Abstract
We demonstrate the efficacy of double-stranded RNA-mediated interference (RNAi) of gene expression in generating “knock-out” phenotypes for specific proteins in several Drosophila cell lines. We prove the applicability of this technique for studying signaling cascades by dissecting the well-characterized insulin signal transduction pathway. Specifically, we demonstrate that inhibiting the expression of the DSOR1 (mitogen-activated protein kinase kinase, MAPKK) prevents the activation of the downstream ERK-A (MAPK). In contrast, blocking ERK-A expression results in increased activation of DSOR1. We also show that Drosophila AKT (DAKT) activation depends on the insulin receptor substrate, CHICO (IRS1–4). Finally, we demonstrate that blocking the expression of Drosophila PTEN results in the activation of DAKT. In all cases, the interference of the biochemical cascade by RNAi is consistent with the known steps in the pathway. We extend this powerful technique to study two proteins, DSH3PX1 and Drosophila ACK (DACK). DSH3PX1 is an SH3, phox homology domain-containing protein, and DACK is homologous to the mammalian activated Cdc42 tyrosine kinase, ACK. Using RNAi, we demonstrate that DACK is upstream of DSH3PX1 phosphorylation, making DSH3PX1 an identified downstream target/substrate of ACK-like tyrosine kinases. These experiments highlight the usefulness of RNAi in dissecting complex biochemical signaling cascades and provide a highly effective method for determining the function of the identified genes arising from the Drosophila genome sequencing project.
Double-stranded RNA (dsRNA)-mediated interference (RNAi) of gene expression has become a widely used method facilitating reverse genetic studies in Caenorhabditis elegans (reviewed in refs. 1–4). Discovery of this phenomenon came from anti-sense RNA experiments, where it was observed that both sense and anti-sense RNAs were effective at producing phenocopies of genetic loss-of-function mutations (1, 5, 6). It was later determined that, if the complimentary strands of a specific RNA are combined, producing dsRNA, there was potent and specific interference of protein expression (6). Originally, dsRNA was injected into the gonad of the worm, but subsequent experiments showed that the dsRNA could be injected anywhere in the worm and still produce mutant phenotypes in the adult. Furthermore, feeding C. elegans bacteria engineered to produce dsRNA (7) or soaking the worms in a dsRNA solution produced mutant phenotypes (8). Surprisingly, this phenomenon can be passed on to the adult worms' progeny, allowing the study of mutant phenotypes throughout all stages of development.
The mechanism by which dsRNA prevents target gene expression is not completely understood. Recent studies suggest that dsRNA probes ranging from 200 to 1000 bp, derived from sequences that are present in the mature transcript but not from those in the promoter or intergenic regions, are able to interfere with gene expression (6). However, some variability of efficacy has been noted for different dsRNA probes that target the same gene (9). Only a few copies of dsRNA per cell are necessary to eliminate protein production, suggesting that the mechanism is catalytic in nature, and that it does not function by titrating endogenous mRNA, as proposed for traditional anti-sense RNA techniques. The targeted gene does not appear to be mutated, nor does transcription of the gene cease in the presence of dsRNA. However, the mRNA produced by the RNAi-targeted gene is absent from the cytoplasm and reduced in the nucleus. These results indicate that the dsRNA exerts its effect during or following RNA processing, but before protein translation (1, 10).
Recently, RNAi has been reported to function in Drosophila as well as in other invertebrates and plants (reviewed in ref. 4). In Drosophila, injection of embryos with dsRNA before cellularization (syncytial blastoderm) successfully phenocopies previously characterized loss-of-function embryonic mutations (11, 12). Because embryos at this stage of development have no cell membranes, it is unclear whether dsRNA is able to cross Drosophila cell membranes. However, the RNAi effect persists throughout development and can be observed in the adult at low penetrance, although transmission of the RNAi effect to progeny has not been observed in Drosophila (12).
