Abstract
One of the greatest bottlenecks in producing recombinant proteins in Escherichia coli is that over-expressed target proteins are mostly present in an insoluble form without any biological activity. DCase (N-carbamoyl-D-amino acid amidohydrolase) is an important enzyme involved in semi-synthesis of β-lactam antibiotics in industry. In the present study, in order to determine the amino acid sites responsible for solubility of DCase, error-prone PCR and DNA shuffling techniques were applied to randomly mutate its coding sequence, followed by an efficient screening based on structural complementation. Several mutants of DCase with reduced aggregation were isolated. Solubility tests of these and several other mutants generated by site-directed mutagenesis indicated that three amino acid residues of DCase (Ala18, Tyr30 and Lys34) are involved in its protein solubility. In silico structural modelling analyses suggest further that hydrophilicity and/or negative charge at these three residues may be responsible for the increased solubility of DCase proteins in E. coli. Based on this information, multiple engineering designated mutants were constructed by site-directed mutagenesis, among them a triple mutant A18T/Y30N/K34E (named DCase-M3) could be overexpressed in E. coli and up to 80% of it was soluble. DCase-M3 was purified to homogeneity and a comparative analysis with wild-type DCase demonstrated that DCase-M3 enzyme was similar to the native DCase in terms of its kinetic and thermodynamic properties. The present study provides new insights into recombinant protein solubility in E. coli.
Keywords: N-carbamoyl-D-amino acid amidohydrolase, directed evolution, negative charge, protein folding, protein solubility
Abbreviations: DCase, N-carbamoyl-D-amino acid amidohydrolase; DCase-M3, a triple DCase mutant A18T/Y30N/K34E; D-HPG, D-p-hydroxyphenylglycine; β-gal, β-galactosidase; IB, inclusion body; IPTG, isopropyl-β-D-thiogalactoside; LB, Luria–Bertani; Ni-NTA, Ni2+-nitrilotriacetate; ONPG, O-nitrophenyl-β-D-galactopyranoside; X-gal, 5-bromo-4-chloroindol-3-yl β-D-galactopyranoside; WT, wild-type
INTRODUCTION
Although various bacterial expression systems have been established for fast and efficient expression of heterologous proteins [1], one common problem of most heterologous expression systems is the poor solubility/misfolding of recombinant proteins. In most cases, target proteins failed to fold into their native states and instead accumulated in inactive IBs (inclusion bodies) when expressed heterologously [2]. Current experimental strategies to form soluble and native protein molecules include: the co-expression of foldases or chaperones [3–5]; the fusion of tags that contain a highly soluble polypeptide or an increased negatively charge polypeptide [6,7]; the use of promoters with different strengths, in order to control the rate of protein synthesis; the expression of the protein at a lower temperature; and the optimization of growth medium and culturing conditions [8]. Although with some success, the major disadvantage of these strategies is that they are very time-consuming. Another successful approach is site-directed mutagenesis of target proteins by modifying the intrinsic folding stability and solubility. Based on structural information, site-directed rational mutation of one or a few amino acids can sometimes result in huge improvement of protein solubility and folding [9]. However, this approach generally requires extensive trial-and-error to find the amino acid substitutions that will result in enhanced solubility of the target proteins, and is therefore not suitable for high-throughput applications [10]. Moreover, the outcomes are not always satisfactory, because it is difficult to predict the necessary changes. Another alternative method is to use the directed evolution technique, which can generate a large number of mutants in a very short period of time. For application of directed evolution techniques, the establishment of an efficient screening method is a prerequisite. Some commonly used high-throughput screening methods use fusion reporter systems, such as the green fluorescent protein folding report method [11], the N-terminal fusion system with chloramphenicol acetyltransferase [12] and LacZα (β-galactosidase α peptide) complementation solubility reporter assay [13].
DCase (N-carbamoyl-D-amino acid amidohydrolase; EC 3.5.1.77) is a rate-limiting enzyme in a two-step reaction system for producing D-HPG (D-p-hydroxyphenylglycine), an important intermediate for semi-synthesis of β-lactam antibiotics such as penicillins and cephalosporins [14]. In the process, a starting substrate DL-5-substituted hydantoin is asymmetrically hydrolysed to N-carbamoyl-D-p-hydroxyphenylglycine by D-hydantoinase (EC 3.5.2.2), followed by stereo-specific transformation of N-carbamoyl-D-p-hydroxyphenylglycine into its corresponding D-HPG by DCase. DCase normally has lower activity relative to that of D-hydantoinase [15,16]. During the past decade, several studies have been reported on the cloning and characterization of DCase encoding genes from a variety of micro-organisms and their overexpression in the Escherichia coli system [17–20]. However, overproduction of DCase proteins in E. coli often results in the formation of biologically inactive IBs. In our previous study [21], a DCase gene was cloned from Burkholderia pickettii. The gene encodes a peptide of 304 amino acids with a calculated molecular mass of 34334 Da. DCase forms a homotetramer and is an intracellular protein without disulfide bonds. Attempts were made to overexpress DCase in E. coli, and the results showed that its activity was very low, because nearly 80% of the recombinant DCase was partitioned into insoluble aggregates. In the present study, the DCase encoding gene from B. pickettii was subjected to random mutation by combining error-prone PCR and DNA shuffling, followed by a simple and efficient screening protocol based on structural complementation between the α- and ω-fragments of β-gal (β-galactosidase) [13]. Using this approach, we have identified three amino acid residues (Ala18, Tyr30 and Lys34) that are related to the protein solubility of DCase. Molecular modelling analysis suggested that enhanced solubility of the DCase proteins attributed mainly to the increases in hydrophilicity and/or negative charge of the substituted amino acids in E. coli. Multiple mutants of DCase were rationally designed and overexpressed in E. coli; among them, DCase-M3 (a triple DCase mutant A18T/Y30N/K34E) was found, which was up to 80% soluble.
