Abstract
MSK1 (mitogen- and stress-activated kinase 1) is a dual kinase domain protein that acts downstream of the ERK1/2 (extracellular-signal-regulated kinase 1/2) and p38 MAPK (mitogen-activated protein kinase) signalling pathways in cells. MSK1, and its related isoform MSK2, phosphorylate the transcription factors CREB (cAMP-response-element-binding protein) and ATF1 (activating transcription factor 1), and the chromatin proteins histone H3 and HMGN1 (high-mobility-group nucleosomal-binding protein 1) in response to either mitogenic stimulation or cellular stress. MSK1 activity is tightly regulated in cells, and activation requires the phosphorylation of MSK1 by either ERK1/2 or p38α. This results in activation of the C-terminal kinase domain, which then phosphorylates further sites in MSK1, leading to the activation of the N-terminal kinase domain and phosphorylation of substrates. Here, we use precursor ion scanning MS to identify five previously unknown sites in MSK1: Thr630, Ser647, Ser657, Ser695 and Thr700. One of these sites, Thr700, was found to be a third site in MSK1 phosphorylated by the upstream kinases ERK1/2 and p38α. Mutation of Thr700 resulted in an increased basal activity of MSK1, but this could be further increased by stimulation with PMA or UV-C radiation. Surprisingly, however, mutation of Thr700 resulted in a dramatic loss of Thr581 phosphorylation, a site essential for activity. Mutation of Thr700 and Thr581 to an alanine residue resulted in an inactive kinase, while mutation of both sites to an aspartic acid residue resulted in a kinase with a significant basal activity that could not be further stimulated. Together these results are consistent with a mechanism by which Thr700 phosphorylation relieves the inhibition of MSK1 by a C-terminal autoinhibitory helix and helps induce a conformational shift that protects Thr581 from dephosphorylation.
Keywords: cAMP-response-element-binding protein (CREB), extracellular-signal-regulated kinase (ERK), mitogen-activated protein kinase (MAPK), p38, p90 ribosomal S6 kinase (RSK), precursor ion scanning MS
Abbreviations: AGC-type kinases, protein kinase A/protein kinase G/protein kinase C-family kinases; ATF, activating transcription factor; CREB, cAMP-response-element-binding; ERK, extracellular-signal-regulated kinase; GST, glutathione S-transferase; LC, liquid chromatography; MAPK, mitogen-activated protein kinase; MEK1/2, MAPK/ERK kinase 1/2; MAPKAPK2/3, MAPK-activated protein kinase-2/3; MNK, MAPK-integrating kinase; MRM, multiple reaction monitoring; MSK, mitogen- and stress-activated kinase; MS/MS, tandem MS; PDK1, phosphoinositide-dependent kinase 1; PKI, protein kinase A inhibitor; RSK, p90 ribosomal S6 kinase
INTRODUCTION
MSK1 (mitogen- and stress-activated kinase 1) and MSK2 are closely related kinases involved in the regulation of gene transcription downstream of mitogenic signalling, pro-inflammatory cytokines and cellular stress. MSKs are able to regulate transcription by the phosphorylation of both transcription factors, including CREB (cAMP-response-element-binding protein) and ATF1 (activating transcription factor) [1–6], and the chromatin-associated proteins HMG-14 (high-mobility group 14) and histone H3 [7–10]. Genes reported to be regulated by MSK downstream of CREB or ATF1 include c-fos [11], junB [4], MUC5A (mucin 5A) [12] and genes encoding the NR4A group of nuclear orphan receptors [13].
The molecular mechanism of MSK activation is complex and requires multisite phosphorylation by upstream kinases [either ERK1/2 (extracellular-signal-regulated kinase 1/2) or p38α] and subsequent autophosphorylation for its activity. MSKs are most closely related to the RSK (p90 ribosomal S6 kinase) family of kinases and, like RSK, MSKs contain two distinct kinase domains within a single polypeptide [14–16]. The N-terminal domain, which is thought to phosphorylate MSK substrates, is a member of the AGC-type kinases (protein kinase A/protein kinase G/protein kinase C-family kinases), while the C-terminal kinase domain is related to the calmodulin-dependent protein kinase family [17]. MSK1 was originally identified [14] through a search for novel AGC kinases that might be regulated by PDK1 (phosphoinositide-dependent kinase 1), a ‘master’ kinase required for the activation of a subset of AGC kinases including RSK. PDK1 acts by phosphorylating a specific residue in the T-loop of its substrates, and the phosphorylation of this site is essential for kinase activation [18–20]. Work using PDK1−/− embryonic stem cells confirmed that PDK1 was required for the activation of RSK1, RSK2 and RSK3 [20]. Surprisingly, however, the use of these knockout cells has also demonstrated that the activation of MSK1 [20] and RSK4 [21] is independent of PDK1, and that T-loop phosphorylation for these kinases occurs via a different mechanism.
In cells, it has been shown that the activation of MSK by mitogens could be blocked by inhibitors of the ERK1/2 cascade, while the activation of MSK by cellular stress was blocked by inhibitors of p38α/β [14–16]. While both p38α and p38β can phosphorylate MSK1 in vitro [14], p38α appears to be the isoform responsible in vivo, as MSK1 activation is greatly reduced in cells from p38α, but not p38β, knockout mice [13,22]. In response to agonists capable of activating both ERK1/2 and p38, such as nerve growth factor or tumour necrosis factor, it was found that inhibitors of both pathways were required to block MSK activation [14–16,23].
