1 being the start of the initiation codon) was cloned by PCR amplification of genomic DNA in four fragments and assembled in PCRII (Invitrogen). The four fragments were: (1) −2261 to +72; (2) +72 to +910; (3) +910 to +1746; and (4) +1746 to +4661. Nucleotide sequence changes were introduced into the PCR-amplified DNA using primers containing the desired changes. The following changes were introduced: (1) at +69 the genomic sequence ACACCTAGGTCGTCGGAA was replaced by ACAGCTAGC TCGTAATCGGAA; (2) at +909 the sequence TTCATGACTC TGGAA was replaced by TTCATGACTCTGTAGGGATAA CAGGGTAATCTGGAA; (3) at +1744 TTCCCGGGACTAAC TCTG was replaced by TTCCCGGGACTATAAACTCTG. Step 2 introduced an I-SceI site, and steps 1 and 3 introduced stop codons to either side of the I-SceI site. The cloned DNA in pCRII was flanked by NotI sites, and the NotI fragment containing the cloned genomic fragment with modifications was transferred to pTV2. This GC donor element was transformed into Drosophila.
The pug donor element
An 8.9-kb fragment containing the pug+ gene and flanking genomic DNA was obtained from the P1 genomic clone DS01137 by digestion with EcoRI. This fragment was cloned into the EcoRI site of pBluescript II SK(+/−). A double-stranded oligonucleotide derived by annealing Nhe-stop-plus (5′-CTAGACTAGTCTAGC-3′) and Nhe-stop-minus (5′-CTAGGCTAGACTAGT-3′) contains stop codons in all three reading frames. It was cloned into the NheI site in the 3′ region of the pug+ coding sequence. The pug+pBS+Nhe-stop was next cut wth NruI, and a Spe Linker (5′-CTAGACTAGTCTAG-3′) from New England Biolabs was ligated into this site. This pug− cassette, with point mutations, was removed as an EcoRI fragment and ligated into pHSS6 at its EcoRI site. Next an I-SceI recognition sequence generated by annealing two oligonucleotides (5′-GGCCTAGGGATAACAGGGTAAT-3′ and 5′-GGC CATTACCCTGTTATCCCTA-3′) was ligated into the ApaI site located in approximately the center of the pug− cassette. The completed donor element was then transferred to the targeting vector pTV2 by cutting at the NotI sites in pHSS6 and pTV2, and transformed into flies by standard techniques.
The p53 donor element
An 8.6-kb genomic fragment that includes the Drosophila p53 gene was PCR-amplified using the genomic P1 clone DS02942 as template, and cloned into pBluescript. A mutation was introduced into the coding region by oligo-directed mutagenesis, which changed the genomic sequence of 5′-CTGCAGGACATTCAGAT-3′ to 5′-CTGTAGG TAATTAAGAT-3′. An I-SceI cut site was introduced by the same method, which changed the genomic sequence of 5′-AAG GTCCAGATCA-3′ to 5′-AAGGTCATTACCCTGTTATCCCT ACAGATCA-3′. The 8.6-kb fragment was then removed as an Acc65I–NotI fragment (with the NotI site from the endogenous p53 sequence), cloned into pTV2, and transformed into Drosophila.
The CG11305 donor
The engineered gene was produced by PCR using the following primers with genomic DNA as a template. The 5′ end of the gene was produced using primer 1 (5′-TGAAGCGGCCGCTTTGGCCAGCAGCCGGATAT-3′, which adds an NotI site) and primer 2 (5′-GGTGAATTCTT TATTCCTAAGCCCTGCGAAATGATGTTG-3′, which adds an in-frame stop codon and an EcoRI site), and was cloned into a modified pBluescript KS vector using NotI and EcoRI sites. The region from the NotI site to an internal XhoI site was then replaced by the NotI- and SalI-cut PCR product of primer 5 (5′-ATTTGCGGCCGCGATGTACACACGGGTTC-3′, adding an NotI site) and primer 6 (5′-GCGTCGACTAGGGATAA CAGGGTAATGGAACTCCTCCACCTGCCG-3′, adding I-SceI and SalI sites). The resulting plasmid, carrying the 5′ end of CG11305, was then cut with EcoRI and HindIII, and the 3′ fragment of the gene was ligated into these sites. The 3′ portion of the gene was generated by PCR using primer 3 (5′-TAG GAATAAAGAATTCACCAAGCATCTAGTTTGCTACACG-3′, providing an EcoRI site that, when ligated to EcoRI from primer2, also causes a shift in the reading frame downstream of the stop codon from primer 2) and primer 4 (5′-GGGGTAC CATTGACTCAAGGGTAATCATT-3′, adding an Acc65I site). This was cloned into pBluescript KS at the EcoRI and EcoRV sites, and then removed by digestion with EcoRI and HindIII for ligation to the 5′ end of the gene, as mentioned. The resulting engineered donor was cloned into pTV2 using NotI and Acc65I sites.