In contrast to C. elegans, there are a number of established Drosophila cell lines offering many experimental advantages for biochemical studies. In this report, we show that dsRNA is capable of specifically blocking the production of targeted proteins in cell culture. First, we demonstrate RNAi efficacy by blocking the expression of two proteins under study in our laboratory. One of these proteins, DSH3PX1, is abundantly expressed in Schneider 2 (S2) cells and is the homologue of the human sorting nexin, SH3PX1 (13, 14). The other protein, Drosophila ACK (DACK), is a protein tyrosine kinase homologous to the family of mammalian activated Cdc42 kinases, ACK (15). Once we optimized the experimental conditions, we used the well-known insulin signaling pathway to establish the validity of RNAi to dissect signal transduction pathways. Importantly, we show that inhibiting the expression of four distinct proteins within the insulin signaling pathway modifies the downstream response in a predictable manner. Finally, we apply RNAi to study the relationship between DSH3PX1 and DACK, concluding that DACK is a kinase upstream of DSH3PX1 phosphorylation.
Materials and Methods
Cell Culture.
S2 cells were propagated in 1× Schneider's Drosophila media (GIBCO) supplemented with 10% FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin in 75-cm2 T-flasks (Sarstedt) at room temperature. S2 cells transfected with 6xHis-tagged Dock SH2 domain were propagated in the same media supplemented with 200 μg/ml hygromycin (Roche Molecular Biochemicals). KC (a gift from Michael Horscht, University of Michigan, Ann Arbor) and BG2-C6 (a gift from Kumiko Ui-Tei, Nippon Medical School, Tokyo) cells were propagated by using the above media.
dsRNA Production.
Individual DNA fragments approximately 700 bp in length, containing coding sequences for the proteins to be “knocked out” were amplified by using PCR. Each primer used in the PCR contained a 5′ T7 RNA polymerase binding site (GAATTAATACGACTCACTATAGGGAGA) followed by sequences specific for the targeted genes. The PCR products were purified by using the High Pure PCR Purification Kit (Roche Molecular Biochemicals). The purified PCR products were used as templates by using a MEGASCRIPT T7 transcription kit (Ambion, Austin, TX) to produce dsRNA. The dsRNA products were ethanol-precipitated and resuspended in water. The dsRNAs were annealed by incubation at 65°C for 30 min followed by slow cooling to room temperature. Six micrograms of dsRNA were analyzed by 1% agarose gel electrophoresis to ensure that the majority of the dsRNA existed as a single band of approximately 700 bp. The dsRNA was stored at −20°C.
Primer sequences used to generate specific dsRNAs were obtained as follows: DSH3PX1, GenBank accession no. AF223381, sense-primer 1124–1139, antisense-primer 1749–1765; DACK, accession no. AF181642, sense-primer 2197–2214, antisense-primer 2807–2824; DSOR1, accession no. D13782, sense-primer 1251–1268, antisense-primer 2200–2220; extracellular signal-regulated kinase (ERK)-A, accession no. M95126, sense-primer 306–325, antisense-primer 1066–1084; DPTEN, accession no. AF144232, sense-primer 205–222, antisense-primer 895–912; CHICO, accession no. AF154826, sense-primer 226–235, antisense-primer 932–940; DPTP61F, accession no. L11253, sense-primer 1127–1146, antisense-primer 1734–1751.
Conditions for RNAi in Drosophila Cell Culture.
Drosophila cell culture cells were diluted to a final concentration of 1 × 106 cells/ml in Drosophila expression system (DES) serum-free medium (Invitrogen). One milliliter of cells was plated per well of a six-well cell culture dish (Corning). dsRNA was added directly to the media to a final concentration of 37 nM. For dsRNAs of approximately 700 nt, this corresponds to 15 μg of dsRNA. This was followed immediately by vigorous agitation. The cells were incubated for 30 min at room temperature followed by addition of 2 ml of 1× Schneider's media containing FBS. The cells were incubated for an additional 3 days to allow for turnover of the target protein.
Extract Preparation, Western Analysis, and Drosophila AKT (DAKT)/Protein Kinase B (PKB) Enzyme Activity Analysis.
Cells were harvested by trituration and pelleted by centrifugation at 1000 × g. Cells were lysed in 400 μl RIPA buffer (50 mM Tris, pH 8.0/150 mM NaCl/1.0% Nonidet P-40/0.5% Deoxycholate/0.1% SDS/0.2 mM NaVO4/10 mM NaF/0.4 mM EDTA/10% glycerol), and 20 μl lysate was added to 20 μl of Laemmli loading buffer followed by electrophoresis on 7.5% SDS polyacrylamide gels. Western analyses were performed as previously described (16). Purification of the 6xHis-tagged-SH2 domain from S2 cells was performed by using nickel agarose as described by the manufacturer (Qiagen, Chatsworth, CA).
