Abstract
Stably integrated transgenes flanked by the chicken β-globin HS4 insulator are protected against chromosomal position effects and gradual extinction of expression during long-term propagation in culture. To investigate the mechanism of action of this insulator, we used bisulfite genomic sequencing to examine the methylation of individual CpG sites within insulated transgenes, and compared this with patterns of histone acetylation. Surprisingly, although the histones of the entire insulated transgene are highly acetylated, only a specific region in the promoter, containing binding sites for erythroid-specific transcription factors, is highly protected from DNA methylation. This critical region is methylated in noninsulated and inactive lines. MBD3 and Mi-2, subunits of the Mi-2/NuRD repressor complex, are bound in vivo to these silenced noninsulated transgenes. In contrast, insulated cell lines do not show any enrichment of Mi-2/NuRD proteins very late in culture. In addition to the high levels of histone acetylation observed across the entire insulated transgene, significant peaks of H3 acetylation are present over the HS4 insulator elements. Targeted histone acetylation by the chicken β-globin insulator occurs independently of gene transcription and does not require the presence of a functional enhancer. We suggest that this acetylation is in turn responsible for the maintenance of a region of unmethylated DNA over the promoter. Whereas DNA methylation often leads to histone deacetylation, here acetylation appears to prevent methylation.
Keywords: Chromatin, histone acetylation, DNA methylation, insulator, methyl-binding domain (MBD) proteins, silencing
The chromosomes of higher eukaryotes are subdivided into discrete functional domains in which gene expression is either repressed or facilitated. Active or potentially active genes are packaged into a chromatin structure, defined as euchromatin, that exhibits increased nuclease sensitivity and reduced amounts of linker histones such as H1 and H5, and is subject to several types of chemical modifications. The inactive chromatin, often described as condensed chromatin, is relatively inaccessible to DNA-modifying reagents, and it contains hypoacetylated histones and methylated DNA. Euchromatic and condensed chromatin regions are interspersed along the chromosome, and boundary elements can mark the border between adjacent domains. These DNA sequences can functionally isolate neighboring genes, thereby blocking their interactions. Several insulators or boundary elements have been described in Drosophila, yeast, mammalian cells, chicken, and Xenopus (Geyer 1997; Bi and Broach 2001; West et al. 2002). They possess at least one of two properties potentially related to boundary formation. First, insulators can block communication between an enhancer and a promoter if they are located between them, thereby preventing inappropriate gene activation. A second activity of some chromatin insulators is their ability to protect a stably integrated gene from local position effects, as well as from gradual extinction of activity during long-term propagation of the transformed cells.
The first insulator described in vertebrates—the chicken β-globin 5′HS4 element (Chung et al. 1993)—has been successfully used to protect transgenes against position effects and prevent their silencing in different cell lines and transgenic animals (Bell et al. 2001). When two copies of the 1.2-kb β-globin boundary element surround an erythroid-specific reporter gene in immature erythroid 6C2 chicken cells, expression is uniform among different lines and is maintained after 80 d of incubation in the absence of selection (Pikaart et al. 1998). In contrast, noninsulated cell lines display varying levels of expression after selection, and silencing of the transgene frequently occurs in less than 80 d. The silent transgene loses several epigenetic hallmarks of active chromatin, including DNA hypomethylation and histone hyperacetylation. This 5′HS4 function has also been confirmed in transgenic mice, rabbits, and cell lines (for review, see Bell et al. 2001). However, the biological mechanisms of action of the chicken β-globin insulator in protection against position-dependent transgene silencing have not yet been established. At least two possible models have been suggested (Pikaart et al. 1998). One model proposes that the insulator can exclude or block access of repressor complexes to the promoter of the flanked transgene. These repressor complexes can include histone deacetylases (HDACs), DNA methyltransferases, methyl-binding domain (MBD) proteins, transcription repressors, and proteins such as HP1 that are associated with condensed chromatin. A second possibility is that insulators can recruit chromatin remodeling complexes and histone acetyltransferases (HATs), although it has been shown that the 5′HS4 insulator does not function independently as a transcription activation complex. It is possible that the above models function simultaneously.
In this paper, we explore the question of how insulators keep the chromatin of insulated genes in an open and transcriptionally active state, and how they prevent gene silencing. To address these questions, we examined the patterns of DNA methylation and histone acetylation across insulated and noninsulated transgenes, and tested for the presence of repressor complexes. We found that the chicken β-globin insulator prevented DNA methylation of specific CpG dinucleotides; this effect was largely confined to a promoter region that is critical for transgene expression. In noninsulated and inactive lines, this region was methylated. We found that MBD3 and Mi-2, which are subunits of the Mi-2/NuRD repressor complex, were bound to silenced noninsulated transgenes in vivo. This was accompanied by histone underacetylation over the entire transgene, except at the β/ɛ enhancer. In the insulated lines, neither MBD3 nor Mi-2 proteins were present, and specific hyperacetylation patterns of histones H3 and H4 were observed across the insulated domains. These and other results reported here indicate that histone acetylation plays a dominant role in the ability of the chicken β-globin insulator to protect against silencing.
Results
The chicken β-globin insulator prevents DNA methylation of specific CpG dinucleotides in the transgene promoter
To determine how the chicken β-globin insulator keeps the chromatin of insulated genes in an open and transcriptionally active state, we applied the model system developed in our laboratory for studying the function of the 5′HS4 insulator (Pikaart et al. 1998). The gene encoding the Tac subunit of the interleukin 2 receptor (IL2R) under the control of the chicken βA-globin promoter and the β/ɛ enhancer (Fig. 1A) was stably integrated into the chicken early erythroid 6C2 cell line. After withdrawal of hygromycin, which was used for selection of the transformed cells, there was a gradual extinction of activity over a period of 20–80 d of growth (data not shown; Pikaart et al. 1998). However, when the transgene was flanked by two copies of the 1.2-kb chicken insulator (Fig. 1B), position effects were suppressed and extinction of expression was prevented (data not shown; Pikaart et al. 1998).
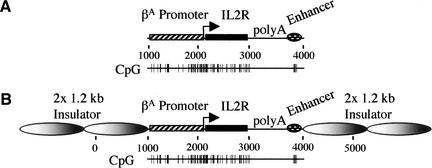
Figure 1.
Insulated and noninsulated IL2R reporters. (A) The IL2R cDNA (black rectangle) and the SV40 poly A signal were linked to the chicken βA-globin promoter (striped rectangle) and a β/ɛ enhancer (hatched oval). (B) Insulated IL2R reporter construct. The expression cassette was flanked by two copies of the 1.2-kb globin insulator (large graded gray ovals). The darker regions in the 1.2-kb insulators indicate the positions of the 250-bp HS4 “core” elements.