Based on the similarity of MSKs to the ERK1/2-regulated kinase RSK, it was suggested that MSK was activated by phosphorylation on two sites by the upstream MAPK (mitogen-activated protein kinase) [14]. One site was in the activation loop of the C-terminal kinase domain (Thr581 in human MSK1), while the other (Ser360) was in the linker region between the two domains. Subsequent work has shown that mutagenesis of these sites in MSK1 and MSK2 prevents the full activation of MSK, and these sites have been shown to be phosphorylated in cells by ERK1/2 or p38 [24,25]. Phosphorylation of MSK1 by ERK1/2 or p38 was found to activate the C-terminal kinase domain, which then autophosphorylates two sites in the linker region (Ser376 and Ser381) and a site in the T-loop of the N-terminal kinase domain (Ser212). Of these three sites, Ser212 and Ser376 appear to be essential for MSK activation as their mutation, to alanine prevents MSK1 activation [24]. These autophosphorylation events activate the N-terminal kinase domain, allowing the phosphorylation of three sites in the C-terminus of MSK1 (Ser750, Ser752 and Ser758), as well as the phosphorylation of MSK substrates such as CREB and ATF1.
Here, we report the use of an Applied Biosystems 4000 Q TRAP MS system to identify further cellular phosphorylation sites in MSK1 and examine the role of these sites in the activation of MSK1.
METHODS
Plasmids
Mammalian expression vectors for FLAG and GST (glutathione S-transferase)-tagged MSK1 have been described previously [14]. Mutagenesis was performed by PCR using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). The MSK1 encoding region of each expression vector was fully sequenced to confirm the presence of the desired mutation. DNA sequencing was carried out by the sequencing service (School of Life Sciences, University of Dundee; http://www.dnaseq.co.uk) using Applied Biosystems Big-Dye Ver3.1 chemistry on a capillary sequencer.
Antibodies
MSK1 antibodies against phospho-Ser360, phospho-Ser376 and phospho-Thr581 were from Cell Signaling Technology, while the phospho-Ser212, phospho-Ser381 phospho-Ser750 and dually phosphorylated Ser750/752 and GST antibodies have been described previously [24]. FLAG antibody was from Sigma. The phospho-Thr700 antibody was raised in a rabbit as described in [26] against the phosphopeptide SNPLM(phospho-T)PDIL (695–704 of MSK1). The resulting antibody was affinity-purified from serum against the immunogenizing phosphopeptide.
Cell culture
HEK-293 cells (human embryonic kidney 293 cells) were cultured in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) fetal bovine serum (Sigma), 2 mM L-glutamine, 50 units/ml penicillin G and 50 μg/ml streptomycin (Invitrogen). HEK-293 cells were transfected using a modified calcium phosphate protocol as described in [14]. Purification of GST–MSK1 from transfected HEK-293 cell lysates was carried out on glutathione–Sepharose as described previously [14].
Q-Trap mass spectrometer phosphorylation-site analysis
GST–MSK1 samples were separated by SDS/PAGE, stained with colloidal Coomassie Blue (Invitrogen) and the gel bands were excised and digested in 50 mM triethylammonium bicarbonate with 5 μg/ml trypsin (Promega) at 30 °C for 18 h. The supernatant was removed, the gel pieces were extracted with 2.5% (v/v) formic acid/50% (v/v) acetonitrile and the combined extracts were dried under vacuum. Digests were reconstituted in 0.1 ml of 1% formic acid in water and analysed by LC (liquid chromatography)–MS on an LC-Packings Ultimate HPLC system interfaced to an Applied Biosystems 4000 Q TRAP system.
Peptides were separated on a 150 mm×0.075 mm PepMapC18 column equilibrated in 0.1% formic acid in water at a flow rate of 350 nl/min and eluted with a discontinuous acetonitrile gradient at the same flow rate. The column eluate was mixed with a sheath liquid of propan-2-ol/water (4:6, v/v) at 300 nl/min using a capillary Mixing Tee (Upchurch Scientific) and the combined flow plumbed into the microionspray head of the 4000 Q TRAP system mass spectrometer fitted with a New Objectives Picotip emitter (FS-360 75 15 N).
Electrospray MS was performed in an automated precursor of 79 duty cycle (6 s total) in negative ion mode (−2300 V), with Q1 masses scanned between 500 and 2000 m/z (3 s), collided with a variable collision energy of −65 to −110 V and daughter ions detected in Q3 after trapping and expelling from the linear ion trap (50 ms fill time). If a daughter ion of PO3− was detected, the polarity at the microionspray head was automatically switched to positive ion mode (+2400 V after 700 ms dwell) and an enhanced resolution scan followed by an enhanced product ion scan [MS/MS (tandem MS)] of the precursors was performed. The polarity was then switched back to −2300 V and the duty cycle repeated. All the MS/MS spectra were searched against local databases using the Mascot search engine (MatrixScience) run on a local server, and sites of phosphorylation were manually assigned from individual MS/MS spectra viewed using Bioanalyst software (MDS-Sciex).
A list of phosphopeptides to be analysed by MRM (multiple reaction monitoring) were generated using the MRM Builder Script supplied by MDS-Sciex as described in [27].
MSK1 kinase assays
For kinase assays, FLAG–MSK1 was immunoprecipitated from 0.2 mg of precleared cell lysate using 2 μg of anti-FLAG antibody coupled with Protein G–Sepharose, or GST–MSK1 pulled down with 10 μl of glutathione–Sepharose. Precipitates were washed twice in 0.5 M NaCl, 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA and 0.1% (v/v) 2-mercaptoethanol and once in 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA and 0.1% 2-mercaptoethanol. Precipitates were then resuspended in 35 μl of reaction buffer [Tris/HCl, pH 7.5, EGTA, PKI (protein kinase A inhibitor) and Crosstide peptide (GRPRTSSFAEG)], and the reaction was started by the addition of 10 μl of 50 mM magnesium acetate, 0.5 mM [32P]ATP and incubated at 30 °C for 15 min. Final concentrations of reagents in the assay were 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA, 0.1% 2-mercaptoethanol, 2.5 μM PKI, 30 μM Crosstide peptide, 10 mM magnesium acetate and 0.1 mM [32P]ATP. Reactions were stopped by transfer on to P81 paper and washing in 75 mM orthophosphoric acid. One unit was defined as the incorporation of 1 nmol of phosphate into the substrate peptide in 1 min.
