Abstract
Global, age-dependent changes in gene expression from rodent models of inherited ALS caused by dominant mutations in superoxide-dismutase 1 (SOD1) were identified by using gene arrays and RNAs isolated from purified embryonic and adult motor neurons. Comparison of embryonic motor neurons expressing a dismutase active ALS-linked mutant SOD1 with those expressing comparable levels of wild-type SOD1 revealed the absence of mutant-induced mRNA changes. An age-dependent mRNA change that developed presymptomatically in adult motor neurons collected by laser microdissection from mice expressing dismutase active ALS-linked mutants was dysregulation of the d/l-serine biosynthetic pathway, previously linked to both excitotoxic and neurotrophic effects. An unexpected dysregulation common to motor neurons expressing either dismutase active or inactive mutants was induction of neuronally derived components of the classic complement system and the regenerative/injury response. Alteration of these mutant SOD1-induced pathways identified a set of targets for therapies for inherited ALS.
Keywords: amyotrophic lateral sclerosis, gene expression profile, laser microdissection
Amyotrophic lateral sclerosis (ALS) is a fatal, adult-onset neurodegenerative disease that selectively kills brain and spinal cord motor neurons (reviewed in ref. 1). Although most ALS cases are sporadic and of unknown origin, some familial instances are caused by acquired toxic properties of dominant missense point mutations in the ubiquitously expressed superoxide dismutase 1 (SOD1) independent of its dismutase activity (2, 3). Because both familial and sporadic forms lead to what is nearly indistinguishable diseases, the analysis of mutant (human or mouse) SOD1-overexpressing rodent ALS models, which develop a fatal, late-onset, progressive ALS-like paralysis (4–7), can provide valuable tools for dissecting the overall ALS disease mechanism.
The central question in understanding ALS in mutant SOD1-expressing mouse and rat models is what are the slowly acting toxic mechanisms, or build-up of toxic products that are responsible for ultimately leading to the degeneration of the most at-risk cell type, the large spinal cord motor neurons (1). Such properties of mutant SOD1 seem likely to induce responses in the genes expressed by motor neurons (their transcriptome) at early disease stages or even before obvious clinical symptoms. Identifying the manner in which motor neurons respond to, or are affected by, ALS-linked mutant SOD1-induced damage could provide key insights either into primary mechanisms of toxicity or to potential therapeutic approaches that may be successful in slowing disease progression in ALS.
High-density oligonucleotide microarrays provide an excellent tool to test on a genome wide level the effects of candidate events on the cellular expression profile. Because certain aspects of ALS are non-cell autonomous and originate from mutant SOD1 accumulation in non-motor neuronal cells (8, 9), cell-type specificity is of high importance when using such an approach. To date, three main gene-expression profiling studies have been reported (10–12), all of which were restricted to analysis of the adult disease phase in one SOD1 mouse line that accumulates a dismutase active mutant to extraordinarily high levels during disease progression [between 10- and 20-fold over the already abundant endogenous mouse SOD1 (4, 13)]. Two studies (10, 11) used the entire spinal cord, thus losing cell-type specificity and masking mRNA changes within motor neurons as a consequence of the much more numerous glial cells and their activation before or during disease progression. A more recent effort (12) used profiling by laser microdissection of motor neurons. The major outcome of this study was an elevation of vimentin beginning in the presymptomatic phase. No comparison was undertaken to evaluate changes due simply to accumulation of high levels of SOD1 protein and activity (e.g., by parallel analysis of similar motor neurons from mice expressing high levels of human wild-type SOD1). In any case, no insights on dysregulated candidates common to SOD1 mutants of divergent biochemical characters can be identified from these earlier efforts.
Using both purified embryonic and adult motor neurons collected by laser microdissection, in combination with Affymetrix GeneChips, (Affymetrix, Inc., Santa Clara, CA) gene dysregulations that arise within the at-risk motor neurons before the overall appearance of clinical symptoms are now identified by the following: (i) analysis of expression profiles from embryonic motor neurons of rats expressing high levels of mutant SOD1G93A as compared with wild-type overexpressing SOD1 controls that do not develop disease; and (ii) analysis of expression profiles at adult stages before appearance of overall clinical symptoms within motor neurons isolated from mice expressing dismutase active (SOD1G37R) or inactive (SOD1G85R) SOD1 mutants, as compared with motor neurons isolated from mice expressing high levels of wild-type SOD1 or normal motor neurons. Two molecular pathways are identified whose dysregulation is common to dismutase active and inactive mutants.
Results
Absence of Mutant SOD1-induced mRNA Dysregulations in Embryonic Motor Neurons.
To test with a global approach whether at embryonic stages mutant SOD1-induced changes were already present in vivo, whole transcriptome gene expression profiling (Affymetrix Rat 230v2.0 GeneChip; 30,000 transcripts) was performed on highly pure spinal cord motor neurons from embryonic day (E) 14 rats expressing similar levels of either mutant (SOD1G93A) or wild-type (SOD1WT) human SOD1 (Fig. 1 and Tables 1 and 2). Independent preparations of motor neurons from four mutant SOD1G93A and three wild-type SOD1WT preparations (see Materials and Methods) were directly isolated without any in vitro culturing and then compared.
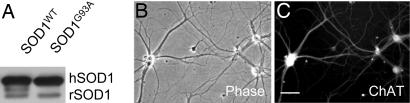
Fig. 1.
Purified embryonic day 14 (E14) rat spinal cord motor neurons used for gene expression profiling. (A) Immunoblot of spinal cord extracts from transgenic mutant (SOD1G93A) and wild-type (SOD1WT) human SOD1 (hSOD1) rats (rSOD1; endogenous rat SOD1). (B and C) Purified embryonic (E14) motor neurons were cultured for 7 days in vitro and stained for choline acetyltransferase (ChAT) to test the viability and purity of the isolated motor neurons. Gene expression profiling was performed on noncultured, freshly isolated embryonic (E14) motor neurons from mutant SOD1G93A and wild-type SOD1wt rats (see Tables 1 and 2). [Scale bar: 20 μm (B and C).]
Table 1.
Purity of noncultured, freshly isolated embryonic (E14) rat motor neurons
| Gene | Expression | Specificity |
|---|---|---|
| Cd11b | Absent | Microglia |
| Iba1 | Absent | Microglia |
| EAAT2 | Absent | Astrocytes |
| EAAT3 | Present | Neurons |
| p75 | Present | Motor Neurons |
| lsl1 | Present | Motor Neurons |
Assessment of motor neuron purity by testing (using microarrays) for the expression (Present) or absence of expression (Absent) of glial and neuronal marker genes.
