Abstract
HATs (histone acetyltransferases) contribute to the regulation of gene expression, and loss or dysregulation of these activities may link to tumorigenesis. Here, we demonstrate that expression levels of HATs, p300 and CBP [CREB (cAMP-response-element-binding protein)-binding protein] were decreased during chemical hepatocarcinogenesis, whereas expression of MOZ (monocytic leukaemia zinc-finger protein; MYST3) – a member of the MYST [MOZ, Ybf2/Sas3, Sas2 and TIP60 (Tat-interacting protein, 60 kDa)] acetyltransferase family – was induced. Although the MOZ gene frequently is rearranged in leukaemia, we were unable to detect MOZ rearrangement in livers with hyperplastic nodules. We examined the effect of MOZ on hepatocarcinogenic-specific gene expression. GSTP (glutathione S-transferase placental form) is a Phase II detoxification enzyme and a well-known tumour marker that is specifically elevated during hepatocarcinogenesis. GSTP gene activation is regulated mainly by the GPE1 (GSTP enhancer 1) enhancer element, which is recognized by the Nrf2 (nuclear factor-erythroid 2 p45 subunit-related factor 2)–MafK heterodimer. We found that MOZ enhances GSTP promoter activity through GPE1 and acts as a co-activator of the Nrf2–MafK heterodimer. Further, exogenous MOZ induced GSTP expression in rat hepatoma H4IIE cells. These results suggest that during early hepatocarcinogenesis, aberrantly expressed MOZ may induce GSTP expression through the Nrf2-mediated pathway.
Keywords: glutathione S-transferase placental form (GSTP), hepatocarcinogenesis, histone acetyltransferase (HAT), MafK, monocytic leukaemia zinc-finger protein (MOZ), nuclear factor- erythroid 2 p45 subunit-related factor 2 (Nrf2)
Abbreviations: AAF, 2-acetylaminofluorene; AML1, acute myeloid leukaemia 1; bZIP, basic region leucine zipper; CREB, cAMP-response-element-binding protein; CBP, CREB-binding protein; DEN, diethylnitrosoamine; DTT, dithiothreitol; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GCN5, positive general control of transcription-5; GST, glutathione S-transferase; GSTP, GST placental form; GPE, GSTP enhancer; HA, haemagglutinin; HAT, histone acetyltransferase; HBO1, HAT binding to ORC1 (origin recognition complex subunit 1); ING, inhibitor of growth; MOZ, monocytic leukaemia zinc-finger protein; MORF, MOZ related factor; MYST, MOZ, Ybf2/Sas3, Sas2 and TIP60; Nrf2, nuclear factor-erythroid 2 p45 subunit-related factor 2; ORF, open reading frame; P/CAF, p300/CBP-associated factor; PH, partial hepatectomy; PHD, plant homeodomain; RT, reverse transcriptase; TIF2, transcriptional intermediary factor 2, TIP60, Tat-interacting protein, 60 kDa; TRE, PMA (‘TPA’)-responsive element
INTRODUCTION
Acetylation is an important post-translational modification known to occur in histones, transcription factors and other proteins [1]. Histone acetylation is catalysed by HATs (histone acetyltransferases), which transfer an acetyl group from acetyl-CoA to lysine residues in histones. Lysine acetylation destabilizes the nucleosome structure and promotes the accessibility of transcription factors to a genetic locus. In agreement with these phenomena, acetylated chromatin has been associated with a transcriptionally activated state [2].
The well-characterized transcriptional co-activators CBP [CREB (cAMP-response-element-binding protein)-binding protein], p300, GCN5 (positive general control of transcription-5), and P/CAF (p300/CBP-associated factor) have intrinsic HAT activity [3–6]. HATs are divided into several groups on the basis of their similarity in homologous regions including acetyl-CoA-binding motifs [1,7]. For example, p300 and CBP, and P/CAF and GCN5 share a remarkable degree of similarity throughout their sequences respectively, and they play distinct but functionally overlapping roles [1,8,9]. Another group of evolutionarily related HATs is the MYST [MOZ (monocytic leukaemia zincfinger protein), Ybf2/Sas3, Sas2 and TIP60 (Tat-interacting protein, 60 kDa)] family. MYST proteins not only contribute to transcriptional activation, but they also have diverse roles in various nuclear processes, including cell-cycle progression, DNA repair, DNA replication and gene silencing [10–16].
Recent studies indicate that some HATs play roles in tumour suppression and that loss or misregulation of these activities may lead to cancer [17]. Development of liver cancer is controlled by several transcriptional factors, such as c-Jun, Foxm1b (forkhead box m1b) and p53 [18,19]. These factors are acetylated by p300 and CBP, which thereby modulate the transcriptional activity of these factors [1,19]. Hyperacetylation of histones in the promoter region of c-Jun is detected [20]. In addition, p53 recruits p300 to nucleosomal histones within the p21 promoter in vitro [21]. These results suggest that HATs may be involved in hepatocarcinogenesis, but the underlying mechanism has not been addressed. Here, we show that MOZ (also known as MYST3), a member of the MYST family of HATs, is induced during hepatocarcinogenesis. MOZ frequently is rearranged in leukaemia [10,22–27], and it regulates transcription mediated by the haemopoietic transcriptional factor AML1 (acute myeloid leukaemia 1) and the MOZ fusion protein, which is generated by translocation, down-regulates in haemopoiesis and leads to leukaemogenesis [28].
GSTP [GST (glutathione S-transferase) placental form] is a Phase II detoxification enzyme and a well-known tumour marker that is specifically elevated during chemical hepatocarcinogenesis in the rat [29]. GSTP gene expression is regulated mainly through GPE (GSTP enhancer), located approx. 2.5 kb upstream from the cap site, and the silencer [30,31]. GPE1, a strong enhancer element in GPE, is responsible for GSTP gene expression during hepatocarcinogenesis in vivo [32,33]. Recently, we showed that a heterodimer comprising Nrf2 (nuclear factor-erythroid 2 p45 subunit-related factor 2) and MafK binds to GPE1 and enhances GSTP promoter activity [34]. Nrf2, a member of bZIP (basic region leucine zipper) family of transcription factors, induces Phase II detoxifying and antioxidative genes [35]. Nrf2 plays a crucial role in early defence against chemical stress and carcinogenesis [36].