MATERIALS AND METHODS
Random mutagenesis combining error-prone PCR and DNA shuffling
Primers, 5′-GGGAATTCCATATGACACGTCAGATGATACTT-3′and 5′-CCCAAGCTTGCGCCAGAACCAGCAGCGGAGCCAGCGGATCCGAGTTCCGCGATCAGACC-3′, were designed to PCR amplify the DCase gene from the plasmid pXZ-total [21]. Of note, we chose to use the DNA fragment 5′-GGATCCGCTGGCTCCGCTGCTGGTTCTGGCGCAAGCTT-3′ coding for an amino acid linker Gly-Ser-Ala-Gly-Ser-Ala-Ala-Gly-Ser-Gly-Ala-Ser between DCase and the α-fragment of β-gal in the latter primer [11]. To enhance the natural mutation rate, the DCase gene was initially amplified by error-prone PCR [22]. The reaction mixture for a 100 μl error-prone PCR sample contained: 7 mM MgCl2, 50 mM KCl, 10 mM Tris/HCl (pH 8.5), 0.01% gelatin, 0.2 mM dATP, 0.2 mM dGTP, 1.0 mM dCTP, 1.0 mM dTTP, 250 pmol of each primer, 500 ng of pXZ-total template DNA, MnCl2 (0.075–0.15 mM final concentration) and 5 units of Taq polymerase (Promega). After denaturation for 6 min at 94 °C, the PCR was run for 30 cycles of 94 °C, 30 s; 58 °C, 30 s; 72 °C, 50 s; with a final extension step at 72 °C for 5 min. PCR products were checked by electrophoresis on a 1.0% (w/v) agarose gel and then purified using a DNA purification kit (Qiagen). Subsequently, the PCR product was randomly cleaved with DNaseI (New England Biolabs). Finally, DNA fragments were reassembled with a primerless PCR and an additional primer PCR as described by Stemmer [23].
Construction and screening of mutant library
The reassembled genes, double digested with NdeI and HindIII, were ligated into the pMAL-c2x vector (New England Biolabs) digested with NdeI and HindIII using the rapid DNA ligase to form an in-frame DCase–α-fragment fusion protein. The ligation mixture was transformed into E. coli DH5α competent cells (Novagen) yielding a library of over 10000 transformants on LB (Luria–Bertani) plates supplemented with 100 μg/ml ampicillin, 80 μg/ml X-gal (5-bromo-4-chloroindol-3-yl β-D-galactopyranoside) and 0.1 mM IPTG (isopropyl-β-D-thiogalactoside) [13]. After the plates were incubated at 37 °C for 18–24 h, colonies showing darker blue than the WT (wild-type) were selected.
Sequencing and analysis
Plasmids isolated from the possible positive colonies were purified using a plasmid mini kit (Qiagen). DNA sequencing was carried out using fluorescent dye terminator sequencing chemistry on an ABI 3730 sequencer (Invitrogen). Nucleotide and amino acid sequence alignment analysis was performed using ClustalW [24]. The secondary structure prediction of protein was provided by ESPript [25].
Measurement of β-gal activity in vitro and solubility analysis
E. coli DH5α cells containing the WT DCase–α-fragment and mutated DCase–α-fragment fusion expression constructs were inoculated into a liquid LB medium supplemented with 100 mg/ml ampicillin and grown at 37 °C until a D600 of 0.6 was reached. The target proteins were induced with 0.3 mM IPTG for 3 h. A 1 ml aliquot of the culture was then transferred into a 1.5 ml Eppendorf tube. After adding 10 μl of X-gal (100 μg/ml), the tube was shaken at 37 °C for 30 min. β-Gal activity was then evaluated by the appearance of a blue colour. Moreover, solubility of the DCase–α-fragment fusion protein was monitored by native PAGE. A 1 ml aliquot of the cell culture was pelleted in a 1.5 ml Eppendorf tube and resuspended in 200 μl lysis buffer A (100 mM Tris/HCl, pH 7.5, 100 mM NaCl and 1 mM EDTA). After sonication on ice (six bursts, 5 s each at 250 W), the lysates were centrifuged at 15000 g for 20 min at 4 °C. The supernatant fractions were analysed using native-PAGE. The gel was stained with buffer Z (10 mM KCl, 2.0 mM MgSO4, 60 mM Na2HPO4, 40 mM Na2PO4, pH 7.0) containing 80 μg/ml X-gal. The activity of β-gal was determined using ONPG (O-nitrophenyl-β-D-galactopyranoside) as a substrate as described previously [13].
Oligonucleotide-directed mutagenesis
Site-directed mutagenesis for 22 single point mutants of DCase, in the three mutation sites: Ala18, Tyr30 and Lys34 (A18L, A18Y, A18E, A18D, A18N, A18K, A18R, Y30L, Y30A, Y30G, Y30E, Y30D, Y30N, Y30K Y30R, K34L, K34L, K34A, K34G, K34H, K34D, K34R), three double mutants (A18T/Y30N, A18T/K34E, and Y30N/K34E) and one triple mutant (A18T/Y30N/K34E) were generated by overlap extension PCR [26]. All the mutations were confirmed by DNA sequencing.
Protein expression and solubility test
All DCase genes, including WT and the mutated DCase by directed evolution as well as mutants by site-directed mutagenesis, were PCR amplified using the forward primer: 5′-GGGAATTCCATATGACACGTCAGATGATACTT-3′; and the reverse primer: 5′-CCCAAGCTTTCAGAGTTCCGCGATCAGACC-3′. After double digestion with NdeI and HindIII, the DCase genes were subcloned into the pET-28a expression vector (Novagen) and heterologously expressed in E. coli strain BL21 (DE3) (Novagen). Cells harbouring the plasmid with WT or the evolved DCase genes were grown to the mid-log phase at 37 °C, induced with IPTG at a final concentration of 0.3 mM at 22 °C. Cells from 1 ml of culture were harvested about 10 h post-induction by centrifugation at 1500 g for 10 min at 4 °C and resuspended in 400 μl lysis buffer A without 1 mM EDTA. The cell suspensions were lysed by sonication on ice (four bursts, 30 s). After removal of insoluble protein and cell debris by centrifugation of the lysates at 15000 g for 10 min at 4 °C, the supernatant fraction and precipitant fraction were subjected to SDS/PAGE analysis (10% gels). Protein solubility was estimated by scanning Coomassie Blue-stained gels in relation to a reference protein.