Table 2.
Gene dysregulations comparing embryonic (E14) motor neurons from mutant SODG93A and wild-type SOD1WT rats.
| ProbeSet | Gene name | Symbol | Fold change | Function |
|---|---|---|---|---|
| 1385969_at | A disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 16 | Adamts16 | 3.1 | Metalloprotease |
| 1368192_at | Chemokine (C-X-C motif) receptor 3 | Cxcr3 | 2.7 | Chemokine receptor |
| 1396014_at | Transcribed locus, similar to Tim8 A (deafness dystonia protein 1 homolog) | LOC680794 | 2.3 | Unknown |
| 1377705_at | Transcribed locus, similar to Ab2-143 (Rattus norvegicus) | BF549971 | 2.2 | Unknown |
| 1389901_at | RNA binding motif protein 8 | Rbm8 | 2.0 | mRNA processing |
| 1382005_at | Transcribed locus, weakly similar to NP_598738.1 transferrin (mouse) | BI280285 | −1.6 | Unknown |
| 1379398_at | Family with sequence similarity 31, member B | Fam31b | −2.0 | Unknown |
| 1395574_at | Transcribed locus | BE118358 | −2.1 | Unknown |
| 1382998_at | RNA (guanine-7-) methyltransferase | Rnmt | −2.2 | mRNA Processing |
| 1390777_at | Sterol-C5-desaturase (fungal ERG3, delta-5-desaturase) homolog (yeast) | Sc5d | −2.4 | Fatty-acid metabolism |
| 1393271_at | Indian hedgehog homolog (Drosophila) | Ihh | −2.9 | Cell-cell signaling |
| 1381407_at | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 (mitochondrial) | Ndufc2 | −3.2 | Oxidative phosphorylation |
Mutant SOD1-induced gene dysregulations (average fold changes) comparing (noncultured, freshly isolated) purified preparations of mutant SOD1G93A (n = 4) and wild-type SOD1WT (n = 3) expressing embryonic motor neurons.
Surprisingly few significant mRNA changes (using high stringency: minimal lowest fold change ≥1.50) were present despite ≈50% of the 30,000 tested transcripts expressed above background levels. Only 12 genes were dysregulated (Table 2). Among them, transcripts up-regulated in mutant SOD1 motor neurons were Cxcr3 (a chemokine receptor), Adamts16 (metallopeptidase) and Rbm8 (RNA binding protein; mRNA processing). Transcripts down-regulated in mutant SOD1 motor neurons were Ndufc2 (mitochondrial NADH dehydrogenase component), Rnmt (mRNA CAP complex; mRNA processing), and Sc5d (sterol-desaturase; fatty-acid metabolism). The chemokine receptor Cxcr3 would be expected to be expressed on microglial rather than neuronal cells (14). However, the high purity of the isolated motor neurons and the absence of several markers for potential microglial contaminants (including CD11b and Iba1) (Table 1) strongly suggested this Cxcr3 induction to reflect a surprising mutant-dependent neuronal mRNA change.
Previous studies had suggested that after culturing in vitro early embryonic motor neurons from ALS model mice were already adversely affected by mutant SOD1 expression. Such changes included altered excitability and altered AMPA receptor responses and an increased sensitivity of the Fas cell-death pathway to exogenous activation (15–18). None of these correlated with mutant SOD1-induced transcriptional alterations in embryonic motor neurons in vivo. There were no mutant-dependent gene dysregulations among the components of the Fas cell-death pathway (including FasL, Fas, Fadd Daxx, p38, nNOS, and Casp8) (Table 2), demonstrating that mutant-dependent increased sensitivity of this network (18) is not based on mRNA dysregulations or requires additional aging/stressing to be revealed.
Major, Age-Dependent Gene Dysregulations in Adult Motor Neurons Long Before Appearance of Clinical Symptoms.
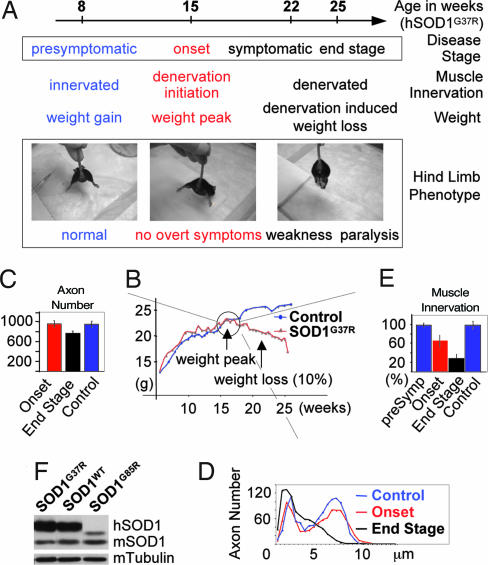
To find gene dysregulations in adult motor neurons of mutant SOD1-expressing mice before the appearance of overall clinical symptoms, we focused initially on the group of dismutase active SOD1s. We used laser microdissection to isolate motor neurons from mice expressing comparable levels of either mutant SOD1G37R or wild-type SOD1WT (Fig. 2). Two presymptomatic time points were chosen (8 weeks and 15 weeks of age), both long before the appearance of overt clinical symptoms (hind limb weakness appears at ≈22 weeks and paralysis at ≈25 weeks) (Fig. 2A). Animal weight has been demonstrated (19) to be a simple, reliable measure for disease “onset” and progression in these mice, with continuing weight gain in the presymptomatic phase (up to 15 weeks) and denervation-induced weight loss in the symptomatic phase (starting at 22 weeks) (Fig. 2B). The inflection point of the weight curve provides a simple definition of disease onset, although it is before obvious symptoms (normal hind limb spread reflex, fully active moving behavior) and with normal number and bimodal caliber distribution of lumbar L5 motor axons (Fig. 2 A, C, and D). Denervation has already initiated at this point (seen, for example, in the gastrocnemius hind limb muscle; Fig. 2E). Continued denervation induced muscle atrophy is reflected in the symptomatic phase by near complete loss of neuromuscular junctions, axonal degeneration, weight loss, hind limb weakness and paralysis (Fig. 2 D and E).
Fig. 2.
Characteristics of the different disease stages in mutant SOD1 mice. (A) Definition of disease stages in mutant SOD1 transgenic ALS mice used for the gene expression profiling of adult laser microdissected mouse spinal cord motor neurons. (B) Typical weight curves of mutant SOD1G37R and control mice. Disease onset is defined as the start of denervation-induced weight loss. (C–E) Progression of motor axonal degeneration [in L5-ventral roots; C (axon number) and D (axonal caliber distribution)] and hind limb muscle (gastrocnemius) denervation (E) in mutant SOD1G37R mice (compared with control, wild-type SOD1WT mice; n = 3 for each line and time point). (F) Levels of transgenic human SOD1 (hSOD1) in the mutant SOD1G37R, SOD1G85R, and wild-type SOD1WT mouse lines determined by immunoblotting.