To account for minor differences in the expression level of different constructs, the value for each sample was normalized relative to MSK expression. MSK1 expression was quantified using immunoblotting for the FLAG tag on the MSK1 constructs. Blots were visualized using the Licor Odyssey system and densitometry of bands was carried out using LI-COR software.
In vitro phosphorylation of MSK1
A GST–MSK1 C-terminal kinase domain–His tag construct, which comprised amino acids 398–802 of the human MSK1 sequence, was expressed in Escherichia coli and purified by affinity chromatography on glutathione–Sepharose and then Ni-NTA (Ni2+-nitrilotriacetate)–Sepharose using standard techniques. Then, 2 μg of MSK1 or myelin basic protein was incubated with 40 m-units of active GST–ERK2 or GST–p38α in 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA, 0.1% 2-mercaptoethanol, 2.5 μM PKI, 10 mM magnesium acetate and 0.1 mM [32P]ATP. After 20 min at 30 °C, the reaction was stopped by the addition of SDS sample buffer.
Immunoblotting
Soluble cell extract (25 μg) was run on 4–12% NuPAGE gels (Invitrogen) and transferred on to nitrocellulose membranes and blotted according to the manufacturer's protocols. For total antibodies, membranes were blocked in 5% (w/v) dried milk in 1×TBS-T [50 mM Tris/HCl, pH 7.6, 150 mM NaCl and 0.1% (v/v) Tween 20] and the primary antibody incubation carried out in the same buffer. For phospho-antibodies, blocking and primary antibody incubations were carried out in Odyssey blocking reagent (LI-COR). Commercial antibodies were used according to the manufacturer's recommendations and in house antibodies were used at 1 μg/ml in the presence of 10 μg/ml of the appropriate dephosphopeptide.
Detection was achieved using secondary antibodies coupled with an appropriate fluorophore and the membranes were scanned using a LI-COR Odyssey scanner. Densitometry of bands was carried out using LI-COR software according to the manufacturer's protocols.
RESULTS
Analysis of MSK1 phosphorylation by precursor ion scanning MS
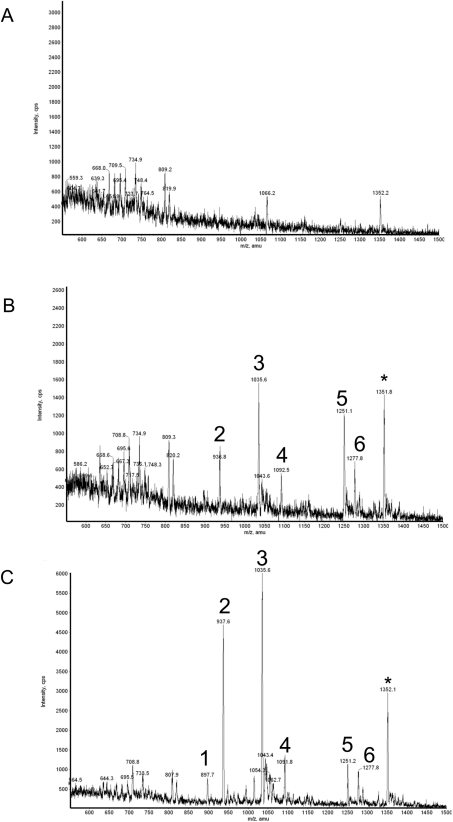
GST–MSK1 was expressed in HEK-293 cells by transient transfection, and the GST–MSK1 was then purified by affinity chromatography from the lysates of cells that had either been serumstarved or stimulated with either PMA or UV-C (Figures 1A–1C). Purified MSK1 was run on SDS/polyacrylamide gels and the MSK1 band was excised and digested with trypsin. The tryptic peptides were analysed on a 4000 Q TRAP system using precursor ion scanning to identify phosphorylated peptides. Potential phosphorylated peptides were then identified by MS/MS. For MSK purified from PMA-stimulated cells, this resulted in the identification of seven peaks, and for six of these MS/MS analysis it was possible to establish that the sequence corresponded to an MSK1 fragment (Figure 1C). The seventh peak corresponded to a peptide from the linker region between GST and MSK1. Two of the peaks corresponded to residues 371–385 of MSK1 that had been phosphorylated on Ser376 or on both Ser376 and Ser381 (Figure 1, peaks 1 and 2). These sites had been previously identified as sites phosphorylated by the C-terminal kinase domain [24]. A peptide corresponding to 210–226 of MSK1 that was phosphorylated on Ser212, another site phosphorylated by the C-terminal kinase domain, was also identified in this analysis (Figure 1, peak 3). The remaining three peptides all corresponded to previously unidentified sites in MSK1. Two of these peptides corresponded to residues 683–716 of MSK1 with either one or two phosphates. The monophosphorylated peptide contained a phosphate at Thr700, while the dually phosphorylated peptide contained phosphate at both Ser695 and Thr700 (Figure 1, peaks 5 and 6). The final peptide corresponded to residues 644–661 of MSK1 and contained phosphates on residues Ser647 and Ser657 (Figure 1, peak 4). Similar peptides were also identified in MSK1 purified from UV-C-stimulated cells (Figure 1B), although at reduced levels compared with PMA-stimulated cells. This reduction is consistent with the weaker activation of MSK1 by UV-C than PMA in this system (see Figure 2). Peak 1 (Ser376) was not detected above the background in the UV-C sample. It is likely however that this reflects a problem of sensitivity rather than a lack of Ser376 phosphorylation for two reasons. UV-C stimulation did result in the detection of peak 2, which corresponds to Ser376 and Ser381 phosphorylation. In addition, UV-C has previously been shown to induce phosphorylation of MSK1 on Ser376, as judged by immunoblotting [24]. The levels of the phospho-MSK1 peaks from unstimulated cells were not detected above background (Figure 1A). The two known MAPK sites, Ser360 and Thr581, were not identified in this analysis. The Thr581 tryptic peptide (residues 571–627 + PO4=6653.19 Da) would generate an ion outside the usable mass range of the 4000 Q TRAP system mass spectrometer. The reason Ser360 phosphorylation was not detected was not clear; however, it could be a result of a trypsin miscleavage resulting in a peptide outside the mass range of the spectrometer.