For experiments involving insulin, dsRNAs were applied to S2 cells as described above. After a 2-day recovery, the cells were serum starved for 2 h, after which they were exposed to 10 μg/ml insulin (human) for 5 min. Extracts were prepared for Western analysis as described above. Extract preparation, immunoprecipitation, and in vitro kinase assays for DAKT/PKB activity were performed as described (17) by using 6 μM Crosstide as the substrate (Upstate Biotechnology, Lake Placid, NY; no. 12-331). Radioactivity incorporated into the peptide was measured by using the manufacturer's suggested protocol.
Antibodies.
Polyclonal antibodies were raised in rabbits against recombinant DSH3PX1, DACK, and Dock (Cocalico, Reamstown, PA). 4G10 anti-phosphotyrosine antibody was purchased from Upstate Biotechnology. Monoclonal antibodies generated against MEK/DSOR1 and phospho-MEK/DSOR1 were purchased from New England Biolabs. Monoclonal antibodies for ERK/ERK-A and phospho-ERK/ERK-A were purchased from Sigma.
Results
RNAi Functions in Drosophila Cell Culture.
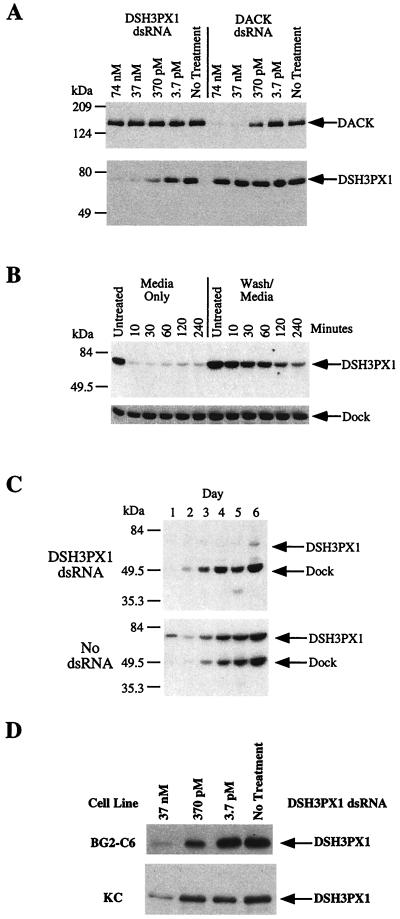
To determine whether RNAi functions in cell culture, we chose to analyze the expression profiles of three proteins under study in our laboratory: DSH3PX1, DACK, and Dock. As previously mentioned, DSH3PX1 resembles a sorting nexin and is comprised of an NH2-terminal SH3 domain and a central phox homology (PX) domain (13, 14), whereas DACK is a tyrosine kinase (15). Dock, the Drosophila homologue of Nck, is an adaptor protein containing three SH3 and one SH2 domains (18, 19). Our laboratory has recently obtained evidence that these proteins work together in a signaling complex (unpublished observations). Here, we demonstrate that specific dsRNA fragments, representing DSH3PX1 and DACK, cause the selective disappearance of the corresponding encoded proteins in S2 cell culture. Briefly, S2 cells were plated in serum-free media, and DSH3PX1 or DACK dsRNA was added to the cultures at concentrations of 74 nM, 37 nM, 370 pM, and 3.7 pM. After 30 min in serum-free media, 2 ml of serum containing media were added per well, and the cells were incubated for 3 days. Western blots of cell extracts show that DSH3PX1 dsRNA specifically reduced DSH3PX1 protein levels in a concentration-dependent manner, whereas DACK dsRNA similarly reduced DACK protein levels (Fig. 1A). A dsRNA concentration of 37 nM was able to maximally inhibit protein synthesis, as addition of 74 nM dsRNA did not result in further inhibition. For both DSH3PX1 and DACK, loss of protein was approximately 95–99% (data not shown). The specificity of RNAi is also apparent from Fig. 1A, as dsRNA corresponding to DACK has no effect on DSH3PX1 protein levels and DSH3PX1 dsRNA does not alter DACK protein levels. To date, we have successfully inhibited the production of DSH3PX1, DACK, Dock, and 12 other proteins, including kinases, phosphatases, adaptor proteins, and receptors, suggesting that the RNAi technique is effective toward a broad range of proteins. Using the conditions described in Materials and Methods, all dsRNAs tested were effective in specifically ablating the production of their respective proteins (data not shown). Interestingly, if the dsRNA is added to S2 cell culture in the presence of serum, the RNAi effect is still apparent but is an order of magnitude less potent (data not shown). The ease and specificity by which proteins can be effectively “knocked-out” of S2 cells makes this an ideal system for dissecting signal transduction pathways and may provide the basis for high throughput screens to determining the functions for specific Drosophila proteins.