From each spinal cord, laser microdissection was used to isolate ≈3,000 motor neurons from the lumbar L4–L6 region, the area most affected in ALS mice (Fig. 3). Two-round linear amplification of total RNA was hybridized to Affymetrix GeneChips (Mouse MOE430A/B GeneChip-Set; 39,000 transcripts). RT-PCR of the isolated RNA revealed an ≈50-fold enrichment of motor neuron mRNAs versus whole spinal cord (Fig. 3G). Because measurement (Bioquant Software) determined that motor neurons in a lumbar spinal cord cross section occupy ≈2% of the total area, this ratio established that essentially all of the RNA was derived from motor neurons.
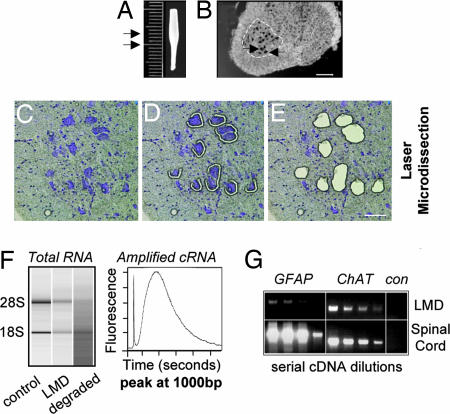
Fig. 3.
Leica laser microdissection-assisted gene expression profiling of adult spinal cord motor neurons from mutant SOD1 mice. (A) L4–L6 lumbar spinal cord region (3 mm) containing the most at-risk motor neurons (arrows) in ALS-like disease in mice. (B) Ethanol-fixed and Nissl-stained 20-μm-thick cryosection containing large (≥25 μm diameter) ventral horn motor neurons (arrows). (C–E) Specificity of laser microdissection. (F) Motor neuron RNA quality after laser microdissection (LMD) as measured with a Bioanalyzer. Total RNA still shows intact 28S and 18S rRNA bands with RNA from fresh cryosections as positive and degraded RNA as negative controls. Size distribution of the amplified cRNA pool (after two-round linear amplification). (G) Purity of laser microdissected (LMD) motor neurons assessed by comparison with signals from total lumbar spinal cord using RT-PCR for a motor neuron marker (ChAT) and an astrocyte marker (GFAP) (con, negative control without reverse transcriptase). [Scale bars: 150 μm (B) and 50 μm (E).]
For the 8- and 15-week time points, both of which are before the appearance of overall symptoms at 22 weeks, we used biological replicate samples of three or four mice per mutant SOD1G37R and three mice per wild-type SOD1WT line (one mouse per MOE430A/B GeneChip-Set; only females were used). Analysis of the motor neuron expression profiles was performed with GCOS v1.2 (Affymetrix) in combination with dChip v1.3 (20). All samples showed an average of ≈50% of all of the 39,000 transcripts to be present (above background level). At the very early presymptomatic 8-week time point, only seven genes were dysregulated (using high stringency: minimal lowest fold change ≥1.50); transcripts up-regulated in mutant SOD1 motor neurons were: Phgdh (d/l-serine-biosynthesis) and Hspa1a (heat-shock protein 1A); whereas transcripts down-regulated in mutant SOD1 motor neurons were: Tnfrsf1b (TNF receptor 2), Hbb (hemoglobin beta-chain), Tmem10 (membrane protein), Dbp (transcription factor; involved in detoxification) and an expressed sequence tag (EST) (EST 1445268_at) [Table 3 and supporting information (SI) Tables 4 and 5].
Table 3.
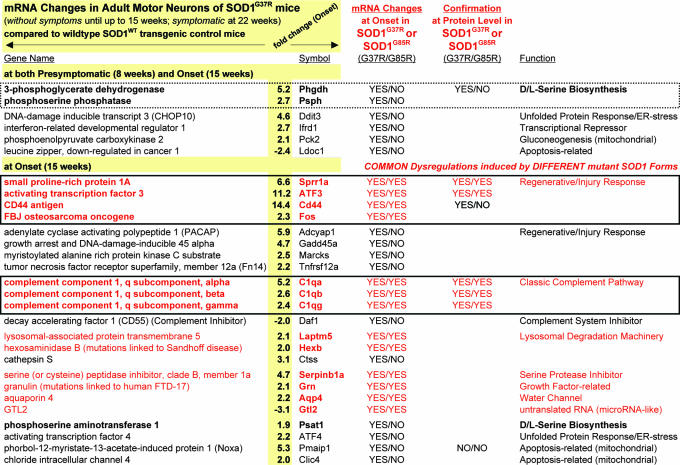
Gene dysregulations appearing before the appearance of overt symptoms and induced by dismutase active and inactive SOD1 mutants within motor neurons of SOD1G37R and SOD1G85R mice
Shown are the 29 most promising candidate genes that were dysregulated in mutant SOD1G37R mice by the 15-week “onset” time point (average fold-changes indicated for onset; for the full list, see SI Tables 4 and 5). Candidates that were also dysregulated in motor neurons of SOD1G85R mutants are indicated in the fourth column and marked in red (for the full list, see SI Tables 4–6). The d/l-serine biosynthesis pathway (highlighted with a dotted box) was early-on induced in dismutase-active SOD1G37R mutants, whereas candidate genes of the neuronal regenerative/injury response and the classic complement pathway (highlighted with solid boxes) were induced in both SOD1G37R and SOD1G85R mice.
However, during the presymptomatic phase between 8 and 15 weeks, a major motor neuron response resulted in the dysregulation by ≥1.50-fold of ≈108 genes by the 15-week onset time point (using high stringency; minimal lowest fold change ≥1.50). Eighty one genes were up-regulated and 27 genes were down-regulated in mutant SOD1 motor neurons (Table 3 and SI Tables 4 and 5). Of these 108 genes, the most promising candidates could be grouped in the following eight functional groups: d/l-serine-biosynthesis (Phgdh, Phsph, Psat), neuronal regenerative/injury response (Sprr1a, CD44, ATF3, Adcyap1, Fos, Fn14, GAP-43, Gadd45a, Marcks), unfolded protein-response/ER-stress (ATF4, Ddit3/CHOP10), neuronal cell-cycle reentry (Ccnd1), lysosomal degradation machinery (Ctss, Laptm5, Hexb), apoptosis-related (Noxa, Nupr1, Clic4, Ldoc1), mitochondrion-associated (Pck2, Noxa, Clic4) and the complement system (C1qa, C1qb, C1qc, Daf1). All of these genes were up-regulated in mutant SOD1 motor neurons, except two (Ldoc1 and Daf1) (Table 3 and SI Tables 4 and 5).
