Abstract
Retinoic acid-inducible gene I (RIG-I) plays a pivotal role in the regulation of cytokine production induced by pathogens. The RIG-I also augments the production of IFN and other cytokines via an amplification circuit. Because the production of cytokines is closely controlled, up- and down-regulation of RIG-I signaling also needs strict regulation. The mechanism of this regulation, however, remains elusive. Here, we found that RIG-I undergoes proteasomal degradation after conjugation to ubiquitin by RNF125. Further, RNF125 conjugates ubiquitin to MDA5, a family protein of RIG-I as well as IPS-1, which is also a downstream protein of RIG-I signaling that results in suppressing the functions of these proteins. Because RNF125 is enhanced by IFN, these functions constitute a negative regulatory loop circuit for IFN production.
Keywords: innate immunity, signal transduction
Upon viral infection, host cells activate the innate immune signaling cascades critical for effective antiviral immune responses (1, 2). The viral components, such as viral DNA or RNA, are recognized by Toll-like receptors (TLRs) that stimulate the production of antiviral factors including cytokines and induce inflammatory and adaptive immune responses (1–8). In addition to TLR-dependent activation of innate immunity, retinoic acid-inducible gene I (RIG-I) and other proteins in this family can also detect viral components or dsRNA, inducing the production of cytokines necessary to activate innate and adaptive immune responses (9, 10). The RIG-I is a DExD/H box RNA helicase with two caspase-recruiting domain (CARD)-like sequences and a helicase domain that is required for its interaction with dsRNA. The CARD-like domains are responsible for activating downstream signaling (9) through interactions with IPS-1/MAVS/VISA/Cardif (11–13).
The rapid induction of type-1 IFN expression is the key event in the initiation of the innate antiviral response, and it requires the pathogen-inducible activation of transcription factors that function synergistically to induce gene expression (9, 14–16). Among members of the IFN regulatory factor family, IRF3 and IRF7 play essential roles in virus-induced type-1 IFN gene activation after infection (9, 13, 17, 18).
Activation of TLRs results in a proinflammatory response necessary to prevent the spread of infection. Limiting TLR signaling, however, is essential to prevent this protective response from causing injury to the host. Several regulatory mechanisms that suppress TLR signaling have been described in earlier reports (19–22). Regulation of the TLR-independent, RIG-I-mediated signaling pathway regulating IRF3; however, it is not well understood. Recently, A20, an NF-κB-inducible ubiquitin-editing protein able to inhibit TLR3- and Sendai virus-induced activation of the ISRE promoter (23) was shown to inhibit this signaling pathway (24).
To clarify the mechanisms underlying the negative regulation of RIG-I signaling, we searched for a Ubl E3 ligase that modifies RIG-I. The result of the search was the identification of RNF125 (Homo sapiens ring-finger protein 125, GenBank accession no. NM_017831), also named TRAC-1 (T cell RING protein identified in activation screen), an E3 ubiquitin ligase serving a positive regulatory role in T cell activation (25), that functioned as an E3 ligase for RIG-I ubiquitin conjugation. Ubiquitin-conjugated RIG-I was degraded in a proteasome-dependent manner. We also observed that RNF125 possessed the ability to conjugate ubiquitin to MDA5, a member of the RIG-I protein family, as well as IPS1/MAVS/VISA/Cardif (referred to as IPS1 hereafter), a protein downstream of RIG-I signaling, which also suppresses RIG-I mediated signaling.
Results
Isolation of RNF125 as an Interacting Protein with UbcH8.
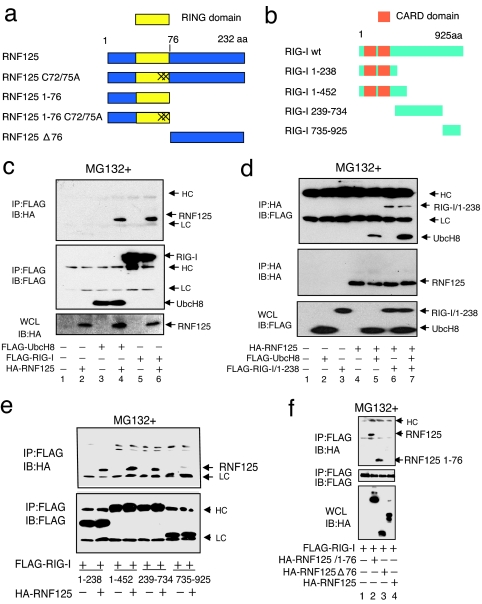
RIG-I is reported to be conjugated (ISGylated) to ISG15 (26). UBE1L and UbcH8 function as E1 and E2 enzymes for ISGylation, respectively. Cotransfection of 293FT cells with plasmids encoding UBE1L, UbcH8, and ISG15 promoted the ISGylation of RIG-I (data not shown). The UbcH8 functions as an E2 enzyme for both ubiquitin and ISG15 conjugation (16, 27–33). We made an attempt to isolate a candidate E3-ligase capable of conjugating ISG15 or ubiquitin to RIG-I. We hypothesized that this candidate E3-ligase would interact with UbcH8 because, under certain circumstances, the interaction of E2 enzymes and E3 ligases can be observed in vitro during the conjugation of ubiquitin-like protein (Ubl) (32, 34). Using a yeast-two hybrid screening, we isolated RNF125 as an interacting protein with UbcH8. RNF125, a RING motif containing proteins, can act as a ubiquitin E3 ligase (25). A schematic diagram of RNF125 and its mutants used in this paper are shown in Fig. 1a. We found that RIG-I interacted with RNF125 as well as UbcH8 (Fig. 1 c and d). We analyzed the regions of RIG-I interacting with RNF125 by using deletion mutants of RIG-I as well as the region of RNF125 interacting with RIG-I (Fig. 1 e and f). From this analysis, the CARD domain of RIG-I as well as the C-terminal region of RIG-I were found to interact with the N-terminal region of RNF125.