To characterize the roles of HATs during hepatocarcinogenesis, we examined their expression profiles and showed that expression of MOZ was induced under these conditions. Further, we found that MOZ acted as a co-activator of the Nrf2–MafK heterodimer and induced expression of GSTP. These results suggest that MOZ induces GSTP expression through the Nrf2-mediated pathway during early hepatocarcinogenesis.
EXPERIMENTAL
Chemical hepatocarcinogenesis of rats
Carcinogenic experiments were performed according to the Solt–Farber protocol [37]. Experiments were initiated by intraperitoneal injection of DEN (diethylnitrosoamine; 200 mg/kg) into 5-week-old Wister rats. After the animals had been fed basal diets for 2 weeks, diets were changed to basal diets containing 0.02% AAF (2-acetylaminofluorene). Three weeks after the DEN injection, a PH (partial hepatectomy) was performed; livers were extirpated 7 weeks after the DEN injection. Control rats were injected with saline and fed basal diets. All animal care and handling procedures were approved by the Animal Care and Use Committee of Osaka University.
Preparation of nuclear extracts, cytosol fractions and RNA from rat liver
Procedures for preparation of nuclear extracts and cytosol fractions from rat liver were described previously [38]. Livers were homogenized in a sucrose-containing buffer, and nuclei were purified by centrifugation. Nuclear proteins were extracted with 0.55 M KCl and centrifuged at 40000 g for 60 min at 4 °C. The supernatants were used for the HAT assay and Western blot analysis. Total RNA was isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, U.S.A.) in accordance with the manufacturer's recommendations.
Western blotting and antibodies
Proteins were resolved using SDS/PAGE, transferred to nitrocellulose or PVDF membrane and detected using the ECL® (enhanced chemiluminescence) Western blotting analysis detection system (Amersham Biosciences, Piscataway, NJ, U.S.A.). For the generation of antibodies against the N- and C-terminal regions of MOZ, nucleotides corresponding to amino acid residues 1–331 and 1717–1998 respectively were cloned into pET-28a (Novagen, Darmstadt, Germany). The resulting His6-tagged fusion poly-peptides were expressed in bacteria and purified over nickel-nitrilotriacetic acid–agarose (Qiagen, Hilden, Germany). These proteins were injected into rabbits, and antibodies were affinity-purified using Protein A–Sepharose (Amersham Biosciences). The anti-P/CAF antibody was a gift from Dr Y. Nakatani (Harvard Medical School, Boston, MA, U.S.A.). The following antibodies were commercially available: anti-p300 (N-15, Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), anti-CBP (A-22, Santa Cruz Biotechnology), anti-GCN5 (N-18, Santa Cruz Biotechnology), anti-TIP60 (Upstate Biotechnology, Lake Placid, NY, U.S.A.), anti-MORF (MOZ-related factor; C-15, Santa Cruz Biotechnology), anti-MYST (Upstate Biotechnology), anti-GSTP (Biotrin, Dublin, Ireland), anti-HA (haemagglutinin) (6B12, Babco, Berkeley, CA, U.S.A.), and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (MAB374, Chemicon, Temecula, CA, U.S.A.).
Plasmid construction
The rat MOZ expression plasmid pCI-MOZ has been described previously [39]. Mutants within the PHD (plant homeodomain) finger and the MYST regions of the Moz gene (pCI-MOZ-PHDmut and pCI-MOZ-MYSTmut) were generated using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.) following the manufacturer's recommended protocols. All mutations were verified by sequencing over the region of change. For construction of Myc-tagged MOZ-expressing plasmids, the MOZ ORF (open reading frame) was subcloned into the EcoRI–NotI site of pCMV-Myc (Clontech, Franklin Lakes, NJ, U.S.A.). For construction of −2.5GST-luciferase, the fragment from −2.5 kb to −91 kb of the GSTP gene [30] was inserted into the SacI site of −91GST-luciferase [38]. To generate −2.15GST-luciferase (the GPE deletion reporter plasmid), −2.5GST-luciferase was digested with SmaI and AccI, blunted with Klenow fragment (Toyobo, Osaka, Japan), and then self-ligated. The Nrf2 expression plasmid (pAβ2-Nrf2), including the human β-actin promoter and enhancer, and GPE1 reporter plasmid (GPE1-luciferase) were described previously in [40]. The HA-tagged rat MafK expression plasmid (pCMV-HA-MafK) was generated by PCR amplification of the MafK ORF [40] using primers that incorporate SalI and NotI at the 5′ and 3′ ends respectively. The PCR product was cloned into the SalI–NotI site of pCMV-HA (Clontech). The non-tagged MafK expression plasmid pRSV-MafK contained MafK cDNA controlled by the Rous sarcoma virus long terminal repeat. For construction of MafK/GEX-KG, MafK cDNA was cloned into pGEX-2T (Amersham Biosciences).
Cell culture
Rat hepatoma H4IIE cells and mouse embryonic carcinoma F9 cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) FBS (fetal bovine serum). HeLa cells were cultured in minimal essential medium supplemented with 10% FBS.
Transfection and reporter gene assays
Transfection of H4IIE and F9 cells was performed using FuGENE™ 6 (Roche, Indianapolis, IN, U.S.A.) in accordance with the manufacturer's instructions. For H4IIE cells, all transfections included 100 ng of the reporter plasmid, with or without 1 μg of the MOZ expression plasmid (pCI-MOZ). The amount of plasmid in the transfection was kept constant by using empty pCI vector. Transfectants were harvested 48 h after transfection. The luciferase assay was performed as described previously [38] and protein concentrations were determined by the method of Bradford. Luciferase activities were normalized to the protein amount. In some experiments, the transfection efficiency was checked by co-transfection with pRSV-GAL, a eukaryotic expression plasmid that contained the Escherichia coli β-galactosidase structural gene controlled by the Rous sarcoma virus long terminal repeat. β-Galactosidase activity was assayed as described in [38]. We confirmed that the variation of transfection efficiency was <15%. Relative luciferase activity was estimated by the luciferase activity from −2.5GST-luciferase in the absence of MOZ.