The 70I-CreI transgene
Two oligonucleotide primers (5′-GTACCCGGATCCATGAATACAAAATATAAT-3′; 5′-GTGACT CGGTCGACTACGGGGACGATTTCTT-3′) were used in the PCR to amplify the I-CreI coding sequence from the plasmid pB-E (Seligman et al. 1997), adding a BamHI site upstream of the start and a SalI site downstream of the stop. To place this under control of the Drosophila hsp70 promoter, the plasmid p70ATG-Bam (Petersen and Lindquist 1989) was digested with BamHI and SalI. The I-CreI fragment was also cut with BamHI and SalI, and it was ligated into p70ATG-Bam, replacing the hsp70 coding sequence with the I-CreI coding sequence, to make 70I-CreI. The gene was removed as an HindIII–EcoRI fragment and cloned into HindIII- and EcoRI-cut pHSS6. The gene was then removed as an NotI–NotI fragment, cloned into the P-element vector pYC1.8 (Fridell and Searles 1991) at the unique NotI site, and transformed into Drosophila.
Genetic procedures
Detailed information about the Drosophila genes and chromosomes mentioned here can be found at http://flybase.bio.indiana.edu/. Crosses were carried out using standard techniques. Mapping and stock-making were accomplished using standard balancer chromosomes. Heat shocks were performed as described (Golic and Lindquist 1989).
Targeting crosses
Two methods were used to screen for targeting. In the first, we screened for mobilization of the marker gene in test crosses, as described by Rong and Golic (2000), except that w+ was the marker rather than y+. In the second, more frequently used method, we screened for a lack of white+/white mosaicism induced by FLP, as previously described (Rong and Golic 2001). This relies on the fact that in a targeted allele the w+ marker is no longer flanked by FRTs and is not subject to excision following FLP expression. It provides a quicker method to proceed from a transformant of the donor P element to the targeted allele. Details of the crosses can be found in the original works.
Reduction to single copy by I-CreI
In a typical crossing scheme, flies carrying a Class II targeted allele (e.g., w1118/Y; p53w+/p53w+ males) are crossed to w1118 70I-CreI; Sb/TM6 females. (Autosomal insertions of 70I-CreI are also available for use in X-linked gene-targeting procedures.) After 2–3 d, these parents are removed from the vials, and the progeny are heat-shocked (36°C, 1 h). Males that eclose (and are typically white+/white mosaics) are individually mated to w1118; Sb/TM6 females. The white-eyed Sb+ sons (w1118/Y; p53reduced/TM6) are collected and mated to w1118; Sb/TM6 females to make stocks of the reduced alleles. Typically, only one son is collected from a given father. Several independently derived stocks are generated and tested for the presence of an introduced mutation.
Verification of targeting
Southern blot analyses were used to verify targeting for all genes except NLaz and were performed as described (Rong and Golic 1998). For verification of NLaz targeting, PCR was used. First, flies were chosen in which the w+ marker gene had moved from its original X-chromosome location to the target chromosome. Genomic DNA was prepared to be used as template. The primers 5′-GAGACCACCTAAAATTGGCA-3′ and 5′-TATCGCGA TGTGCATACAGA-3′ were used to amplify a 1.3-kb fragment from the NLaz gene that spanned the location of the I-SceI site and both mutations (which were marked by unique SpeI and PvuI sites). We reasoned that this amplified fragment would be cut with SpeI or PvuI only if the donor had integrated at the target locus and retained at least one of the mutations: the wild-type NLaz alleles would be amplified but not cut, and nontargeted integration would not be a substrate for amplification because the integrated ends would, most likely, be facing apart.
Verification of the incorporation of point mutations
For NLaz, the same PCR reaction that was used to verify targeting was also used to detect single-copy reduction alleles that retained the SpeI-marked mutation. Genomic DNA from homozygous stocks was used as template, and we looked for complete cutting of the amplified fragment with SpeI to verify that the mutant allele was the only one present, also providing additional confirmation of the initial targeting.
For GC, allele-specific PCR was used to verify the incorporation of mutations. For the upstream mutation, two primers (5′-CACAAAGTAACAGCTAGCTCGTAA-3′ and 5′-AGGATTC CCGGCTTGGAA-3′) were used that allowed amplification of a 1.2-kb fragment only from the mutant allele. For the downstream mutation, two primers (5′-CGAGATGAAGTTGGT CAGAGTTTA-3′ and 5′-GACGTGGAATAACCACAGCTA -3′) were used that also gave a 1.2-kb fragment only from the mutant allele.