Fig. 1.
Association of RNF125 with UbcH8 and RIG-I. (a and b) Schematic structure of RNF125, RIG-I, and their derivatives used in this work. “X” indicates site of cysteine residue substitution with alanine at the 72nd and 75th residues in RNF125. (c and d) Coimmunoprecipitation experiments. The 293FT cells were transfected as indicated. Thirty-six hours after transfection, protein associations were analyzed either by coimmunoprecipitation using anti-FLAG antibody, followed by Western blot using anti-HA antibody (c), or by coimmunoprecipitation using anti-HA antibody, followed by anti-FLAG antibody (d). (e) Analysis of the RIG-I domain that interacts with RNF125. Plasmids expressing various deletion mutants, including amino acids 1–238, 1–452, 239–734, and 735–925 of RIG-I fused to a FLAG epitope tag (b), were transfected into 293FT cells with or without a plasmid encoding HA-RNF125. Complex formation was examined by immunoprecipitation with an anti-FLAG antibody followed by immunoblotting using the anti-HA antibody (Upper). The amount of each RIG-I mutant in the complex is indicated (Lower). (f) Analysis of the association of RIG-I with RNF125 mutants. Plasmids as indicated were transfected into 293FT cells. The lysates were immunoprecipitated by using an anti-FLAG antibody, followed by blotting with the anti-HA antibody (Top). The quantity of FLAG-RIG-I in the immunocomplexes as well as the RNF125 and mutants in whole-cell lysates are also shown in Middle and Bottom, respectively. HC and LC indicate the heavy and light chains of human immunoglobulins, which are also indicated in the following figures. All cells in c–f were treated with MG132.
RNF125 Is an E3-Ligase for Conjugation of Ubiquitin to RIG-I.
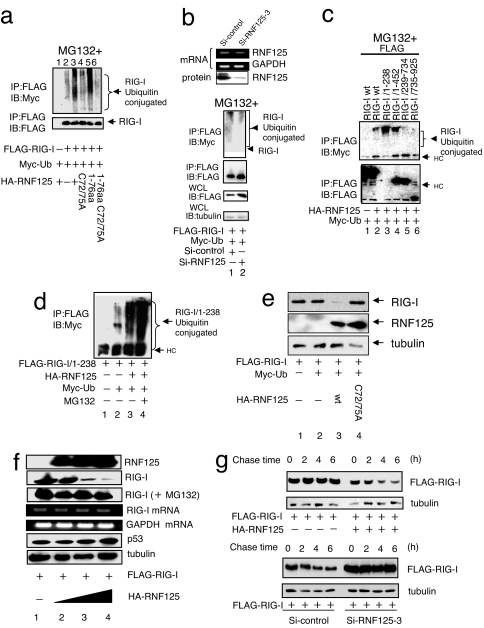
We analyzed whether the E3-ligase activity of RNF125 is responsible for the ubiquitination of RIG-I. When FLAG-RIG-I, HA-RNF125, and Myc-Ub were expressed in 293FT cells, ubiquitin conjugation to RIG-I was observed (Fig. 2a). The level of ubiquitination was low in the absence of ectopic RNF125 expression. The fact that RNF125 is the ubiquitin E3-ligase for RIG-I was further confirmed by the suppression of endogenous RNF125 using siRNA. Transfection of cells with siRNF125–3 strongly suppressed the production of RNF125 mRNA, leading to a substantial reduction in RNF125 protein levels in 293FT cells (Fig. 2b Upper). In cells treated with siRNF125–3, with the exception of those treated with control siRNA, the levels of RIG-I polyubiquitination were substantially reduced (Fig. 2b Lower).
Fig. 2.