For F9 cells, all transfections contained 100 ng of reporter plasmid (GPE1-luciferase) and 5 ng of Renilla luciferase plasmid phRL-tk (Promega, Madison, WI, U.S.A.) as an internal control to normalize for transfection efficiency, with or without 1 μg of the MOZ expression plasmid (pCI-MOZ) in the presence or absence of 5 ng of the Nrf2 expression plasmid (pAβ2-Nrf2). The amount of plasmid in the transfection was kept constant by using empty pCI vector. At 48 h after transfection, cells were harvested and assayed for luciferase activity using the Dual-luciferase Reporter Assay System (Promega) in accordance with the manufacturer's recommendations. Reported values are relative to the activity of GPE1-luciferase without transfection of Nrf2 and MOZ. All experiments were repeated at least three times with two or three different preparations of DNA.
GST pull-down assay
The recombinant plasmid was transformed into BL21(DE3)pLysS cells. Transformants were grown overnight at 30 °C in Luria–Bertani medium containing 100 μg/ml ampicillin. The culture then was diluted 25-fold and grown to an attenuance (D600) of 0.4; at that time, isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 0.1 mM. The cells were allowed to grow for an additional 4.5 h and then were harvested by centrifugation; resuspended in a buffer containing 0.15 M KCl, 50 mM Tris (pH 8.0), 10% (v/v) glycerol, 0.1% Tween 20, 1 mM DTT (dithiothreitol) and 1 mM PMSF, and disrupted by sonication. After centrifugation at 7000 g for 10 min, the supernatant was cross-linked to glutathione–Sepharose 4B with dimethylpimelimidate. 35S-labelled MOZ proteins were produced using pCI-MOZ, pCI-MOZ-PHDmut and pCI-MOZ-MYSTmut as templates by in vitro transcription–translation with the TNT T7-coupled reticulocyte lysate system (Promega). A 5 μl aliquot of the reticulocyte lysate reaction containing 35S-labelled MOZ proteins was incubated for 3 h at 4 °C in a buffer containing 0.15 M KCl, 50 mM Tris (pH 7.6), 10% (v/v) glycerol, 1 mM DTT and 1 mM PMSF with GST fusion proteins. After extensive washes, bound proteins were separated by SDS/PAGE and detected by autoradiography.
Immunoprecipitation assay
Myc-tagged MOZ expression plasmid (pCMV-Myc-MOZ) was co-transfected into HeLa cells with HA-tagged MafK (pCMV-HA-MafK) or non-tagged MafK (pRSV-MafK) by the calcium phosphate co-precipitation method [41]. The cells were harvested 48 h after transfection, and nuclear extracts from the transfected HeLa cells were prepared as described in [38]. Nuclear extracts were diluted by adding nuclear lysis buffer containing 20 mM Hepes (pH 7.9), 1 mM EDTA, 0.5 mM spermidine, 1 mM DTT, 10% glycerol, 1 mM PMSF, 1 μg/ml pepstatin A and 1 μg/ml leupeptin (final KCl concentration, 0.15 M). To immunoprecipitate HA-tagged protein, we incubated extracts with anti-HA antibody immobilized on Sepharose beads overnight at 4 °C. For control experiments, control mouse IgG coupled with Sepharose was used. After extensive washes, bound proteins were separated by SDS/PAGE and detected by Western blotting.
Induction of endogenous GSTP expression by MOZ in rat hepatoma H4IIE cells
Rat hepatoma H4IIE cells were transfected with various amounts of the MOZ expression plasmid pCI-MOZ by using the FuGENE™ 6 reagent in 35 mm plates. The total amount of plasmid DNA was adjusted by supplementing with empty pCI vector to 1 μg. After 36 h, cell lysates were prepared with a buffer comprising 25 mM Tris phosphate (pH 7.8), 2 mM cyclohexane-1,2-diaminetetra-acetic acid, 10% glycerol, 2 mM DTT and 1% Triton X-100. Cell lysates were separated by SDS/PAGE (15% gel), and expression of endogenous GSTP and GAPDH was detected by Western blotting.
RESULTS
Activity and expression profiles of HATs during hepatocarcinogenesis
To evaluate the activity and expression profiles of HATs during hepatocarcinogenesis, we performed chemical carcinogenesis in the rat liver in accordance with the Solt–Farber protocol [37]. This model of liver chemical carcinogenesis is a widely used system for the study of molecular and cellular processes leading to cancer. In this protocol, rats were fed a combination of DEN and AAF and then underwent PH. At the end of 7 weeks, the livers had large numerous hyperplastic nodules, and the rats were killed (Figure 1A). We prepared four types of control experiments: rats underwent saline injection; were injected with DEN; underwent AAF feeding; underwent PH but were not treated with DEN and AAF. We checked the reproducibility of the carcinogenic experiments. Western blotting analysis of the cytosol fractions with an anti-GSTP antibody revealed that GSTP was induced at 7 weeks after combined treatment with DEN, AAF and PH, but no GSTP was detected in any of the control rats (Figure 1B).
Figure 1. Expression profiles of HAT during hepatocarcinogenesis.
(A) The Solt–Farber protocol for chemically induced hepatocarcinogenesis in rats [37]. BD, basal diet; S, times at which rats were killed. (B) Expression profiles of HATs were investigated in control livers and those with hyperplastic nodules. Nuclear extracts were prepared from livers of various rats, and immunoblot analysis was performed with specific antibodies, as described in the Experimental section. GSTP in the cytosol fraction was also detected (bottom). The fractions shown in lanes 1 and 2 were from control rats; lanes 3–6, rats having livers with hyperplastic nodules; lane 7, rat treated with DEN only; lane 8, rat treated with AAF only; lane 9, rat underwent PH only.
