Abstract
In Saccharomyces cerevisiae, heterochromatin-like regions are found near telomeres and at the silent mating-type loci, where they can repress genes in an epigenetic manner. Several proteins are involved in telomeric heterochromatin structure including Rap1, Sir2, Sir3, Sir4, yKu70 (Hdf1), yKu80 (Hdf2), and the N termini of histones H3 and H4. By recognizing cis-acting DNA-binding sites, Rap1 is believed to recruit Sir and other silencing proteins and determine where heterochromatin forms. The integrity of heterochromatin also requires the binding of Sir proteins to histones that may form a scaffold for Sir protein interactions with chromatin. In this study we describe how the heterochromatin complex may form initially and how it differs from the complex that spreads along the chromosome. We found that close to the telomere end, Sir4 can bind Rap1 independently of Sir2, Sir3, yKu70/yKu80, and the intact H4 N terminus. In contrast, Sir4 binding requires all of the silencing factors further along telomeric heterochromatin. These data indicate that Sir4 binding to Rap1 initiates the sequential association of Sir and other proteins, allowing the subsequent spreading of the heterochromatin proteins along the chromosome.
Keywords: Heterochromatin, Sir, telomere, Rap1, initiation, spreading
In complex eukaryotes, such as Drosophila melanogaster, heterochromatin at centromeres and telomeres is cytologically condensed throughout the cell cycle, located near the nuclear periphery, and represses gene activity in an epigenetic manner. Such repression occurs by the spreading of the heterochromatin complex into adjacent genes (Henikoff 1990; Wakimoto 1998). Yeast, Saccharomyces cerevisiae, also contains heterochromatin-like regions that include telomeres and the silent mating-type loci (HMLα and HMRa). These regions, that are found near the nuclear periphery, use a number of known proteins that include Rap1; Silencing Information Regulators Sir2, Sir3, Sir4; and histones H3 and H4 to form heterochromatin (Laurenson and Rine 1992; Grunstein 1997; Cockell and Gasser 1999). Among these proteins Rap1 has a special role in the initiation of heterochromatin because it directly binds cis-acting DNA elements such as C1–3A repeat sequences at telomeres and the E and I silencers at the silent HM mating-type loci. Because of the interaction of Rap1 with both Sir3 and Sir4 in two-hybrid experiments and in vitro (Moretti et al. 1994), and interactions in cell extracts and in vitro among Sir3–Sir4–Sir2 (Moazed et al. 1997; Strahl-Bolsinger et al. 1997), it has been proposed that Rap1 may initiate the formation of heterochromatin by recruiting the Sir complex (Moretti et al. 1994; Hecht et al. 1996). The yKu70/yKu80 heterodimer, the yeast homolog of mammalian Ku70/Ku80 that binds to the ends of double-stranded DNA with high affinity (Mimori and Hardin 1986), is also present at the telomeres and is required for the integrity of telomeric heterochromatin (Laroche et al. 1998; Mishra and Shore 1999). Recent studies have shown that the yKu70/yKu80 heterodimer participates in silencing by counteracting Rif1, a Rap1-binding factor that prevents Rap1–Sir4 interaction (Mishra and Shore 1999). Although yKu70, like Rap1, interacts with Sir4 in two-hybrid experiments (Tsukamoto et al. 1997) it is unknown whether the yKu70/yKu80 heterodimer can directly recruit Sir proteins or whether it functions solely by antagonizing Rif1.
The N termini of histones H3 and H4 may serve as a platform for the interaction of Sir proteins with chromatin. This has been postulated as a result of genetic and direct binding data. The histone N-terminal domains containing sequences (H3 residues 4–20; H4 residues 16–29) that interact directly with Sir3 and Sir4 (Hecht et al. 1995) are also those uniquely required for gene repression by heterochromatin in vivo (Johnson et al. 1990, 1992; Thompson et al. 1994). In fact, as shown by chromatin immunoprecipitation (ChIP) assay, a single mutation at H4 lysine 16 to glutamine (H4 K16Q) can disrupt Sir3 binding and spreading of the Sir complex at all sites examined between 0.5 kb and 2.8 kb from the telomeric end of Chromosome VI-R (Hecht et al. 1996). Because mutations in RAP1 or any of the SIR genes have a similar effect as H4 K16Q, it has been proposed that the proteins of heterochromatin form an interdependent complex that must interact with histone tails at these sites (Hecht et al. 1996). Whether the interdependent heterochromatin complex is involved in both initiation and spreading of heterochromatin is unknown.
Several clues suggest that different Sir proteins have unique roles and that these roles must be accounted for in understanding the initiation and spreading of heterochromatin. Only SIR3, when overexpressed, can extend gene repression from the telomere (Telomere Position Effect, or TPE) farther than 3 kb at Chromosome VI-R. The overproduction of Sir3 from a 2μ high-copy plasmid causes TPE to extend as far as 17 kb in a manner that is dependent on SIR2 and SIR4 (Renauld et al. 1993; Hecht et al. 1996). However, extended TPE is not caused by the spreading of the entire Sir complex as in the first 3 kb. Instead, it is mainly Sir3 that spreads in extended telomeric heterochromatin (Strahl-Bolsinger et al. 1997) by interacting with the hypoacetylated histone H4 N termini (Braunstein et al. 1993; Carmen et al. 2001). This finding supports the proposal that Sir3 is especially important in the spreading of TPE (Renauld et al. 1993). Sir2 is an NAD-dependent histone deacetylase (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000) with specificity in vitro for K16 of histone H4 and K9, K14 of H3 (Imai et al. 2000). Hence, Sir2 has been proposed to deacetylate histone H4 K16 to allow Sir3 to bind and spread. Finally, overexpression of SIR4 but not SIR3 suppresses the silencing defect of a yku70Δ strain (Mishra and Shore 1999), indicating that Sir4 has functions distinct from those of Sir3. Furthermore, in one-hybrid experiments, Sir4 fused to a transcription activation domain can activate an HIS marker at ∼50 bp from the left arm of Chromosome VII in the absence of Sir3 (Bourns et al. 1998). However, although it has been proposed on the basis of these indirect experiments that Sir4 may be important in initiation, the unique role of Sir4 protein in this regard is not clear. This is especially so given that both Sir4 and Sir3 interact with Rap1 in two-hybrid experiments and GST-fusion direct-binding studies (Moretti et al. 1994; Moretti and Shore 2001).