Comparing these two (symptom-free) time points revealed that two of the 7 genes dysregulated already at 8 weeks were increasingly (and in the same direction) also dysregulated at 15 weeks: Phgdh (d/l-serine-biosynthesis) was up-regulated (2.9- to 5.2-fold) and a gene previously identified only as an EST (EST-1445268_at) was down-regulated (1.9- to 3.2-fold). Seven of the 108 that were dysregulated at 15 weeks (fold changes between 1.8 and 5.2) were already dysregulated in the same direction, but less strongly at 8 weeks (using low stringency; minimal average fold change ≥1.5). These genes included Psph (d/l-serine-biosynthesis), Ddit3/CHOP10(unfolded protein response/ER-stress), Pck2(mitochondrial, gluconeogenesis), Ldoc1 (apoptosis-related), Ifrd1(transcriptional repressor), Ddr2 and Chac1 (all up-regulated, except Ldoc1) (Table 3 and SI Tables 4 and 5). None of these genes dysregulated in the SOD1G37R mice were overlapping with dysregulated genes found in mutant SOD1-expressing E14 embryonic motor neurons (Table 2 and SI Tables 4 and 5).
Dysregulation of the d/l-Serine Biosynthetic Pathway.
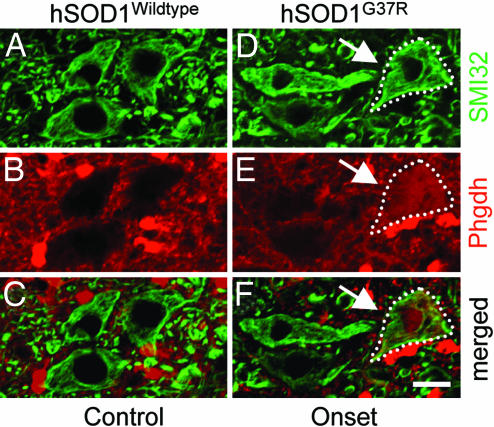
Inspection of the 113 genes dysregulated by 15 weeks revealed the up-regulation in motor neurons of a pair of mRNAs encoding products functioning in the d/l-serine biosynthetic pathway (21). By 8 weeks both Phgdh (3-Phosphoglycerate dehydrogenase) and Psph (Phosphoserine phosphatase) were induced (by 2.9- and 1.7-fold, respectively) in mutant SOD1 motor neurons. By 15 weeks, induction of both (Phgdh and Psph) increased and Psat1, the third of the three major enzymes known to be involved in the synthesis of d/l-serine, was also up-regulated (1.9-fold; Table 3 and SI Tables 4 and 5). Immunohistochemistry was used to confirm the mRNA induction at a protein level. Phgdh, although known to be constitutively expressed in glial cells (21), was clearly induced in a subset of large lumbar ventral horn motor neurons in 15-week-old (onset) mutant SOD1G37R mice, but was completely absent in motor neurons of age-matched wild-type SOD1WT control mice (Fig. 4).
Fig. 4.
Immunohistochemical confirmation of induction of Phgdh (involved in d/l-serine biosynthesis) at onset within a subset of motor neurons before appearance of symptoms in SOD1G37R mutant mice. Double-immunohistochemistry for Phgdh (B and E; red) and SMI32 (an antibody against neurofilaments to detect large ventral horn neurons; A and D; green) on lumbar spinal cord sections of mutant SOD1G37R (D–F) and control wild-type SOD1WT mice (A–C). (E) Induction of Phgdh on a protein-level at disease onset (15 weeks; arrows) in mutant SOD1G37R mice. (Scale bar: 20 μm.)
In addition to Phgdh, Noxa (phorbol-12-myristate-13-acetate-induced protein 1, Pmaip1; apoptosis-related) and CD44(CD44 antigen; neuronal regenerative/injury response; ref. 22), two other candidates which showed strong mRNA induction at 15 weeks (onset) (5-fold and 14-fold, respectively) were chosen for confirmation at the protein level. Noxa was specifically found within large ventral horn motor neurons, albeit no difference in the level of protein accumulation could be detected between mutant and wild-type SOD1WT mice either at onset or symptomatic stages (SI Fig. 7 and data not shown). However, immunohistochemistry clearly confirmed the up-regulation of CD44 within a subpopulation of large ventral horn lumbar motor neurons in 15-week-old (onset) mutant SOD1G37R mice whereas it was absent from age-matched wild-type SOD1WT motor neurons (SI Fig. 7).
Dysregulation of the Complement Cascade Within Motor Neurons.
A very unexpected finding was that components of the classic pathway of the complement cascade were expressed and dysregulated in mutant SOD1 motor neurons. By disease onset (15 weeks), mRNAs for all of the three subunits of the C1q protein [C1qa, C1qb, C1qc (complement component 1; q subcomponent; α, β, and γ polypeptides)] were strongly induced in mutant SOD1-expressing motor neurons (Table 3 and SI Tables 4 and 5). C1q, a secreted, extracellular polypeptide which can bind antibody aggregates, is the main initiating factor for the classic complement pathway (23). Although in the central nervous system complement components would be expected to be produced by the resident immune cells rather than by motor neurons, markers like CD11b and Iba1 (both measures for potential microglial contaminants) were not detectable by GeneChip analysis (“Absent”) in the mRNA pool of laser microdissected motor neurons. In addition, one of the major cellular complement cascade inhibitors, Daf1 (decay-accelerating factor 1; CD55; GPI-attached extracellular membrane protein), was down-regulated at this same onset time point, further demonstrating mutant SOD1-induced dysregulation of neuronally derived complement components.
Gene Dysregulations Common to Dismutase Active and Inactive Mutants.
To assess the significance of the identified gene dysregulations in motor neurons of mutant SOD1G37R mice as candidate transcriptional changes involved in a common mechanism of toxicity arising from SOD1 mutants of divergent biochemical character, we expanded our gene expression profiling to motor neurons from mice that develop ALS-like motor neuron disease from expression of the dismutase inactive mutant SOD1G85R, which causes disease in mice when accumulated to levels below that of endogenous mouse SOD1 (levels 5- to 50-fold below that needed for disease from dismutase active mutants) (5).