For pug, we used PCR to determine which mutations were present in the chromosome subsequent to targeting and reduction. In one procedure (as shown in Fig. 7) the primers ProxH5′ (5′-GGAAATTGCGTATGCCAGCA-3′) and MTH4826u (5′-CTCAATTTCTCCGGTTTCCGTATTCAGA-3′) were used to amplify a 3.7-kb segment of the pug gene that spanned the sites of both point mutations. The point mutations introduce SpeI recognition sequences. The amplified product was digested with SpeI: bands of 0.5 and 3.2 kb indicate the presence of the more upstream mutation at NruI (designated U in Fig. 7); bands of 2.6 and 1.1 kb indicate the presence of the downstream mutation engineered at the NheI site (designated D in Fig. 7); and the presence of three bands of 0.5, 2.1, and 1.1 kb indicates both mutations in a single-copy reduction allele. A single 3.7-kb band after SpeI digestion indicates neither mutation is present.
To confirm the presence of the mutation introduced to the NruI site, PCR was carried out with three primers: ProxH5′, NruSpeId (5′-CCGTTAGATCCATTCGCGA-3′), and Test4 (5′-ACCTTCGTTAACCGTGTGCAA-3′). These prime amplification of a 1005-bp fragment from wild-type and mutant alleles, and a 490-bp fragment specific to the mutant. The presence of both bands (as shown in Fig. 5) indicated that the mutation was present. A similar reaction was used to verify presence or absence of the mutation at the NheI site. The oligonucleotides pugseq1 (5′-TCAGTGTTGGAGCGTCTGAA-3′), Nhestopminusd (5′-ACTTCGGGATAGTGCTAGC-3′), and MTH4275U (5′-CAGAGCAGCTTTTTGACCA-3′) prime amplification of a 1233-bp fragment from wild-type and mutant alleles and a 666-bp fragment specific to the mutant allele.
For p53, we used allele-specific PCR to determine which targeted and reduction alleles carried the point mutation. Three primers were used: (1) 5′-GTTCGCCTGGATCTTAATTA-3′; (2) 5′-GTTCGCCTGGATCTGAATGT-3′; and (3) 5′-AATCGC TGCATGCGGTAGTA-3′. Primers 1 and 3 generate a 1.3-kb fragment specifically from the mutant allele. Primers 2 and 3 generate a 1.3-kb fragment specifically from the wild-type allele.
For CG11305, Southern blotting of EcoRI-digested genomic DNA was used to ascertain the presence of the point mutation.
Statistical procedures
Statistical analyses were performed with the aid of GraphPad Instat version 3.0 for Macintosh (GraphPad Software).
For testing the difference between male and female germ-line targeting, a contingency test of homogeneity was performed for all pug and p53 donors that had been tested in both males and females and that gave any targeting events (these are, as listed in Table 1: first, third, and fourth for pug; and second and third for p53). The heterogeneity χ2 was not significant (3.79, 4 d.f., 0.25 < P < 0.5), and the results obtained for all these donors were added, with the sums used in a 2 × 2 contingency test of female versus male targeting efficiency.
To test the significance of the difference between targeted and nontargeted insertions recovered from females versus males, we used a 2 × 2 contingency test of total targeted and nontargeted events recovered from each sex. Contingency tests were also used to test whether different insertions of the same donor construct targeted with different efficiencies, and to test whether targeting efficiencies for GC and CG11305 were significantly different.
The coefficient of correlation (and corresponding one-tailed P value) between the frequencies of targeted and nontargeted events was determined using the unweighted average targeting (or nontargeting) frequency of each gene in females.