To date, investigators have detected no differences in the protein requirements for initiation or spreading of yeast telomeric heterochromatin. As expected, proteins required for initiation are also required for spreading. Sir2, Sir3, and Sir4 do spread along telomeric heterochromatin in the wild-type cell when examined by ChIP (Hecht et al. 1996; Strahl-Bolsinger et al. 1997). However, even Rap1 appears to spread away from its C1–3A repeat binding sites as far as Sir proteins spread (2.8 kb at Chromosome VI-R). This can be explained by data indicating that the telomere folds back onto itself (Strahl-Bolsinger et al. 1997; de Bruin et al. 2000, 2001). This would allow Rap1 to contact telomere distal sites without actually spreading. Nevertheless, ChIP data do argue that Rap1 initiates the formation of heterochromatin. A mutation at the C terminus of Rap1 (rap1-21) that prevents silencing (Liu and Lustig 1996) and the interaction of Rap1 with Sir3 does not dislodge Rap1 from its cis-acting DNA sites (Strahl-Bolsinger et al. 1997). Rap1 is also the only heterochromatin protein known to remain at its binding sites in the absence of Sir proteins, which suggests that Rap1 does initiate heterochromatin from telomeric C1–3A repeats. In this paper we investigate whether proteins other than Rap1 are involved in the initiation of telomeric heterochromatin. To do so, we have used ChIP to examine Sir protein interactions as close as 70 bp from the C1–3A repeats. This is much closer to the telomeric end than previously analyzed by ChIP. These interactions were compared with those at telomere distal regions, 0.5 kb and beyond. We have found that Rap1 interacts with Sir4 at the telomere proximal site in the absence of other Sir proteins and other heterochromatin factors examined. In contrast, Sir4 binding at telomere distal heterochromatin is strongly dependent on the other heterochromatic factors. These data support the argument that by interacting with Rap1, Sir4 initiates the sequential assembly of the Sir complex at the telomeric end.
Results
Sir4 binding to the telomeric end is independent of Sir2 and Sir3
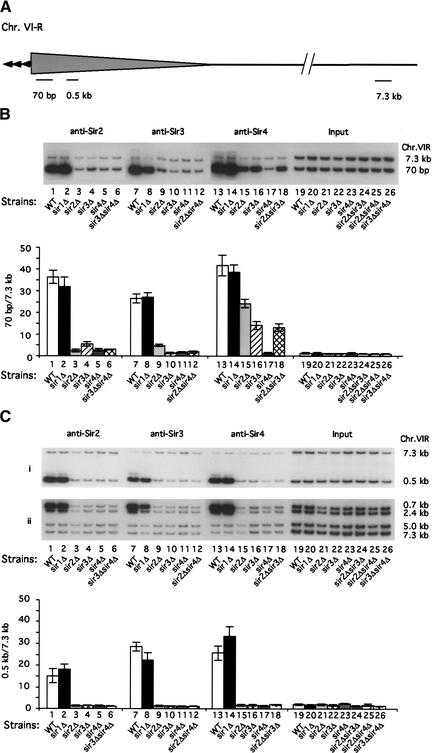
Previous analyses of Sir protein binding at subtelomeric sites focused on regions that were detected by primer sets that started at a number of sites (from 0.7 kb to as far as 2.8 kb from the end of Chromosome VI-R), beyond which little Sir binding was observed (Hecht et al. 1996; Strahl-Bolsinger et al. 1997). At the Sir protein binding sites, mutations in RAP1, SIR3, and histone H4 all caused disruption of the Sir–histone complex. This implied the formation of an interdependent heterochromatin complex at these subtelomeric sites. To ask whether all Sir proteins are required for the formation of the Sir complex even closer to the telomere end, we examined the binding of Sir2, Sir3, and Sir4 proteins by ChIP, using primer sets that start at 70 bp from the C1–3A repeats (Fig. 1A). These data were compared with Sir protein binding in heterochromatin that starts at 0.5 kb and a more telomere distal region at 7.3 kb, where Sir binding is no longer found (Hecht et al. 1996). Binding of Sir2, Sir3, and Sir4 was compared between wild-type (WT) and sir mutant strains to investigate the interdependent binding of the Sir proteins. In addition, we examined the effect of sir1Δ as a control because Sir1 is important for HM but not for telomeric silencing. We find that anti-Sir2 antibody immunoprecipitates 30–35-fold more DNA at 70 bp than at 7.3 kb, in the wild-type or sir1Δ strains (Fig. 1B, lanes 1,2). We also detect Sir2 enrichment at 0.5 kb (Fig. 1Ci, lanes 1,2). As the distance from the telomere is increased to 0.7 and 2.4 kb, Sir2 binding decreases (Fig. 1Cii, lanes 1,2), confirming previous observations of Sir2 distribution in telomeric heterochromatin (Strahl-Bolsinger et al. 1997). However, we find that the level of Sir2 binding is reduced dramatically at all distances from the telomere when SIR2, SIR3, or SIR4 is disrupted (Fig. 1B,C, lanes 3–5). Sir3 binding is similarly affected by deletions in SIR2, SIR3, and SIR4 (Fig. 1B,C, lanes 9–11). Therefore, the binding of Sir2 and Sir3 at 70 bp, 0.5 kb, and 2.4 kb from the telomeric C1–3A repeats is dependent on the presence of the entire Sir complex.
Figure 1.
Sir4 binds to a region 70 bp from the telomeric end in the absence of Sir2 and Sir3. (A) Schematic representation of telomeric primer sets used for ChIP in this study. Primers specific to subtelomeric sequences starting at 70-bp, 0.5-kb, and 7.3-kb regions from the C1-3A repeats at the right end of Chromosome VI (Chr. VI-R) are shown. The 7.3-kb region was used as a background control. The shaded area represents the distribution of the Sir complex (Strahl-Bolsinger et al. 1997). (B) Sir2, Sir3, and Sir4 binding at 70-bp region. (C) Sir2, Sir3, and Sir4 binding at 0.5-kb (i) and more distal regions (ii) from the telomere end. The ChIP assay was performed as described in Materials and Methods. Protein–DNA complexes were immunoprecipitated using anti-Sir2 (lanes 1–6), anti-Sir3 (lanes 7–12), and anti-Sir4 (lanes 13–18) antibodies. DNA samples before immunoprecipitation are shown as Input (lanes 19–26). Telomeric DNA fragments associated with the precipitated proteins were amplified by PCR in the presence of [α-32P]ATP using primers starting at 70-bp, 0.5-kb, 0.7-kb, 2.4-kb, 5.0-kb, and 7.3-kb regions (Materials and Methods). PCR reaction products were then subjected to SDS-PAGE. Individual fragments were quantified using PhosphorImager; data are shown below the gel as fold enrichment at 70 bp and 0.5 kb compared with that of 7.3 kb in the indicated strains. All experiments have been performed independently at least three times, with error bars shown for standard deviations. Binding of individual Sir proteins was assayed in wild-type (WT; JRY4012; lanes 1,7,13,19), sir1Δ (JRY4620; lanes 2,8,14,20), sir2Δ (JRY4587; lanes 3,9,15,21), sir3Δ (JRY4604; lanes 4, 10,16,22), sir4Δ (JRY4579; lanes 5,11,17, 23), sir3Δ sir4Δ (KLY103; lanes 6,26), sir2Δ sir4Δ (KLY102; lanes 12,25), and sir2Δ sir3Δ (KLY101; lanes 18,24) strains.
In contrast, 35%–60% of the Sir4 binding that takes place in the wild-type strain at the telomeric end is retained when SIR2 or SIR3 is disrupted (Fig. 1B, lanes 13–16). This represents ∼10–18-fold more Sir4 bound at 70 bp in the sir2Δ and sir3Δ mutants than in the sir4Δ mutant control. Even when both SIR2 and SIR3 are disrupted in the same strain, there is similar binding of Sir4 as when SIR3 alone is deleted (Fig. 1B, lanes 16,18). This independent binding of Sir4 at 70 bp does not occur at telomere distal regions such as 0.5 kb and beyond, where Sir2, Sir3, and Sir4 binding are all similarly reduced when any of the SIR genes is deleted (Fig. 1Ci,ii; lanes 3–5, 9–11, 15–17). Therefore, Sir2 or Sir3 binding at the telomeric end is strongly dependent on SIR4; however, significant Sir4 binding at this region is independent of SIR2 and SIR3. This contrasts with the interdependent binding of the Sir proteins within more telomere-distal heterochromatin.