One well studied mutant SOD1G85R mouse line (5, 24) reaches end stage at ≈12–13 months of age, a time significantly later than the SOD1G37R mice (≈25 weeks). The later disease onset in this line produces more variable timing of disease stages than is seen in the SOD1G37R mice, with the inflection point of the weight curve (onset) between 7.5 and 9 months, whereas overall clinical symptoms (including hind limb weakness) appear at ≈11–12 months and end stage (defined by hind limb paralysis) is reached at ≈12–13 months. Overall behavior of the SOD1G85R mice (hind limb spread reflex, overall activity) and the integrity of the motor neuron unit (axon number, axon caliber distribution, muscle innervation) were very similar to the corresponding onset time point (15 weeks) in SOD1G37R mice (Fig. 2 and SI Fig. 8). Because in SOD1G37R mice dysregulation of 108 genes appeared at onset, we chose this time point for laser microdissection recovery of motor neurons from SOD1G85R mice, followed by gene expression profiling. Because SOD1G85R accumulates to less than endogenous mSOD1 (Fig. 2), nontransgenic littermates were chosen as the most appropriate comparison animals. To compensate for the spread of the onset in SOD1G85R mice, two time points, 7.5 and 9 months, were analyzed and candidates with an average fold change ≥1.5 (low stringency) in either of them were selected (Table 3 and SI Tables 4 and 5; for high stringency data see SI Table 6). For each time point, four mutant and three control mice were used (as for the SOD1G37R line, only females were taken).
A common set of 21 genes was found to be dysregulated in motor neurons of both the SOD1G37R and the SOD1G85R mice by disease onset (Table 3 and SI Tables 4–6). The most promising candidates fell into three groups: neuronal regenerative/injury response (ATF3, CD44, Fos, Sprr1a), the classic complement system (C1qa, C1qb, C1qc) and lysosomal degradation machinery (Laptm5, Hexb [HEXB-mutations are linked to the neurodegenerative Sandhoff-disease (25)]). Additional genes dysregulated in common between dismutase active and inactive mutants included Serpinb1a (a serine protease inhibitor), Aqp4 (a water-channel), Grn [granulin; growth-factor related; GRN-mutations are linked to frontotemporal-dementia (FTD-17) (26)] and Gtl2 (untranslated regulatory RNA) (all of the transcripts were up-regulated in mutant SOD1 motor neurons, except Gtl2, which was down-regulated).
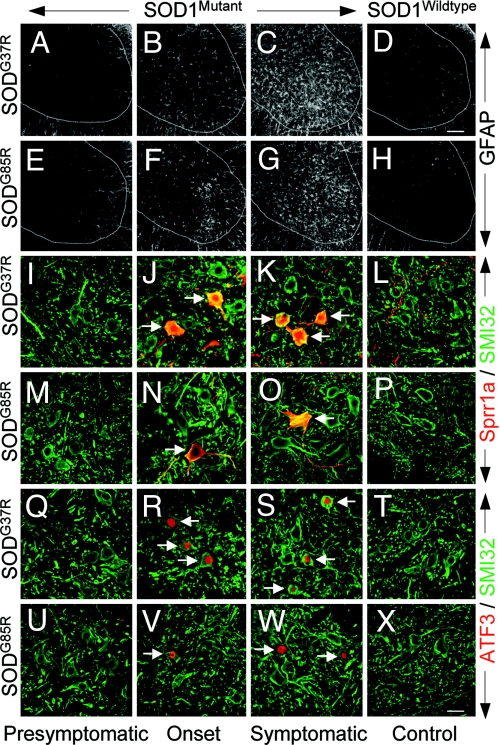
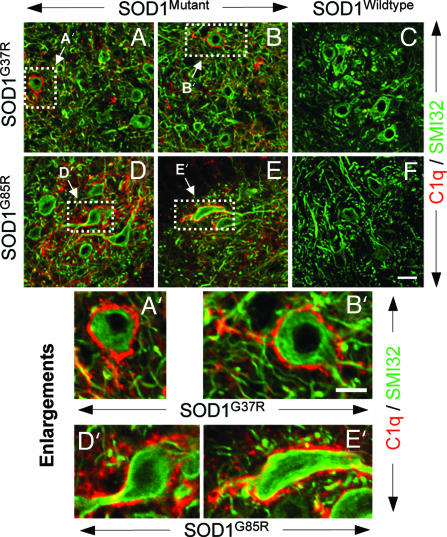
Immunohistochemistry with available antibodies was used to confirm that the mRNA dysregulations were reflected in changes in accumulated proteins within motor neurons. At onset (at 15 weeks in SOD1G37R and at 9 months in SOD1G85R mice), both Sprr1a and ATF3 were clearly selectively induced in a subset of large lumbar ventral horn motor neurons. Parallel analysis of spinal cords from age-matched mice expressing high levels of wild-type human SOD1 or nontransgenic mice showed no detectable staining in any cell-type (Fig. 5). Induction of Sprr1a and ATF3 was further increased in the symptomatic phase (at ≈22 weeks in SOD1G37R and at ≈11–12 months in SOD1G85R mice), although still only in a subset of motor neurons (Fig. 5). Glial activation in the vicinity of the affected neurons was found only after onset, indicating that the induction/dysregulation of Sprr1a and ATF3 is an early event in the motor neuron response to mutant SOD1 toxicity (Fig. 5). A subset of lumbar ventral horn motor neurons of both SOD1G37R and SOD1G85R mice also showed immunoreactivity for complement (C1q) (Fig. 6), whereas age-matched control animals did not, confirming the induction of complement in motor neurons.
Fig. 5.
Immunohistochemical confirmation of induction of Sprr1a and ATF3 (both involved in neuronal regenerative/injury response) at onset within a subset of motor neurons before appearance of symptoms in both dismutase active SOD1G37R and inactive SOD1G85R mutant mice. (A–H) Significant astrocytosis (seen by GFAP immunoreactivity) initiates after onset in mutant SOD1 mice but is absent in age-matched control wild-type SOD1WT or nontransgenic mice. (I–X) Double immunohistochemistry for (I–P) Sprr1a (red) or (Q–X) ATF3 (red) in combination with SMI32 (I–X; to detect large neurons; green) on lumbar spinal cord sections of mutant SOD1G37R (I–K and Q–S), mutant SOD1G85R (M–O and U–W), and age-matched control wild-type SOD1WT (L and T) or nontransgenic mice (P and X). Sprr1a and ATF3 were induced in both mutant SOD1G37R and SOD1G85R mice by disease onset in a subset of lumbar spinal cord ventral horn motor neurons. No staining was detected in motor neurons of age-matched (to symptomatic stages) control mice. [Scale bar in D: 100 μm (for A–H).] [Scale bar in X: 25 μm (for I–X).]