Figure 1.
Specificity and duration of RNAi in S2 cell culture. (A) S2 cells were incubated with the indicated concentrations of DSH3PX1 or DACK dsRNAs. Cellular extracts were prepared after 3 days and subjected to Western analysis as described in Materials and Methods. (B) S2 cells were incubated with 37 nM DSH3PX1 dsRNA for 10–240 min. (Left) Samples undergoing the standard recovery procedure. (Right) Samples rinsed to remove the unincorporated dsRNA before addition of serum containing media. Western analysis on extracts prepared after a 3-day recovery period was performed as described using antibodies generated against DSH3PX1 and Dock. (C) S2 cells were exposed to 37 nM DSH3PX1 dsRNA as described in Materials and Methods. Cellular extracts were prepared on the indicated days and subjected to Western analysis using antibodies against DSH3PX1 and Dock. (D) The specified amounts of dsRNAs corresponding to DSH3PX1 coding sequence were added to either BG2-C6 or KC cells. Western analysis was performed as described using antibodies directed against DSH3PX1. In each panel, an arrow indicates the relative mobility of DACK, DSH3PX1, or Dock.
RNAi Kinetics, Duration, and General Applicability in Drosophila Cell Culture.
After the discovery that the addition of dsRNA to cell culture media was sufficient to abolish targeted protein production in S2 cells, we determined the optimal incubation time. S2 cells were treated with 37 nM DSH3PX1 dsRNA in serum-free media for increasing lengths of time. After this incubation period, either serum-containing media was added as previously described or the cells were rinsed to remove the dsRNA before the addition of serum-containing media. All cultures were incubated for 3 days before examination of DSH3PX1 and Dock levels. At all time points examined, the cells exposed to the dsRNA showed a loss of DSH3PX1 production as expected (Fig. 1B). However, when the dsRNA was removed from the media after a 10- to 60-min incubation period, there was little or no loss of DSH3PX1 protein expression (Fig. 1B). Inhibition of DSH3PX1 production was apparent when the cells were exposed to dsRNA for 2 h and was further enhanced when the dsRNA was left on the cells for 4 h. Maximal inhibition of protein production at this concentration of dsRNA may take as long as 8 h of continuous dsRNA exposure (data not shown).
The duration of the RNAi effect was examined by monitoring DSH3PX1 and Dock protein levels over the course of 6 days. dsRNA for DSH3PX1 was added to cells by using the previously described optimal conditions. After the addition of serum-containing media, the cells were exposed to the dsRNA for 1–6 days, after which they were harvested and analyzed for DSH3PX1 and Dock protein expression levels (Fig. 1C). In the untreated cells, both DSH3PX1 and Dock protein levels increase with increasing cell density. When DSH3PX1 dsRNA is present, the reduction of DSH3PX1 is apparent as early as day 1 and persists through day 6. This demonstrates both the selectivity and longevity of the RNAi effect.
Several Drosophila cell lines were also tested to determine whether the RNAi effect was limited to S2 cells. KC and BG2-C6 cells, originally isolated from different embryonic cell types, were all shown to express DSH3PX1 (20). DSH3PX1 dsRNA was applied to these cells as described above, and DSH3PX1 protein levels were examined. Expression of DSH3PX1 was blocked in all cell lines tested (Fig. 1D). This demonstrates that RNAi selectively blocks protein expression in several different Drosophila cell lines. This observation significantly broadens the application of this methodology because it is likely that these cell lines express distinct proteins depending on their origin.