Purification of WT and mutant DCase-M3
Recombinant WT and the evolved DCase-M3 fused to a His6 tag were purified from E. coli by a Ni2+-affinity column (Qiagen). Briefly, the procedure of protein purification was as follows. (i) The cell pellets from 200 ml of cell culture medium were harvested by centrifugation and resuspended in 6 ml of lysis buffer A containing 10 mM imidazole. After sonication at 4 °C (for 4 min in 5 s pulses), the lysates were centrifuged at 15000 g for 20 min at 4 °C. (ii) Supernatant fractions of the cleared lysates were mixed completely with 1 ml of Ni-NTA (Ni2+-nitrilotriacetate)–agarose on ice by shaking for 1 h, and were loaded on to a 5 ml column. (iii) The Ni-NTA–agarose was cleared using 10 ml of wash buffer (50 mM Tris/HCl, pH 7.5, 100 mM NaCl and 20 mM imidazole). (iv) The target proteins were eluted by elution buffer (50 mM Tris/HCl, pH 7.5, 100 mM NaCl and 250 mM imidazole). Protein purity was assessed by SDS/PAGE followed by staining of the gel with Coomassie Blue. Protein concentration was determined by the method of Bradford [27], using BSA as a standard.
Kinetic analysis of WT DCase and DCase-M3
The kinetic parameters of WT DCase and DCase-M3 were determined as described previously [28]. The substrate concentration of N-carbamoyl-D-p-hydroxyphenylglycine was varied from 3.3 to 20 mM. One unit of enzyme activity was defined as the amount of enzyme that catalyses the formation of D-HPG at the rate of 1 μmol/min under the assay conditions.
In vitro equilibrium denaturation measurement
Native proteins (6 μg of purified WT DCase or DCase-M3) were added into 1 ml of a Tris buffer (20 mM Tris/HCl, pH 7.5 and 50 mM NaCl) with varying concentrations of urea, and the fluorescence intensity of each sample was recorded after equilibrium. The urea-induced unfolding or refolding of the target proteins was monitored by the increase/decrease of the intrinsic fluorescence of tryptophan. Fluorescence measurements were performed using a Hitachi F-4010 fluorescence spectrophotometer with an excitation wavelength of 295 nm and an emission wavelength from 300–400 nm. The excitation and emission bandwidths were 3 nm and 5 nm respectively. All fluorescence experiments were performed at 25 °C. The experimental data were used to calculate the thermodynamic parameters according to the two-state unfolding model [29].
Structural homology modelling
Structural models of the WT DCase and the DCase-M3 proteins of B. pickettii were constructed based on the crystal structure of DCase from Agrobacterium sp. KNK712 from the Protein Data Bank (PDB accession code 1ERZ) [30]. The amino acid replacements were generated using the rotamer library approach of the SCWRL program [31]. The program RasMol was used for display of the protein structure [32]. Charge distribution of protein was generated with GRASP [33].
RESULTS
Directed evolution of soluble DCase variants
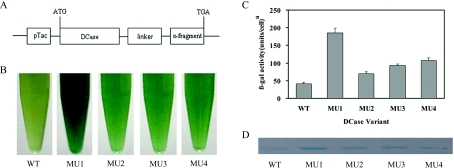
The directed evolution technique involves two key steps: first, the construction of mutant library with enough variations; and secondly, high-throughput screening. In the present study, attempts were made to increase the mutation rate. A variety of DCase gene fragments were first created by error-prone PCR, the fragments were then randomly fragmentized with DNase I before the steps of primerless PCR and primer PCR of DNA shuffling. Through such modifications a higher frequency of mutations, 0.8% at nucleotide level and approx. 3 amino acid changes per enzyme variant, was achieved (data based on sequence analysis of 15 randomly selected clones). Overall, a library of mutated DCase with over 10000 colonies was obtained. The mutated DCase genes were cloned into the pMAL-c2x fusion expression vector (Figure 1A). In addition, we also introduced a colorimetric screening system based on structural complementation between the α- and ω-fragments of β-gal in E. coli to identify DCase variants with improved solubility. The solubility can be visualized by the intensity of blue colour on IPTG/X-gal-treated indicator plates. The method is very high throughput and reproducible. From the initial screening, four mutants (MU1–MU4) with increased solubility were isolated from the library. The relative intensities of blue colour in cells expressing WT DCase–α-fragment or mutated DCase–α-fragment were compared in liquid medium containing X-gal (Figure 1B). The results showed that cells harbouring four mutated DCase–α-fragments displayed blue colouration that was much darker than that of WT DCase–α-fragment. Increased β-gal activities were also observed in MU1–MU4, especially in MU1 where it had a more than 3-fold increase of the β-gal activity compared with that of WT (Figure 1C). Supernatant fractions of these fusion proteins were analysed on native-PAGE to compare further the solubility of WT and MU1–MU4. As expected, more soluble proteins were observed for MU1–MU4 than for WT (Figure 1D).
Figure 1. A high-throughput colorimetric screening procedure.