Sir2 and Sir3 binding at the telomeric end require the enzymatic activity of Sir2
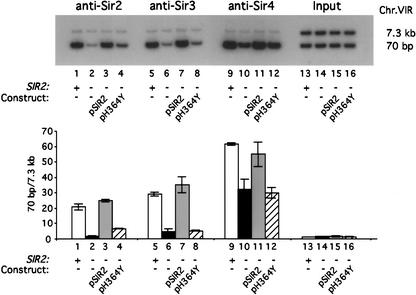
Sir2 is an NAD-dependent deacetylase that has been shown to have specificity for H4 K16 in vitro. A mutation, SIR2-H364Y, that disrupts both Sir2 NAD-dependent deacetylase and ADP-ribosyltransferase activities does not prevent binding of Sir2 to Sir4 in vivo (Tanny et al. 1999). To determine if the enzymatic activity of Sir2 is required for Sir2 or Sir3 binding at the 70-bp region, we examined their interaction in a mutant strain containing SIR2-H364Y. Binding of Sir2 was compared in strains that are wild type, sir2Δ, as well as sir2Δ strains containing plasmid-borne SIR2-H364Y and plasmid-borne wild-type SIR2 genes. As shown in Figure 2, at 70 bp SIR2-H364Y causes a dramatic loss of Sir2 binding relative to wild-type SIR2 (Fig. 2; lane 4 vs. lanes 1 and 3). Similarly, Sir3 binding at 70 bp was nearly abolished in the cells carrying the mutant SIR2 (Fig. 2; lane 8 vs. lanes 5 and 7). Similar to the SIR2 gene disruption studies (Fig. 1), loss of Sir2 enzymatic activity leaves ∼55% of Sir4 bound at 70 bp (Fig. 2; lane 12 vs. lanes 9 and 11). These data support the argument that although Sir2-H364Y can interact with Sir4 as well as wild-type Sir2, such interaction is insufficient for Sir2 recruitment at 70 bp. Sir2 and Sir3 binding at 70 bp from the telomeric end requires the enzymatic activity of Sir2.
Figure 2.
The deacetylase activity of Sir2 is required for Sir2 and Sir3, but not Sir4, to bind to the region 70 bp from the telomeric end. Sir protein binding to the region starting at 70 bp from the C1–3A repeats was assayed in the wild-type (WT) strain (Y1195; lanes 1,5,9,13); the sir2Δ strain containing vector pRS315 (Y1201; lanes 2,6,10,14); the sir2Δ strain containing pSIR2 (Y1203; lanes 3,7,11,15), and the sir2Δ strain containing pH364Y (Y1205; lanes 4,8,12,16). ChIP and PhosphorImager quantitations were performed as described in Figure 1 and are shown as fold enrichment at 70 bp compared with that at 7.3 kb.
Sir4 recruitment to the telomeric end is independent of histone H4 lysine 16 and the yKu70/yKu80 proteins
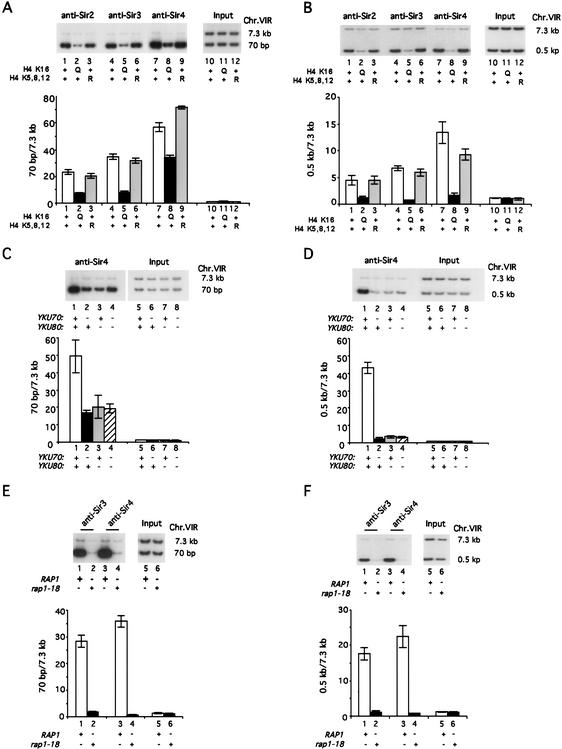
Given that Sir2 and Sir3 are not essential for recruiting Sir4 to the very end of the chromosome, other proteins may be involved in recruiting Sir4. These may include histones, the yKu70/yKu80 heterodimer, and Rap1, each of which is involved in silencing and associates directly with DNA in heterochromatin. Histone H4 K16 plays a crucial role in telomeric and HM silencing, as mutation of this site (H4 K16Q) abolishes the binding of Sir3 from both telomeres and HML (Hecht et al. 1996). To test the possibility that H4 K16 is required for the interaction of Sir4 with the telomeric end, we examined Sir4 binding at 70 bp in a strain containing H4 K16Q. This was compared with Sir4 binding in the wild-type and in a mutant strain in which three acetylatable lysines, K5, K8, and K12, are mutated to arginine (H4 K5,8,12R). We found that the H4 K16Q mutation significantly reduced binding of Sir2 and Sir3 at both 70 bp and 0.5 kb as compared with wild-type or the H4 K5,8,12R mutation (Fig. 3A, lanes 1–6; Fig. 3B, lanes 1–6). However, as shown in Figure 3A (cf. lane 8 to lanes 7 and 9), the strain containing H4 K16Q retains 50%–65% of Sir4 binding at 70 bp as compared with the wild-type or H4 K5,8,12R mutation. Therefore, as with SIR2 and SIR3 disruptions, H4 K16Q does not abolish Sir4 binding at 70 bp. In contrast, H4 K16Q decreases Sir4 binding to ∼10% of wild-type levels at 0.5 kb, as shown in Figure 3B (cf. lane 8 to lanes 7 and 9). We also find that deletions yku70Δ , yku80Δ, and yku70Δ yku80Δ still allow ∼20-fold enrichment of Sir4 binding at 70 bp as compared with 7.3 kb (i.e., 35%–40% of Sir4 bound in the wild-type isogenic strain; Fig. 3C, cf. lanes 2–4 to lane 1). In contrast, yku70Δ and yku80Δ deletions disrupt Sir4 binding almost completely at 0.5 kb (Fig. 3D, cf. lanes 2–4 to lane 1). We conclude that considerable binding of Sir4 close to the telomeric end is independent of histone H4 K16 and yKu70/yKu80 proteins.
Figure 3.