Ubiquitin conjugation to RIG-I. (a) Ubiquitin conjugation to RIG-I by WT and mutant RNF125. Plasmids encoding FLAG-RIG-I and Myc-Ub were cotransfected into 292FT cells with a plasmid encoding either WT or mutant RNF125. The RIG-I ubiquitination was monitored by immunoprecipitation. All cells were treated with MG132. (b) Inhibition of RIG-I ubiquitin conjugation after suppression of endogenous RNF125. The siRNF125–3, an siRNA that down-regulates RNF125 mRNA, or a control siRNA were transfected into the 293FT cells. The mRNA and protein levels of RNF125 were monitored by RT-PCR or Western blot, respectively, 36 h after transfection (Upper). The 293FT cells transfected with plasmids encoding FLAG-RIG-I and Myc-Ub were simultaneously treated with control siRNA or siRNF125–3; RIG-I ubiquitination in cell lysates was then analyzed by coimmunoprecipitation. All cells were treated with MG132. The quantity of RIG-I present in the immunoprecipitants was monitored by immunoblot with an anti-FLAG antibody. The total RIG-I present in an aliquot of whole-cell lysate is also shown. Tubulin serves as the control in this analysis (Lower). (c) Analysis of the region for ubiquitin conjugation in RIG-I. Plasmids encoding HA-RNF125 and Myc-Ub were cotransfected into 293FT cells with WT or deletion mutants of FLAG-RIG-I. We analyzed cell lysates harvested 36 h after transfection for interactions between RNF125 and WT or mutant RIG-I by immunoprecipitation (note that the expression level of 1–238 was very low; despite the low levels, however, the interaction with RIG-I was clearly observed). All cells were treated with MG132. (d) Ubiquitin conjugation stimulates RIG-I degradation in a proteasome-dependent manner. Plasmids encoding a FLAG-tagged version of the RIG-I N terminus, FLAG-RIG-I/1–238, HA-RNF125, and Myc-Ub were transfected into 293FT cells as indicated. After culturing with or without MG132, the levels of ubiquitination were analyzed by immunoprecipitation. (e) RNF125 stimulates RIG-I degradation. Plasmids encoding FLAG-RIG-I and Myc-Ub were cotransfected into 293FT cells with WT or a point mutant of HA-RNF125. Cells were harvested 36 h after transfection and were analyzed for proteins by Western blot. (f) RIG-I was degraded by RNF125 in a dose-dependent manner. The 293FT cells were transfected with a plasmid encoding FLAG-RIG-I (0.5 μg) and varying doses of a plasmid encoding HA-RNF125 (0, 0.5, 1, and 2 μg). Half of each cell aliquot was treated with MG132. The levels of both RIG-I mRNA and protein were examined in cells harvested 48 h after transfection. “RIG-I (+MG132)” indicates analysis of the RIG-I protein in cells treated with MG132. As a control, the cellular proteins p53 and tubulin and GAPDH mRNA analyses are shown. (g) RIG-I was degraded by ectopic and endogenous RNF125. A plasmid encoding FLAG-RIG-I with or without a plasmid encoding HA-RNF125 was transfected into 293FT cells. Thirty-six hours after transfection, the cells were treated with CHX (final concentration, 50 μg/ml), and were analyzed by Western blot (Upper). Plasmids encoding FLAG-RIG-I and RNA for si-control or siRNF125–3 were transfected into 293FT cells. Twenty-four hours after transfection, these cells were treated with Poly I:C for 12 h, followed by addition of CHX. Cells were harvested at the indicated times after addition of CHX and analyzed by Western blot (Lower).
Analysis of the E3-Ligase Domain of RNF125 and a Target Region in RIG-I for Ubiquitination.
To verify the role of the RNF125 RING domain in ubiquitin conjugation to RIG-I, we performed a mutational analysis. Although RIG-I could be conjugated to ubiquitin by WT RNF125 (Fig. 2a, lane 3), an RNF125 mutant [in which the 72nd and 75th cysteine residues were substituted with alanine (C72/75A)] was unable to mediate RIG-I ubiquitination (Fig. 2a, lane 4). A peptide encompassing the first 76 residues of RNF125 (1–76 aa), a region containing the intact RING domain, mediated ubiquitin conjugation similar to the WT RNF125. The mutant Cys-72 and Cys-75 in this peptide (1–76 aa C72/75A), however, completely abrogated its ubiquitin-conjugating activity (Fig. 2a, lane 6).
To verify the region of RIG-I conjugated to ubiquitin, we analyzed ubiquitin conjugation to FLAG-RIG-I mutants in cells coproducing HA-RNF125 and Myc-Ub (Fig. 2c). Peptides containing CARD domain of RIG-I, amino acids 1–238 (RIG-1/1–238) or 1–452 (RIG-I/1–452), were ubiquitinated at higher levels than intact RIG-I. In contrast, C-terminal RIG-I peptides, RIG-I/239–734 and RIG-I/735–925, were only weakly conjugated. Using the N-terminal region of RIG-I (RIG-I/1–238) as a substrate, we confirmed that RNF125, and not Efp (estrogen-responsive finger proteins), another ubiquitin E3 ligase, efficiently conjugated ubiquitin (data not shown), further supporting the conclusion that RNF125 acts as an E3 ligase for RIG-I.
Degradation of RIG-I Is Enhanced by the Expression of RNF125.
Polyubiquitin was primarily conjugated to the N-terminal CARD-containing region of RIG-I (Fig. 2c). Steady-state levels of the ubiquitin-conjugated peptide containing the N-terminal region, RIG-I/1–238, were increased in cells treated with MG132, a proteasome inhibitor (Fig. 2d). These results suggest that RNF125-dependent RIG-I ubiquitination precedes proteasome-dependent degradation. Full-length RIG-I was also degraded with ectopic expression of RNF125 but not with the mutant C72/75A in 293FT cells (Fig. 2e). The levels of RIG-I were reduced as increasing amounts of RNF125 were expressed (Fig. 2f). Under these conditions, mRNA levels of RIG-I, GAPDH, p53, and tubulin remained unchanged. When cells were treated with MG132, the degradation of RIG-I was suppressed. These results clearly demonstrate that RNF125 acts as a ubiquitin E3-ligase regulating the cellular levels of RIG-I through proteasomal degradation. Proteasomal degradation of RIG-I was also observed in other cell types such as HeLa and HepG2 (data not shown).