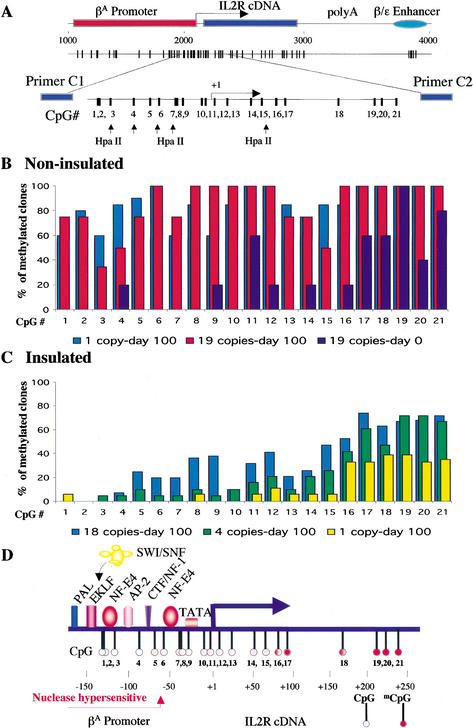
Whereas it has been shown previously that the silent transgene loses several epigenetic hallmarks of active chromatin, including DNA hypomethylation and histone hyperacetylation (Pikaart et al. 1998), there was no clear correlation between the presence of the insulators, transcriptional activity, and DNA methylation of three detectable HpaII restriction sites over the promoter. Some insulated cell lines after long-term growth in culture, even with high expression levels of the IL2R cell surface marker, showed resistance to HpaII digestion due to DNA methylation at these three sites (data not shown; Pikaart et al. 1998). To resolve this apparent paradox and determine precisely how the DNA methylation pattern contributes to the function of the chicken β-globin insulator, we used the bisulfite genomic sequencing method, which, unlike the HpaII digestion method, reveals the methylation state of every CpG. We amplified a region covering the main CpG cluster of the βA-globin promoter, including the transcription start site and part of the IL2R cDNA (Fig. 2A). For each cell line, 20 clones from five individual PCR reactions were sequenced to estimate statistically the level of methylation of each CpG site. The patterns of CpG methylation among the 20 clones were similar from clone to clone (see Supplemental Fig. 1 at http://www.genesdev.org). The noninsulated multicopy cell line 013 was almost free of methylation at CpG dinucleotides 1–10 at day 0 after removal of selection (Fig. 2B), and this was correlated with a high level of IL2R gene expression. The CpGs present downstream of the transcription start site (CpGs 11–21) were partially methylated and obviously did not interfere with transcription. However, this noninsulated cell line at day 100, when it was completely inactive, showed an increase in DNA methylation levels over the entire region, including CpGs 1–10, which were methylated in 40%–100% of sequenced clones (Fig. 2B). The same pattern was also found in a single-copy noninsulated cell line (Fig. 2B). A different picture was observed with insulated cell lines on the same late day in culture. The chicken β-globin insulator prevented the methylation of specific CpG dinucleotides upstream of the transcription start site. Three different cell lines are presented in Figure 2C: line 829 with a single copy of the insulated transgene, line 809 with four copies, and line 804 with 18 copies. In the single-copy and 4-copy lines, CpG sites 1–10 were essentially free of methylation (<10%) after 100 d in culture. It is interesting that this small proximal region of the promoter (−150 to +50 relative to the transcription start site) coincides with binding sites for known factors that specifically regulate the βA-globin promoter in erythroid cells (Fig. 2D; Emerson et al. 1985; Gallarda et al. 1989). In the 18-copy line, the promoter clearly was still protected, but there was a little methylation at sites 5–7 and moderate methylation at sites 8 and 9, possibly related to incomplete protection of some of the copies. The methylation-free region was also shown previously to be hypersensitive to nucleases (Emerson et al. 1985). Therefore, hypomethylation of this region is strongly correlated with gene activity.
Figure 2.
Bisulfite genomic sequencing analysis. (A) The distribution of CpG dinucleotides over the transgene. CpG dinucleotides 1–21 (amplified by primers C1/C2) and the HpaII restriction sites are indicated. (B) CpG methylation over the promoter of noninsulated cell lines 013 (19 copies) and 008 (1 copy). Genomic DNA from day 0 (line 013) and day 100 (lines 013 and 008) was analyzed. The percentage of methylated clones for each CpG is plotted. (C) Distribution of CpG methylation over the insulated promoter. Insulated cell lines 829 (single copy of the transgene), 809 (4 copies), and 804 (18 copies) were analyzed. (D) Methylation map of the transgene flanked by chicken β-globin insulators. The white circles indicate CpGs that are protected from DNA methylation. The partially methylated CpGs are indicated by red-filled circles. The nuclease hypersensitive region protected from DNA methylation corresponds to a part of the chicken βA-globin promoter where transcription factors (colored rectangles and ovals) are known to bind.
We noted that the HpaII sites at CpGs 7 and 15, which were partly resistant to digestion in cell line 804 (Pikaart et al. 1998), were also not methylation-free by the bisulfite-sequencing assay (Fig. 2C). In general, a gradual increase in methylation was seen in CpGs downstream of the transcription start site in insulated transgenes (CpGs 11–21, Fig. 2C), as was observed for the noninsulated transgene at day 0 (see above, Fig. 2B). In both multicopy lines, methylation at sites 16 and higher was greater than 40%.
Methylation of the transgene body and hypomethylation of the enhancer do not correlate with gene activity or insulator presence
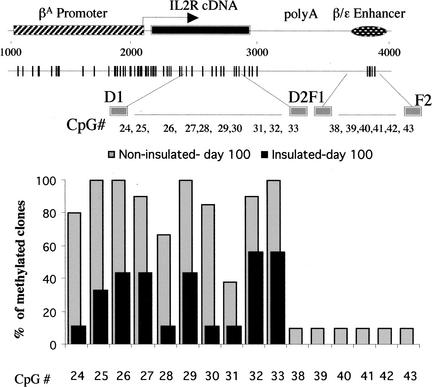
We extended our bisulfite genomic sequencing further into the transgene body and to the β/ɛ enhancer (Fig. 3; Supplemental Fig. 2). We found that CpGs 24–33 within the gene body were methylated to some extent in both insulated and noninsulated transgenes (Fig. 3), and we obtained similar methylation patterns for these transgenes at day 0 and day 100 (data not shown). Therefore, DNA methylation of the gene body, including the CpGs immediately downstream of the transcription start site (CpGs 12–21, Fig. 2C), appeared not to be a determining factor for gene silencing. However, the extent of methylation over the body of the gene was greater in noninsulated transgenes (Fig. 3). We detected little or no CpG methylation in the enhancer fragment, irrespective of gene activity or the presence of the insulators (CpGs 38–43, Fig. 3). Only two of 20 sequenced clones from noninsulated cells showed methylation of the β/ɛ enhancer, despite the fact that the reporter gene was not expressed. This result was not surprising since it confirmed other observations of DNA hypomethylation and chromatin remodeling at enhancers (see below; Lichtenstein et al. 1994; Schübeler et al. 2000).
Figure 3.