Figure 1. Precursor ion scan analysis of MSK1.
GST-tagged MSK1 was expressed in HEK-293 cells by transient transfection. Cells were then serum-starved for 16 h and then left unstimulated (control; A) or stimulated with UV-C (200 J/m2 followed by 30 min at 30 °C; B) or PMA (400 ng/ml, 10 min; C). MSK1 was purified on glutathione–Sepharose, samples were run on polyacrylamide gels and the MSK1 band was excised and digested with trypsin. The tryptic digests were analysed by LC–MS with precursor ion scanning on a 4000 Q TRAP system as described in the Methods section. The sum of the peptide masses detected by precursor ion scanning in the negative ion mode across the 45 min HPLC separation are shown. Stimulation with UV-C gave similar results to stimulation with PMA; however, the peak heights were smaller compared with the PMA sample. Peptides 1–6 were identified from the MS/MS fragmentation spectra by database searching and manual inspection of the spectra as: 1=LFQGY(pS)FVAPSILFK (phospho-Ser376), 2=LFQGY(pS)FVAP(pS)ILFK (phospho-Ser376/Ser381), 3=AY(pS)FCGTIEYMAPDIVR (phospho-Ser212), 4=GDF(pS)FEGEAWKNV(pS)QEAK (phospho-Ser647/Ser657), 5=YNEWLQDGSQLSSNPLM(pT)PDILGSSGAAHTCVK (phospho-Thr700), 6=YNEWLQDG(pS)QLSSNPLM(pT)PDILGSSGAAHTCVK (phospho-Ser695/Thr700). The peptide ion denoted with an asterisk (*) was found to be a phosphopeptide derived from the GST–FLAG tag used to generate this fusion protein.
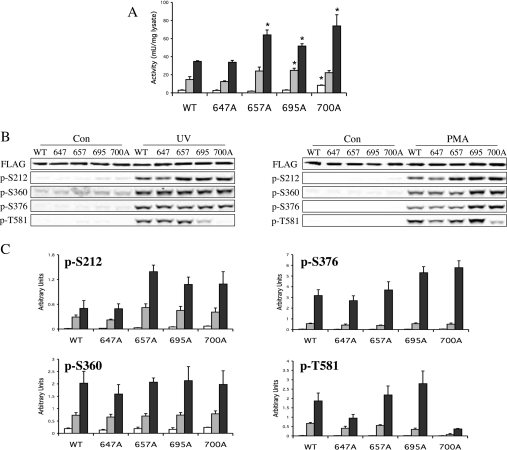
Figure 2. Effect of phosphorylation site mutagenesis on MSK1 activation.
Ser647, Ser657, Ser695 or Thr700 were changed to alanine by site-directed mutagenesis of a FLAG–MSK1 expression vector. Wild-type (WT) or mutant protein was then overexpressed in HEK-293 cells by transient transfection. Once transfected, cells were serum-starved for 16 h and then left unstimulated (white bars) or stimulated with UV-C (200 J/m2 followed by incubation at 30 °C for 30 min; grey bars) or PMA (400 ng/ml, 10 min; black bars). Cells were then lysed and MSK1 activity was determined by immunoprecipitation kinase assays as described in the Methods section (A). One unit was defined as the incorporation of 1 nmol of phosphate into the substrate peptide in 1 min. The results of the different mutations were compared with the wild-type protein from either control, UV-C or PMA stimulations by t tests. A P value of less than 0.05 is indicated by an asterisk. Soluble protein (25 μg) was run on SDS/polyacrylamide gels and immunoblotted using antibodies against FLAG, phospho-Ser360, phospho-Ser376 and phospho-Thr581 (B). To analyse Ser212 phosphorylation, MSK1 was first immunoprecipitated using an anti-FLAG antibody and the immunoprecipitates were blotted for Ser212 phosphorylation. Immunoblots were quantified using an Odyssey LI-COR scanner and the phospho-signal was calculated relative to the FLAG loading control (C). Error bars represent the S.E.M. for three separate stimulations.
Ser647, Ser657 and Ser695 phosphorylation is not required for MSK activation
The requirement for phosphorylation on Ser212, Ser376 or Ser381 for MSK1 activity has been investigated previously by site-directed mutagenesis [24]. To examine if the novel sites, Ser647, Ser657, Ser695 and Thr700, identified in the MS analysis were required for MSK1 activation, each site was mutated to alanine in a FLAG-tagged MSK1 construct. Mutated constructs were transfected into HEK-293 cells, which were then starved for 16 h. The cells were then stimulated by PMA (400 ng/ml for 10 min) or UV-C (200 J/m2 followed by 30 min at 37 °C) and MSK1 activity was measured by immunoprecipitation kinase assays. Mutation of any of these sites did not greatly affect the level of MSK1 expression, as judged by immunoblotting for the FLAG tag.
Mutation of Ser647 to alanine did not affect the ability of PMA or UV-C to activate MSK1 when compared with the wild-type protein (Figure 2A). Consistent with this, the mutation of Ser647 did not affect the phosphorylation of Ser376, Ser360 and Ser212, sites shown previously to be involved in the activation of MSK1 in response to UV-C or PMA (Figures 2B–2C). Surprisingly, the PMA-, and, to a lesser extent, UV-C-induced phosphorylation of Thr581 was reduced by this mutation.
Mutation of Ser657 to alanine resulted in a modest increase in MSK1 activity after stimulation with either UV-C for 30 min or PMA for 10 min compared with wild-type MSK1 (Figure 2A). These increases were, however, less pronounced at later time points, suggesting that this mutation is affecting the rate of activation rather that the specific activity of the activated protein (results not shown). The increased activation at early time points correlated with increases in the phosphorylation of the N-terminal T-loop site, Ser212, in the S657A mutant (Figures 2B–2C). Mutation of Ser657 to alanine did not greatly affect the phosphorylation of Ser360, Ser376 or Thr581 induced by either PMA or UV-C.