We first investigated the HAT activity of nuclear extracts during hepatocarcinogenesis. The assay using core histones or nucleosome histones as substrates revealed that HAT activity in livers with hyperplastic nodules was indistinguishable from that in control rat livers (results not shown). For determination of the expression profiles of HATs during hepatocarcinogenesis, we performed Western blot analysis using nuclear extracts and specific antibodies to each of the HATs (Figure 1B). The HATs best characterized as transcriptional co-activators are p300, CBP, P/CAF and GCN5. The expression levels of P/CAF and GCN5 showed no change during hepatocarcinogenesis, but expression of both p300 and CBP decreased. Next, we observed the expression levels of the MYST-type acetyltransferases, which are involved in a wide range of regulatory functions [1]. Expression of TIP60 was unchanged during hepatocarcinogenesis, whereas MOZ expression increased. MORF was not detected (results not shown). Among those we assayed, MOZ was the sole HAT whose expression was positively correlated with GSTP expression during hepatocarcinogenesis.
Induction of the intact form of MOZ during hepatocarcinogenesis
MOZ belongs to the MYST family of HATs and frequently is rearranged in leukaemia [10]. MOZ fusion partners include CBP, p300 and TIF2 (transcriptional intermediary factor 2); all of these proteins are also known to be transcriptional co-activators [10,24–26]. MOZ is a transcriptional regulator in haemopoiesis, and MOZ fusion proteins antagonize MOZ function and lead to leukaemogenesis [26,28]. Using Western blotting and RT (reverse transcriptase)–PCR analyses, we assessed whether MOZ was translocated and thus fused with these transcriptional co-activators during hepatocarcinogenesis (Figure 2 and results not shown). We generated specific antibodies against the N- and C-terminal regions of rat MOZ, and we also used the anti-MYST antibody, which recognizes a motif (amino acids 671–685) in the MYST region of MOZ. These three antibodies recognize different parts of MOZ. Western blot analysis revealed that MOZ induced in livers with hyperplastic nodules and recognized by the three different antibodies were all the same size (Figure 2A), as was the less-abundant MOZ in the control rat liver. We characterized additional fusion partners, including p300, CBP and TIF2. The sizes of these proteins in livers with hyperplastic nodules were the same as those in control livers (Figures 1B and 2B). These results suggested that the intact form of MOZ was induced and that translocation of MOZ did not occur during chemical hepatocarcinogenesis in rats. To confirm these results, we performed RT–PCR with three sets of primers spanning the MOZ regions in which rearrangement occurred frequently [10,24–26]. Sequencing of PCR products revealed that MOZ rearrangement did not occur during hepatocarcinogenesis (results not shown). We further examined the MOZ–CBP chimaeric transcript by hemi-nested PCR, but the amplification product was not detected (results not shown). These results indicate that the intact form of MOZ was induced and that MOZ translocation did not occur during the early stages of hepatocarcinogenesis.
Figure 2. Induction of the intact form of MOZ during hepatocarcinogenesis.
(A) Nuclear extracts were prepared from control (lanes 1, 4 and 7) and livers with hyperplastic nodules (lanes 2, 3, 5, 6, 8 and 9), separated by SDS/PAGE (7.5% gel), and immunoblotted using polyclonal antibodies against the N- (lanes 1–3) or C- (lanes 7–9) terminal region of MOZ or the anti-MYST antibody (lanes 4–6). (B) The putative MOZ fusion partner, TIF-2, was detected with the anti-TIF-2 antibody. The fraction shown in lane 1 is from control; those in lanes 2 and 3 were from livers with hyperplastic.
Activation of GSTP promoter activity by MOZ through the GPE
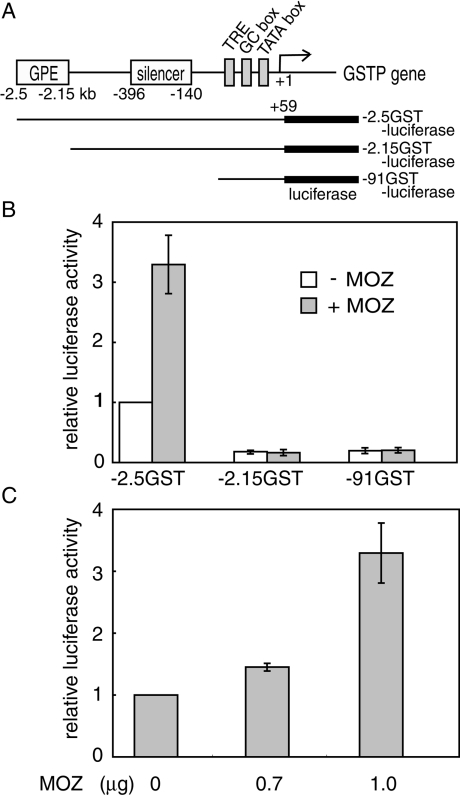
MOZ functions as a transcriptional co-activator and participates as a mediator in haemopoiesis [28,42]. To characterize the effect of MOZ on hepatocarcinogenesis-specific gene expression, we asked whether exogenous MOZ would enhance GSTP promoter activity. GSTP is strongly and specifically expressed during chemical hepatocarcinogenesis and is considered to be an excellent tumour marker [29]. The transcriptional regulatory region of the rat GSTP gene includes enhancer and silencer elements [30,31]. To examine the effect of MOZ on GSTP promoter activity, −2.5GST-luciferase (which has the entire GSTP regulatory region and promoter) was co-transfected with MOZ expression plasmid or control empty vector into rat hepatoma H4IIE cells (Figure 3A). MOZ enhanced GSTP promoter activity (Figure 3B). Luciferase activity in the presence of various concentrations of MOZ was assayed, and MOZ demonstrated dose-dependent enhancement of GSTP promoter activity (Figure 3C). To more closely define the MOZ response element, we used two reporter plasmids: −2.15GST-luciferase, which lacked the GPE, and −91GST-luciferase, which lacked both the GPE and silencer regions (Figures 3A and 3B). These reporter plasmids were not transactivated, thereby suggesting that MOZ activates GSTP promoter activity through the GPE.
Figure 3. MOZ activates the GSTP promoter activity through the GPE.