Degradation of RIG-I is enhanced by the presence of RNF125. The level of RIG-I was reduced after addition of cycloheximide (CHX), when RNF125 was ectopically expressed, but no such reduction in RIG-I was observed in cells without ectopic expression of RNF125 (Fig. 2g Upper). Such degradation of RIG-I in cells treated with poly I:C was not observed in cells expressing RNF125 specific siRNA, siRNA125–3, although substantial amount of RIG-I degradation in cells treated with control siRNA could be observed (Fig. 2g).
E2 Enzymes Involved in Ubiquitin Conjugation to RIG-I.
RNF125 interacts with several E2 enzymes (25), and one of these enzymes most likely acts as an E2 for ubiquitin conjugation to RIG-I. To identify the E2 acting upstream of RNF125 in RIG-I conjugation, we performed an in vitro assay examining ubiquitin conjugation to RIG-I [see supporting information (SI) Fig. 6]. We identified that UbcH1, UbcH5a, UbcH5b, and UbcH5c functioned as an E2 enzyme. The enzymes UbcH1 and UbcH5a-c conjugated ubiquitin to RNF125 and RIG-I via K48 (data not shown).
Further, by analyzing ubiquitin conjugation to RIG-I in 293FT cells by ectopic expression of the E2 enzymes, we observed only UbcH5c, and not other E2 enzymes, showing enhanced ubiquitin conjugation to RIG-I, suggesting that UbcH5c is the major E2 enzyme functioning in vivo (data not shown).
Expression of RNF125 Suppresses the Activation of IRF3.
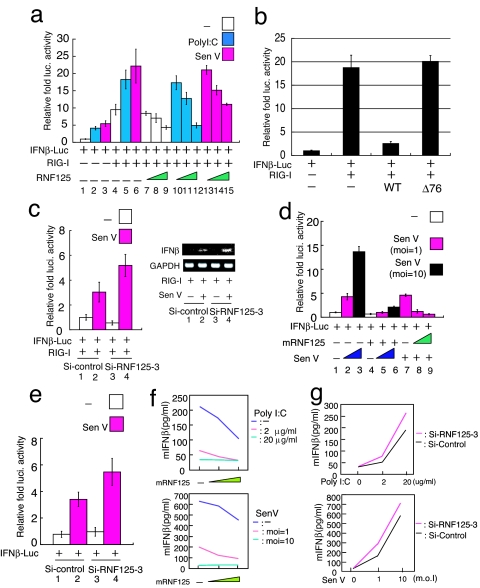
The ability of RNF125 to modulate RIG-I protein levels suggests that RNF125 may play an important role in the regulation of IRF3 activity. We analyzed this possibility by using a luciferase reporter gene driven by the IFNβ promoter, IFNβ-luc, to examine the effect of RNF125 on IRF3 activity after stimulation with poly I:C or Sendai virus infection (Fig. 3a). The 293FT cells expressing IFNβ-Luc, RIG-I, and RNF125 were treated with poly I:C or Sendai virus infection. We observed a marginal increase in luciferase activity in those cells lacking ectopic RIG-I and RNF125 expression (Fig. 3a, lanes 1–3). Luciferase activity was enhanced by the ectopic expression of RIG-I alone and further augmented by treatment with poly I:C or Sendai virus (Fig. 3a, lanes 4–6). Cells expressing RNF125, however, exhibited reduced reporter activity, and this decreased in a dose-dependent manner (Fig. 3a, lanes 7–15). The enhanced luciferase activity seen after polyIC treatment or Sendai virus infection was almost completely abrogated by high levels of RNF125 expression (Fig. 3a, lanes 12 and 15). Suppressive activity of IFNβ-luc was not observed by the mutant RNF125 that lacks the conjugation activity and ability to interact with RIG-I (Fig. 3b). Furthermore, we observed that ectopic RNF125 expression inhibited nuclear localization of IRF3 by poly I:C treatment (SI Fig. 7).
Fig. 3.