Acknowledgments
We gratefully acknowledge the work of many yeast and mouse researchers who pioneered target gene-modification techniques that we have freely borrowed; and Ray Monnat for the I-CreI encoding pB-E and for analysis of cut sites in Drosophila rDNA. This work was supported by grants GM60700 and GM65604 from the National Institutes of Health, by the University of Utah Research Foundation, and by the Stowers Institute for Medical Research. M. Brodsky was supported by a fellowship from the American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kgg@stowers-institute.org; FAX (816) 926-2065.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.986602.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Anonymous. In search of a function. Nat Cell Biol. 2000;2:E137–E138. doi: 10.1038/35019607. [DOI] [PubMed] [Google Scholar]
- Argast GM, Stephens KM, Emond MJ, Monnat RJ. I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J Mol Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- Bentley A, MacLennan B, Calvo J, Dearolf CR. Targeted recovery of mutations in Drosophila. Genetics. 2000;156:1169–1173. doi: 10.1093/genetics/156.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag RJ, Waldman AS, Liskay RM. Homologous recombination in mammalian cells. Ann Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Carroll D. Homologous genetic recombination in Xenopus: Mechanism and implications for gene manipulation. Prog Nucleic Acids Res Mol Biol. 1996;54:101–125. doi: 10.1016/s0079-6603(08)60361-x. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: From yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels WR. Reversal of fortune for Drosophila geneticists? Science. 2000;288:1973–1975. doi: 10.1126/science.288.5473.1973. [DOI] [PubMed] [Google Scholar]
- Fridell Y-WC, Searles LL. vermillion as a small selectable marker gene for Drosophila transformation. Nucleic Acids Res. 1991;19:5082. doi: 10.1093/nar/19.18.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/s0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- Gloor GB. Gene-targeting in Drosophila validated. Trends Genet. 2001;17:549–551. doi: 10.1016/s0168-9525(01)02419-2. [DOI] [PubMed] [Google Scholar]
- Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist SL. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Golic MM, Golic KG. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics. 1996;143:385–400. doi: 10.1093/genetics/143.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Honigberg SM. Effect of chromosomal locus, GC content and length of homology on PCR-mediated targeted gene replacement in Saccharomyces. Nucleic Acids Res. 2001;29:5156–5162. doi: 10.1093/nar/29.24.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Yang CT, Jin D, Stafford DW. Identification of a Drosophila vitamin K-dependent γ-glutamyl carboxylase. J Biol Chem. 2000;275:18291–18296. doi: 10.1074/jbc.M001790200. [DOI] [PubMed] [Google Scholar]
- Lin FL, Sperle K, Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990;10:103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Maryon E, Carroll D. Involvement of single-stranded tails in homologous recombination of DNA injected into Xenopus laevis oocyte nuclei. Mol Cell Biol. 1991;11:3268–3277. doi: 10.1128/mcb.11.6.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U. Ten years of gene targeting: Targeted mouse mutants, from vector design to phenotype analysis. Mech Dev. 1999;82:3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- Ollmann M, Young LM, Di Como CJ, Karim F, Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A, et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Papadopoulou B, Dumas C. Parameters controlling the rate of gene targeting frequency in the protozoan parasite Leishmania. Nucleic Acids Res. 1997;25:4278–4286. doi: 10.1093/nar/25.21.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RB, Lindquist S. Regulation of HSP70 synthesis by messenger RNA degradation. Cell Regul. 1989;1:135–149. doi: 10.1091/mbc.1.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG. Dominant defects in Drosophila eye pigmentation resulting from a euchromatic–heterochromatic fusion gene. Genetics. 1998;150:1551–1566. doi: 10.1093/genetics/150.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- ————— A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Rudin N, Haber JE. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1998;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D, Ganfornina MD, Torres-Schumann S, Speese SD, Lora JM, Bastiani MJ. Characterization of two novel lipocalins expressed in the Drosophila embryonic nervous system. Intl J Dev Biol. 2000;44:349–359. [PubMed] [Google Scholar]
- Seifert HS, Chen EY, So M, Hefron F. Shuttle mutagenesis: A method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci. 1986;83:735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman LM, Stephens KM, Savage JH, Monnat RJ. Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- te Riele H, Maandag ER, Berns A. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc Natl Acad Sci. 1992;89:5128–5132. doi: 10.1073/pnas.89.11.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Yuan X, Kudlicki W, Herrin DL. Cleavage and recognition pattern of a double-strand-specific endonuclease (I-CreI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene. 1992;119:247–251. doi: 10.1016/0378-1119(92)90278-w. [DOI] [PubMed] [Google Scholar]
- Torres RM, Kuhn R. Laboratory protocols for conditional gene targeting. Oxford, UK: Oxford University Press; 1997. [Google Scholar]
- Walker CS, Shetty RP, Clark K, Kazuko SG, Letsou A, Olivera BM, Bandyopadhyay PK. On a potential global role for vitamin K-dependent γ-carboxylation in animal systems: Evidence for a γ-glutamyl carboxylase in Drosophila. J Biol Chem. 2001;276:7769–7774. doi: 10.1074/jbc.M009576200. [DOI] [PubMed] [Google Scholar]
- Wang S, Hazelrigg T. Implications for bcd mRNA localization from spatial distribution of exu protein in Drosophila oogenesis. Nature. 1994;369:400–403. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- Yanez RJ, Porter AC. Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic Acids Res. 2002;3:740–748. doi: 10.1093/nar/30.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]