(A) Diagram of the 5′-flanking region of the rat GSTP gene and the reporter constructs for observing the effect of MOZ on the promoter activity of the GSTP gene. (B) We co-transfected 100 ng of the reporter plasmid with (grey columns) or without (white columns) 1 μg of MOZ expression plasmid (pCI-MOZ) into H4IIE rat hepatoma cells. All transfection assays were repeated at least three times. The relative luciferase activity was calculated from mean values relative to the activity of −2.5GST-luciferase in the absence of MOZ. Each error bar indicates ±S.D. (C) Dose-dependent transactivation of −2.5GST-luciferase by MOZ. Relative luciferase activities are shown as in (B).
MOZ interacts with MafK both in vitro and in vivo
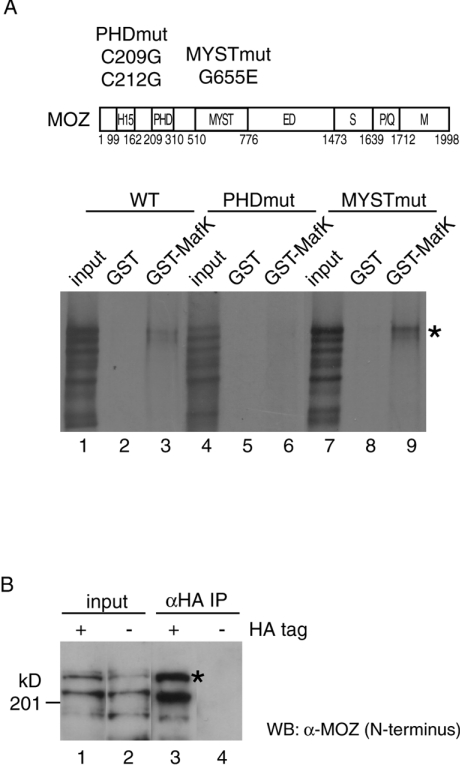
The GPE1 element in GPE is a key control element responsible for GSTP expression in preneoplastic tissue. GPE1 is similar in sequence to ARE (antioxidant-response-like element), MARE (Maf recognition element) and TRE [PMA (‘TPA’)-responsive element] [30,33,34]. A recent study showed that the Nrf2–MafK heterodimer binds to GPE1 and regulates GSTP promoter activity [34]. To determine the mechanism of the MOZ-associated enhancement of GSTP promoter activity, we tested whether MOZ could bind Nrf2 and MafK. We previously showed that MOZ interacted with c-Jun through the bZIP domain in vitro [39]; Nrf2 and MafK also have bZIP domains. To determine MOZ binding partners, we performed an in vitro pull-down assay using 35S-labelled full-length MOZ. We fused the Nrf2 DNA-binding domain to maltose-binding protein, incubated it with 35S-labelled MOZ, and precipitated it with amylose resin, but interaction between MOZ and the Nrf2 DNA-binding domain was not detected (results not shown). Next, we evaluated the interaction between MOZ and MafK using a GST pull-down assay and found that 35S-labelled MOZ interacted with GST–MafK but not with GST alone (Figure 4A, lanes 1–3). Unique structural domains are identified in MOZ [28]. To identify the region required for the interaction between MOZ and MafK, two MOZ derivatives with double and single point mutations in the PHD zinc-finger (C209G and C212G) and the MYST (G655E) regions respectively, were generated. The PHD zinc-finger and the MYST regions are important for binding to specific nuclear protein partners and HAT activity, respectively [43,44]. The mutant in the PHD zinc-finger region was not able to interact with GST–MafK, whereas the mutation in the putative acetyl-CoA-binding site in the MYST region did not affect the binding to MafK (Figure 4A, lanes 4–9). These results suggest that MOZ interacts with MafK in the absence of the heterodimer partner, Nrf2, mediated by the PHD zincfinger region of MOZ.
Figure 4. MOZ interacts with MafK in vitro and in vivo.
(A) Structural domains of MOZ were indicated as follows: H15, histones H1- and H5-like module; MYST, MYST acetyltransferase domain; ED, glutamic acid/aspartic acid-rich acidic regions; S, serine-rich domain; P/Q, proline/glutamine-stretch; and M, methionine-rich domain. Also shown are the mutation positions in the PHD finger and MYST regions. Indicated wild-type and mutated in vitro-translated [35S]MOZ proteins were incubated with GST (lanes 2, 5 and 8) or GST–MafK (lanes 3, 6 and 9). MOZ protein retained on the GST-conjugated beads after extensive washing was analysed by SDS/PAGE and autoradiography. The amount of input (lanes 1, 4 and 7) is equivalent to 10% of the reaction volume in the assay. [35S]MOZ proteins are indicated by asterisks (*). (B) MOZ expression plasmid was co-transfected with HA-tagged MafK (lanes 1 and 3) or nontagged MafK (lanes 2 and 4) into HeLa cells, and nuclear extracts were prepared. Immunoprecipitation (IP) experiments were performed with anti-HA antibody. Immunoprecipitates (lanes 3 and 4) and 5% of input (lanes 1 and 2) were resolved by SDS/PAGE (7.5% gel) and detected by Western blotting using anti-N-terminal MOZ antibody. MOZ proteins are indicated by asterisks (*).
To evaluate the interaction between MOZ and MafK under physiological conditions, we attempted to detect immunoprecipitated MOZ, but endogenous MOZ in nuclear extracts from H4IIE and HeLa cells could not be detected. Therefore we next introduced the MOZ expression plasmid with HA-tagged or non-tagged MafK into HeLa cells, and nuclear extracts were prepared. MOZ was immunoprecipitated only in nuclear extracts expressing HA-tagged MafK (Figure 4B). Some degraded MOZ proteins were detected in nuclear extracts and these proteins were also immunoprecipitated. GST pull-down and immunoprecipitation experiments suggest that MOZ may interact with the MafK moiety of the Nrf2–MafK heterodimer in vivo.