Suppression of RIG-I function by ubiquitination. (a) The RNF125 suppressed the IFNβ-driven luciferase activity, activated by RIG-I (at the left). The 293FT cells were transfected with plasmids encoding RIG-I (50 ng) and IFNβ-luc (50 ng) with varying amounts of a plasmid encoding RNF125 (10, 50, and 100 ng). Twenty-four hours after transfection, cells were treated with polyIC (blue) or infected with Sendai virus (pink). Cells were harvested 12 h after the treatment; luciferase activity in the lysates was then measured. The RNF125 suppressed the production of IFNβ mRNA. (b) The 293FT cells transiently expressing IFNβ-luc, RIG-I, and RNF125WT or Δ76 were analyzed for luciferase activity 36 h after transfection. (c) Effect of siRNF125–3, a small inhibitory RNA specific for RNF125 mRNA, on IFNβ-luc activity and IFNβ level. The 293FT cells were transfected with plasmids encoding IFNβ-luc and RIG-I and treated with control siRNA or siRNF125–3. An aliquot of cells was then infected with Sendai virus. Twenty-four hours after infection, luciferase activity (Left) and IFNβ mRNA levels, assessed by RT-PCR (Right), were measured. (d) Effect of mouse RNF125 on endogenous RIG-I signaling in primary MEFs. The MEFs were transfected with plasmids encoding mouse RNF125 (100 ng in lanes 4, 5, 6, and 8 or 200 ng in lane 9) and IFNβ-luc (50 ng) in combination as indicated. Twelve hours after transfection, cells were infected with Sendai virus at the indicated multiplicity of infection (MOI) or mock infected. Luciferase activity in cell lysates prepared 24 h after treatment was measured. White box, mock infected; pink box, Sendai virus infected with MOI 1; black box, Sendai virus with MOI of 10. (e) The MEFs were transfected with plasmids encoding IFNβ-luc and treated with control siRNA or siRNF125–3 (final concentration of 8 nM). Cells were then infected with Sendai virus at MOI of 1. Twenty-four hours after infection, luciferase activity was measured. (f) The level of IFNβ in culture medium in cells was decreased in an RNF125 dose-dependent manner. Cells transfected with different amounts of plasmids encoding RNF125 were treated with 0 (green), 2 (pink), and 20 (blue) μg/ml of poly I:C (Upper) and with 0 (green), 1 (pink), and 10 MOI (blue) of Sendai virus (Upper). Twelve hours after the treatment, IFNβ in the culture medium was measured by ELISA system. (g) The level of IFNβ was increased in MEFs treated with siRNA for RNF125-specific mRNA. Cells treated with control siRNA (black bars) or siRNA125–3 (pink bars) were treated with different doses of poly I:C or Sendai virus infection and were analyzed for IFNβ in culture medium.
We next examined the effect of endogenous RNF125 on IRF3 activation by knocking down RNF125 mRNA by using siRNA. The 293FT cells treated with siRNF125–3 were subsequently transfected with plasmids encoding IFNβ-luc and RIG-I. Cells were then infected with Sendai virus or mock infected. Sendai virus-induced luciferase activity was substantially greater in cells in which RNF125 expression was suppressed by siRNF125–3 (Fig. 3c Left, lanes 2 and 4). Additionally, endogenous IFNβ mRNA production was also increased in those cells (Fig. 3c Right).
To examine whether the ability of RNF125 to suppress IRF3 activation in 293FT cells was physiologically relevant, an experiment was performed by using primary mouse embryonic fibroblasts (MEFs). RIG-I signaling is essential for virus-induced cytokine secretion in conventional dendritic cells (DCs) and MEFs (35). The MEFs transfected with plasmids encoding mouse RNF125 (mRNF125) and IFNβ-luc were infected with Sendai virus or mock infected. Induced induction of luciferase activity was directly correlated with the degree of Sendai virus infection (Fig. 3d). However, the induced luciferase activity was strongly suppressed by the ectopic expression of mRNF125 (Fig. 3d, lanes 7–9). Furthermore, transfection of MEFs with siRNF125–3 led to reduced mRNF125, mRNA, and protein levels (data not shown) and somewhat enhanced luciferase production after Sendai virus infection (Fig. 3e).
We further confirmed the suppression of RIG-I signaling by RNF125 by examining the release of IFNβ in the culture medium. In cells treated with poly I:C as well as infected with Sendai virus, IFNβ released was significantly reduced by the expression of RNF125 in a dose-dependent manner (Fig. 3f). Furthermore, MEFs treated with si-RNF125–3 showed substantial increase of IFNβ release in the culture medium upon polyI:C and Sendai virus infection (Fig. 3g).
RNF125, UbcH5, and RIG-I Are Up-Regulated by IFN-α and Poly I:C.
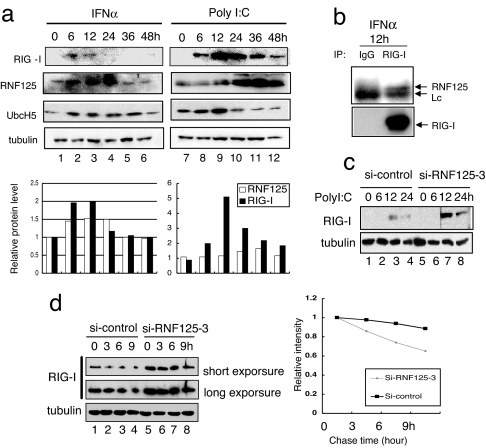
The production of proteins related to Ubl conjugation is often regulated by IFNs (36). Thus, the modification of RIG-I by ubiquitin is most likely regulated by the coordinated induction of Ubl-conjugating proteins. We analyzed the levels of RIG-I, RNF125, and UbcH5 in HepG2 cells at various times after treatment with IFNα and poly I:C (Fig. 4a). Low levels of RNF125 were present before IFNα and Poly I:C treatment, but its expression peaked 12–36 h after induction. Although RIG-I was barely detectable at the time of induction, it became detectable at 6 h and peaked at 12 h. The decreases in RIG-I expression correlated temporally with the increases in RNF125 and UbcH5 (Fig. 4a), suggesting that RNF125 decreased the levels of RIG-I after IFN induction. Quantitative analysis indicated that RIG-I level showed reverse correlation of RNF125 (Fig. 4a, graph). The fact that constitutive association of endogenous RIG-I and RNF125 (Fig. 4b) supports the possibility that RNF125 conjugates ubiquitin to RIG-I to promote proteasomal degradation. We confirmed this hypothesis by treating cells with RNF125 siRNA. In cells treated with control siRNA, RIG-I expression increased at 12 and 24 h after poly I:C treatment, but, in cells treated with siRNF125–3, RIG-I expression was higher, and it sustained longer than treated control cells (Fig. 4c, compare lanes 3 and 7 and 4 and 8). To measure a half-life of RIG-I, we treated HepG2 cells expressing siRNA specific to RNF125 or control siRNA with IFNα for 12 h and then added CHX. Cells harvested at the indicated time after CHX treatment were analyzed for RIG-I by Western blot and quantified the intensity of the bands (Fig. 4d). Calculated half-life of RIG-I in cells treated with siRNA specific to RNF125 was 16 h and 8 h in cells treated with control siRNA.