Mutation of Ser695 to alanine also resulted in small increases in the UV-C- or PMA-induced activation of MSK1 (Figure 2A), and again this correlated with increases in Ser212 phosphorylation, as well as slight increases in the PMA-induced level of Ser376 and Thr581, but not Ser360, phosphorylation compared with wild-type protein (Figures 2B–2C). Mutation of Ser647, Ser657 or Ser695 to an aspartic acid residue had similar effects as mutation to alanine on the activation of MSK1 in cells (results not shown).
Thr700 is a new MAPK site involved in the regulation of MSK1 activity
Mutation of Thr700 resulted in a moderate increase in the PMA- and UV-C-stimulated level of MSK1 activity; however, for this mutant an increase in the basal level of activity was also observed (Figure 2A). This correlated with an increase in the phosphorylation of Ser212. Surprisingly however, phosphorylation of Thr581 was greatly reduced in the Thr700 mutant compared with wild-type MSK1 after either UV-C or PMA stimulation (Figures 2B–2C). This was unexpected, as the mutation of Thr581 to alanine prevents the activation of MSK1 [24].
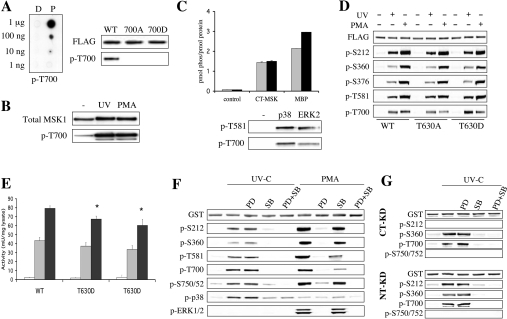
Interestingly, Thr700 is followed by a proline residue suggesting that it may be a new MAPK site. A phosphospecific antibody was therefore raised against Thr700. This antibody was specific for phospho-Thr700 as judged by both dot-blotting Thr700 phospho- and dephospho-peptides, and immunoblotting of UV-C-stimulated HEK-293 cells expressing wild-type or T700A MSK1 (Figure 3A). To confirm that this site was phosphorylated in endogenous MSK1, MSK1 was immunoprecipitated from serum-starved, UV-C- or PMA-stimulated HEK-293 cells and immunoblotted. Both UV-C and PMA were found to stimulate the phosphorylation of Thr700 in endogenous MSK1 (Figure 3B).
Figure 3. Thr700 of MSK1 is phosphorylated by ERK1/2 and p38.
(A) To allow analysis of Thr700 phosphorylation in MSK1 an anti-(phospho-Thr700) antibody was raised in rabbits. To analyse the specificity of this antibody, it was tested in dot-blots against the peptides SNPLMTPDIL (D, dephospho) and SNPLM (phosphoT)PDIL (P, phospho), which correspond to the sequence around Thr700 in MSK1 (A, left panel). The antibody was tested for its ability to recognize wild-type (WT), T700A or T700D MSK1 expressed in HEK-293 cells that had been PMA-stimulated before lysis (A, right panel). (B) Endogenous MSK1 was immunoprecipitated from 1 mg of soluble lysate from serum-starved HEK-293 cells or cells stimulated with UV-C (200 J/m2 followed by 30 min at 37 °C) or PMA (400 ng/ml, 10 min). Immunoprecipitates were then immunoblotted for total and phospho-Thr700 MSK1. (C) A 2 μg portion of recombinant C-terminal kinase domain of MSK1 (CT-MSK1), purified from E. coli, was phosphorylated by using 40 m-units of ERK2 (grey bars) or p38α (black bars) for 20 min at 30 °C as described in the Methods section. As controls, MSK was incubated without ERK or p38 (control), and ERK or p38α was used to phosphorylate the known MAPK substrate myelin basic protein (MBP). The protein was then run on SDS/polyacrylamide gels and, after Coomassie Blue staining, the MSK1 bands were excised and the 32P incorporation was determined by Cerenkov counting (C, upper panel). Samples from the same reactions were also immunoblotted using phospho-Thr581 and phospho-Thr700 antibodies (C, lower panel). (D) FLAG-tagged wild-type, T630A or T630D MSK1 was expressed in HEK-293 cells by transient transfection. Following transfection, cells were serum-starved for 16 h and then either left unstimulated or stimulated with UV-C (200 J/m2 followed by 30 min at 37 °C) or PMA (400 ng/ml, 10 min). Lysates were then immunoblotted for phospho-Ser212, -Ser360, -Ser376, -Thr581 and - Thr700 MSK1 and for GST as a loading control. (E) As (D) except that anti-FLAG antibodies were used to immunoprecipitate MSK1 and kinase activity was measured as described in the Methods section. The kinase activity from unstimulated cells is shown by white bars, UV-C-stimulated cells by light grey bars and PMA-stimulated cells by dark grey bars. Error bars represent the S.E.M. for three independent stimulations. One unit was defined as the incorporation of 1 nmol of phosphate into the substrate peptide in 1 min. The results of the different mutations were compared with the wild-type protein from either control, UV-C or PMA stimulations by t tests. A P value of less than 0.05 is indicated by an asterisk. (F) Wild-type GST-tagged MSK1 was expressed in HEK-293 cells by transient transfection. Following transfection, cells were serum-starved for 16 h and then incubated for a further 1 h with either no inhibitor, 2 μM PD 184352 (PD), 5 μM SB 203580 (SB) or both PD 184352 and SB203580 (PD + SB). Cells were then left unstimulated or stimulated with UV-C (200 J/m2 followed by incubation at 30 °C for 30 min) or PMA (400 ng/ml, 10 min). Soluble protein (25 μg) was run on SDS/polyacrylamide gels and immunoblotted using antibodies against GST, phospho-Ser360, phospho-Ser376, phospho-Thr581, phospho-Thr700 or phospho-Ser750/752. To analyse Ser212 phosphorylation, MSK1 was first pulled down using GSH–Sepharose and the pull-downs blotted for Ser212 phosphorylation. (G) as (F), except that cells were transfected with either C-terminal kinase dead (CT-KD, top panel) or N-terminal kinase dead (NT-KB, lower panel) GST-tagged MSK1, and cells were stimulated with UV-C only.