MOZ functions as a co-activator of the Nrf2–MafK heterodimer
MOZ preferentially interacted with MafK and up-regulated GSTP promoter activity through GPE, which contains the binding site for the Nrf2–MafK in the reporter assay (Figures 3 and 4). These data suggest that MOZ is a potential co-activator of Nrf2–MafK heterodimer. To test this hypothesis, we investigated whether MOZ could stimulate Nrf2–MafK-mediated transactivation (Figure 5). We have previously reported that Nrf2 simulates GPE1-mediated transactivation in F9 cells, which are considered to lack AP1 (activator protein 1) activity and to express excess amounts of small Maf proteins, including MafK [34]. MOZ or Nrf2 expression plasmid was co-transfected with the reporter plasmid GPE1-luciferase (which includes GPE1 and the GSTP promoter) into F9 cells. MOZ and Nrf2 slightly enhanced the activity of the reporter construct. As expected, MOZ, when in the presence of Nrf2, dose-dependently stimulated GPE1-mediated GSTP promoter activity. MOZ did not stimulate the promoter activity of reporter plasmids including the mutated Nrf2 binding site (results not shown).
Figure 5. MOZ is a co-activator of Nrf2.
Nrf2-mediated transactivation by MOZ was examined in mouse embryonic carcinoma F9 cells. We co-transfected 100 ng of the reporter plasmid (GPE1-luciferase, in the panel) and 5 ng of Renilla luciferase plasmid (phRL-tk) with 0, 0.3, 0.7 and 1 μg MOZ expression plasmid (pCI-MOZ) in the absence (–) or presence (+) of Nrf2 expression plasmid (pAβ2-Nrf2, 5 ng). The luciferase activity was normalized to Renilla luciferase activity. Relative luciferase activity was calculated from the mean values relative to the activity of GPE1-luciferase without Nrf2 and MOZ. Each error bar indicates ±S.D.
Induction of endogenous GSTP expression by MOZ in rat hepatoma cells
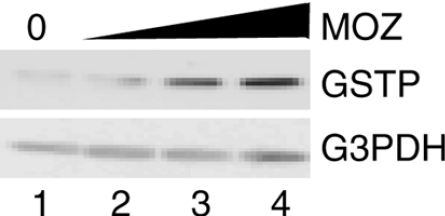
As we described above, MOZ stimulated the GSTP promoter activity mediated by Nrf2–MafK. We then assessed the effects of MOZ overexpression on GSTP expression in H4IIE cells. Transiently overexpressed MOZ induced expression of endogenous GSTP but not GAPDH (Figure 6). The induction of GSTP protein was dependent on the exogenous MOZ expression. These results suggest that MOZ functions as a co-activator of the Nrf2–MafK heterodimer and may stimulate GSTP gene expression during hepatocarcinogenesis.
Figure 6. MOZ induces endogenous GSTP expression.
H4IIE cells were transfected with 0, 0.4, 0.7 and 1.0 μg of MOZ expression plasmid (pCI-MOZ, lanes 1–4), and cell lysates were prepared. Endogenous GSTP and GAPDH (‘G3PDH’) were detected by immunoblotting.
DISCUSSION
HATs contribute to tumour suppression, and loss or dysregulation of these activities may be linked to tumorigenesis [45]. To gain insight into the roles of HATs in liver cancer, we analysed the expression profiles of HATs during hepatocarcinogenesis and evaluated their roles in hepatocarcinogenic-specific gene expression.
We have shown that MOZ expression was up-regulated during hepatocarcinogenesis. MOZ functions as a co-activator of AML1-mediated transcription, and the AML1–MOZ complex might play a role in cell differentiation [28,42]. MOZ frequently is rearranged in leukaemia, and the MOZ fusion protein antagonizes MOZ function in haemopoiesis [26,28]. Even though MOZ rearrangement does not occur during hepatocarcinogenesis, we documented an anomalous increase of MOZ. Because HATs regulate global gene expression [1,46], dysregulation of MOZ may induce unusual gene expression, leading to hepatocarcinogenesis.
Recently, the MOZ–MORF complex including BRPF (bromo-domain- and PHD finger-containing) 1/2/3 paralogue and ING5 (inhibitor of growth 5; tumour suppressor) was purified [47]. MORF was not detected in nuclear extracts from livers with hyperplastic nodules (results not shown), so that MOZ, but not MORF, complex may regulate GSTP expression. ING5 is also included in HBO1 [HAT binding to ORC1 (origin recognition complex subunit 1)] complex. Interestingly, ING4, another member of ING family proteins, exists in HBO1 complex, but not MOZ complex. AML1-dependent promoter activity is stimulated by ING5, but not ING4 [47]. This raises a possibility that overexpressed MOZ may affect regulation of AML1-dependent gene expression. ING5 tumour suppressor is included in both HBO1 and MOZ complexes, which are important for DNA synthesis [47]. Overexpressed MOZ might trap ING5 and generate partial complexes, and further, HBO1 complex would be affected with the change of ING5 level. Thus aberrantly expressed MOZ during hepatocarcinogenesis may disturb the tumour suppressor function of ING5 complexes and DNA synthesis, which lead to tumorigenesis.
We also found that the expression of p300 and CBP were decreased during hepatocarcinogenesis. Although AAF blocks the proliferation of hepatocytes, GSTP-expressing cells escape from the growth inhibition and continuously grow in the Solt–Farber model. Trautwein et al. [48] reported that AAF blocks cell-cycle progression after PH by inducing the cyclin-dependent kinase inhibitor p21. Expression of p21 is regulated mainly by the tumour-suppressor protein p53, and full transcriptional activity of p53 requires the co-activators p300/CBP [49–51]. Down-regulation of p300 and CBP reduces p53 activity and leads to cell-cycle progression of GSTP-expressing cells, suggesting that p300 and CBP may be considered tumour suppressors, and their loss of function may be a link to hepatocarcinogenesis.