(A) Fusion expression construct with DCase–α-fragment in the pMAL-c2x vector. The linker between DCase and the α-fragment of β-gal is Gly-Ser-Ala-Gly-Ser-Ala-Ala-Gly-Ser-Gly-Ala-Ser. (B) Photograph of E. coli colonies expressing genes encoding the WT and evolved DCases in liquid medium. (C) Assay of β-gal activity using the substrate ONPG in vitro. A unit of β-gal activity is defined as the amount of enzyme required to hydrolyse 1 μmol of ONPG to O-nitrophenol and D-galactose per min. (D) Native PAGE of soluble fractions of the WT and evolved DCases expressed with α-fragment fusion tag at 37 °C. MU1–MU4, DCase mutants 1–4 as indicated in the main text.
Determination of amino acid sites related to solubility in DCase
To identify the molecular basis of protein solubility change, plasmids harbouring evolved DCase genes were extracted for DNA sequencing. Only one substitution was found in MU1 and MU2, K34E and A18T respectively. Whereas MU3 and MU4 had the same substitution of Y30N, additional analysis showed that three other substitutions in MU3 or MU4 (Q23H, R278C and L300Q) did not affect the solubility of DCase (results not shown). The results suggested that the three amino acid residues (Ala18, Tyr30 and Lys34) were related to protein solubility of DCase. The sequence alignment of six amidohydrolases is listed in Figure 2 and the characteristics related to the three mutations are summarized in Table 1.
Figure 2. Sequence alignment of six homologous amidohydrolases.
1, DCase from B. pickettii; 2, DCase from Agrobacterium sp. KNK712; 3, DCase from Pseudomonas sp. KNK003A; 4, nitrilase from Polaromonas sp. JS666; 5, aliphatic amidase from Saccharopolyspora spinosa; 6, β-alanine synthase from Brevibacillus agri. The Figure was generated using the ClustalW program and drawn with ESPript. The secondary structure elements of DCase are shown above the sequences. Identical residues are boxed in black. Glu47, Lys127 and Cys172, the catalytically important residues, are indicated by stars. The three mutations in DCase, A18T, Y30N and K34E, are indicated by closed circles (●).
Table 1. Randomly evolved mutations leading to improved solubility of DCase.
| Amino acid substitution | Base change (codon) | Secondary structure at position | Location | Conserved residue | Hydrophilicity change* |
|---|---|---|---|---|---|
| A18T | GCG→ACG | Turn | Surface | No | +2.5 |
| Y30N | TAC→AAC | α-Helix | Surface | No | +2.2 |
| K34E | AAA→GAA | α-Helix | Surface | Yes | −0.4 |
*Increase or decrease of hydrophilicity of the mutated sites are indicated by ‘+’, ‘−’respectively. Hydropathy indices are obtained by the method of Kyte and Doolittle [34].
Effects of site mutations on DCase solubility
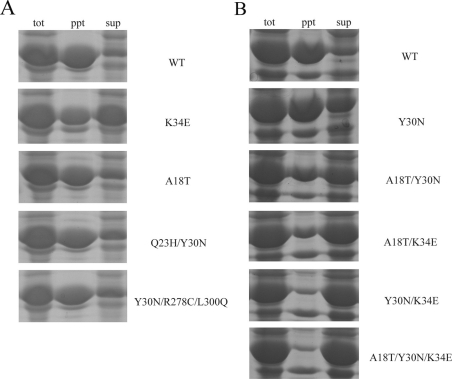
Due to the low expression level of DCase from the pMAL-c2x vector and the purification problems, the pET system was used to overexpress the target proteins. SDS/PAGE analysis of proteins from whole cells showed that overall expression levels of the WT DCase and the evolved DCase proteins were at the same level, approx. 70% of total protein (results not shown). Fractionation of the cells by sonication and centrifugation showed that more of the evolved DCase proteins remained soluble than the WT DCase protein (Figure 3A). Of the three single mutations (A18T, Y30N and K34E), the A18T and Y30N mutants were solubly expressed in approx. 35% of the total DCase proteins, whereas the mutant K34E produced up to approx. 50% soluble expression. To check whether cumulative effects are present among the three point mutations on protein solubility, three double mutants (A18T/Y30N, A18T/K34E and Y30N/K34E) and one triple mutant (A18T/Y30N/K34E) were constructed using site-directed mutagenesis. Solubility testing indicated that more soluble proteins could be obtained by any combination of more than one mutation (Figure 3B). The triple mutant DCase-M3 (A18T/Y30N/K34E) showed the highest solubility in all mutants, approx. 3-fold higher than that of the WT DCase. In comparison with the WT DCase, solubility of the evolved DCase mutants had all been improved from 10% to 60%, with the highest for DCase-M3 where more than 80% of the target proteins were soluble.
Figure 3. Solubility analysis of target proteins that were expressed using the pET expression system at 22 °C.
The mutations are listed on the right-hand side of the panels. (A) Solubility analysis of WT DCase and evolved recombinant DCases. (B) Expression test of a single mutant (Y30N), three double mutants (A18T/Y30N; A18T/K34E; and Y30N/K34E) and a triple mutant (A18T/Y30N/K34E). tot, total cell; ppt, precipitant fraction; sup, supernatant fraction.