Dissection of Insulin Signal Transduction Pathways in S2 Cells.
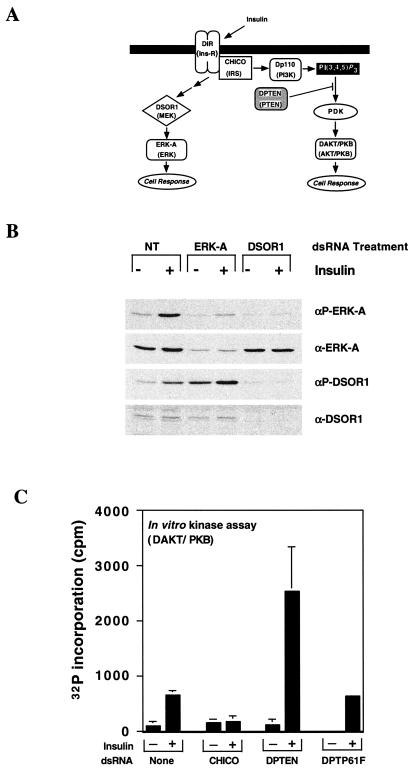
We then assessed the suitability of RNAi for dissecting signal transduction cascades using the well-characterized insulin signaling pathway. Similar to the mammalian insulin receptor, activation of the Drosophila insulin receptor triggers the activation of various intracellular effectors, including the mitogen-activated protein kinase (MAPK) and phosphoinositol-3 kinase (PI3K) pathways (Fig. 2A) (21–24).
Figure 2.
Dissection of the insulin signal transduction pathway in S2 cell culture. (A) Schematic representation of the ERK-A and PI3K branches of the insulin signal transduction pathway. The mammalian homologues of the Drosophila proteins are in parentheses. (B) S2 cells were incubated in the absence (NT) or presence of dsRNAs representing DSOR1 or ERK-A (dsRNA DSOR1/ERK-A). After 3 days, the cells were either left untreated or treated with 10 μg/ml insulin for 5 min. Cellular extracts were prepared and subjected to Western analysis using antibodies directed against phospho-ERK-A (αP-ERK-A), ERK-A (αERK-A), phospho-DSOR1 (αP- DSOR1), or DSOR1 (α- DSOR1). (C) S2 cells were treated with dsRNAs directed against the expression of CHICO, PTEN, and DPTP61F as indicated. Cellular extracts were prepared and DAKT/PKB activity measured as described in Materials and Methods.
Activation of the MAPK pathway by insulin results in the increased phosphorylation and activity of downstream effectors, including DSOR1, which subsequently phosphorylates and activates ERK-A (Fig. 2A). Applying dsRNA corresponding to DSOR1-coding sequences results in the loss of both phosphorylated and unphosphorylated DSOR1 from the cells (Fig. 2B). As predicted, the lack of DSOR1 precludes the activation of ERK-A after insulin stimulation (Fig. 2B). Similarly, applying dsRNA corresponding to ERK-A coding sequences results in the loss of both activated and inactivated ERK-A. Interestingly, removal of ERK-A results in activation of DSOR1 both in the absence and presence of insulin (Fig. 2B). This confirms several reports reviewed by Schaeffer and Weber (25) that implicates ERK-A in the down-regulation of the MAPK pathway.
We have further characterized RNAi applicability to another branch of the insulin signaling cascade. In Drosophila, genetic analysis of Chico, the homologue of the vertebrate insulin receptor substrate (IRS) family, demonstrates that the insulin signal transduction pathway controls cell size (26). This branch of insulin receptor signaling involves phosphorylation of CHICO, which activates PI3K-mediated increased phosphatidylinositol (3,4,5)-trisphosphate [PI(3,4,5)P3] levels (Fig. 2A). Increases in this lipid result in DAKT/PKB activation. PTEN is a phosphatase that has been shown to hydrolyze PI(3,4,5)P3 (27). Consistently, mouse PTEN knockouts display high levels of cellular PI(3,4,5)P3, leading to constitutive activation of AKT/PKB (reviewed in ref. 28). Therefore, we anticipate that removal of CHICO and DPTEN will regulate DAKT/PKB activity in opposite ways.