GSTP is a Phase II detoxification enzyme involved in the metabolism of carcinogens, and it plays a protective role during chemical hepatocarcinogenesis [52]. The Nrf2–MafK heterodimer is important for the GSTP expression during early hepatocarcinogenesis, but it is difficult to explain the markedly increased expression of GSTP in livers with hyperplastic nodules solely on the basis of the increased quantity of Nrf2 [34]. We found that the expression of MOZ was well correlated with GSTP expression during hepatocarcinogenesis; MOZ also functioned as a co-activator of the Nrf2–MafK heterodimer. We reported that the binding activity of Nrf2–MafK heterodimer to GPE1 is much stronger than that of MafK homodimer. Further Nrf2 alone could not bind to GPE1, and the Nrf2 mRNA level is increased in cells from hyperplasic nodules when compared with those from normal livers [34]. Histones H3 and H4 are acetylated in both GPE1 and in the promoter regions of the GSTP gene in the H4IIE hepatoma cell line but not normal liver [34]. This acetylation coincides with the activation of GSTP expression. MOZ may contribute acetylation of histones in the regulatory region of the GSTP gene. Elevation of both MOZ and Nrf2 expression may be required for the dramatically increased gene expression of GSTP observed during hepatocarcinogenesis in vivo. To understand the molecular mechanism of the GSTP induction mediated by Nrf2–MafK heterodimer and MOZ, we proceeded to identify the regions of MOZ and MafK required for the GSTP expression in exogenously Nrf2-expressed H4IIE cells.
The activation mechanism of GSTP expression is classified into two types: specific induction in livers with hyperplastic nodules by chemical carcinogens, and non-specific induction by non-carcinogenic agents such as antioxidants [29,53]. The former induction may require both Nrf2 and MOZ, but only Nrf2 may be necessary for the latter. Preneoplastic foci and nodules are derived from GSTP-positive single cells [54]. The mechanism of the generation of the GSTP-positive single cell is unclear, and specific induction of GSTP has not been reproduced in cell lines by using chemical carcinogens. The use of transgenic or MOZ knockout animals would probably enable us to demonstrate the mechanism of chemical carcinogen-associated GSTP induction during hepatocarcinogenesis.
Acknowledgments
This research was supported in part by grants from a Sasakawa Scientific Research Grant from the Japan Science Society, Sankyo Foundation of Life Science, the LRI (Long-range Research Initiative) of the JCIA (Japan Chemical Industry Association), the Japanese Ministry of Education, Culture, Sports, Science, and Technology and JSPS (Japan Society for the Promotion of Science). We are grateful to Dr Yoshihiro Nakatani (Harvard Medical School, Boston, MA, U.S.A.) for kindly providing the anti-P/CAF antibody. We also thank the staff of the Radioisotope Research Center, Osaka University (Osaka, Japan). We thank Mitsumasa Kurita and Kiyoto Kageyama for their helpful discussions.
References
- 1.Sterner D. E., Berger S. L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebbes T. R., Thorne A. W., Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brownell J. E., Allis C. D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X. J., Ogryzko V. V., Nishikawa J., Howard B. H., Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A. J., Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 7.Carrozza M. J., Utley R. T., Workman J. L., Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- 8.Xu W., Edmondson D. G., Evrard Y. A., Wakamiya M., Behringer R. R., Roth S. Y. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 9.Yamauchi T., Yamauchi J., Kuwata T., Tamura T., Yamashita T., Bae N., Westphal H., Ozato K., Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borrow J., Stanton V. P., Jr, Andresen J. M., Becher R., Behm F. G., Chaganti R. S., Civin C. I., Disteche C., Dube I., Frischauf A. M., et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat. Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 11.Reifsnyder C., Lowell J., Clarke A., Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat. Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 12.Smith E. R., Eisen A., Gu W., Sattah M., Pannuti A., Zhou J., Cook R. G., Lucchesi J. C., Allis C. D. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3561–3565. doi: 10.1073/pnas.95.7.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iizuka M., Stillman B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 1999;274:23027–23034. doi: 10.1074/jbc.274.33.23027. [DOI] [PubMed] [Google Scholar]
- 14.Ikura T., Ogryzko V. V., Grigoriev M., Groisman R., Wang J., Horikoshi M., Scully R., Qin J., Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 15.Osada S., Sutton A., Muster N., Brown C. E., Yates J. R., III, Sternglanz R., Workman J. L. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 2001;15:3155–3168. doi: 10.1101/gad.907201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howe L., Auston D., Grant P., John S., Cook R. G., Workman J. L., Pillus L. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 2001;15:3144–3154. doi: 10.1101/gad.931401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiltz R. L., Nakatani Y. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta. 2000;1470:M37–M53. doi: 10.1016/s0304-419x(99)00037-2. [DOI] [PubMed] [Google Scholar]
- 18.Eferl R., Ricci R., Kenner L., Zenz R., David J. P., Rath M., Wagner E. F. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/s0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- 19.Kalinichenko V. V., Major M. L., Wang X., Petrovic V., Kuechle J., Yoder H. M., Dennewitz M. B., Shin B., Datta A., Raychaudhuri P., et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomson S., Clayton A. L., Mahadevan L. C. Independent dynamic regulation of histone phosphorylation and acetylation during immediate-early gene induction. Mol. Cell. 2001;8:1231–1241. doi: 10.1016/s1097-2765(01)00404-x. [DOI] [PubMed] [Google Scholar]
- 21.Barlev N. A., Liu L., Chehab N. H., Mansfield K., Harris K. G., Halazonetis T. D., Berger S. L. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell. 2001;8:1243–1254. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 22.Carapeti M., Aguiar R. C., Goldman J. M., Cross N. C. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- 23.Liang J., Prouty L., Williams B. J., Dayton M. A., Blanchard K. L. Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood. 1998;92:2118–2122. [PubMed] [Google Scholar]