Fig. 4.
Induction of RIG-I and the Ubls by IFNα or poly I:C treatment. (a) HepG2 cells were harvested at the indicated times after treatment with IFNα (103 units/ml) or Poly I:C (2 μg/ml) and were analyzed for protein levels by Western blot. As a control, the level of tubulin is shown. The protein level of each band is quantified and graphed. (b) Endogenous association of RIG-I and RNF125. Jurkat cells were incubated with IFNα (1,000 units/ml) and MG132 (final 10 μM). After 12 h, whole-cell lysates were subjected to immunoprecipitation assay using anti-RIG-I antibody or control antibody, followed by immunoblotting for detection of RNF125 and RIG-I. (c) Influence of siRNA specific to RNF125 for RIG-I levels. Cells treated with siRNA were further treated with poly I:C and then harvested at the indicated times after poly I:C treatment. The lysates were analyzed for the levels of endogenous RIG-I. Tubulin acts as a control. (d) HepG2 cells treated with siRNA were further treated with IFNα for 12 h and then harvested at the indicated times after CHX treatment. The lysates were analyzed for the levels of endogenous RIG-I. Tubulin acts as a control. The intensity of each band is quantified and graphed.
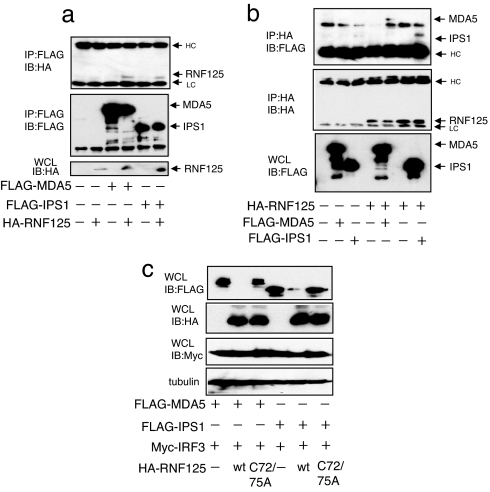
RNF125 Exhibits Ubiquitin-Conjugating E3-Ligase Activity for MDA5 and IPS1.
MDA5 is a member of the RIG-I protein family and contains CARD domain. The IPS1, a CARD-containing protein, downstream of RIG-I signaling interacts with RIG-I through its CARD domain. These proteins are associated with RNF125 (Fig. 5 a and b). The concentrations of MDA5 and IPS1 were observed to reduce in cells expressing RNF125 ectopically (Fig. 5c). Furthermore, we noticed that these proteins were ubiquitin-conjugated by RNF125, and not by the RNF125 mutant, although the levels of conjugation differed between the substrates (SI Fig. 8 a and b). Suppression of conjugation of ubiquitin to these proteins was observed by treatment with siRNA specific to RNF125 (SI Fig. 8 a and b Right). Bringing all these data together, RNF125 serves as a ubiquitin E3-ligase for MDA5 and IPS1 as well as for RIG-I.
Fig. 5.
Association of RNF125 with MDA5 and IPS-1. A plasmid encoding FLAG-MDA5 or FLAG-IPS1 was transfected into 293FT cells in the presence or absence of a plasmid encoding HA-RNF125. (a and b) Protein–protein interactions were monitored by coimmunoprecipitation using the anti-FLAG antibody, followed by Western blot with the anti-HA antibody (a) and coimmunoprecipitation using the anti-HA antibody, followed by Western blot with the anti-FLAG antibody (b). All cells were grown in medium containing MG132. (c) MDA5 and IPS1 were also degraded by RNF125. A plasmid encoding Myc-IRF3 and FLAG-MDA5 or FLAG-IPS1 were transfected into 293FT cells with or without a plasmid encoding HA-RNF125 or HA-RNF125 C72/75A. Thirty-six hours after transfection, cells were harvested and were analyzed for protein levels by Western blot.
Analyzing a candidate E2 enzyme for ubiquitin conjugation to MDA5 and IPS1 by RNF125 in an in vitro assay, we observed that UbcH1 and UbcH5a-c were also involved in the process (SI Fig. 8c).
Signaling from MDA5 as Well as from IPS1 Was Suppressed by RNF125 Ubiquitin Conjugation-Dependent Manner.