Bisulfite genomic sequencing analysis of the gene body and the enhancer in insulated and noninsulated transgenes. For the transgene body, bisulfite-treated genomic DNA was amplified with primers D1/D2 and was analyzed for methylation of CpGs 24–33. Primer set F1/F2 was used to analyze methylation of CpGs 38–43 in the β/ɛ enhancer. Combinations of data for insulated (black bars) and noninsulated (gray bars) cells are plotted.
The chicken β-globin insulator prevents binding of MBD3 and Mi-2 subunits of the Mi-2/NuRD repressor complex to transgenes in vivo
One possible model of insulator action is that insulators protect against transgene silencing by excluding methyl-CpG-binding proteins, histone deacetylases, or other components required for the formation of a repressive chromatin conformation. To investigate this possibility, we used the chromatin immunoprecipitation (ChIP) strategy for MBD3 and the Mi-2 SWI/SNF nucleosomal ATPase, which are subunits of the Mi-2/NuRD repressor complex. It was shown previously that this large multiprotein complex contains histone deacetylases as well as nucleosome remodeling factors, and it has been suggested that this complex could repress transcription by targeting deacetylation of the core histones, either through recognition of regions of methylated DNA (Wade et al. 1999; Zhang et al. 1999; for review, see Bird and Wolffe 1999) or of DNA-binding proteins such as Ikaros (Kim et al. 1999).
To ascertain whether these proteins are present in chicken, we performed Western blots with chicken 6C2 cell nuclear extract (Fig. 4A) using specific antibodies against MBD3 and Mi-2 (Wade et al. 1999). The specific reactions of the antibodies on the blots indicated that these proteins exist in chicken. As controls, the same reactions were done with human HeLa cell nuclear extract (Fig. 4A).
Figure 4.
Presence of MBD3 and Mi-2 proteins in chicken and their binding to silenced transgenes in vivo. (A) Western blot analyses of MBD3 and Mi-2 in chicken 6C2 cell nuclear extract. As a control, immunoblotting with human HeLa cell nuclear extract was also performed. (B) ChIP of a noninsulated cell line. Noninsulated cells that actively transcribe the IL2R gene (day 0 after hygromycin selection) and cells with inactive transgenes (day 100) were analyzed. ChIP was performed with antibodies specific to MBD3, Mi-2, histones H4 and H1, and nonimmune IgG as a control. The values from MBD3, Mi-2, H4, and H1 ChIPs were corrected by subtraction of the nonspecific signal derived from the nonimmune IgG ChIP. The fold enrichments of the analyzed proteins are plotted. (C) ChIP assays showing fold enrichments of MBD-3 and Mi-2 in a single-copy noninsulated cell line at day 100 versus a single-copy insulated cell line at day 100.
Cells that actively transcribed the IL2R gene (noninsulated line 013, day 0) and cells with completely inactive and methylated transgenes (same line, day 100) were analyzed for the presence of MBD3 and Mi-2 proteins (Fig. 4B). After ChIP, the bound fraction was analyzed for the presence of transgene sequences by the fluorescence-based real-time PCR (TaqMan) method. This allowed the precise quantitation of the amount of each immunoprecipitated DNA (Litt et al. 2001b). Each individual bar in Figure 4B and C represents the ratio of the abundance of the probed sequence in the immunoprecipitated fraction to its abundance in the input fraction before ChIP, and was compared with the same ratio for the chicken ovalbumin gene as an internal control. Inactive cell lines showed between four- and 12-fold enrichment of the transgene in MBD3- and Mi-2-bound fractions (Fig. 4B,C). This demonstrated that these proteins are bound to methylated and silenced transgenes in vivo. In a control ChIP experiment with both active and inactive cells, we did not find any difference in the distribution of histone H4 over the transgene (Fig. 4B). However, association of the linker histone H1 was about twofold less with the active IL2R gene, compared to the inactive gene (Fig. 4B). This observation confirms that linker histones are depleted from transcriptionally active chromatin in chicken erythrocytes, as described previously (Kamakaka and Thomas 1990).
We showed in the previous section that the insulators were not able to protect the entire transgene from DNA methylation. Such regions might be targets for Mi-2/NuRD repressor complex (Fig. 2C). To determine whether this occurred, we performed ChIP experiments with insulated cell lines carrying a single copy of the IL2R transgene, and we looked for the presence of MBD3 and Mi-2 (Fig. 4C). In contrast to the behavior of the noninsulated line at day 100 (Fig. 4C), the insulated cell line (line 829) did not show any enrichment of these proteins very late in culture. The chicken β-globin insulator thus directly or indirectly prevents recruitment of the MBD3 and Mi-2 proteins, and consequently of the associated histone deacetylases that might otherwise contribute to inactivation.
High levels of histone acetylation exist over the entire insulated but not noninsulated transgene, with significant peaks of H3 acetylation over the insulator elements
The second model for insulator action suggests that the chicken β-globin insulator recruits transcription activation complexes carrying histone acetyltransferase activity and preserves the insulated transgene in an open chromatin conformation. Consistent with this idea, Litt et al. (2001b) recently showed that there is a strong peak of histone acetylation near the endogenous 5′HS4 element during erythroid development. It was speculated that the natural function of the HS4 insulator in the chicken β-globin locus is to serve as a chain terminator to block spreading of the upstream condensed chromatin region (Litt et al. 2001b).
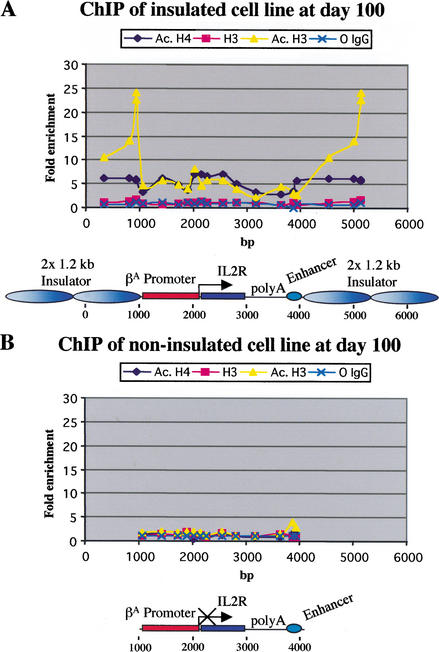
In earlier work (Pikaart et al. 1998), it was shown that the histones in exon 2 of insulated IL2R transgenes are hyperacetylated. To better understand insulator function, it was necessary to determine whether the histone modification was localized at the insulator, or whether this modification was spread over the entire insulated domain. We addressed this question by performing ChIP assays across the entire transgene, including the 1.2-kb insulator elements, using antibodies specific for the acetylated forms of histones H3 and H4. For high resolution, we designed primer sets with TaqMan probes spanning the region at intervals of 200–300 bp. We also used a low formaldehyde concentration, which allowed shearing of chromatin to small-sized fragments. Multicopy cell lines were chosen to preferentially amplify the transgene signal over the endogenous signal. Under these circumstances, the chicken βA-globin promoter, the β/ɛ enhancer, and the HS4 “core” in the endogenous locus of erythroid 6C2 cells should not contribute significantly to the acetylation signal (data not shown; Litt et al. 2001b).