- 24.Chaffanet M., Gressin L., Preudhomme C., Soenen-Cornu V., Birnbaum D., Pebusque M. J. MOZ is fused to p300 in an acute monocytic leukemia with t(8;22) Genes Chromosomes Cancer. 2000;28:138–144. doi: 10.1002/(sici)1098-2264(200006)28:2<138::aid-gcc2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 25.Kitabayashi I., Aikawa Y., Yokoyama A., Hosoda F., Nagai M., Kakazu N., Abe T., Ohki M. Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia. 2001;15:89–94. doi: 10.1038/sj.leu.2401983. [DOI] [PubMed] [Google Scholar]
- 26.Deguchi K., Ayton P. M., Carapeti M., Kutok J. L., Snyder C. S., Williams I. R., Cross N. C., Glass C. K., Cleary M. L., Gilliland D. G. MOZ–TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–271. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 27.Imamura T., Kakazu N., Hibi S., Morimoto A., Fukushima Y., Ijuin I., Hada S., Kitabayashi I., Abe T., Imashuku S. Rearrangement of the MOZ gene in pediatric therapy-related myelodysplastic syndrome with a novel chromosomal translocation t(2;8)(p23;p11) Genes Chromosomes Cancer. 2003;36:413–419. doi: 10.1002/gcc.10172. [DOI] [PubMed] [Google Scholar]
- 28.Kitabayashi I., Aikawa Y., Nguyen L. A., Yokoyama A., Ohki M. Activation of AML1-mediated transcription by MOZ and inhibition by the MOZ–CBP fusion protein. EMBO J. 2001;20:7184–7196. doi: 10.1093/emboj/20.24.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato K. Glutathione transferase as markers of preneoplasia and neoplasia. Adv. Cancer Res. 1989;52:205–255. doi: 10.1016/s0065-230x(08)60214-6. [DOI] [PubMed] [Google Scholar]
- 30.Sakai M., Okuda A., Muramatsu M. Multiple regulatory elements and phorbol 12-O-tetradecanoate 13-acetate responsiveness of the rat placental glutathione transferase gene. Proc. Natl. Acad. Sci. U.S.A. 1988;85:9456–9460. doi: 10.1073/pnas.85.24.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imagawa M., Osada S., Okuda A., Muramatsu M. Silencer binding proteins function on multiple cis-elements in the glutathione transferase P gene. Nucleic Acids Res. 1991;19:5–10. doi: 10.1093/nar/19.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimura S., Suzuki T., Hochi S., Yuki A., Nomura K., Kitagawa T., Nagatsu I., Imagawa M., Muramatsu M. Trans-activation of glutathione transferase P gene during chemical hepatocarcinogenesis of the rat. Proc. Natl. Acad. Sci. U.S.A. 1993;90:2065–2068. doi: 10.1073/pnas.90.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki T., Imagawa M., Hirabayashi M., Yuki A., Hisatake K., Nomura K., Kitagawa T., Muramatsu M. Identification of an enhancer responsible for tumor marker gene expression by means of transgenic rats. Cancer Res. 1995;55:2651–2655. [PubMed] [Google Scholar]
- 34.Ikeda H., Nishi S., Sakai M. Transcription factor Nrf2/MafK regulates rat placental glutathione S-transferase gene during hepatocarcinogenesis. Biochem. J. 2004;380:515–521. doi: 10.1042/BJ20031948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itoh K., Chiba T., Takahashi S., Ishii T., Igarashi K., Katoh Y., Oyake T., Hayashi N., Satoh K., Hatayama I., et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 36.Kwak M. K., Wakabayashi N., Kensler T. W. Chemoprevention through the Keap1–Nrf2 signaling pathway by phase 2 enzyme inducers. Mutat. Res. 2004;555:133–148. doi: 10.1016/j.mrfmmm.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 37.Solt D., Farber E. New principle for the analysis of chemical carcinogenesis. Nature. 1976;263:701–703. [Google Scholar]
- 38.Osada S., Takano K., Nishihara T., Suzuki T., Muramatsu M., Imagawa M. CCAAT/enhancer-binding proteins α and β interact with the silencer element in the promoter of glutathione S-transferase P gene during hepatocarcinogenesis. J. Biol. Chem. 1995;270:31288–31293. doi: 10.1074/jbc.270.52.31288. [DOI] [PubMed] [Google Scholar]
- 39.Ohta K., Osada S., Nishikawa J., Nishihara T. Cloning and characterization of a cDNA encoding the histone acetyltransferase MOZ (monocytic leukemia zinc finger protein) in the rat. J. Health Sci. 2005;51:253–256. [Google Scholar]
- 40.Ikeda H., Serria M. S., Kakizaki I., Hatayama I., Satoh K., Tsuchida S., Muramatsu M., Nishi S., Sakai M. Activation of mouse Pi-class glutathione S-transferase gene by Nrf2 (NF-E2-related factor 2) and androgen. Biochem. J. 2002;364:563–570. doi: 10.1042/BJ20011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bristow C. A., Shore P. Transcriptional regulation of the human MIP-1α promoter by RUNX1 and MOZ. Nucleic Acids Res. 2003;31:2735–2744. doi: 10.1093/nar/gkg401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Utley R. T., Cote J. The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- 45.Timmermann S., Lehrmann H., Polesskaya A., Harel-Bellan A. Histone acetylation and disease. Cell. Mol. Life Sci. 2001;58:728–736. doi: 10.1007/PL00000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krebs J. E., Fry C. J., Samuels M. L., Peterson C. L. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell. 2000;102:587–598. doi: 10.1016/s0092-8674(00)00081-7. [DOI] [PubMed] [Google Scholar]
- 47.Doyon Y., Cayrou C., Ullah M., Landry A. J., Cote V., Selleck W., Lane W. S., Tan S., Yang X. J., Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Trautwein C., Will M., Kubicka S., Rakemann T., Flemming P., Manns M. P. 2-Acetaminofluorene blocks cell cycle progression after hepatectomy by p21 induction and lack of cyclin E expression. Oncogene. 1999;18:6443–6453. doi: 10.1038/sj.onc.1203045. [DOI] [PubMed] [Google Scholar]
- 49.Gu W., Shi X. L., Roeder R. G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 50.Gu W., Roeder R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 51.Liu L., Scolnick D. M., Trievel R. C., Zhang H. B., Marmorstein R., Halazonetis T. D., Berger S. L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 53.Satoh K., Kitahara A., Soma Y., Inaba Y., Hatayama I., Sato K. Purification, induction, and distribution of placental glutathione transferase: a new marker enzyme for preneoplastic cells in the rat chemical hepatocarcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 1985;82:3964–3968. doi: 10.1073/pnas.82.12.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satoh K., Hatayama I., Tateoka N., Tamai K., Shimizu T., Tatematsu M., Ito N., Sato K. Transient induction of single GST-P positive hepatocytes by DEN. Carcinogenesis. 1989;10:2107–2111. doi: 10.1093/carcin/10.11.2107. [DOI] [PubMed] [Google Scholar]