Effect of conjugation of ubiquitin to MDA5 and IPS1 by RNF125 on their downstream signaling was analyzed by luciferase assay. Cells were transfected with plasmids expressing IFNβ-luc, MDA5, or IPS1 together with that expressing RNF125 or its mutant. The luciferase activities in cells expressing MDA5 and IPS1 were severely impaired by the expression of RNF125, and not by Δ76 (SI Fig. 9 a and b). Alternatively, those cells treated with siRNF125–3 showed increased activity of luciferase in the cells bearing both MDA5 and IPS1 (SI Fig. 9 a and b). Luciferase activity driven by p53-binding promoter was not affected in cells expressing ectopic p53 and RNF125 (SI Fig. 9c).
Discussion
We demonstrated that RIG-I is ubiquitinated before its proteasome-dependent degradation. The RNF125 functioned as an E3-ligase for this conjugation process, because ectopic expression of RNF125 enhanced (Fig. 2a) and suppression of endogenous RNF125 by siRNA severely impaired the ubiquitin conjugation of RIG-I (Fig. 2b). The RNF125, a RING finger-containing protein (25), also known as TRAC-1, exerts a positive regulatory role on T cell activation. Expression of RNF125/TRAC-1 is higher in lymphoid tissues but is also expressed in a variety of other tissues. The role of RNF125 described here appears to be distinct from that previously reported.
It can be observed that the N-terminal region of RIG-I, the region containing the CARD domain, was conjugated to ubiquitin, by minimal incorporation of ubiquitin to the C-terminal half of RIG-I (Fig. 2c). Conjugation was enhanced after the deletion of the C-terminal portion of RIG-I, which contained the helicase domain, a result that is consistent with the earlier postulation that the C terminus of RIG-I may mask the function of the N-terminal region (9).
The MDA5, an RIG-I family member, and IPS1, the downstream signaling proteins that interact with RIG-I via the RIG-I CARD, interacted with RNF125 and were also ubiquitin conjugated (Fig. 5 a and b and SI Fig. 8 a and b). These results suggest that RNF125 is a ubiquitin E3-ligase with activity against proteins containing CARD domains. These two ubiquitin-conjugated proteins also underwent proteasomal degradation (Fig. 5c). We noticed that MDA5 was ubiquitin-conjugated less than RIG-I by RNF125, but the level of this proteins decreased substantially (Fig. 5c). We do not know the reason for this, but there may be the possibility that a small amount of ubiquitination to MDA5 may be sufficient to trigger the degradation or that there is a pathway other than ubiquitin-dependent proteasomal degradation for MDA5 degradation. These observations suggest that ubiquitination of RIG-I/MDA5 and IPS1 inhibits RIG-I signaling by shunting these proteins toward proteasomal degradation. Given that the production of cytokines affects a wide variety of responses in innate and acquired immunity, activation of RIG-I signaling by viral pathogens is quite likely to be strictly controlled. Suppression of endogenous RNF125 levels by siRNA enhanced luciferase activity driven by the IFNβ promoter and up-regulated IFNβ expression (Fig. 3c), which clearly demonstrates that RNF125 functions as a negative regulator of RIG-I signaling. Importantly, the negative regulatory function of RNF125 is inversely related to IFN signaling in cells, a factor most likely contributing to the down-regulation of IFN levels as the viral infection resolves.
The suppression of IRF3 signaling by RNF125 was not limited to cultured cell lines, but was also seen in primary MEFs (Fig. 3d), in which RIG-I is the primary factor activating IRF3 during RNA virus infection (35). These results clearly indicate that RNF125 plays an important role in suppressing RIG-I signaling in physiologically immunocompetent cells.
The IFNα and Poly I:C treatment transiently up-regulated RIG-I levels, which peaked at 12 h after the treatment of HepG2 cells (Fig. 4a). Reductions in RIG-I expression correlated with the induction of RNF125 and UbcH5, suggesting that these proteins may enhance the degradation of RIG-I by ubiquitin conjugation. This result is found to be consistent with the in vitro experiments (Fig. 2 a and b).
Materials and Methods
Cell Culture, Transfection, and Luciferase Reporter Assays.
For details on cell culture, transfection, and luciferase reporter assays, see SI Methods.
Yeast Two-Hybrid Screening.
Yeast two-hybrid screening was performed as described in SI Methods.
Construction of cDNA Expression Plasmids.
Expression constructs for RIG-I, MDA5, and IPS-1 have been described (9). Plasmids expressing WT and mutant ubiquitin were obtained from K. Nakayama (Division of Cell Biology, Medical Institute of Bioregulation, Kyushu University, Fukuoka, Kyushu, Japan). WT RNF125 (amino acid 1–232) and mouse RNF125 were cloned into the pCAG vector. To generate point mutants, alanine was substituted for the targeted residues by PCR. The N- and C-terminal truncation mutants (RNF125 1–76 and RNF125Δ76) were generated by standard PCR. The RNF125 and RIG-I were cloned into the pGEX-6P-1 vector (Amersham, Piscataway, NJ) in-frame with an N-terminal GST.
siRNA and Measurement of mRNA.
siRNA duplex sequences (siRNF125–3, 5′-CCGUGUGCCUUGAGGUGUU-3′) were custom synthesized by Proligo (Boulder, CO). A control nucleotide, si-control, was purchased from Dharmacon (Lafayette, CO) (nonspecific control duplex IV). mRNA was measured by RT-PCR.