Several cell lines were tested, and a typical profile of histone acetylation over the insulated transgene is presented in Figure 5A. Histones were acetylated across the entire transgene and the insulators. Both the promoter and the IL2R gene body showed high levels of acetylation, with a peak around the transcription start site. The SV40 splice/polyadenylation signal element marked a decrease in acetylation levels, whereas the β/ɛ enhancer showed another peak. The histone H4 levels of hyperacetylation were quite uniform across the transgene, though these were slightly higher in the gene body compared to the acetylated H3. Of particular note were the very large peaks of H3 acetylation over the 1.2-kb insulators.
Figure 5.
Patterns of histone acetylation. (A) Pattern of histone acetylation over the insulated transgene. After a long period in culture, cells with multicopy insulated transgenes were used for ChIP with antibodies against acetylated histones H3 and H4, unmodified histone H3, and nonimmune IgG. Primer sets with TaqMan probes spanning every 200–300 bp of the transgene were used to amplify the bound and input DNA. As a reference, we used the chicken ovalbumin gene. The graph shows the fold enrichment of analyzed proteins in the bound fraction versus input chromatin. (B) Histone acetylation across a noninsulated IL2R transgene at day 100. Inactive cells carrying a multicopy transgene without insulators were processed as described above.
Because bound Mi-2/NuRD repressor complex can induce histone deacetylation, we also carried out a similar experiment with a noninsulated cell line (line 013) at day 100, when the gene is completely off and the promoter is heavily methylated. Very low levels of H3 and H4 acetylation were found throughout much of the noninsulated transgene (Fig. 5B), and this result confirmed that the binding of MBD proteins to DNA is correlated with a wave of deacetylation. The notable exception was the β/ɛ enhancer, which showed about fivefold enrichment of acetylated histones (Fig. 5B), particularly in the case of histone H3; this was similar to the enrichment seen in the transcriptionally active state (Fig. 5A). The acetylation of this enhancer also correlated with the hypomethylated status of its DNA in inactive transgenes (Fig. 3), and this more “open” configuration is consistent with the fact that this enhancer is present as a DNase I hypersensitive site at its endogenous location in 6C2 cells, where the β-globin genes are not expressed (Boyes and Felsenfeld 1996), but where transcription factors are bound.
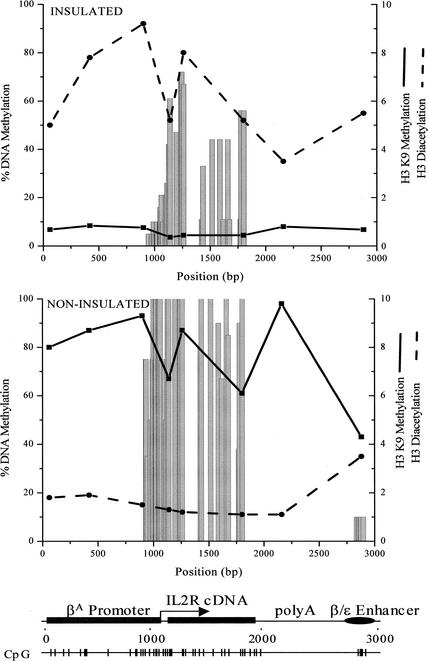
We compared the pattern of DNA methylation with the histone acetylation profile over the insulated gene (Fig. 6). Since CpG islands usually include the promoters and the 5′ transcribed regions of housekeeping genes, they represent a model for the “active” chromatin structure, which is marked by acetylated histones and DNA hypomethylation (Tazi and Bird 1990). Correlation between acetylation and DNA hypomethylation was found for the small promoter region where the erythroid-specific transcription factors are bound (Fig. 2D), but not for the rest of the CpG cluster, including the gene body, where high levels of acetylation were seen even though the DNA was methylated. In addition, the partially methylated region near the transcription start site (Fig. 2C) was also associated with hyperacetylated histones (Fig. 6).
Figure 6.
Comparison of DNA methylation with patterns of histone modifications over the IL2R transgene. After 100 d in culture, cells with insulated and noninsulated transgenes were used for ChIP with antibodies against diacetylated histone H3 and dimethyl-histone H3 (Lys 9). The patterns of these modifications are shown in comparison with DNA methylation. The left y-axis indicates the percentage of methylated clones for each CpG tested. To plot H3 Lys 9 methylation and H3 di-acetylation together, enrichments of 4- and 10-fold, respectively, are set to 10 (right y-axis).
We also determined the pattern of histone H3 lysine 9 (Lys 9) methylation across the transgene in both insulated and noninsulated cell lines (Fig. 6). As expected from previous studies (Litt et al. 2001a), levels of Lys 9 methylation were inversely related to levels of H3 acetylation. Histone methylation at Lys 9 was uniformly low in the insulated transgene, and high in the noninsulated, silenced transgene that had been kept in culture for 100 d. In insulated cells, there was no obvious correlation between DNA methylation over the coding region and either Lys 9 methylation or H3 acetylation, whereas in noninsulated cells, high DNA methylation levels correlated with high levels of Lys 9 methylation at the promoter and over the gene body.
Histone acetylation associated with the chicken β-globin insulator is independent of transgene transcription and the presence of a functional enhancer
To investigate whether transcription is required for targeted histone acetylation by the HS4 element, we abolished IL2R expression by using a nonfunctional enhancer. Three different IL2R insulated constructs, one with a mutation of the NF-E2 site in the β/ɛ enhancer (Fig. 7A), a second with mutations of the two GATA-1 sites in the enhancer (Fig. 7B), and a third with a completely deleted enhancer (Fig. 7C) were stably integrated in chicken 6C2 cells. For each mutation, only a small number of clones expressed the IL2R surface marker (data not shown; Pikaart et al. 1998), in agreement with previous transient transfections in chicken red blood cells (Reitman and Felsenfeld 1988).
Figure 7.
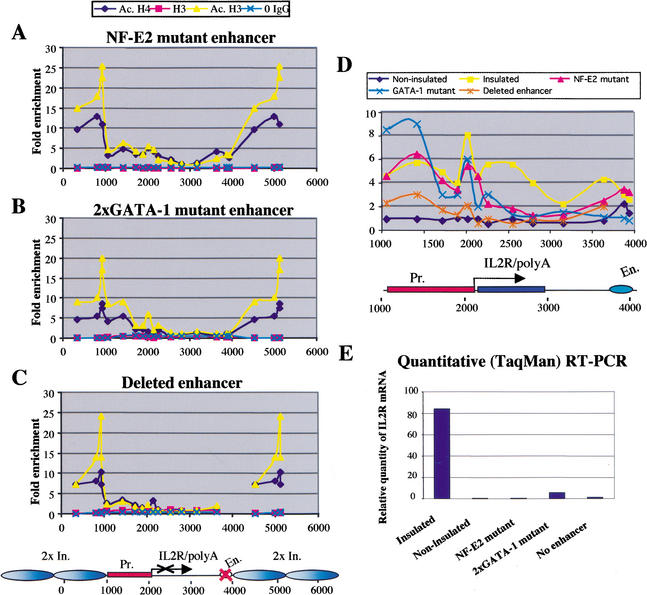
IL2R expression and histone acetylation patterns of insulated transgenes with nonfunctional enhancers. (A) Pattern of histone acetylation over an insulated transgene with an NF-E2 mutant enhancer. ChIP assays with an inactive cell line were performed as described in Figure 5. (B) Histone acetylation pattern over an insulated transgene with a mutation of the two GATA-1 binding sites in the β/ɛ enhancer. (C) Pattern of histone acetylation across an insulated transgene with a deleted enhancer. (D) High-resolution comparison of patterns of H3 acetylation across insulated transgenes with nonfunctional enhancers (insulator acetylation not shown). Insulated and noninsulated cell lines contain a transgene with a wild-type enhancer. (E) Quantitative (TaqMan) RT–PCR of the same cell lines. Each bar on the graph represents the amount of IL2R mRNA relative to the folate receptor mRNA per copy of the transgene.