Comparative characterization of the WT DCase and DCase-M3
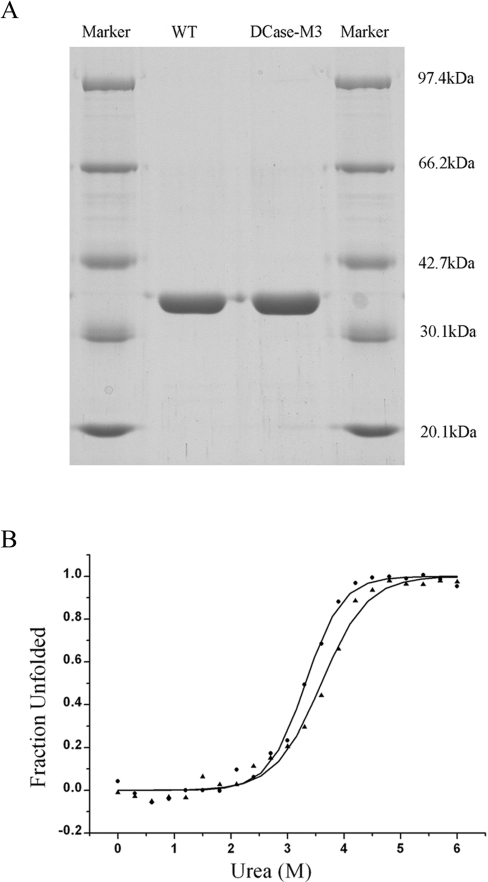
In order to evaluate the effect of three mutations (A18T, Y30N and K34E) on the enzyme properties of DCase-M3 and WT DCase, biochemical characterisation was performed. Both the WT DCase and DCase-M3 enzymes were produced at a purity of above 95% on Ni2+-affinity columns, as determined by SDS/PAGE (Figure 4A). We comparatively analysed two categories of steady-state characteristics of the enzymes. The first was the kinetic properties of enzymes, such as Km, kcat, and kcat/Km. Analytical results showed that DCase-M3 had similar kcat and Km values compared with WT DCase (Table 2). The second was the thermodynamic stability of the enzymes. The equilibrium unfolding transitions of the proteins by urea were carried out by monitoring fluorescence emission at 340 nm. The curves of the fluorescence change versus the increase of urea concentration are shown in Figure 4(B). Profiles of the WT DCase and the DCase-M3 showed almost identical and typical sigmoidal curves, indicative of a two-state unfolding model. According to the unfolding model, the Gibbs free energy changes (ΔGH2O) were calculated (Table 3). The ΔGH2O of the WT DCase (5.72±0.41 kcal/mol; 1 cal=4.184 J) was similar to that for Dcase-M3 within the experimental error (6.13±0.60 kcal/mol). Furthermore, values of Cm and m were only slightly changed (Table 3). Taken together, the results suggested that the kinetic properties and thermodynamic parameters of DCase-M3 were essentially identical with those of the WT DCase.
Figure 4. Equilibrium denaturation curves of WT and Dcase-M3.
(A) SDS/PAGE of purified WT DCase and DCase-M3. (B) Urea-induced equilibrium transition curves for the unfolding of WT DCase (▲) and DCase-M3 (●). Curves of urea-induced denaturation were monitored by the fluorescence of tryptophan at 340 nm.
Table 2. Kinetic parameters of WT DCase and DCase-M3.
The values for kinetic parameters are means±S.E.M. for three independent experiments.
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (mM−1·s−1) |
|---|---|---|---|
| WT | 0.82±0.1 | 5.4±0.3 | 6.6 |
| DCase-M3 | 0.80±0.1 | 5.9±0.2 | 7.4 |
Table 3. Thermodynamic properties of WT and DCase-M3 by urea denaturation (25 °C).
Unfolding of WT and DCase-M3, induced by the addition of urea, was monitored by the intrinsic fluorescence of tryptophan. The data containing the fraction of protein in the folded and unfolded state were calculated as the two-state unfolding model (ΔG=−RT·lnKu). ΔGH2O is the Gibbs free energy in the absence of denaturant, ΔG=ΔGH2O−m[urea]. Cm represents denaturant concentration at the midpoint of the denaturant transition. The m value represents the dependence of the free energy charge of unfolding on denaturant concentration, that is, the slope of plots shown in Figure 4(B).
| Protein | ΔGH2O (kcal/mol) | Cm (M) | m (kcal/mol per M) |
|---|---|---|---|
| WT | 5.72±0.41 | 3.56±0.03 | 1.61±0.10 |
| DCase-M3 | 6.13±0.60 | 3.21±0.10 | 1.91±0.13 |
Additional site-directed mutation of the three residues: Ala18, Tyr30 and Lys34
To explore further the molecular mechanism for increased soluble expression, three amino acid residues of DCase were mutated using overlap extension PCR. Replacements for the amino acids Ala18, Tyr30 and Lys34 were primarily selected on the basis of the hydrophobicity/hydrophilicity of the amino acids [34], and secondarily on their classification [35]. All mutant proteins were expressed in E. coli. Both soluble and insoluble fractions were fractionized and analysed by SDS/PAGE. Solubility tests of various point mutants of DCase are listed in Table 4. For the Ala18 residue, five mutant proteins, A18Y, A18T, A18E, A18D and A18N, showed a marked increase in their solubility, whereas the other muteins did not increase in solubility compared with the WT DCase. For the Tyr30 residue, three mutant proteins, Y30E, Y30D and Y30N, were generated and they were all significantly more soluble than the WT DCase. For the Lys34 residue, two mutated DCases, K34E and K34D, had higher solubility than the WT DCase (Table 4).
Table 4. Solubility test of point mutants of DCase.
Hydrophilicity of amino acids is from weak to strong: L, A, G, T, Y, H, E, D, N, K, and R. Solubility of proteins was analysed after cell lysis by ultrasonication, and then supernatant fractions were resolved by SDS/PAGE. The intensity of the target protein band of muteins was compared with that of wild-type DCase. −, no increase in solubility. Total expression of the target protein (DT) was calculated by adding the integrated density of the soluble (DS) and insoluble fractions (DI), and the soluble fraction was defined as SF=DS/DT [11]. ΔSF=(SF of mutein−SF of WT)/SF of WT.