Only Rap1 is required for Sir4 recruitment at the region 70 bp from the telomeric end. ChIP assay was performed as described in Materials and Methods and Figure 1. (A,B) Sir4 recruitment to the 70-bp region from the telomeric end does not depend on H4 K16. Sir2, Sir3, and Sir4 binding at 70 bp (A) and 0.5 kb (B) in wild-type (WT; JTY34U; lanes 1,4,7,10), H4 K16Q (JTY502U; lanes 2,5,8,11), and H4 K5,8,12R (JTY503U; lanes 3,6,9,12) strains are shown. (C,D) Sir4 binding to the 70-bp region does not depend on YKU70 or YKU80. The strains used are wild-type (WT; AJL275–2a-VIIL-URA; lanes 1,5), yku70Δ (KLY16; lanes 2,6), yku80Δ (KLY17; lanes 3,7), and yku70Δ yku80Δ (KLY161; lanes 4,8). Sir4 binding at 70-bp (C) and 0.5-kb regions (D) is shown. (E,F) rap1-18 abolishes Sir4 binding at the 70-bp region. Sir3 and Sir4 binding at the 70-bp region (E) and the 0.5-kb region (F) were assayed in wild-type (WT; AJL275–2a-VIIL-URA; lanes 1,3,5) and rap1-18 (AJL399–4b; lanes 2,4,6) strains. PhosphorImager quantitations are shown as fold enrichment at 70 bp compared with that at 7.3 kb.
Genetic and biochemical evidence for the direct recruitment of Sir4 by Rap1 to the telomeric end
The rap1-18 mutant protein has a truncation in its C-terminal domain (Δ678–827) that is required for Rap1 association with Sir3/Sir4 in vitro and in two-hybrid studies as well as for TPE (Kyrion et al. 1992). Therefore, to determine whether Rap1 recruits Sir4 preferentially, we asked whether rap1-18 interferes with Sir3 and Sir4 binding at 70 bp. Binding was compared with the telomere-distal regions at 0.5 kb and 7.3 kb. As shown in Figure 3, E and F (lanes 1–4), both Sir3 and Sir4 binding were completely abolished at 70 bp and at 0.5 kb in the rap1-18 mutant. Therefore, of all the heterochromatin protein sequences examined for their requirement in Sir4 binding to the 70-bp site, only the C-terminal region of Rap1 is essential for Sir4 binding to the telomeric end.
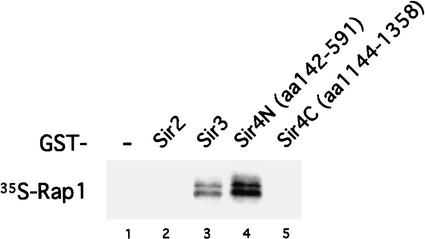
Previous in vitro studies have shown that Rap1 interacts directly with a large fragment (residues 737–1358, ∼50% of the total protein) at the C terminus of Sir4 (Moretti and Shore 2001). To determine whether Rap1 also interacts with the N terminus of Sir4, we examined the binding in vitro of Rap1 with a GST–Sir4 N-terminal fusion protein as compared with GST alone. We found that Rap1 can interact with the N-terminal region of Sir4 (residues 142–591; Fig. 4, lane 4) but not with a shorter C-terminal domain (residues 1144–1358; Fig. 4, lane 5). Therefore, Rap1 interacts directly with regions contained in N-terminal residues 142–591 and C-terminal residues 737–1358. Our data also confirm that Rap1 interacts with Sir3, but not Sir2 (Fig. 4, lanes 3 and 2). These data are consistent with earlier observations showing that both N and C termini of Sir4 are required for Sir4 function in vivo (Marshall et al. 1987). These direct binding data and the genetic requirement of Rap1 for Sir4 binding at 70 bp support the argument that Rap1 recruits Sir4 to the telomeric end.
Figure 4.
Rap1 binds Sir4 in vitro. In vitro binding of Rap1 and GST–Sir fusion proteins. GST-fusion protein experiments were performed as described in Materials and Methods. Full-length Rap1 was labeled in vitro with 35S-Methionine and incubated with GST (lane 1), GST–Sir2 (amino acids 275–562, lane 2), GST–Sir3 (amino acids 503–970, lane 3), GST–Sir4N (amino acids 142–591, lane 4), and GST–Sir4C (amino acids 1144–1358, lane 5). Interacting proteins were analyzed by SDS-PAGE and fluorography.
The recruitment of Sir4 by Rap1, independently of other silencing proteins, is a feature that is specific for telomeric heterochromatin
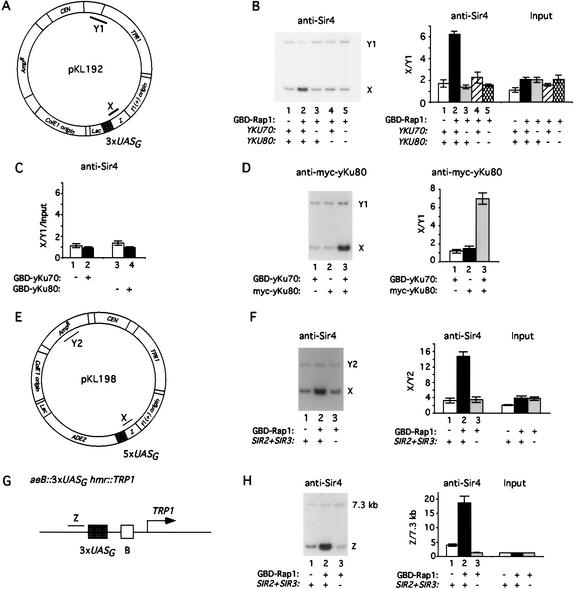
To determine whether Rap1 may recruit Sir4 at a nontelomeric episomal site, we introduced a GBD (Gal4 DNA binding domain)–Rap1 fusion construct into a strain carrying a plasmid (pKL192) containing three copies of UASG sites (Gal4-DNA-binding sites; Fig. 5A). Sir4 binding was examined using ChIP with primer sets to the region (X) adjacent to the UASG sites and comparison to a sequence (Y1) that is distant from the UASG sites in pKL192. Using anti-Sir4 antibody we found that Sir4 was enriched at the UASG sites of pKL192 in cells that carry GBD–Rap1 but not GBD alone (Fig. 5B, cf. lanes 2 and 1). Therefore, Rap1 is likely to recruit Sir4 to the UASG sites of pKL192.
Figure 5.