The amount of IL2R cell-surface marker was analyzed by FACS and compared with the profiles obtained from an active insulated and an inactive noninsulated cell line (data not shown). We also performed quantitative (TaqMan) RT–PCR with the IL2R-negative lines to ensure that the β/ɛ enhancer mutations had abolished transgene transcription (Fig. 7E). For each cell line, the level of IL2R mRNA was compared to the level of folate receptor mRNA, which is expressed in 6C2 cells (Prioleau et al. 1999). This experiment confirmed that IL2R transcription occurs only in the insulated cell line with a wild-type enhancer (Fig. 7E). We analyzed the level of histone acetylation across the insulated transgenes with nonfunctional enhancers. A significant peak of H3 acetylation was found over the HS4 element at both ends of the construct in all analyzed inactive cell lines, even when the enhancer was completely deleted (Fig. 7A–C). Surprisingly, even in the absence of transcription these cell lines show regions of elevated H3 and H4 acetylation within the reporter. In lines carrying deletions of either the NF-E2 or GATA-1 binding sites (Fig. 7A,B), considerable acetylation was seen over the promoter, comparable to that seen in the wild-type construct (Fig. 7D). Lines with a deleted enhancer also showed increased levels of promoter acetylation relative to the levels seen in noninsulated lines (Fig. 7C,D; data not shown). The deletions had their greatest effect on acetylation over the body of the IL2R gene, which was decreased, especially at points more distal to the promoter (Fig. 7D). In all of these mutant lines, the promoter region was also largely hypomethylated, as seen in active cell lines, except that the TATA box region (CpGs 8 and 9) was hypermethylated in the case of the NF-E2 and deleted enhancer mutants (data not shown). We conclude that the insulators permit maintenance of significant levels of acetylation over the promoter in the absence of transcription. Furthermore, the high levels of acetylation seen over HS4 are independent of the transcriptional activity of the adjacent gene, the presence of the enhancer, or the site of insertion into the genome.
The histone deacetylase inhibitor TSA can prevent transcriptional silencing of noninsulated transgenes
If the main function of the β-globin insulator is to recruit HAT activity and maintain the positive balance between histone acetylation and histone deacetylation over the insulated transgene, it should be possible to prevent the extinction of noninsulated transgene activity by inhibiting the process of histone deacetylation. In fact, the level of acetylation is a result of dynamic equilibrium between competing HATs and HDACs. We therefore cultured noninsulated cell lines in the presence of nanomolar concentrations of the histone deacetylase inhibitor trichostatin A (TSA) from the first day after selection, and we analyzed IL2R expression by FACS every 5 d (Fig. 8A). After day 10, a small fraction of cells became inactive (20%–25%), but the rest of the cells maintained a high level of expression throughout long-term growth. It is interesting that the ratio of active versus inactive cells at day 10 was preserved almost constantly for at least 60 d of growth (Fig. 8A). This effect was not because TSA prevented silencing due to a reduced cell proliferation rate. We tested this by starving the cells with reduced amounts of fetal serum in the medium to decrease cell growth. The serum-starved cells showed doubling times similar to the TSA-treated cells, but only the TSA-treated cells expressed the transgene after 20 d in culture (data not shown). When noninsulated cells were cultured under normal conditions in the absence of TSA, very rapid extinction of expression was observed (Fig. 8A). In addition, when we removed TSA from the media after 40 d of growth (data not shown), all of the cells rapidly became inactive, suggesting that maintenance of the stably active state requires constant inhibition of histone deacetylation.
Figure 8.
Effect of TSA and 5-azaC on noninsulated IL2R transgene expression in 6C2 cells. (A) Cells carrying a noninsulated IL2R transgene (line 21) were cultured in the presence (+TSA) or absence of TSA (−TSA) after removal of hygromycin selection. Transgene activity was estimated by FACS, and the number of IL2R-negative and -positive cells was calculated using CellQuest software (Becton Dickinson). The percentages of active and inactive cells are plotted. Nontransfected 6C2 cells were used as an IL2R-negative control. (B) Percentages of active and inactive cells in the presence and absence of 5-azaC. A cell line carrying a noninsulated transgene (line 21) was constantly cultured in the presence of the DNA methylation inhibitor 5-azaC. Experiments with three different concentrations of 5-azaC are presented.
The DNA methylation inhibitor 5-aza-2‘-deoxycytidine (5-azaC) is another drug widely used for reactivation of silenced genes. We showed in the previous section that IL2R transgene silencing is accompanied by heavy DNA methylation over the critical region in the βA-globin promoter. It was therefore interesting to investigate whether 5-azaC alone would stop the extinction of gene expression. We used three different concentrations of 5-azaC to treat the cells from the first day after selection, and we analyzed IL2R activity by FACS analysis. The ratio of active versus inactive cells is presented in Figure 8B. In a dose-dependent manner, 5-azaC somewhat delayed transcription extinction at days 5 and 10, but the majority of cells became inactive after 20 d in culture.
These results show that the histone deacetylase inhibitor TSA, but not the DNA methylation inhibitor 5-azaC, can prevent transcriptional silencing of noninsulated transgenes during long-term growth. Since this scenario is analogous to the behavior seen with insulated transgenes, these experiments further support the notion that the chicken β-globin insulator prevents transgene silencing by inhibiting HDACs and/or by recruiting HATs.