Treatment of S2 cells with insulin results in a 4-fold increase in DAKT/PKB activity (Fig. 2C). Cells exposed to dsRNAs for CHICO are no longer able to activate DAKT/PKB. Conversely, cells treated with dsRNA corresponding to PTEN, the negative regulator of this pathway, demonstrate a 19-fold increase in DAKT/PKB activity on insulin treatment. This effect is specific for PTEN, as addition of dsRNA directed against another phosphatase expressed in S2 cells, DPTP61F (16, 29), does not increase the activity of DAKT/PKB in response to insulin. In addition, the increase in DAKT/PKB activity can also be blocked by treating cells with dsRNAs corresponding to the Drosophila homologues of PI3K (Dp110) and phosphoinositide-dependent kinase (data not shown).
DSH3PX1 Phosphorylation Is Downstream of DACK Function.
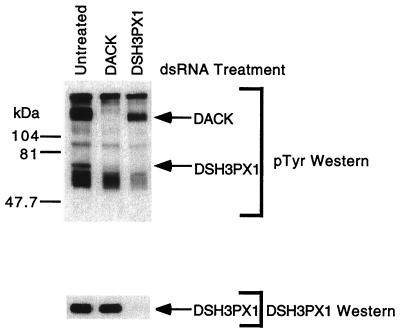
After having demonstrated the usefulness of RNAi to dissect the previously characterized signaling pathways described above, we analyzed a signaling complex composed of DSH3PX1, Dock, and DACK (unpublished observations). Examination of DSH3PX1 by Western blotting using the anti-phosphotyrosine antibody 4G10 reveals that it is tyrosine-phosphorylated. To test the hypothesis that DACK is responsible for the observed tyrosine phosphorylation of DSH3PX1, S2 cells expressing a 6xHis-tagged version of the Dock SH2 domain were treated with DACK or DSH3PX1 dsRNA. The Dock SH2 domain was purified on a Ni-agarose column, and the bound protein complex including DACK and DSH3PX1 were detected by 4G10 anti-phosphotyrosine Western analysis (Fig. 3). In the cells treated with DACK dsRNA, DACK is no longer detectable as a tyrosine phosphorylated protein. Interestingly, in cells that lack DACK, tyrosine phosphorylation of DSH3PX1 is greatly diminished although the amount of DSH3PX1 present in the Dock SH2-associated complex remains the same (Fig. 3). This experiment brings up two interesting points: (i) that the interaction between DSH3PX1and Dock's SH2 domain may be indirect, and (ii) that DSH3PX1 tyrosine phosphorylation is downstream of DACK, making DSH3PX1 the first effector/substrate for DACK. Additional experiments are in progress to establish the relationship between DACK and DSH3PX1. This simple experiment highlights the usefulness of RNAi in unraveling the complexity associated with these signaling molecules.
Figure 3.
DACK is upstream of DSH3PX1 phosphorylation. S2 cells stably expressing the 6xHis-tagged SH2 domain of Dock were treated with dsRNAs representing DACK and DSH3PX1. The protein complex associated with Dock's SH2 domain was purified as described in Materials and Methods and subjected to Western blot analysis using antibodies recognizing phosphotyrosine (pTyr) or DSH3PX1. The relative mobilities of DACK and DSH3PX1 are indicated.
Discussion
The use of RNAi in cell culture to dissect signal transduction pathways has several advantages over methods requiring the introduction of DNA into cells. Transfection experiments can lead to nonphysiological concentrations of the recombinant protein, often making the interpretation of results difficult. Likewise, transfection experiments often result in up-take of the DNA into significantly less than 100% of the cells. Our results with RNAi suggest that all of the cells effectively take up the dsRNA because 95–99% of the protein disappears from the cultured cells.
The use of dsRNA in S2 cells is technically simple, requiring only the production of dsRNA from a PCR product that has T7 RNA polymerase binding sites at each end. The method is also quick, in that results of the protein “knock-out” experiment can be obtained within 2–3 days. This contrasts sharply with the time necessary to produce selective gene “knock-outs” in mammalian cells. Likewise, the method appears to be highly reproducible, working on a wide range of proteins in several Drosophila cell lines. We suggest that RNAi will greatly facilitate the study of intracellular signaling processes.