| Ala18 | Tyr30 | Lys34 | |||
|---|---|---|---|---|---|
| Mutation | ΔSF (%) | Mutation | ΔSF (%) | Mutation | ΔSF (%) |
| A18L | − | Y30L | − | K34L | − |
| A18T | ∼34 | Y30A | − | K34A | − |
| A18Y | ∼40 | Y30G | − | K34G | − |
| A18E | ∼47 | Y30E | ∼81 | K34H | − |
| A18D | ∼44 | Y30D | ∼122 | K34E | ∼81 |
| A18N | ∼42 | Y30N | ∼55 | K34D | ∼37 |
| A18K | − | Y30K | − | K34R | − |
| A18R | − | Y30R | − | ||
Structural analysis of the engineering designed DCase-M3
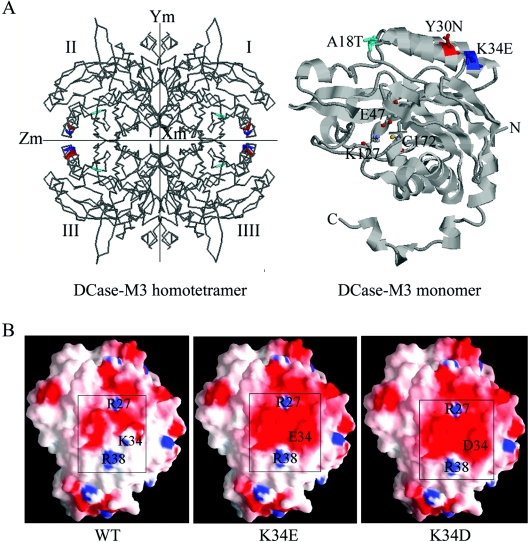
Crystal structures of DCase from several sources have been solved [30,36]. The protein from Agrobacterium sp. KNK712, solved at 1.7 Å (1 Å=0.1 nm) resolution, was employed to model the WT DCase and the DCase-M3. Sequence identity between the DCase from B. pickettii and the DCase from Agrobacterium sp. KNK712 was approx. 99% in the optimal alignment. The DCase-M3 model was constructed by replacing the 18–39 residue fragment containing the three mutated sites: A18T, Y30N and K34E using the SCWRL program [31]. In the structural models of the DCase-M3 homotetramer and the DCase-M3 monomer (Figure 5A), the three residues were distributed on the surface of the target protein, and were located far from the catalytic sites (Glu47, Lys127 and Cys172). The results suggested that the mutation of the three residues might not directly affect the function and structure of DCase, which is consistent with the results of the kinetic characterization. The structure model also showed that the residue at position 18 was situated at a turn between a β-sheet and an α-helix, and the amino acids at position 30 and 34 were located in an α-helix (Figure 5A). Replacement of the residues at positions 18 and 30 increased the hydrophilicity of DCase, whereas mutation of residue 34 decreased the hydrophilicity slightly (Table 1). Moreover, according to the charge distribution on protein surface generated by GRASP [33], charge distribution of the region near the amino acid at position 34 had a notable change on the molecular surface of the DCase-M3 model relative to that of the WT DCase (Figure 5B).
Figure 5. Model of DCase based on its similarity with DCase of Agrobacterium sp. KNK712.
(A) The three-dimensional structure of the DCase-M3 homotetramer and monomer. In the DCase-M3 homotetramer (left-hand panel), the three replacements are coloured: A18T (cyan), Y30N (red) and K34E (blue). In the DCase-M3 monomer (right-hand panel), the three mutations in DCase-M3 are indicated as space-filling models including A18T, Y30N and K34E, which are located on the surface of the protein. The catalytically important residues (Glu47, Lys127 and Cys172) are displayed as ball and stick model, and the N- and C-termini are also labelled in the structure. The Figure was generated using RasMol. (B) Charge distribution of the region near the amino acid at position 34 on the molecular surface of modelled WT and evolved DCases (K34E and K34D). The electrostatic potential is coloured: uncharged amino acids (white), negative amino acids (red) and positive amino acids (blue). All Figures were generated using GRASP.
DISCUSSION
Due to low solubility and improper folding, proteins often aggregate when over-expressed in heterologous expression systems. Directed evolution has been demonstrated as a powerful and efficient tool to overcome the difficulties of expressing target proteins in a soluble form [10,13,37,38]. In the present study, we have applied a directed evolution approach to improve the solubility of DCase in E. coli. The success of this approach was attributed in a large part to an efficient and high-throughput colorimetric screening system for monitoring protein solubility/misfolding in vivo using structural complementation of β-gal. This system greatly simplified the screening procedure and allowed visual inspection of colonies. Consistent with a previous report [13], the detailed analysis of β-gal activity and native PAGE showed that there was clear correlation between β-gal activity and solubility/folding of the evolved target proteins, and it was completely feasible to apply this method to screen clones with more soluble DCase (Figure 1).
The causes of IB formation have been widely investigated. The major factors that have been have been suggested include, the protein's amino acid composition in E. coli, its hydrophobicity and its overall net charge [39]. Interestingly, when comparing the amino acid sequences of the WT DCase with the evolved DCase-M3, two prominent differences were the hydrophobicity and charge properties of the mutated residues. The replacement by a hydrophilic residue may be able to counteract the hydrophobic site, resulting in improved solubility [39,40]. By mutation of Ala18 and Tyr30 with selected amino acids (Table 4), solubility tests revealed that increases in hydrophilicity of the residues at positions 18 and 30 improved its solubility. However, when proteins were substituted by positively charged residues (lysine or arginine), which had the strongest hydrophilic effect, no increase of solubility was observed. It was speculated that the positive effect of hydrophilicity (increase in solubility) and the negative effect of positive charge (decrease in solubility) might counteract with each other. Moreover, there is an obvious charge change in mutation K34E. Therefore, in this case the major force to reduce aggregation seems to be the increased negative charge. Additional replacement at the Lys34 position showed that mutated proteins replaced with negative charges (glutamate or aspartate) were more soluble than the WT DCase (Table 4). Although a reduced pI has been reported in several soluble muteins [7,9], no change of pI was observed for the A18T or Y30N single muteins and double mutein (A18T/Y30N). However, the single mutant K34E and triple mutant DCase-M3 (A18T/Y30N/K34E) had lower pI (6.57 and 6.57) values than WT DCase (pI 6.84). Although more proof is still needed, the results suggested that the pI change may be not the major factor affecting solubility in DCase, and it could still be partially involved in the increase in solubility of some of the muteins.