To determine if ERK2 or p38α could phosphorylate this site in vitro, the C-terminal kinase domain (residues 398–804) of MSK1 was expressed in E. coli and used as a substrate for these kinases. Both ERK2 and p38α were able to phosphorylate this domain on the Thr581 and Thr700 MSK1 sites, although p38α appeared to be slightly more effective than ERK2 at the phosphorylation of Thr700 in vitro, as judged by immunoblotting using antibodies against these sites (Figure 3C). The presence of Thr700 phosphorylation in the ERK2-phosphorylated MSK1 domain was further confirmed using precursor ion scanning MS (results not shown). Surprisingly, this also identified a further site (equivalent to Thr630 in full-length MSK1) as being phosphorylated. Re-analysis of the MS data on HEK-293-cell-expressed MSK1 using MRM techniques suggested that this site may also be phosphorylated in cells; however, mutation of this site to alanine or aspartic acid in full-length MSK1 did not affect the phosphorylation of the other sites (Figure 3D) or the activation of MSK1 (Figure 3E).
In HEK-293 cells both UV-C and PMA were able to stimulate the phosphorylation of Thr700 in wild-type MSK1. The phosphorylation of Thr700 downstream of UV-C stimulation was blocked by the p38α/β inhibitor SB203580, but unaffected by the MEK1/2 (MAPK/ERK kinase 1/2) inhibitor PD184352, suggesting that p38 can directly phosphorylate this site in cells (Figure 3F). In response to PMA the MEK1/2 inhibitor greatly decreased the phosphorylation of Thr700, suggesting that, in response to PMA, ERK1/2 are the major kinases for this site (Figure 3F). Surprisingly, the p38 inhibitor SB203580 was also able to decrease the phosphorylation of Thr700 in response to PMA, although the effect was not as great as for PD184352. As PMA did not significantly activate p38α, as judged by immunoblotting for phospho-p38 (Figure 3F), this may be due to a role for basal p38 activity in this phosphorylation, or represent a non-specific effect of the inhibitor. Similar results were obtained for Ser360 and Thr581, the other MAPK sites in MSK1 (Figure 3F). To confirm that Thr700 was not an autophosphorylation site, its phosphorylation was examined after UV-C treatment of cells expressing either N- or C-terminal kinase dead forms of MSK1. As expected, Thr700 was phosphorylated after UV-C treatment in both the N- and C-terminal kinase dead MSK1, although phosphorylation of the appropriate autophosphorylation sites (Ser212 for the C-terminal kinase domain and Ser750/752 for the N-terminal kinase domain) was blocked in the kinase dead mutants (Figure 3G).
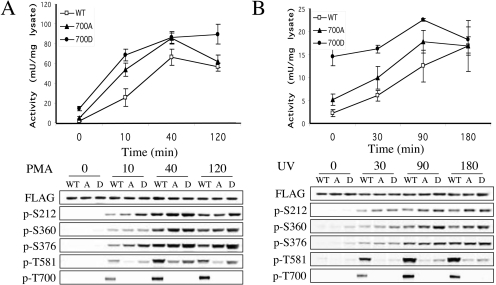
To further examine the role of Thr700 in MSK activation, Thr700 was also mutated to an aspartic acid in an attempt to mimic phosphorylation. Wild-type, T700A or T700D MSK1 were expressed in HEK-293 cells and their activation was measured over a time course of PMA and UV-C stimulation. As seen in Figure 4, mutation of Thr700 to alanine resulted in a 2-fold increase in the basal level of MSK1 activity in the transfected cells. The basal MSK1 activity was even higher (∼7-fold increase compared with wild-type) for the T700D mutant. UV-C and PMA were however able to further stimulate the activities of both these mutants, as well as the wild-type protein (Figures 4A and 4B). At 10 and 40 min of PMA treatment or 30 and 60 min after UV-C treatment the mutant proteins were still more active than their wild-type controls; however, this was less pronounced at longer time points (120 min for PMA or 180 min for UV-C). The level of Ser212, Ser360 or Ser376 phosphorylation was either unchanged or slightly increased in the T700A or T700D mutants after PMA or UV-C stimulation compared with wild-type protein. In contrast, phosphorylation of Thr581, a critical site for MSK1 activation, was greatly reduced in both Thr700 mutants compared with wild-type protein in response to UV-C treatment. In response to PMA stimulation, phosphorylation of Thr581 was also significantly reduced in the T700A mutant in response to PMA; however, this phosphorylation was only slightly reduced in the T700D MSK1 compared with wild-type protein.
Figure 4. Effect of Thr700 mutagenesis on MSK1 activation.
Wild-type (open squares), T700A (closed triangles) or T700D (closed circles) MSK1 was overexpressed in HEK-293 cells by transient transfection. Once transfected, cells were serum-starved for 16 h and then left unstimulated or stimulated with 400 ng/ml PMA (A) or 200 J/m2 UV-C followed by incubation at 37 °C (B) for the times indicated. Cells were then lysed and MSK1 activity was determined by immunoprecipitation kinase assays as described in the Methods section (upper panels). One unit was defined as the incorporation of 1 nmol of phosphate into the substrate peptide in 1 min. Soluble protein (25 μg) was run on SDS/polyacrylamide gels and immunoblotted using antibodies against FLAG, phospho-Ser360, phospho-Ser376, phospho-Thr581 and phospho-Thr700. To analyse Ser212 phosphorylation, MSK1 was first immunoprecipitated using an anti-FLAG antibody and the immunoprecipitates were blotted for Ser212 phosphorylation (lower panels).