Fig. 6.
Immunohistochemical confirmation of mRNA induction on a subset of motor neurons of both SOD1G37R and SOD1G85R mice of C1q (of the classic complement system). (A–F) Double-immunohistochemistry for C1q (red) in combination with SMI32 (to detect large neurons; green) on lumbar spinal cord sections of mutant (A and B) SOD1G37R, mutant (D and E) SOD1G85R and age-matched control WT SOD1WT (C) or nontransgenic (F) mice. C1q (a secreted polypeptide, whose mRNA was already induced at onset) was detectable at a protein level on ventral horn motor neurons of symptomatic SOD1G37R and SOD1G85R mice. No C1q could be detected on motor neurons of age-matched control mice. (A′–E′) Enlargements showing individual C1q-positive motor neurons. [Scale bar in F: 25 μm (for A–F).] [Scale bar in B′: 10 μm (for A′–E′).]
Discussion
Cell-type specific gene expression profiling of the at-risk spinal cord motor neurons in mutant SOD1-expressing ALS rodent models has revealed three important findings.
First, there are no inherent mutant SOD1-induced mRNA changes at the developmental birth of embryonic motor neurons. Although embryonic motor neurons have been argued to be already adversely affected by mutant SOD1 expression (15–18), it is now clear that these effects are not the result of mutant SOD1-induced transcriptional changes in vivo. Rather, because deficits in embryonic motor neurons are seen only after culture in vitro, this additional stress is required to induce/reveal mutant-dependent changes.
Second, mutant SOD1 toxicity establishes its effects on mRNA dysregulations in an age-dependent manner and marked transcriptional changes arise in adult motor neurons well before the appearance of overt clinical symptoms. Indeed, major mutant SOD1-induced gene dysregulations develop during the presymptomatic postnatal adult lifespan of the at-risk motor neurons, underlining the need for an age-dependency for the overall toxic process (8, 9).
Third, a common set of gene dysregulations originate from dismutase active and inactive mutants. Indeed, a set of 21 genes were dysregulated in dismutase active and inactive mutants before the appearance of the overall symptomatic phase, three of which (Sprr1a, ATF3, C1q) we confirmed at a protein level. Thus, motor neuron specific induction/dysregulation of these proteins is common to SOD1 mutants of divergent biochemical characters.
The significance of these findings is amplified by our analysis of the data from an earlier gene array study (12) with an additional dismutase active SOD1 mutant (SOD1G93A). The analysis revealed a 43% overlap (49 genes) with our own dismutase active mutant SOD1G37R data set, including all our major functional candidate groups (SI Tables 4 and 5). None of these 49 genes, with the exception of vimentin, were initially recognized as promising candidates, perhaps because of masking from the use of nontransgenic littermates as the comparison animals instead of the more appropriate wild-type SOD1 transgenic mice.
Most importantly, inspection of genes dysregulated within motor neurons in all three mutant lines of SOD1 mice produced a set of 16 dysregulated genes common to all of the mutant lines, including our two most promising functional groups of neuronal regenerative/injury response (ATF3, Sprr1a, CD44) and neuronally derived components of the classic complement pathway (all three subunits of C1q) (SI Tables 4 and 5). Candidates of the neuronal regenerative/injury response (Sprr1a/ATF3/CD44) are known to be strongly induced by axonal injury in affected neurons (22, 27, 28) (SI Tables 4 and 5) and ATF3 (together with another of our candidates Ddit3/CHOP10) have been shown, on a protein level, to be induced in SOD1G93A mice (29). In light of a report of mutant SOD1G93A-dependent impairment of the axonal regenerative response (30), the simplest view is that induction of this pathway is a compensatory response.
The unexpected induction of mRNAs of the classic complement pathway (C1qa, C1qb, C1qc) long before appearance of obvious clinical symptoms and before major neuroinflammation suggests that mutant SOD1-induced up-regulation of motor neuron derived complement components is a likely aspect of a toxicity developed within motor neurons that contributes to neurodegeneration. C1q is a secreted, extracellular polypeptide which can bind antibody aggregates and is the main initiating factor for the classic complement pathway which is used to clear/lyse (antibody-marked) debris, pathogens and injured or degenerating cells (23). Complement activation has been associated with several neurodegenerative diseases, including ALS, Parkinson's and Alzheimer's (31–35) and analysis of entire spinal cords from mutant SOD1G93A mice showed transcriptional up-regulation of complement components at early symptomatic stages (10). C1q components have also previously been suggested to be produced by injured or stressed neurons, especially in Alzheimer's disease (36–40). Furthermore, deletion of C1q has been shown to reduce pathological signs in APP-transgenic Alzheimer-mice (41). In light of the recent finding that a proportion of mutant SOD1 may be secreted (42), as well as liberated by cell death, and therefore could act from outside the motor neurons, it is tempting to speculate that the induced complement system might recognize such extracellular, misfolded mutant SOD1 and mark the motor neuron for attack by immune cells.
Thirty-three genes were also identified to be specifically induced/dysregulated by a pair of dismutase active mutant SOD1s (SOD1G37R, this study; SOD1G93A (12); SI Tables 4 and 5). Among the most provocative candidate pathways was the early dysregulation of the d/l-serine biosynthetic pathway (SI Tables 4 and 5). With both a function as a trophic factor (l-serine; ref. 21) and as a NMDA-receptor coagonist (d-serine; refs. 43 and 44), this pathway could be involved in protective neurotrophic functions as well as in detrimental excitotoxic actions.
Future studies are needed to assess whether modulating the neuronal regenerative/injury candidate genes found in our study (Sprr1a/ATF3/CD44) can increase the neuronal regenerative response and slow disease in ALS, whether systemic activation of the complement system in ALS and also the herein suggested role of neuron-specific induction of components of the complement pathway is detrimental or beneficial, and whether induction of the d/l-serine pathway is protective to stressed neurons.
Materials and Methods
Animals.
Transgenic human SOD1-expressing mice, wild-type SOD1WT, line 76 (6), mutant SOD1G37R, line 42 (6) and mutant SOD1G85R, line 148 (5), were on a pure C57BL/6 genetic background, whereas transgenic human SOD1-expressing rats, wild-type SOD1WT (45) and mutant SOD1G93A (7), were on a Sprague–Dawley genetic background. Genotyping was performed as described (46).
Embryonic Rat Motor Neurons.