Western Blotting and Immunoprecipitation.
Western blotting and immunoprecipitation were done as reported in SI Methods.
ELISA.
For details on ELISA, see SI Methods.
Antibodies and Reagents.
Antibodies to FLAG (anti-Flag; M2), HA (12CA5) and HA (3F10) were purchased from Sigma (St. Louis, MO) and Roche (Indianapolis, IN), respectively. Anti-Myc (9E10), anti-IRF3 (FL425) antibody, and anti-ubiquitin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-UbcH5, which reacts with UbcH5a-c, and anti-α tubulin were acquired from Chemicon (Temecula, CA) and Oncogene Research Products (San Diego, CA), respectively. Anti-GST antibodies were purchased from Amersham. Anti-RNF125 polyclonal antibodies were generated in rabbits by using the RNF125 peptide from 215 to 232 aa. Anti-RIG-I monoclonal antibody was described (9). Anti-RIG-I polyclonal antibodies were generated in rabbits by using the RIG-I recombinant protein from 1 to 238 aa. CHX was purchased from Nacalai Tesque (Kyoto, Japan). Poly (I:C) and MG132 were purchased from Amersham and Peptide Institute (Osaka, Japan), respectively.
The GenBank accession number for mRNF125 is AB259692.
Supplementary Material
Acknowledgments
We thank Dr. K. I. Nakayama for providing the plasmids expressing the ubiquitin mutants. This work was supported by grants-in-aid for cancer research and for the second-term comprehensive 10-year strategy for cancer control from the Ministry of Health, Labour and Welfare as well as by Grant-in-Aid for Scientific Research on Priority Areas “Integrative Research Toward the Conquest of Cancer” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations
- CARD
caspase-recruiting domain
- CHX
cycloheximide
- MEF
mouse embryonic fibroblast
- TLR
Toll-like receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611551104/DC1.
References
- 1.Iwasaki A, Medzhitov R. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 2.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Akira S. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R, Janeway CA., Jr Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 5.Janeway CA, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 6.Akira S, Takeda K, Kaisho T. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 7.Takeda K, Kaisho T, Akira S. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 8.Le Bon A, Tough DF. Curr Opin Immunol. 2002;14:432–436. doi: 10.1016/s0952-7915(02)00354-0. [DOI] [PubMed] [Google Scholar]
- 9.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 10.Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 12.Seth RB, Sun L, Ea CK, Chen ZJ. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 14.Malakhov MP, Kim KI, Malakhova OA, Jacobs BS, Borden EC, Zhang DE. J Biol Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz DC, Hochstrasser M. Trends Biochem Sci. 2003;28:321–328. doi: 10.1016/S0968-0004(03)00113-0. [DOI] [PubMed] [Google Scholar]
- 16.Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. Proc Natl Acad Sci USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M., Jr J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Proc Natl Acad Sci USA. 2004;101:17705–17710. doi: 10.1073/pnas.0407974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sweet MJ, Leung BP, Kang D, Sogaard M, Schulz K, Trajkovic V, Campbell CC, Xu D, Liew FY. J Immunol. 2001;166:6633–6639. doi: 10.4049/jimmunol.166.11.6633. [DOI] [PubMed] [Google Scholar]
- 20.Chuang TH, Ulevitch RJ. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr, Medzhitov R, Flavell RA. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa R, Naka T, Tsutsui H, Fujimoto M, Kimura A, Abe T, Seki E, Sato S, Takeuchi O, Takeda K, et al. Immunity. 2002;17:677–687. doi: 10.1016/s1074-7613(02)00449-1. [DOI] [PubMed] [Google Scholar]
- 23.Wang YY, Li L, Han KJ, Zhai Z, Shu HB. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 24.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, Li CC, Pardo J, Chu PC, Liao CX, Huang J, Dong JG, Zhou X, Huang Q, Huang B, et al. J Immunol. 2005;174:5288–5297. doi: 10.4049/jimmunol.174.9.5288. [DOI] [PubMed] [Google Scholar]
- 26.Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Proc Natl Acad Sci USA. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar S, Kao WH, Howley PM. J Biol Chem. 1997;272:13548–13554. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- 28.Chin LS, Vavalle JP, Li L. J Biol Chem. 2002;277:35071–35079. doi: 10.1074/jbc.M203300200. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K, Suzuki T, Chiba T, Shimura H, Hattori N, Mizuno Y. J Mol Med. 2001;79:482–494. doi: 10.1007/s001090100242. [DOI] [PubMed] [Google Scholar]
- 30.Niwa J, Ishigaki S, Doyu M, Suzuki T, Tanaka K, Sobue G. Biochem Biophys Res Commun. 2001;281:706–713. doi: 10.1006/bbrc.2001.4414. [DOI] [PubMed] [Google Scholar]
- 31.Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. Nature. 2002;417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 32.Moynihan TP, Ardley HC, Nuber U, Rose SA, Jones PF, Markham AF, Scheffner M, Robinson PA. J Biol Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Proc Natl Acad Sci USA. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. Mol Cell Biol. 2002;22:5222–5234. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Dastur A, Beaudenon S, Kelley M, Krug RM, Huibregtse JM. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.