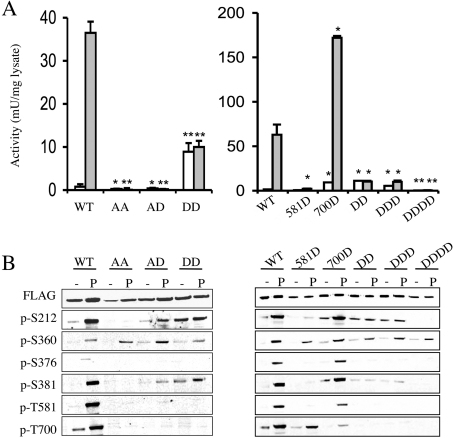
The reduction in Thr581 phosphorylation that occurs when Thr700 was mutated is surprising given that the Thr700 mutations were active, as Thr581 phosphorylation has previously been shown to be critical for activity. We therefore analysed the effects of combinations of Thr581 and Thr700 mutations (Figure 5). Mutation of both Thr581 and Thr700 to alanine resulted in an inactive kinase that was not stimulated by either PMA (Figure 5A) or UV-C (results not shown) and was not phosphorylated on either of the Ser212 and Ser376 MSK1 autophosphorylation sites (Figure 5B), consistent with what had previously been reported for the single T581A mutation [24]. The T581A/T700A double mutation did not, however, prevent the interaction of the mutant with its upstream kinases ERK1/2 or p38, as the MAPK site Ser360 was phosphorylated normally in response to PMA or UV-C in this mutant (Figure 5B). Mutation of Thr581 to alanine and Thr700 to aspartic acid also resulted in an inactive kinase, reinforcing the critical role of Thr581 (Figure 5A). To further investigate the interaction between Thr581 and Thr700, Thr581 was mutated to an aspartic acid to try and mimic phosphorylation. A single T581D mutation was found to have extremely low activity, indicating that this mutation was not completely successful at replicating the effects of phosphorylation (Figure 5A). The T581D mutant was, however, phosphorylated on the other MAPK sites, Ser360 and Thr700, in response to PMA stimulation, and a small increase in Ser212 phosphorylation was also observed (Figure 5B). Mutation of both Thr581 and Thr700 to aspartic acid resulted in a kinase with an elevated basal activity in unstimulated cells, similar to what was seen for the single T700D mutant. However, in contrast with the T700D single mutant, the activity of the double mutant could not be further stimulated by PMA (Figure 5A) or UV-C (results not shown). Consistent with the elevated basal activity of the T581D/T700D mutant, an elevated level of Ser212 phosphorylation that was not significantly enhanced by PMA treatment was also observed (Figure 5B).
Figure 5. Analysis of MSK1 Thr581/Thr700 mutants.
Wild-type or mutant MSK1 was expressed in HEK-293 cells by transient transfection. After transfection cells were serum-starved for 16 h and then left unstimulated (white bars) or stimulated by PMA (400 ng/ml, 10 min; grey bars). Cells were then lysed and MSK1 activity was determined by immunoprecipitation kinase assays as described in the Methods section (A). Activity was normalized to expression based on anti-FLAG blots. Error bars represent the S.E.M. for three stimulations. One unit was defined as the incorporation of 1 nmol of phosphate into the substrate peptide in 1 min. The results of the different mutations were compared with the wild-type protein from either control or PMA stimulations by t tests. A P value of less than 0.05 is indicated by an asterisk, and a value of less than 0.01 by double asterisks. Soluble protein (30 μg) was run on SDS/polyacrylamide gels and immunoblotted using antibodies against FLAG, phospho-Ser360, phospho-Ser376, phospho-Ser381, phospho-Thr581 and phospho-Thr700. To analyse Ser212 phosphorylation, MSK1 was first immunoprecipitated using an anti-FLAG antibody and the immunoprecipitates were blotted for Ser212 phosphorylation (B). Analysis of wild-type (WT) as well as T581D (581D), T700D (700D), T581D/T700D (DD), S376D/T581D/T700D (DDD), S212D/S376D/T581D/T700D (DDDD), T581A/T700D (AA) and T581A/T700D (AD) mutants are shown.
Previous studies have suggested that mutation of Ser376 to Asp successfully mimics phosphorylation of this residue [24]. We therefore tested if mutation of Ser376 to aspartic acid could further increase the activity of the T581D/T700D mutant. The triple mutation of Ser376, Thr581 and Thr700 to aspartic acid, however, had a similar activity and phosphorylation pattern to that of the double mutant (Figure 5). Mutation of all the critical phosphorylation sites in MSK1, Ser212, Ser276, Thr581 and Thr700, to an aspartic acid resulted in an inactive kinase. This is consistent with the previous finding that mutation of Ser212 to an acidic residue in the N-terminal kinase domain T-loop was unable to activate this domain [28].
DISCUSSION
We have previously studied the activation mechanism of MSK1 and demonstrated that ERK1/2 or p38α phosphorylate two sites in MSK1, Ser360 and Thr581, that are required for its activation [24]. These phosphorylation events activate the C-terminal kinase domain of MSK1, which then autophosphorylates Ser212, Ser376 and Ser381, resulting in the activation of the N-terminal kinase domain. Here, we describe the identification of five new phosphorylation sites in MSK1: Thr630, Ser647, Ser657, Ser695 and Thr700. One of these sites, Thr700 lies in a consensus MAPK phosphorylation sequence (ΨXS/TP where Ψ is a proline or aliphatic residue), and was found to be phosphorylated by both ERK and p38α both in vitro and in cells. The remaining sites were not direct targets of MAPKs and represent autophosphorylation sites, or sites for an as yet undetermined kinase. All of these sites lie in the C-terminal domain of MSK1, which is most closely related to MAPKAPK2/3 (MAPK-activated protein kinase-2/3), MNK1/2 (MAPK-integrating kinase 1/2) and the C-terminal domain of RSK [29]. The equivalent site to Ser647 in MSK1 is a threonine residue in RSK1; however, it is replaced by a glutamic acid residue in MNK and MAPKAPK2, while the Ser657 site in MSK1 is conserved in RSK, MSK and MAPKAPK2. In contrast, the Thr630 and Ser695 sites are not conserved in the other kinases. The MAPK site Thr700 is not conserved in RSK; however, it is conserved in MNK1 and MNK2. A threonine residue is present at this position in MAPKAPK2; however, it is not followed by a proline residue making it an unlikely MAPK target. Interestingly, MAPKAPK2 does contain a potential p38 phosphorylation site four residues N-terminal to this position (Figure 6).