The effects of the mutations on soluble expression of DCase can be partially explained by homology structural modelling. There were three to four hydrophilic residues at intervals in the N-terminus of the WT DCase from the primary structure of protein, and molecular modelling predicted that both α1 and α2 had comparatively typical amphiphilic helixes, that is, one surface of each helix is mainly hydrophilic side-chains that interact favourably with the solvent, while the opposite surface is mainly hydrophobic side-chains to satisfy the core [41]. Notably, one of the most hydrophobic cores was formed in this region, which is composed of five secondary structure elements (α1, α2, β1, β2 and β3). According to the ‘hydrophobic collapse model’, the native polypeptide conformation forms by rearrangement of a compact collapsed structure during protein folding [42], and formation of α1 and α2 may be a very significant initiating process. Hence, the three mutations, A18T, Y30N and K34E, located on this domain could issue profound effects on folding or formation of the native polypeptides. In addition, the other mechanism may contribute to the delay in folding of passenger proteins by increasing negative charge. Recent evidence shows that protein aggregation can also be decreased by peptide extensions with large net negative charge; the net negatively charged peptide extensions could prolong the time that nascent polypeptides remain in productive folding pathways by increasing electrostatic repulsion between nascent polypeptides [7]. In the present study, when the residue at position 34 was replaced by amino acids with a negative charge, the solubility of muteins was increased (Table 4). This conclusion was further supported by analysing the charge distribution of region near the amino acid at position 34 on the molecular surface of modelled WT DCase and evolved DCases (K34E and K34D; Figure 5B). Significantly, the monomer interface of the evolved DCases showed a more favourable charge distribution relative to the WT protein. In the evolved K34D or K34E proteins, Asp34 or Glu34 balanced the positive potential of Arg27 and Arg38 from the symmetry-related subunit. In contrast, in the model of WT DCase, Arg27, Lys34 and Arg38 displayed an asymmetric negative and positive charge distribution. The favourable charge distribution will help proper folding of proteins by suppressing formation of non-specific off-pathway aggregates [37].
In summary, by combining error-prone PCR and DNA shuffling with a high-throughput colorimetric screening assay, we have developed a powerful directed evolution approach to obtain a DCase with better solubility when expressed in E. coli. There are two key parameters that may correlate with DCase solubility/misfolding in E. coli, which are charge average and hydrophilicity. The present study provides new insights into protein solubility/folding in E. coli and is also important for various rational design studies in improving protein solubility of similar types of proteins.
Acknowledgments
This work was supported by a grant from the Knowledge Innovation Program of the Chinese Academy of (SciencesKSCX2-YW-G-018, 007) and a grant from the Ministry of Science and Technology of China (National Basic Research Program of China, 2007CB707803). We thank Professor Haiyan Liu (Key Laboratory of Structural Biology, University of Science and Technology of China, Chinese Academy of Science, Hefei, China) and Professor Hongyu Hu (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China) for their kind help in molecular modelling and determination of thermodynamic parameters respectively.
References
- 1.Kleman G. L., Strohl W. R. Developments in high cell density and high productivity microbial fermentation. Curr. Opin. Biotechnol. 1994;5:180–186. doi: 10.1016/s0958-1669(05)80033-3. [DOI] [PubMed] [Google Scholar]
- 2.Wetzel R. For protein misassembly, it's the ‘‘I’’ decade. Cell. 1996;86:699–702. doi: 10.1016/s0092-8674(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 3.Goloubinoff P., Gatenby A. A., Lorimer G. H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 4.Roman L. J., Sheta E. A., Martasek P., Gross S. S., Liu Q., Masters B. S. S. High-level expression of functional rat neuronal nitric oxide synthase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S. C., Olins P. O. Effect of overproduction of heat shock chaperones GroESL and DnaK on human procollagenase production in Escherichia coli. J. Biol. Chem. 1992;267:2849–2852. [PubMed] [Google Scholar]
- 6.Chopra A. K., Brasier A. R., Das M., XU X. J., Petersoir J. W. Improved synthesis of Salmonella typhimurium enterotoxin using gene fusion expression systems. Gene. 1994;144:81–85. doi: 10.1016/0378-1119(94)90207-0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y. B., Howitt J., McCorkle S., Lawrence P., Springer K., Freimuth P. Protein aggregation during overexpression limited by peptide extensions with large net negative charge. Protein Expression Purif. 2004;36:207–216. doi: 10.1016/j.pep.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Georgiou G., Valax P. Expression of correctly folded proteins in Escherichia coli. Curr. Opin. Biotechnol. 1996;7:190–197. doi: 10.1016/s0958-1669(96)80012-7. [DOI] [PubMed] [Google Scholar]
- 9.Dale G. E., Broger C., Langen H., D'Arcy A., Stuber D. Improving protein solubility through rationally designed amino acid replacements: solubilization of the trimethoprim-resistant type S1 dihydrofolate reductase. Protein Eng. 1994;7:933–939. doi: 10.1093/protein/7.7.933. [DOI] [PubMed] [Google Scholar]