Sir4 recruitment by Rap1 depends on YKU70/YKU80, SIR2, and SIR3 at nontelomeric sites. (A–D) yKu70/yKu80 are required for Sir4 recruitment by GBD–Rap1 on a plasmid. Plasmid pKL192 carrying 3× UASG sites (Gal4 DNA-binding sites) and PCR amplification products used for ChIP are shown schematically in A. PCR fragment X is immediately next to the UASG sites. Fragment Y1, which is 1.4 kb away from the UASG sites, served as a background control. (B) Sir4 recruitment by GBD–Rap1 is abolished in yku70Δ and yku80Δ mutants. ChIP was carried out as described in Figure 1. The GBD–Rap1 fusion construct was cotransformed with pKL192 into wild-type (WT; AJL275–2a-VIIL-URA; lanes 1,2), yku70Δ (KLY16; lane 3), yku80Δ (KLY17; lane 4), and yku70Δ yku80Δ (KLY161; lane 5) strains. Sir4 association with pKL192 was analyzed. The ratio of enrichments at X versus at Y1 was plotted. (C) GBD–yKu70 and GBD–yKu80 do not recruit Sir4 to pKL192. GBD alone (lane 1,3), GBD–yKu70 (lane 2), and GBD–yKu80 (lane 4) constructs were cotransformed with pKL198 into wild-type (WT; AJL275–2a-VIIL-URA), respectively. Sir4 association with pKL192 was analyzed as in B. The ratio of enrichments at X versus at Y1 was plotted after normalization with Input DNA. (D) yKu70 and yKu80 interact at pKL192. GBD–yKu70 was transformed with pKL192 into a strain carrying a myc-tagged yKu80 (GA1009). Antibody specific to the myc tag was used to examine myc-yKu80 binding on pKL192 in strains carrying wild-type yKu80 (GA426) and GBD–yKu70 (lane 1); myc-yKu80 (GA1009) and GBD alone (lane 2); and myc-yKu80 (GA1009) and GBD–yKu70 (lane 3). (E,F) Sir2 and Sir3 are required for Sir4 recruitment by GBD–Rap1 to plasmid pKL198. Plasmid pKL198 carrying 5× UASG sites is shown schematically in E with regions amplified by PCR. PCR fragment X is immediately next to the UASG sites. Fragment Y2 is 2.75 kb away from the UASG sites and served as a background control. GBD (lanes 1) and GBD–Rap1 fusion (lanes 2,3) plasmids were cotransformed with pKL198 into wild-type (WT; RS1001; lanes 1,2) and sir2Δ sir3Δ (KLY111; lane 3) strains. Sir4 binding on pKL198 was analyzed as shown in F. The ratio of enrichments at X versus Y2 is plotted. (G,H) Sir2 and Sir3 are required for Sir4 recruitment by GBD–Rap1 to the HMR locus. The mutated hmr locus is shown schematically in G (Chien et al. 1993). PCR fragment Z is immediately next to the 3× UASG sites. The subtelomeric region at 7.3 kb from the right end of Chromosome VI served as a background control. GBD (lane 1) and GBD–Rap1 fusion (lanes 2,3) plasmids were transformed into wild-type (WT; RS1001; lanes 1,2) and sir2Δ sir3Δ (KLY111; lane 3) strains. Sir4 binding analyzed by ChIP is shown in H. The ratio of enrichments at Z versus 7.3 kb is plotted.
We also asked whether yKu70 and yKu80 are required for GBD–Rap1 recruitment of Sir4 by examining Sir4 binding to GBD–Rap1 in yku70Δ and yku80Δ strains. We found that GBD–Rap1 was not able to recruit Sir4 to the UASG sites of pKL192 in either yku70Δ or yku80Δ strains (Fig. 5B, lanes 3,4). Therefore, although yKu70 and yKu80 proteins are not essential for Sir4 recruitment at the telomeric end, these proteins strongly participate in GBD–Rap1 recruitment of Sir4 at episomal UASG sites.
To test if yKu70 or yKu80 can recruit Sir4 directly, we also tethered either GBD–yKu70 or GBD–yKu80 fusion proteins to the UASG sites of pKL192. As shown in Figure 5C, neither GBD–yKu70 nor GBD–yKu80 fusion proteins were able to recruit Sir4 to the UASG sites. This is the case even though myc–yKu80 protein associates with GBD–yKu70 at the UASG sites, supporting the argument that the yKu70/yKu80 heterodimer forms at the UASG sites bound by GBD–yKu70 (Fig. 5D). We conclude that unlike Rap1, yKu70 and yKu80 do not recruit Sir4 to the UASG sites but are required for Rap1 recruitment of Sir4 to these nontelomeric sites.
We also asked whether GBD–Rap1 can recruit Sir4 independently of SIR2 and SIR3 to 5× UASG sites of pKL198 (Fig. 5E). We observed that Sir4 is recruited by GBD–Rap1 in wild-type cells (Fig. 5F, lane 2 vs. lane 1). However, such recruitment could not be detected in the absence of Sir2 and Sir3 (Fig. 5F, lane 3). We observed similar results when we targeted GBD–Rap1 to a mutated hmr locus on Chromosome III (Fig. 5G) bearing 3× UASG sites at aeB hmr::TRP1 (Chien et al. 1993). GBD–Rap1 was able to recruit Sir4 in the wild-type strain but not in the sir2Δ sir3Δ strain (Fig. 5H, cf. lanes 2 and 3). From these data we conclude that Rap1, when tethered artificially to a nontelomeric site, is capable of recruiting Sir4. However, GBD–Rap1 recruitment of Sir4 at these nontelomeric sites is dependent on the other proteins of heterochromatin, yKu70, yKu80, Sir2, and Sir3.
Discussion
Differences in Sir4 interaction with chromatin at telomere-proximal and more distal sites in heterochromatin
We have shown that Sir4 has a unique role in the formation of telomeric heterochromatin. Only Sir4 interacts with the telomeric end in the absence of other Sir and yKu70/yKu80 proteins or the intact histone H4 N terminus. Our data also support the argument that Rap1 recruits Sir4 directly to the Rap1 DNA-binding sites at the telomeric end. First, a mutation in RAP1 (rap1-18) that allows Rap1 binding to DNA but disrupts TPE also prevents Sir4 binding to the telomeric end. Second, Rap1 and Sir4 interact directly in vitro. Rap1–Sir4 interactions at the telomeric end contrast with Sir4 binding at telomere-distal heterochromatin sites (0.5 kb and beyond), at which all the proteins of heterochromatin analyzed are essential for Sir4 binding.
Nevertheless, Sir4 recruitment by Rap1 even at the telomeric end appears to be partially reduced (on average 55%) by mutations in other heterochromatin factors. This reduction may be explained by the following technical reasons. The PCR primer set that is used to amplify the immunoprecipitated chromatin by anti-Sir4 antibodies generates a DNA fragment that starts at 70 bp and extends to 340 bp from the telomeric C1–3A sequences. This primer set not only detects adjacent Rap1-binding sites at the telomeric end but also a telomere-distal region that contains one to two nucleosomes over which telomeric heterochromatin may spread. Sir4 presence in this distal region may be dependent on other heterochromatin proteins. The inevitable inclusion in the ChIP of the telomere-distal region would make it appear that other heterochromatin proteins are required for Sir4 recruitment by Rap1 even at the telomeric end. This apparent requirement of other proteins would be further exaggerated by the immunoprecipitation (in ChIP) of overlapping, randomly sheared, DNA fragments sonicated to 300–500 bp. Therefore, Sir4 recruitment by Rap1 to the telomeric end is likely to be even more independent of other heterochromatin proteins than our data would suggest. This independent Sir4 recruitment by Rap1 helps explain why the overexpression of SIR4 (but not SIR3) can suppress the negative effect of Rif1, the Rap1-binding factor that inhibits Rap1–Sir4 interaction at telomeres (Mishra and Shore 1999). Given our data and the one-hybrid data of Bourns et al. (1998), Sir4 recruited to Rap1 sites independently of other proteins of heterochromatin is likely to initiate the sequential assembly of telomeric heterochromatin.