Embryonic motor neurons were purified from 14 day old (E14) heterozygous wild-type SOD1WT or mutant SOD1G93A rat embryos as described (47) by using a 6% Optiprep (Sigma, St. Louis, MO) gradient separation followed by immunopanning using an anti-p75 (low-affinity NGF receptor) antibody (Chemicon, Temecula, CA). Of each motor neuron preparation (five to seven hSOD1WT or hSOD1G93A positive embryos) an average of ≈100,000 highly pure large motor neurons were obtained. Cells were frozen at −80°C and ≈750 ng of total RNA could be extracted (Absolutely RNA-MicroPrep Kit; Stratagene, La Jolla, CA; with DNase treatment). On the same day, both one mutant and one wild-type SOD1 motor neuron preparation were prepared. This step was repeated four times to give biological replicates. Additional procedures are described in SI Materials and Methods.
Laser Microdissection of Adult Mouse Motor Neurons.
For both mutant SOD1G37R and SOD1G85R and wild-type SOD1WT (or nontransgenic control) mouse lines, three to four animals were used per time point (females only). Lumbar spinal cords were rapidly recovered, rinsed and frozen in −40°C Isopentane in HistoPrep OCT compound (Fisher Scientific International, Hampton, NH), cryosectioned (20 μm) and sections mounted on RNase-free PEN-foil covered glass slides (Leica, Bannockburn, IL). Sections were fixed for 60 s in 75% EtOH, stained for 30 s in 4% cresyl-violet-acetate/0.3% acetic-acid (Sigma) to identify motor neurons, dehydrated in graded solutions of ethanol (70%, 95%, 100%) followed by 45 s in xyelene and dehydrated for 1 h before laser microdissection (Leica AS/LMD-20). From one spinal cord ≈3,000 large ventral horn motor neurons (diameter ≥25 μm) of the lumbar L4–L6 region were directly collected in lysis buffer and the lysate stored at −80°C. Approximately 150 ng of total RNA was recovered per 3,000 laser microdissected motor neurons [Absolutely RNA-NanoPrep Kit, Stratagene; with DNase treatment; quantified with a 20-μl SubMicro Quartz Spectrophotometer Cell (Starna Cells, Atascadero, CA)] and RNA integrity was assessed on a Bioanalyzer 2100 (Agilent, Santa Clara, CA).
RNA Amplification and GeneChip Hybridization.
For both laser microdissected and embryonic motor neurons, 100 ng of total RNA was used for two-round linear amplification (Eukaryotic Small Sample Target Labeling Assay Version II; Affymetrix, Santa Clara, CA) and per sample one Affymetrix Mouse MOE 430 A/B or Rat 230v2.0 GeneChip was hybridized with 15 μg of biotinylated cRNA. All GeneChip hybridizations were performed either at the VA GeneChip Core Facility (University of California at San Diego) or the TGen Microarray Core Facility (Phoenix, AZ) as part of the National Institutes of Health Neuroscience Microarray Consortium according to standard Affymetrix protocols.
GeneChip Data Analysis.
CEL files obtained by using GCOS v1.2 (Affymetrix) were analyzed with dChip v1.3 (20), whereas “Present/Absent” calls were calculated with GCOS. In dChip, all mutant and control CEL files per time point were normalized and “model-based expression-indexes” were calculated by using the match/mismatch model (20). Comparison analysis between three to four mutant and three control samples was performed with dChip by using genes that were called “Present” on at least 50% of the chips in either the control or the mutant group per time point. Selection criteria were a minimal lowest fold change of ≥1.50 (high stringency) or a minimal average fold change of ≥1.5 (low stringency). For all gene names, NetAffx Annotation Update R21 (Nov 28, 2006) was used (Affymetrix). GeneChip raw data (CEL-files) were deposited at http://arrayconsortium.tgen.org (lobsi-affy-mouse-193446 and lobsi-affy-rat-194438) and will be available through the Gene Expression Omnibus (GEO) Database, www.ncbi.nlm.nih.gov/geo.
Immunohistochemistry, assessment of the integrity of the motor neuron unit, RT-PCR, immunoblotting, and additional details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ms. Rebecca Halperin (TGen, Phoenix, AZ) and Ms. Lutfunnessa Shireen [University of California at San Diego (UCSD)] for GeneChip hybridizations, Dr. Jean Lozach (UCSD) for help with data analysis, Drs. Ben Barres and Beth Stevens (Stanford University, Stanford, CA) for helpful discussions regarding the complement system, Dr. Andy Lee and Craig Rappaport (Leica) for help with the LMD system, Ms. Janet Folmer (The John Hopkins University, Baltimore MD) for help with root preparations, Mr. Brendan C. Brinkman (UCSD) for help with confocal analysis, Dr. Pak Chan (Stanford University) for his kind gift of hSOD1WT rats, Drs. Stephen Strittmatter (Yale University, New Haven, CT) and Masahiko Watanabe (Hokkaido University, Hokkaido, Japan) for their kind gift of antibodies, and Ms. Melissa McAlonis-Downes, Ms. Seung-Joo Chun, and Ms. Gabi Szwajkowski for help with genotyping. This work was supported by National Institutes of Health Grant NS 27036 (to D.W.C.). Salary support for D.W.C is provided by the Ludwig Institute for Cancer Research. C.S.L. was supported, in part, by a fellowship from the Swiss National Science Foundation and S.B., in part, by a Fondation pour la Recherche Medical fellowship, an INSERM fellowship, and a developmental grant from the Muscular Dystrophy Association.
Abbreviations
- SOD1
Cu/Zn superoxide dismutase 1
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
Data deposition: GeneChip raw data (CEL-files) were deposited at http://arrayconsortium.tgen.org (accession nos. lobsi-affy-mouse-193446 and lobsi-affy-rat-194438) and are available through the Gene Expression Omnibus (GEO) Database, www.ncbi.nlm.nih.gov/geo.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702230104/DC1.