Discussion
The mechanism by which insulators interfere with chromosomal position effects and the silencing of integrated transgenes is not well understood. In this paper, we have asked how transgene expression is regulated and how the chicken β-globin insulator can influence it. Methylation is a mechanism that can be used to silence foreign DNA or to defend against parasitic sequences that are integrated in the host genome (Yoder et al. 1997). We therefore began with a detailed analysis of the DNA methylation pattern of the transgene. We found that although noninsulated transgenes had mostly methylation-free promoters and were transcribed at day 0 after selection, silencing occurred usually in less than 80 d in culture and was accompanied by methylation of the promoter as well as the coding region of the gene. The insulated lines showed a different methylation pattern. As for noninsulated transgenes, methylation over the coding region at day 100 was relatively high. However, insulation did protect CpGs 1–10 in the promoter against methylation. The correlation between expression and low levels of promoter methylation strongly suggests that these sites are critical for gene expression. The fact that CpG sites distributed downstream of the transcription start site and in the gene body were methylated in both active and inactive cells suggests that DNA methylation does not interfere with transcription elongation. This is in agreement with previous studies showing that some genes can be transcribed despite heavy methylation within their coding sequences (Gonzalgo et al. 1998; Jones and Takai 2001). Usually, transgenes are integrated in the host genome in multicopy arrays, which may result in repeat-induced silencing, the mechanism of which is still unknown (Dorer and Henikoff 1997). To investigate whether DNA methylation is somehow related to repeat-induced transgene silencing, we tested cells with a single-copy IL2R transgene. The same profile of CpG methylation was observed, showing that this is not the case.
Promoter methylation is commonly involved in gene repression through at least two possible mechanisms. The most direct effect of cytosine methylation is to prevent binding of transcription factors to their recognition sequence (Tate and Bird 1993). The second mechanism involves recruitment by methyl-CpG-binding proteins of transcriptional repressor complexes to methyl-CpG-enriched regions (Bird and Wolffe 1999; Wade 2001; Magdinier and Wolffe 2001). In our transgene system, DNA methylation of the coding region is not sufficient to induce histone deacetylation and gene repression.
To determine whether complexes involved in transgene silencing were recruited to both insulated and noninsulated transgenes, we used the ChIP strategy to detect the presence of MBD3 and the Mi-2 nucleosomal ATPase. MBD3 was shown previously to be a component of the NuRD or Mi-2 histone deacetylase and nucleosome remodeling complex, purified from both HeLa cells and Xenopus egg extracts (Wade et al. 1999; Zhang et al. 1999). This complex appears to be the most abundant macromolecular form of histone deacetylase found in Xenopus eggs and in mammalian cells (Bird and Wolffe 1999). However, its repressor effect in vivo remains unclear. The Mi-2 subunits of these complexes belong to the CHD (chromo-helicase/ATPase-DNA binding) family of proteins (Woodage et al. 1997). The Mi-2 nucleosomal ATPase presumably remodels nucleosome structure and facilitates the deacetylation of core histones. The noninsulated and inactive cell lines that we analyzed showed 4–12-fold enrichment of the transgene in MBD3- and Mi-2-bound fractions, which demonstrates that these proteins are bound to silenced transgenes in vivo. We were not able to map precisely the binding sites for these proteins over the transgene, because a higher concentration of formaldehyde was required to cross-link these proteins to DNA, and the average chromatin fragment size was 1 kb, which decreased the resolution of the ChIP assay. Nonetheless, the absence of MBD3 and Mi-2 from the insulated transgenes suggests that local DNA methylation is not a sufficient recognition signal for these proteins.
We have described at least three contributions to the inactivation of the noninsulated IL2R reporter gene when it is integrated in the host genome: DNA methylation of specific CpGs in the promoter, binding of the Mi-2 repressor complex to the transgene, and remodeling of the chromatin by deacetylation of core histones, presumably leading to chromatin condensation (not necessarily occurring in that order). How do insulators flanking the reporter gene antagonize these layers of epigenetic modifications and prevent gene silencing? First, histone acetylation appears to be used by the β-globin insulator as one layer of transcriptional control. This is evident from the high peak of histone H3 di-acetylation that was found over the insulator elements in the insulated transgene, as well as over the endogenous 5′HS4 element in the chicken β-globin locus in both erythroid and nonerythroid cells (Litt et al. 2001b). It should be noted that individual acetylation sites on histone H4 are also elevated over the endogenous HS4 element, but tetra-acetylated H4, though enriched, is not as abundant (as in the case of the IL2R transgene). These elevated acetylation levels suggest that histone acetyltransferase activity is recruited (and/or HDAC activity is inhibited), and it constantly keeps the insulated region in an accessible chromatin conformation that facilitates transcription. Interestingly, this does not depend on transcriptional activity or on the presence of a functional enhancer. A similar effect has been described in yeast, where the histone acetyltransferase Gcn5 is targeted by the transcription activator Gcn4 to a specific promoter independently of transcription (Kuo et al. 2000). Another effect of insulator acetylation could be to provide a boundary against the spreading of condensed chromatin at unfavorable transgene integration sites. This is consistent with a report showing that targeting of HAT activity between the budding yeast heterochromatic HMR-E silencer and the al gene is sufficient to prevent silencing extending from the HMR-E element (Donze and Kamakaka 2001). In vertebrates, condensed chromatin is marked not only by decreased histone acetylation, but also by an increase in histone H3 methylation at Lys 9 and a decrease at Lys 4 (Litt et al. 2001a). It has been suggested (Litt et al. 2001a) that the competing acetylation reaction at the HS4 insulator, which also modifies Lys 9, may block the advance of a processive Lys 9 methylation (Bannister et al. 2001) and thus confer protection against further chromatin condensation and consequent silencing.
Because the action of the insulator appeared to involve maintenance of histone acetylation over the transgene, we asked whether another method of maintaining high levels of acetylation would have the same effect. We therefore grew cell lines carrying the noninsulated transgene in the presence of the histone deacetylase inhibitor TSA. The results in Figure 8 show that in the constant presence of TSA, IL2R activity is to a large extent maintained, suggesting that maintenance of acetylation is indeed sufficient to sustain activity. These results must be compared to earlier studies, which showed that TSA treatment after partial extinction of expression of noninsulated IL2R transgenes could result in partial reactivation (Pikaart et al. 1998). Other evidence (Lorincz et al. 2000) suggests that the incompleteness of reactivation reflects DNA methylation of crucial CpG sites within the locus, with the methylation density increasing with time in culture. Interestingly, inhibition of DNA methylation by 5-azaC alone was not sufficient in our experiments to stop transcriptional extinction in long-term cultures (Fig. 8B). It did however delay extinction up to 10 d in culture, consistent with previous studies showing partial reactivation of early-stage noninsulated transgenes during short-term culture in the presence of 5-azaC (Pikaart et al. 1998). Thus, maintenance of acetylation appears to be the critical event in preventing extinction in long-term cultures.