Independent recruitment of Sir4 by Rap1 is specific to the telomere
Interestingly, the independent recruitment of Sir4 by Rap1 is a feature that is specific to the telomere in our experiments. The reasons for this specificity are unknown. However, they may reflect the presence of considerably more Rap1 available for Sir4 binding at telomeres. Telomeres contain ∼300–350 bp of C1–3A repeat sequence that form ∼15–18 Rap1-binding sites (Gilson et al. 1993). In contrast, the nontelomeric sites examined contain 3–5 Gal4-binding sites. It is possible that increased cooperative interactions of Rap1 and Sir4 at telomeres generate a complex that is more stable and therefore more independent. We tested this possibility by examining Rap1–Sir4 interactions at ∼900 bp of C1–3A repeat sequence integrated at a nontelomeric site of Chromosome II (Stavenhagen and Zakian 1994). We find that Rap1 can recruit Sir4 in the absence of Sir2 and Sir3 at this site (data not shown). This helps explain why Rap1–Sir4 interactions are less stable in the absence of yKu proteins on plasmids that lack a large number of Rap1 sites. These data also suggest that factors other than Rap1 may contribute to stable Sir4 recruitment at the HM mating-type loci that contain a single Rap1 site. Sir1, the HM-specific silencing factor, may serve this function (Triolo and Sternglanz 1996) because GBD–Sir1 that is targeted to the same mutated hmr as used for the GBD–Rap1 experiment (Fig. 5G) can recruit Sir4 independently of SIR2 (data not shown). Therefore, the recruitment of Sir4 may be an initial event in the formation of heterochromatin even at the silent HM mating loci.
Association of Sir2 and Sir3 at telomere-proximal and -distal regions
The recruitment of Sir2 and Sir3 both at the telomeric end and in telomere-distal heterochromatin is dependent on the presence of Rap1 and Sir4 (Fig. 1). Given that Sir4–Sir2 and Sir4–Sir3 interact directly in vitro (Moazed et al. 1997; Strahl-Bolsinger et al. 1997), it appears likely that Sir4 then recruits the remaining Sir proteins to help form heterochromatin. A problem in interpreting ChIP data is that we cannot exclude the possibility that Sir2 and Sir3 can be recruited to the Rap1 sites but are less easily detectable in the absence of histone interactions. Nevertheless, this secondary recruitment of Sir2/Sir3 may involve an interaction with chromatin for the following reasons. The histone deacetylase activity of Sir2 is required for stable binding of Sir2 and Sir3 at 70 bp (Fig. 2). Moreover, the histone H4 mutation H4 K16Q reduces the binding of Sir2 and Sir3 to the 70-bp region much more than Sir4 binding to the same site (Fig. 3). Therefore, in contrast to the Rap1–Sir4 interaction, the association of Sir2/Sir3 with the 70-bp region requires a likely interaction of these proteins with histone H4. This is true not only at the telomere end but also at telomere-distal sites where the spreading of heterochromatin takes place.
A model for the initiation and spreading of telomeric heterochromatin
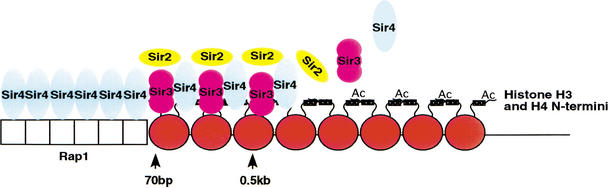
The data presented above and previous findings allow us to suggest the following steps in the initiation and spreading of telomeric heterochromatin (Fig. 6). Sir4 is the first of the Sir proteins that is recruited to Rap1-binding sites and helps determine the subsequent association of heterochromatin proteins. Sir4 then recruits Sir2, an interaction that requires histone H4, at the nucleosome adjacent to the Rap1 DNA-binding sites, for stable binding. Sir2 deacetylates histone H4 and histone H3 (Imai et al. 2000; Smith et al. 2000), enabling Sir3 and Sir4 binding to the histone tails. The presence of Sir2 and Sir3 starts the process of heterochromatin elongation as Sir3 recruits more Sir4, which, in turn, binds Sir2, enabling additional Sir3 and Sir4 binding to chromatin. Given that Sir proteins also interact in a homotypic manner with each other, cooperative interactions between Sir proteins may allow the spreading of an increasingly more stable Sir complex along the histone tails increasingly further from the telomeric end. Telomeric heterochromatin is further stabilized by two higher-order events. These include the folding back of the telomere (Strahl-Bolsinger et al. 1997; de Bruin et al. 2000, 2001), allowing additional Rap1–Sir and Sir–Sir interactions (Strahl-Bolsinger et al. 1997), and the association of the telomere with the Sir-rich nuclear periphery (Maillet et al. 1996).
Figure 6.
Model for the initiation and spreading of telomeric heterochromatin. Sir4 is the first Sir protein recruited to the telomere end by Rap1 to initiate the formation of the silencing complex. Sir4 recruits Sir2 and Sir3. Sir2 deacetylates histone tails to allow Sir3 binding to nucleosome(s) adjacent to the Rap1 sites. Sir3 recruits more Sir4 onto nucleosomes. The process is repeated as Sir4 recruits Sir2 and Sir3 and the complex propagates along the chromosome. The complex that initiates heterochromatin (detected at 70 bp from the telomeric end) differs from that which spreads (detected at 0.5 kb and beyond) in telomeric heterochromatin.
Materials and methods
Yeast transformation
All plasmid and DNA fragments were transformed into yeast cells using the LiAc/SS-DNA/PEG method as described in Agatep et al. (1998).
Plasmids and strains
Strains are as described in Table 1. Strain KLY101 was obtained by transforming the HindIII fragment of plasmid pES28 (sir2::URA3; Chien et al. 1993) into strain JRY4604 (Fox et al. 1995). KLY102 and KLY103 were obtained by transforming the HindIII fragment of plasmid pCTC77 (sir4::URA3; Chien et al. 1993) into strains JRY4587 and JRY4604 (Fox et al. 1995), respectively. KLY111 was obtained as follows: Primers 5′-GAG GAG GAG AAA ACG TAT TTC TGA AAA GAA TTT CTG ATG GCG TAC GCT GCA GGT CGA C-3′ and 5′-CCC ACT AAA TCA TTC ACT ACA CTT ATC CAA GAA AAT TTC TAT CGA TGA ATT CGA GCT CG-3′ were used in PCR to generate the sir3::kanMX4 fragment. This fragment was then transformed into RS1042 (Chien et al. 1993) to generate KLY111 (sir2::URA3, sir3::kanMX4). The yku mutants were obtained as follows: The yku70::kanMX4 fragment was obtained by PCR from plasmid pFA6a-kanMX4 (Wach et al. 1994) with primers 5′-ACT GTA ACA GAG AGG ATG CCA AGG AAG GTA TCT ATG AAC TTT CGT ACG CTG CAG GTC GAC-3′ and 5′-GTG GGT TTT TGA AAT CGG ATG GGT TAT ATC CAT CCC TCA AGA TCG ATG AAT TCG AGC TCG-3′. Primers 5′-GTC CCG TAT CTG AGA ATT CCC AGG AAA TAC CAA ATG TGT TCC CGT ACG CTG CAG GTC GAC-3′ and 5′-GAG TCA GGA AGC TCT GGG ATT TCA AAT TCC CTG GAA TCA TTG ATC GAT GAA TTC GAG CTC G-3′ were used in PCR to amplify the yku80::kanMX4 fragment. These fragments were then transformed into AJL275–2a-VIIL-URA (Kyrion et al. 1993) to make KLY16 (yku70::kanMX4) and KLY17 (yku80::kanMX4). The cells were selected on YPD plates with 200 mg/L Geneticin, and mutants were verified by PCR. The yku70::kanMX4, yku80::LEU2 strain was made as follows: The complete YKU80 gene sequence was amplified from yeast genomic DNA using PCR primers 5′-CCG AAG CTT GCC ACC ATG TCA AGT GAG TCA ACA ACT TTC-3′ and 5′-CGC GAG CTC TTA ATT ATT GCT ATT GTT TGG ACT TC-3′. The PCR product was subsequently cloned into HindIII and SacI sites of pAHe240.1, a pBluescript derivative vector, to produce pKL178. The LEU2 marker was isolated from pJJ252 by digesting the plasmid with BamHI and SphI, and the BamHI–SphI fragment was then cloned into pKL178 with the same restriction sites to generate pKL185. The HindIII–SacI fragment of pKL185 was transformed into KLY16 to yield KLY161. The cells were selected on SD-LEU plates, and mutants were verified by PCR.