References
- 1.Boillee S, Vande Velde C, Cleveland DW. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 3.Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. Proc Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 5.Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, Borchelt DR, Price DL, et al. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- 6.Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, Sisodia SS, Cleveland DW, Price DL. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
- 7.Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J, DeVito L, Psaltis G, et al. Proc Natl Acad Sci USA. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillee S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, et al. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 9.Boillee S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 10.Olsen MK, Roberds SL, Ellerbrock BR, Fleck TJ, McKinley DK, Gurney ME. Ann Neurol. 2001;50:730–740. doi: 10.1002/ana.1252. [DOI] [PubMed] [Google Scholar]
- 11.Yoshihara T, Ishigaki S, Yamamoto M, Liang Y, Niwa J, Takeuchi H, Doyu M, Sobue G. J Neurochem. 2002;80:158–167. doi: 10.1046/j.0022-3042.2001.00683.x. [DOI] [PubMed] [Google Scholar]
- 12.Perrin FE, Boisset G, Docquier M, Schaad O, Descombes P, Kato AC. Hum Mol Genet. 2005;14:3309–3320. doi: 10.1093/hmg/ddi357. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Lillo C, Jonsson PA, Vande Velde C, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, et al. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Rappert A, Bechmann I, Pivneva T, Mahlo J, Biber K, Nolte C, Kovac AD, Gerard C, Boddeke HW, Nitsch R, et al. J Neurosci. 2004;24:8500–8509. doi: 10.1523/JNEUROSCI.2451-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pieri M, Albo F, Gaetti C, Spalloni A, Bengtson CP, Longone P, Cavalcanti S, Zona C. Neurosci Lett. 2003;351:153–156. doi: 10.1016/j.neulet.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Spalloni A, Albo F, Ferrari F, Mercuri N, Bernardi G, Zona C, Longone P. Neurobiol Dis. 2004;15:340–350. doi: 10.1016/j.nbd.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Raoul C, Estevez AG, Nishimune H, Cleveland DW, deLapeyriere O, Henderson CE, Haase G, Pettmann B. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 18.Raoul C, Buhler E, Sadeghi C, Jacquier A, Aebischer P, Pettmann B, Henderson CE, Haase G. Proc Natl Acad Sci USA. 2006;103:6007–6012. doi: 10.1073/pnas.0508774103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, Shinobu LA, Ward CM, Young D, Cleveland DW. J Neurochem. 2005;93:875–882. doi: 10.1111/j.1471-4159.2005.03054.x. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Wong WH. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuya S, Watanabe M. Arch Histol Cytol. 2003;66:109–121. doi: 10.1679/aohc.66.109. [DOI] [PubMed] [Google Scholar]
- 22.Raivich G, Bohatschek M, Da Costa C, Iwata O, Galiano M, Hristova M, Nateri AS, Makwana M, Riera-Sans L, Wolfer DP, et al. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Kishore U, Reid KB. Immunopharmacology. 2000;49:159–170. doi: 10.1016/s0162-3109(00)80301-x. [DOI] [PubMed] [Google Scholar]
- 24.Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- 25.Neufeld EF. J Biol Chem. 1989;264:10927–10930. [PubMed] [Google Scholar]
- 26.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 27.Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 28.Bonilla IE, Tanabe K, Strittmatter SM. J Neurosci. 2002;22:1303–1315. doi: 10.1523/JNEUROSCI.22-04-01303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vlug AS, Teuling E, Haasdijk ED, French P, Hoogenraad CC, Jaarsma D. Eur J Neurosci. 2005;22:1881–1894. doi: 10.1111/j.1460-9568.2005.04389.x. [DOI] [PubMed] [Google Scholar]
- 30.Pun S, Santos AF, Saxena S, Xu L, Caroni P. Nat Neurosci. 2006;9:408–419. doi: 10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 31.Bonifati DM, Kishore U. Mol Immunol. 2007;44:999–1010. doi: 10.1016/j.molimm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Donnenfeld H, Kascsak RJ, Bartfeld H. J Neuroimmunol. 1984;6:51–57. doi: 10.1016/0165-5728(84)90042-0. [DOI] [PubMed] [Google Scholar]
- 33.Fonseca MI, Kawas CH, Troncoso JC, Tenner AJ. Neurobiol Dis. 2004;15:40–46. doi: 10.1016/j.nbd.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Goldknopf IL, Sheta EA, Bryson J, Folsom B, Wilson C, Duty J, Yen AA, Appel SH. Biochem Biophys Res Commun. 2006;342:1034–1039. doi: 10.1016/j.bbrc.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 35.Grewal RP, Morgan TE, Finch CE. Neurosci Lett. 1999;271:65–67. doi: 10.1016/s0304-3940(99)00496-6. [DOI] [PubMed] [Google Scholar]
- 36.Fan R, Tenner AJ. J Neuroinflammation. 2005;2:1. doi: 10.1186/1742-2094-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Y, Li R, McGeer EG, McGeer PL. Brain Res. 1997;769:391–395. doi: 10.1016/s0006-8993(97)00850-0. [DOI] [PubMed] [Google Scholar]
- 38.Terai K, Walker DG, McGeer EG, McGeer PL. Brain Res. 1997;769:385–390. doi: 10.1016/s0006-8993(97)00849-4. [DOI] [PubMed] [Google Scholar]
- 39.Huang J, Kim LJ, Mealey R, Marsh HC, Jr, Zhang Y, Tenner AJ, Connolly ES, Jr, Pinsky DJ. Science. 1999;285:595–599. doi: 10.1126/science.285.5427.595. [DOI] [PubMed] [Google Scholar]
- 40.Afagh A, Cummings BJ, Cribbs DH, Cotman CW, Tenner AJ. Exp Neurol. 1996;138:22–32. doi: 10.1006/exnr.1996.0043. [DOI] [PubMed] [Google Scholar]
- 41.Fonseca MI, Zhou J, Botto M, Tenner AJ. J Neurosci. 2004;24:6457–6465. doi: 10.1523/JNEUROSCI.0901-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urushitani M, Sik A, Sakurai T, Nukina N, Takahashi R, Julien JP. Nat Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 43.Kartvelishvily E, Shleper M, Balan L, Dumin E, Wolosker H. J Biol Chem. 2006;281:14151–14162. doi: 10.1074/jbc.M512927200. [DOI] [PubMed] [Google Scholar]
- 44.Panatier A, Theodosis DT, Mothet JP, Touquet B, Pollegioni L, Poulain DA, Oliet SH. Cell. 2006;125:775–784. doi: 10.1016/j.cell.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 45.Chan PH, Kawase M, Murakami K, Chen SF, Li Y, Calagui B, Reola L, Carlson E, Epstein CJ. J Neurosci. 1998;18:8292–8299. doi: 10.1523/JNEUROSCI.18-20-08292.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lobsiger CS, Garcia ML, Ward CM, Cleveland DW. Proc Natl Acad Sci USA. 2005;102:10351–10356. doi: 10.1073/pnas.0503862102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pennica D, Arce V, Swanson TA, Vejsada R, Pollock RA, Armanini M, Dudley K, Phillips HS, Rosenthal A, Kato AC, et al. Neuron. 1996;17:63–74. doi: 10.1016/s0896-6273(00)80281-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.