Our observation that the insulator prevents the methylation of specific CpG dinucleotides in the promoter revealed DNA methylation as a second layer of insulator regulation. The small hypomethylated promoter proximal region (−150 to +50 relative to the transcription start site, Fig. 2D) in insulated cell lines contrasts with the rest of the transgene and with silenced noninsulated genes, where higher CpG methylation levels exist. It is significant that this promoter region corresponds to binding sites for factors that specifically regulate the βA-globin promoter in erythroid cells (Fig. 2D; Emerson et al. 1985; Gallarda et al. 1989). These factors include NF-E4, AP-2, CTF/NF-1, and the erythroid-specific EKLF factor, which can target an SWI/SNF-related chromatin remodeling complex to the human β-globin gene promoter (Armstrong et al. 1998). We therefore propose a model (Fig. 9) for the role of this region in protection against silencing that involves histone acetylation and transcription factor binding: (1) The insulators are able to recruit HAT activity and to maintain an open chromatin conformation across the insulated domain, even after an extended period in cell culture. (2) This more accessible chromatin structure subsequently facilitates the binding of regulatory proteins at the promoter and ultimately, the recruitment of RNA polymerase. (3) It has been shown that the occupation of a promoter region by bound regulatory proteins prevents the access of DNA methyltransferases, thus inhibiting DNA methylation. Promoter demethylation, dependent on replication, has been found at sites where transcription factors are bound to DNA (Matsuo et al. 1998). Recently, with the use of the E. coli lac repressor-operator system, it was shown that site-specific binding of proteins protects DNA sites on an episome and in the genome from de novo methylation in human cells (Lin et al. 2000; Han et al. 2001). Our results thus suggest that there is a second relationship between histone acetylation and DNA methylation in addition to the familiar chain of events in which DNA methylation induces binding of methyl-DNA specific complexes, which in turn recruit histone deacetylases that lead to gene inactivation (Bird and Wolffe 1999).
Figure 9.
A schematic representation of the proposed mechanism by which the β-globin insulator protects the IL2-R promoter from inactivation by DNA methylation. We suggest that because the insulator elements maintain a high level of histone acetylation over the protected region they surround, the promoter is more accessible to transcription factors. The binding of these factors in turn protects the promoter DNA from methylation. The coding region, though it also carries acetylated histones, does not bind such factors and is therefore not protected against methylation.
There is recent evidence for a dependence of DNA methylation at CpG sites in Neurospora, or CpNpG sites in Arabidopsis, upon enzymatic activities responsible for histone H3 methylation at Lys 9 (Tamaru and Selker 2001; Jackson et al. 2002). This behavior may reflect mechanisms in which methylated histones locally recruit DNA methylases. Our results, however, do not show a straightforward correlation between the patterns of H3 Lys 9 methylation and DNA methylation. As shown in Figure 6, the coding region DNA of the insulated IL2R transgene is highly methylated despite the fact that H3 Lys 9 methylation levels are low over that same region. This shows either that DNA methylation does not always require H3 Lys 9 methylation, or that the events establishing these patterns occurred at some earlier point after transgene integration when Lys 9 methylation patterns were different.
It has been suggested (Bird 2002) that the protection against methylation observed in CpG islands is a consequence of expression of the adjacent genes during embryonic development. Although it is unlikely that CpG islands could be protected against methylation solely by binding of transcription factors, it may be that the presence of histones modified by acetylation and other markers of transcriptional activity provide additional signals that inhibit DNA methylation (Bird 2002). Such a model, in which histone modification prevents DNA methylation, would be consistent with our observations. Bird also suggests that DNA methylation at previously active genes may be a secondary event that follows inactivation of gene expression through other mechanisms. The pathway we propose involves histone deacetylation as the primary determinative event in inactivation, with DNA methylation a subsequent event, perhaps not essential to the inactivation. The data we have presented suggest that in insulated lines the maintenance of gene activation depends on local acetylation of the promoter with consequent prevention of DNA methylation. The additional finding that high levels of acetylation within the insulated domain do not depend on having a transcriptionally active gene provides support for models in which insulators act as the primary elements in maintenance of acetylation. These data are also consistent with the observation that insulators appear to maintain (rather than alter) whatever levels of expression are characteristic of the internal promoter and enhancers they surround.
Materials and methods
Plasmid constructs, cell culture, transfections, and FACS analysis
The insulated and noninsulated IL2R reporter constructs carrying the wild-type β/ɛ enhancer or the enhancer with mutated NF-E2 or GATA-1 sites were described previously (Pikaart et al. 1998). The reporter construct with a deleted enhancer is identical to the wild-type version, except that the β/ɛ enhancer fragment downstream of the IL2R gene is missing. 6C2 cells were grown as described (Boyes and Felsenfeld 1996). For the starving experiment, the amount of fetal serum was reduced from 10% to 0.5% and the cells were incubated at 32°C. For histone deacetylase inhibitor experiments, TSA (Wako Pure Chemical Industries) was added at a final concentration of 1–10 nM. The drug and the medium were replenished regularly. For experiments with a DNA methylation inhibitor, 5-azaC (Sigma) at a 0.1–50 μM final concentration was administered daily due to its short half-life in aqueous solution.
6C2 cells were stably transfected with the reporters as described (Boyes and Felsenfeld 1996). IL2R expression was monitored by FACS analysis as described (Pikaart et al. 1998). Cells were maintained in log-phase growth for all experiments.
Bisulfite genomic sequencing
Bisulfite conversion of DNA was carried out by the method developed by Clark et al. (1994) with minor modifications. Two micrograms of purified genomic DNA was denatured by adding freshly prepared NaOH to a final concentration of 0.2 N in a 50-μL reaction volume, with incubation for 10 min at 37°C. Fresh solutions of hydroquinone (Sigma) and sodium bisulfite (Sigma) (pH 5.0) were prepared and added to the denatured DNA at final concentrations of 0.5 mM and 3.1 M, respectively, in a final volume of 600 μL. The reactions were mixed, overlaid with mineral oil, and incubated in the dark for 18 h at 50°C. The DNA was desalted followed by a desulfonation step with 0.3 N NaOH for 10 min at room temperature. The samples were neutralized and the DNA was recovered by ethanol precipitation. The converted DNA was resuspended in 50 μL of 10 mM Tris (pH 8.0). Next, 1.5 μL of each sodium bisulfite-treated DNA sample (50 μL final volume) was subjected to PCR amplification using one of the three primer sets: C1/C2, D1/D2, or F1/F2 (see Results and Supplemental Table). Platinum Taq DNA polymerase (Invitrogen) was used for amplification as follows: 95°C for 5 min; 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final incubation at 72°C for 10 min. The PCR products were purified, subcloned, and individual clones were sequenced using the BigDye terminator cycle sequencing kit (PE Applied Biosystems) and an ABI PRISM 310 DNA sequencer (Perkin Elmer).
Western blot analysis
Nuclear extracts from 6C2 cells were generated as described (Evans at al. 1988). HeLa cell nuclear extracts were purchased from Upstate Biotechnology. The proteins were separated on a 4%–12% NuPAGE bis-tris gel (Novex) and transferred to nitrocellulose membrane (Novex) in NuPAGE transfer buffer (Novex) containing 10% MeOH, 0.01% SDS at 25 V for 1.25 h. The membranes were probed with MBD3 serum (1:1000 dilution, provided by P. Wade) or Mi-2 antibody (1:3000 dilution, Wade et al. 1999) according to the protocol described (Mutskov et al. 1998).