Table 1.
Yeast strains
| Strain
|
Genotype
|
Reference
|
|---|---|---|
| JRY4012 | MATaura3 lys2 his3-11,15 leu2-3,112 trpl-l can1-100 | Fox et al. 1995 |
| JRY4620 | same as JRY4012, sir1::TRP1 | Fox et al. 1995 |
| JRY4587 | same as JRY4012, sir2::TRP1 | Fox et al. 1995 |
| JRY4604 | same as JRY4012, sir3::TRP1 | Fox et al. 1995 |
| JRY4579 | same as JRY4012, sir4::TRP1 | Fox et al. 1995 |
| KLY101 | same as JRY4604, sir2::URA3 sir3::TRP1 | This study |
| KLY102 | same as JRY4587, sir2::TRP1 sir4::URA3 | This study |
| KLY103 | same as JRY4604, sir3::TRP1 sir4::URA3 | This study |
| JTY34U | MATa ade2-101(och) ura3-52 his3-200 lys2-801 (amb) trp1-901 hh1,hhf1::LEU2 hht2,hhf2::HIS3 adh4::URA3-TEL plus pJT34 (H3/H4-LYS2) | Thompson et al. 1994 |
| JTY502U | Same as JTY34U, except with pJT4Q16 (H4K16Q-LYS2) | Thompson et al. 1994 |
| JTY503U | Same as JTY34U, except with pJT43R (H4 3K->R-LYS2) | Thompson et al. 1994 |
| GA426 | MATa ade2::hisG ura3-52 his3-11 leu2 trpl canl::hisG VR::ADE2-TEL | Martin et al. 1999 |
| GA1009 | Same as GA426, YKU80-myc(::kanMX4) | Martin et al. 1999 |
| AJL275-2a-VIIL-URA | MATα ade2-1 his3 leu2-3,112 trpl ura3-1 VIIL::URA3 | Kyrion et al. 1993 |
| AJL399-4b | Same as AJL275-2a-VIIL-URA, rapl-18 | Kyrion et al. 1993 |
| KLY16 | Same as AJL275-2a-VIIL-URA, yku70::kanMX4 | This study |
| KLY17 | Same as AJL275-2a-VIIL-URA, yku80::kanMX4 | This study |
| KLY161 | Same as AJL275-2a-VIIL-URA, yku70::kanMX4, yku80::LEU2 | This study |
| W303-1B | MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trpl-1 can1-100 | |
| RS1001 | same as W303-1B, aeB::3xUASG hmr(a)::TRP1 gal4::LEU2 | Chien et al. 1993 |
| RS1042 | same as RS1001, sir2::URA3 | Chien et al. 1993 |
| KLY111 | same as RS1001, sir2::URA3 sir3::kanMX4 | This study |
| Y235 | MATa ura3-52 lys2-801 ade2-1010 his3-200 leu2-1 trpl TEL adh4::URA3 | |
| Y1195 | Same as Y235, pRS315 | Tanny et al. 1999 |
| Y1201 | Same as Y235, sir2Δ, pRS315 | Tanny et al. 1999 |
| Y1203 | Same as Y235, sir2Δ, pSIR2-LEU2 | Tanny et al. 1999 |
| Y1205 | Same as Y235, sir2Δ, pH364Y-LEU2 | Tanny et al. 1999 |
The GBD–Rap1 (pSB136) and GBD–yKu70 (pE306) constructs were obtained from Mishra and Shore (1999). GBD–yKu80 was made as follows: The complete YKU80 gene sequence was amplified from yeast genomic DNA using PCR primers 5′-CCG AGA TCT GCC ACC ATG TCA AGT GAG TCA ACA ACT TTC-3′ and 5′-CGC GTC GAC TTA ATT ATT GCT ATT GTT TGG ACT TC-3′. The PCR product was subsequently digested with BglII and SalI and then cloned into BamHI–SalI sites of pGBDC-1 (James et al. 1996) to make pKL186. An XhoI–SalI fragment of pKL186 was transferred into pMA424 (Chien et al. 1993) to generate pKL189. This GBD–yKu80 fusion protein is able to establish silencing when tethered to HMR (Chien et al. 1993).
3× UASG sites plasmid pKL192
DNA oligonucleotides 5′-CGG GAT CCG TCG ACG GAG GAC TGT CCT CCG TCG ACT GCA GTT TT-3′ and 5′-AAA ACT GCA GTC GAC GGA GGA CAG TCC TCC GTC GAC GGA TCC CG-3′ were annealed and digested with BamHI and PstI. 5′-AAA ACT GCA GTC GAC GGA GGA CTG TCC TCC GTC GAT CGA CGG AGG ACT GTC CTC CGT CGA CGG AAT TCC G-3′ and 5′-CGG AAT TCC GTC GAC GGA GGA CAG TCC TCC GTC GAT CGA CGG AGG ACA GTC CTC CGT CGA CTG CAG TTT T-3′ were annealed and digested with PstI and EcoRI. These two fragments were ligated to the BamHI–EcoRI fragment of pRS414 to make pKL192. Insertion was verified by restriction digest and sequencing.
5× UASG sites plasmid pKL198
DNA 5′-TCC CCC GGG CGA AAA GTT GCC TAG TTT CAT G-3′ and 5′-TCC CCC GGG GGA CAC CTG TAA GCG TTG AT-3′ were used to amplify the ADE2 marker gene from yeast genomic DNA by PCR. The PCR product was subject to SmaI digestion and cloned into pRS414 to make pKL197. pFRluc (Stratagene) was digested with PstI and KpnI to isolate the 5× UASG sites. The fragment was cloned into pKL197 to generate pKL198. Insertion was verified by restriction digestion and sequencing.