- 10.Yang J. K., Park M. S., Waldo G. S., Suh S. W. Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:455–460. doi: 10.1073/pnas.0137017100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldo G. S., Standish B. M., Berendzen J., Terwilliger T. C. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell K. L., Mittermaier A. K., Forman-Kay J. D., Davidson A. R. A simple in vivo assay for increased protein solubility. Protein Sci. 1999;8:1908–1911. doi: 10.1110/ps.8.9.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wigley W. C., Stidham R. D., Smith N. M., Hunt J. F., Thomas P. J. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat. Biotechnol. 2001;19:131–136. doi: 10.1038/84389. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa J., Shimizu S. Industrial microbial enzymes: their discovery by screening and use in large-scale production of useful chemicals in Japan. Curr. Opin. Biotechnol. 2002;13:367–375. doi: 10.1016/s0958-1669(02)00331-2. [DOI] [PubMed] [Google Scholar]
- 15.Chao Y. P., Fu H., Lo T. E., Chen P. T., Wang J. J. One-step production of D-p-hydroxyphenylglycine by recombinant Escherichia coli strain. Biotechnol. Prog. 1999;15:1039–1045. doi: 10.1021/bp9901163. [DOI] [PubMed] [Google Scholar]
- 16.Kim G. J., Kim H. S. Optimization of the enzymatic synthesis of D-p-hydroxyphenylglycine from DL-5-substituted hydantoin using D-hydantoinase and N-carbamoylase. Enzyme Microb. Technol. 1995;17:63–67. [Google Scholar]
- 17.Nanba H., Ikenaka Y., Yamda Y., Yajima K., Takano M., Takahashi S. Isolation of Agrobacterium sp. strain KNK712 that produces N-carbamyl-D-amino acid amidohydrolase, cloning of the gene for this enzyme, and properties of the enzyme. Biosci. Biotechnol. Biochem. 1998;62:875–881. doi: 10.1271/bbb.62.875. [DOI] [PubMed] [Google Scholar]
- 18.Grifantini R., Pratesi C., Galli G., Grandi G. Topological mapping of the cysteine residues of N-carbamyl-D-amino-acid amidohydrolase and their role in enzymatic activity. J. Biol. Chem. 1996;271:9326–9331. doi: 10.1074/jbc.271.16.9326. [DOI] [PubMed] [Google Scholar]
- 19.Moller A., Syldatk C., Schulze M., Wagner F. Stereo- and substrate-specificity of a D-hydantoinase and a N-carbamoyl-D-amino acid amidohydrolase of Arthrobacter crystallopoietes AM 2. Enzyme Microb. Technol. 1988;10:618–625. [Google Scholar]
- 20.Ogawa J., Max Ching-Ming C., Hida S., Yamada H., Shimizu S. Thermostable N-carbamoyl-D-amino acid amidohydrolase: screening, purification and characterization. J. Biotechnol. 1994;38:11–19. doi: 10.1016/0168-1656(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 21.Xu Z., Jiang W. H., Jiao R. S., Yang Y. L. Cloning, sequencing and high expression in Escherichia coli of D-hydantoinase gene from Burkholderia pickettii. Chin. J. Biotechnol. 2002;18:149–154. [PubMed] [Google Scholar]
- 22.Cadwell R. C., Joyce G. F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 23.Stemmer W. P. C. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouet P., Courcelle E., Stuart D. I., Metoz F. ESPript: multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 26.Aiyar A., Xiang Y., Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 1996;57:177–191. doi: 10.1385/0-89603-332-5:177. [DOI] [PubMed] [Google Scholar]
- 27.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Ikenaka Y., Nanba H., Yajima K., Yamada Y., Takano M., Takahashi S. Thermostability reinforcement through a combination of thermostability-related mutations of N-carbamyl-D-amino acid amidohydrolase. Biosci. Biotechnol. Biochem. 1999;63:91–95. doi: 10.1271/bbb.63.91. [DOI] [PubMed] [Google Scholar]
- 29.Pace C. N., Hirs C. H. W., Timasheff S. N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–228. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 30.Nakai T., Hasegawa T., Yamashita E., Yamamoto M., Kumasaka T., Ueki T., Nanba H., Ikenaka Y., Takahashi S., Sato M., Tsukihara T. Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases. Structure. 2000;8:729–737. doi: 10.1016/s0969-2126(00)00160-x. [DOI] [PubMed] [Google Scholar]
- 31.Bower M. J., Cohen F. E., Dunbrack R. L. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J. Mol. Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein H. J. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 2000;25:453–455. doi: 10.1016/s0968-0004(00)01606-6. [DOI] [PubMed] [Google Scholar]
- 33.Nicholls A., Sharp K. A., Honig B. Protein folding and association – insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 34.Kyte J., Doolittle R. F. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 35.Timberlake K. C. 5th Edn. New York: Harper-Collins; 1992. Chemistry. [Google Scholar]
- 36.Wang W. C., Hsu W. H., Chien F. T., Chen C. Y. Crystal structure and site-directed mutagenesis studies of N-carbamoyl-D-amino-acid amidohydrolase from Agrobacterium radiobacter reveals a homotetramer and insight into a catalytic cleft. J. Mol. Biol. 2001;306:251–261. doi: 10.1006/jmbi.2000.4380. [DOI] [PubMed] [Google Scholar]
- 37.Pedelacq J. D., Piltch E., Liong E. C., Berendzen J., Kim C. Y., Rho B. S., Park M. S., Terwilliger T. C., Waldo G. S. Engineering soluble proteins for structural genomics. Nat. Biotechnol. 2002;20:927–932. doi: 10.1038/nbt732. [DOI] [PubMed] [Google Scholar]
- 38.Esteban O., Zhao H. M. Directed evolution of soluble single-chain human class II MHC molecules. J. Mol. Biol. 2004;340:81–95. doi: 10.1016/j.jmb.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 39.Mosavi L. K., Peng Z. Y. Structure-based substitutions for increased solubility of a designed protein. Protein Eng. 2003;16:739–745. doi: 10.1093/protein/gzg098. [DOI] [PubMed] [Google Scholar]
- 40.Wurth C., Guimard N. K., Hecht M. H. Mutations that reduce aggregation of the Alzheimer's Aβ42 peptide: an unbiased search for the sequence determinants of Aβ amyloidogenesis. J. Mol. Biol. 2002;319:1279–1290. doi: 10.1016/S0022-2836(02)00399-6. [DOI] [PubMed] [Google Scholar]
- 41.Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 42.Dill K. A., Bromberg S., Yue K., Fiebig K. M., Yee D. P., Thomas P. D., Chan H. S. Principles of protein folding – a perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]