In vivo formaldehyde cross-linking and ChIP
In vivo protein–DNA cross-linking was performed by incubating cells (∼1.5 × 108 cells at a density of 1.5 × 106 cells/mL) with formaldehyde at a final concentration of 1%, as described by Orlando and Paro (1993). For histone acetylation studies, 0.3% of formaldehyde was used. The chromatin isolation and the purification of covalently-linked protein–DNA complexes by CsCl isopyknic centrifugation were done essentially as described (Orlando and Paro 1993), except for the sonication procedure. Three parts of nuclei suspension were mixed with one part of 0.5 mm glass beads (Biospec Products) and sonicated at 4° using a sonicator microtip (Ultrasonic liquid processor, Misonix) at 50%–60% maximum power. Ten pulses of 30 sec each were performed, separated by cooling on ice for 30 sec. All buffers for the chromatin isolation were supplemented with protease inhibitors (1 mM PMSF, 1 μg/mL pepstatin A, 1 μg/mL leupeptin). In some experiments, instead of CsCl gradient purification, nuclei were lysed in a 1% SDS final concentration for 10 min on ice, and the chromatin was directly used for immunoprecipitation. ChIPs, reversal of cross-links, and DNA purifications were performed according to Upstate Biotechnology’s protocol. For each ChIP, 10–20 μL of one of the following polyclonal antibodies were added to the chromatin solution: MBD3 serum, Mi-2 antibody, histone H1 antibody (provided by Iliya Pashev, Bulgarian Academy of Sciences), histone H4 antibody (provided by Iliya Pashev), acetylated histone H3 antibody (Upstate Biotechnology), acetylated histone H4 antibody (Upstate Biotechnology), dimethyl-histone H3 (Lys 9) antibody (Upstate Biotechnology), histone H3 antibody (Upstate Biotechnology), or normal rabbit IgG (Upstate Biotechnology).
Real-time PCR using TaqMan probes
DNA from input and antibody-bound chromatin were analyzed by real-time PCR using the TaqMan Universal PCR Master Mix protocol (PE Applied Biosystems) and an ABI Prism 7700 sequence detector, as described (Litt et al. 2001b). Each PCR was carried out in triplicate to control for PCR variation, and the Ct values were collected. The Ct is the number of PCR cycles necessary to reach a predetermined fluorescence intensity and is a function of the amount of target DNA in the analyzed samples. Quantification was determined by applying the comparative Ct method, as described by Litt et al. (2001b). Primers and TaqMan probes used (Supplemental Table) were selected from the IL2R transgene, including the 1.2-kb chicken β-globin insulator, using PE Applied Biosystems Primer Express software.
Quantitative TaqMan RT–PCR assay
Total RNA was isolated using the RNeasy mini kit (QIAGEN) and was then treated for 15 min at room temperature with DNase I (Invitrogen) at a final concentration of 0.1 U/μL. One microgram of RNA was reverse transcribed with random hexamers (Promega) and Superscript II RNase H− reverse transcriptase (Invitrogen). Subsequently, the cDNA products were amplified by real-time PCR using specific primers and TaqMan probes for the IL2R transgene (see Supplemental Table, IL2R2800) and the folate receptor gene (see Supplemental Table, FolateExon4).
Acknowledgments
We thank Michael Pikaart for providing IL2R transgenic constructs and cell lines, Iliya Pashev for anti-histone H1 and H4 antibodies, Michael Litt for advice on real-time PCR (TaqMan) methods, and Tommy Brodigan for DNA sequencing. We also acknowledge members of the Felsenfeld laboratory for useful discussions and critical reading of this manuscript. The authors note with great regret the death of our co-author and colleague, Alan Wolffe, during the course of this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gary.felsenfeld@nih.gov; FAX (301) 496-0201.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.988502.
References
- Armstrong JA, Bieker JJ, Emerson BM. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- Bi X, Broach JR. Chromosomal boundaries in S. cerevisiae. Curr Opin Genet Dev. 2001;11:199–204. doi: 10.1016/s0959-437x(00)00179-9. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;1:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 2001;20:520–531. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–1190. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson BM, Lewis CD, Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult β-globin gene: Nature of the binding domain. Cell. 1985;41:21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallarda JL, Foley KP, Yang ZY, Engel JD. The β-globin stage selector element factor is erythroid-specific promoter/enhancer binding protein NF-E4. Genes & Dev. 1989;3:1845–1859. doi: 10.1101/gad.3.12a.1845. [DOI] [PubMed] [Google Scholar]
- Geyer PK. The role of insulator elements in defining domains of gene expression. Curr Opin Genet Dev. 1997;7:242–248. doi: 10.1016/s0959-437x(97)80134-7. [DOI] [PubMed] [Google Scholar]
- Gonzalgo ML, Hayashida T, Bender CM, Pao MM, Tsai YC, Gonzales FA, Nguyen HD, Nguyen TT, Jones PA. The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res. 1998;58:1245–1252. [PubMed] [Google Scholar]
- Han L, Lin IG, Hsieh CL. Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol Cell Biol. 2001;21:3416–3424. doi: 10.1128/MCB.21.10.3416-3424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jones P, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kamakaka RT, Thomas JO. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990;9:3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kuo MH, vom Baur E, Struhl K, Allis CD. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol Cell. 2000;6:1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: A novel role for the intronic kappa chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- Lin IG, Tomzynski TJ, Ou Q, Hsieh CL. Modulation of DNA binding protein affinity directly affects target site demethylation. Mol Cell Biol. 2000;20:2343–2349. doi: 10.1128/mcb.20.7.2343-2349.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001a;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Recillas-Targa F, Prioleau M-N, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001b;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Schübeler D, Goeke SC, Walters M, Groudine M, Martin DI. Dynamic analysis of proviral induction and de novo methylation: Implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol Cell Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdinier F, Wolffe AP. Selective association of the methyl-CpG binding protein MBD2 with the silent p14/p16 locus in human neoplasia. Proc Natl Acad Sci. 2001;98:4990–4995. doi: 10.1073/pnas.101617298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Silke J, Georgiev O, Marti P, Giovannini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V, Gerber D, Angelov D, Ausio J, Workman J, Dimitrov S. Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol Cell Biol. 1998;18:6293–6304. doi: 10.1128/mcb.18.11.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes & Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau M-N, Nony P, Simpson M, Felsenfeld G. An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Felsenfeld G. Mutational analysis of the chicken β-globin enhancer reveals two positive-acting domains. Proc Natl Acad Sci. 1988;85:6267–6271. doi: 10.1073/pnas.85.17.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Lorincz MC, Cimbora DM, Telling A, Feng YQ, Bouhassira EE, Groudine M. Genomic targeting of methylated DNA: Influence of methylation on transcription, replication, chromatin structure, and histone acetylation. Mol Cell Biol. 2000;20:9103–9112. doi: 10.1128/mcb.20.24.9103-9112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bird A. Alternative chromatin structure at CpG islands. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Wade PA. Methyl CpG binding proteins: Coupling chromatin architecture to gene regulation. Oncogene. 2001;20:3166–3173. doi: 10.1038/sj.onc.1204340. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: Many functions, many mechanisms. Genes & Dev. 2002;1:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng H-H, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]