The discovery that RNAi functions in Drosophila cell culture will undoubtedly aid in the elucidation of the mechanism that allows dsRNA to inhibit target protein synthesis and may serve as a stepping stone for adaptation of this technique to mammalian systems. This procedure is akin to having inhibitors for specific proteins in a wide range of signal transduction cascades. By using RNAi, we have the potential to rapidly accelerate the characterization of biochemical pathways in Drosophila cell lines.
Acknowledgments
We thank Matt Wishart and Kim Orth for critical review of this manuscript. This work was supported by Grants DK18024 and 18849 from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health (to J.E.D.) and from the Walther Cancer Institute. M.M. is a recipient of Human Frontier Science Program Organization Long-Term Fellowship.
Abbreviations
- DACK
Drosophila ACK
- dsRNA
double-stranded RNA
- RNAi
RNA-mediated interference
- S2
Schneider 2
- ERK
extracellular signal-regulated kinase
- DAKT
Drosophila AKT
- IRS
insulin receptor substrate
- PX
phox homology
- MAPK
mitogen-activated protein kinase
- PI3K
phosphoinositol-3 kinase
- PI(3,4,5)P3
phosphatidylinositol (3,4,5)-triphosphate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110149597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110149597
References
- 1.Montgomery M K, Fire A. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 2.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 3.Hunter C P. Curr Biol. 1999;9:R440–R442. doi: 10.1016/s0960-9822(99)80276-0. [DOI] [PubMed] [Google Scholar]
- 4.Sharp P A. Genes Dev. 1999;13:139–141. [PubMed] [Google Scholar]
- 5.Guo S, Kemphues K J. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Timmons L, Fire A. Nature (London) 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 8.Tabara H, Grishok A, Mello C C. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 9.Powell-Coffman J A, Knight J, Wood W B. Dev Biol. 1996;178:472–483. doi: 10.1006/dbio.1996.0232. [DOI] [PubMed] [Google Scholar]
- 10.Montgomery M K, Xu S, Fire A. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 12.Misquitta L, Paterson B M. Proc Natl Acad Sci USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard L, Nelson K K, Maciewicz R A, Blobel C P. J Biol Chem. 1999;274:31693–31699. doi: 10.1074/jbc.274.44.31693. [DOI] [PubMed] [Google Scholar]
- 14.Kurten R C, Cadena D L, Gill G N. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- 15.Yang W, Cerione R A. J Biol Chem. 1997;272:24819–24824. doi: 10.1074/jbc.272.40.24819. [DOI] [PubMed] [Google Scholar]
- 16.Clemens J C, Ursuliak Z, Clemens K K, Price J V, Dixon J E. J Biol Chem. 1996;271:17002–17005. doi: 10.1074/jbc.271.29.17002. [DOI] [PubMed] [Google Scholar]
- 17.Andjelkovic M, Jones P F, Grossniklaus U, Cron P, Schier A F, Dick M, Bilbe G, Hemmings B A. J Biol Chem. 1995;270:4066–4075. doi: 10.1074/jbc.270.8.4066. [DOI] [PubMed] [Google Scholar]
- 18.Garrity P A, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky S L. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- 19.Desai C J, Garrity P A, Keshishian H, Zipursky S L, Zinn K. Development (Cambridge, UK) 1999;126:1527–1535. doi: 10.1242/dev.126.7.1527. [DOI] [PubMed] [Google Scholar]
- 20.Echalier G. Drosophila Cells in Culture. New York: Academic Press; 1997. [Google Scholar]
- 21.Biggs W H, III, Zipursky S L. Proc Natl Acad Sci USA. 1992;89:6295–6299. doi: 10.1073/pnas.89.14.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, DeFea K, Roth R A. J Biol Chem. 1999;274:9351–9356. doi: 10.1074/jbc.274.14.9351. [DOI] [PubMed] [Google Scholar]
- 23.Ogg S, Ruvkun G. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 24.Paradis S, Ruvkun G. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaeffer H J, Weber M J. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss B F, Beckingham K, Hafen E. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 27.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 28.Maehama T, Dixon J E. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 29.McLaughlin S, Dixon J E. J Biol Chem. 1993;268:6839–6842. [PubMed] [Google Scholar]