In vitro protein binding assay
The GST-fusion protein assay for direct binding was described earlier (Strahl-Bolsinger et al. 1997). The constructs used were pGEX2TK (GST, Pharmacia), pAHe238 (GST–Sir2), pKL104 (GST–Sir3), pKL106 (GST–Sir4N), pSB28a (GST–Sir4C), and pAHe250.2 (Rap1).
Chromatin immunoprecipitation (ChIP) assay
The chromatin immunoprecipitation (ChIP) assay was as described (Suka et al. 1998). Cells were grown in 50 mL of YPD or selective media to OD600 = 0.5–1 and then cross-linked with formaldehyde. Chromatin was fragmented by sonication to a size of 300–500 bp. Anti-Sir2, anti-Sir3, and anti-Sir4 antibodies were used to immunoprecipitate protein–DNA complexes. Precipitated telomere or plasmid DNA fragments were amplified in the presence of [α-32P]ATP by PCR using specific primers and separated by PAGE. DNA samples before immunoprecipitation are shown as Input. All PCR reactions were performed in the linear range for PCR as described before (Hecht et al. 1996). Results were scanned by PhosphorImager and visualized and quantitated using ImageQuant software. Quantitation data was analyzed by Microsoft Excel.
In this study, we investigated Sir complex formation at the right end of Chromosome VI. Whereas other telomeres have conserved Y‘ and X elements, the right end of Chromosome VI is unique and does not contain the conserved sequences that other telomeres have. In our ChIP assay of the telomeric end, a 270-bp PCR product was generated from 70 bp to 340 bp from the C1–3A telomeric repeat. Given the size of sonicated DNA fragments (300–500 bp), we believe that this fragment detects sites involved in the initiation of heterochromatin at Rap1 sites as well as adjacent telomere-distal sites where nucleosomes form immediately distal to the C1–3A repeat (Wright et al. 1992). Primers used to detect regions starting at 0.7 kb, 2.4 kb, and 7.3 kb from the telomeric end were described earlier as 0.77 kb, 2.5 kb, and 7.5 kb (Hecht et al. 1996). The 0.5-kb PCR product extends from 477 bp to 614 bp from the C1–3A telomeric repeat. The 0.7-kb PCR product extends from 728 bp to 1131 bp. The 2.4-kb PCR product extends from 2424 bp to 2803 bp. The 5.0-kb PCR product was generated from 5014 bp to 5348 bp. The 7.3-kb PCR product extends from 7284 bp to 7593 bp. The subtelomeric region starting at 7.3 kb from the C1–3A repeat was used as a background control, because no silencing or silencing complex has been detected there (Strahl-Bolsinger et al. 1997). The sequences of each primer pairs are as follows: 70 bp, 5′-CAT GAC CAG TCC TCA TTT CCA TC-3′ and 5′-ACG TTT AGC TGA GTT TAA CGG TG-3′; 0.5 kb, 5′-CTC GTT AGG ATC ACG TTC GAA TCC-3′ and 5′-GCG TAA CAA AGC CAT AAT GCC TCC-3′; 0.7 kb, 5′-GCG CCT AGT GCA ACT AGT GCA TAT-3′ and 5′-GGA CAG ATC CTT TCG CAT TCC TAC-3′; 2.4 kb, 5′-CTG GAT GAT AAA TGG ACC TGT CCT TC-3′ and 5′-GGC GAG CTG AAT CCC GAA CTT TGT-3′; 5.0 kb, 5′-AAC CCA GTA TTC ATG TCC GGG ACA-3′ and 5′-CGC CAC TCT TTT TTG ATG CTC ATG C-3′; 7.3 kb, 5′-GAC GTG AAA GTT CAG CGC AAC AAG-3′ and 5′-GTA CAT GTC GGC CCT TCT ACG GAT-3′.
Acknowledgments
We are grateful to A.J. Lustig, D. Moazed, J. Rine, D. Shore, and R. Sternglanz for the gift of plasmids and yeast strains. We also thank the members of the Grunstein lab for helpful discussions and their critical comments on this manuscript. This work was supported by Public Health Service grant GM42421 of the National Institutes of Health. M.A.V.-P. received financial support from the Spanish Ministry of Science and Technology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mg@mbi.ucla.edu; FAX (310) 206-9073.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.988802.
References
- Agatep, R., Kirkpatrick, R.D., Parchaliuk, D.L., Woods, R.A., and Gietz, R.D. 1998. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online ( http://tto.trends.com).
- Bourns BD, Alexander MK, Smith AM, Zakian VA. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N-terminus regulates its binding to heterochromatin protein Sir3. J Biol Chem. 2001;277:4778–4781. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D. Targeting of Sir1 protein establishes transcription silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Cockell M, Gasser SM. Nuclear compartments and gene regulation. Curr Opin Genet Dev. 1999;9:199–205. doi: 10.1016/S0959-437X(99)80030-6. [DOI] [PubMed] [Google Scholar]
- de Bruin D, Kantrow SM, Liberatore RA, Zakian VA. Telomere folding is required for the stable maintenance of telomere position effects in yeast. Mol Cell Biol. 2000;20:7991–8000. doi: 10.1128/mcb.20.21.7991-8000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin D, Zaman Z, Liberator RA, Ptashne M. Telomere looping permits gene activation by a downstream UAS in yeast. Nature. 2001;409:109–113. doi: 10.1038/35051119. [DOI] [PubMed] [Google Scholar]
- Fox C, Loo S, Dillin A, Rine J. The origin recognition complex has essential functions in transcriptional silencing and chromosomal replication. Genes & Dev. 1995;9:911–924. doi: 10.1101/gad.9.8.911. [DOI] [PubMed] [Google Scholar]
- Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser SM. Distortion of the DNA double helix by Rap1 at silencers and multiple telomeric binding sites. J Mol Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with Sir3 and Sir4 proteins: A molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor Sir3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990;6:422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Kayne PS, Kahn ES, Grunstein M. Genetic evidence for an interaction between Sir3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1990;87:6286–6290. doi: 10.1073/pnas.87.16.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Fisher-Adams G, Grunstein M. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992;11:2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Boakye KA, Lustig AJ. C-terminal truncation of Rap1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrion G, Liu K, Liu C, Lustig AJ. Rap1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes & Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein Sir2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lustig AJ. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics. 1996;143:81–93. doi: 10.1093/genetics/143.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: A role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Marshall M, Mahoney D, Rose A, Hicks JB, Broach JR. Functional domains of Sir4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and Sir proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by Rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: A Sir2/Sir4 complex and evidence for a regulatory domain in Sir4 that inhibits its interaction with Sir3. Proc Natl Acad Sci. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of Sir proteins interacts with the silencer and telomere-binding protein Rap1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by Sir3 dosage. Genes & Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen JB, Zakian VA. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes & Dev. 1994;8:1411–1422. doi: 10.1101/gad.8.12.1411. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Suka N, Carmen AA, Rundlett SE, Grunstein M. The regulation of gene activity by histones and the histone deacetylase Rpd3. Cold Spring Harb Symp Quant Biol. 1998;63:391–399. doi: 10.1101/sqb.1998.63.391. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and Sir1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Phlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Wright JH, Gottschling DE, Zakian VA. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes & Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]