Figure 6. Sequence alignment of kinase domains.
The C-terminal kinase domains human MSK1, MSK2 and RSK1, as well as the kinase domains of MAPKAPK2, MNK1 and MNK2, are aligned. Phosphorylation sites in MSK1 are indicated by asterisks.
Mutation of Ser647, Ser657, Ser695 or Thr700 to alanine (Figure 1) or aspartic acid (results not shown) did not prevent the activation of MSK1 in response to UV-C or PMA. Mutation of Thr700 to alanine slightly increased the basal activity of MSK1, while mutation to aspartic acid increased the basal activity still further. A similar result has been shown for mutagenesis of the equivalent site of MSK2 [30], although this study did not directly establish that this residue could be phosphorylated. Mutation of the equivalent site in MNK1 or MNK2 has also been reported to give similar results [31,32]; however, while this site is phosphorylated in vitro by ERK [33], it was unclear from these studies if this was an in vivo phosphorylation site. Interestingly the equivalent site (Thr338) is phosphorylated in MAPKAPK2, although this probably occurs via autophosphorylation rather than by p38 or ERK1/2 [34]. A MAPK site (Thr334) does exist in MAPKAPK2 four residues N-terminal to the Thr338 autophosphorylation site. Mutation of the Thr334 in MAPKAPK2 to alanine does not prevent the activation of MAPKAPK2, while mutation to an acidic residue increased the basal activity. Mutation of both Thr344 and the T-loop phosphorylation site in MAPKAPK2 to acidic residues resulted in a kinase that is constitutively active [34,35]. In combination with structural information from crystallographic studies [36,37] this has suggested a model for MAPKAPK2 activation in which phosphorylation of Thr334 promotes the dissociation of an autoinhibitory C-terminal helix from the substrate-binding groove of the kinase domain, while phosphorylation of the activation loop on Thr222 stabilizes its structure and promotes substrate binding [35,37].
Although the C-terminal regions of MSK1 and MAPKAPK2 are not well conserved, activation of the MSK1 C-terminal kinase domain may occur via a similar mechanism, with phosphorylation of Thr700 in MSK1 promoting the dissociation of an autoinhibitory sequence from the kinase domain (Figure 7). Consistent with this, C-terminal truncation of MSK2 has been reported to increase its basal activity [30], although this truncation also deletes nuclear localization and MAPK docking sequences. Such a mechanism would explain the increased basal activity of the Thr700 MSK1 mutations; however, it would not explain why Thr581 phosphorylation is reduced in these mutants. Thr581 phosphorylation is still required for MSK1 activation, as mutation of Thr700 to aspartic acid does not rescue the activity of a T581A mutation. One possibility is that mutation of Thr700 does not prevent Thr581 phosphorylation but instead reduces the stability of the Thr581 phosphorylation. In such a model, Thr581 would be phosphorylated, allowing the activation of MSK1; however, its prolonged phosphorylation would require interaction of the phospho-Thr581 with another part of MSK1 in a way that protects it from dephosphorylation. Mutation of Thr700 would interrupt this interaction and therefore result in accelerated dephosphorylation of Thr581. Interestingly, reduced Thr581 phosphorylation has also been observed in other MSK1 mutants, such as those that inactivate the C-terminal or N-terminal kinase domains [24], in line with the suggestion that this site can be easily dephosphorylated if the correct active conformation of MSK1 cannot be formed. The idea that the active conformation of a kinase can protect it from dephosphorylation has been suggested previously for other kinases. For instance, binding of the phosphorylated hydrophobic motif of PKB, RSK and S6K (S6 kinase) to a phosphate-binding pocket in the kinase has been suggested to protect the hydrophobic motif from dephosphorylation [18,38,39].
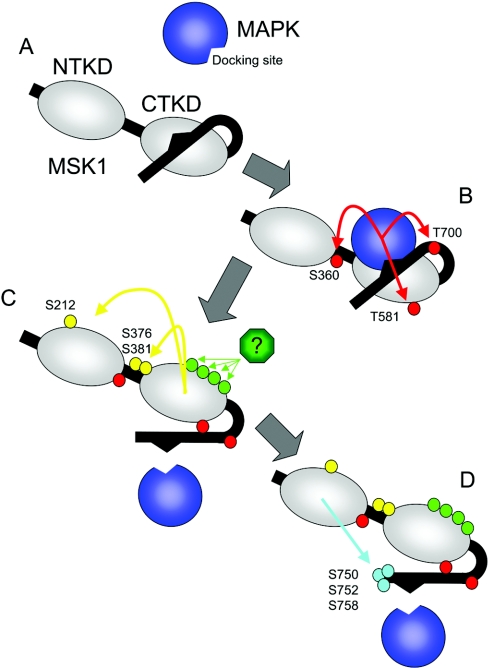
Figure 7. Activation model for MSK1.
MSK1 consists of two kinase domains, joined by a short linker region, and a C-terminal extension containing a MAPK kinase docking domain after the second kinase domain. In the inactive state (A), MSK1 is unphosphorylated and the C-terminus of the protein may fold back and act as an inhibitor of the C-terminal kinase domain, as has been described for the related kinase MAPKAPK2. Active ERK1/2 or p38 MAPK is able to bind to the docking motif in the C-terminal region of MSK1, resulting in the phosphorylation of three sites, Ser360, Thr581 and Thr700 in MSK1 (red circles; B). Thr581 is in the activation loop of the C-terminal kinase domain and its phosphorylation may be required to stabilize the active confirmation of the activation loop. Thr700 is proposed to lie in a hinge region between the C-terminal kinase domain and the inhibitory C-terminal sequence and phosphorylation of this residue would promote the dissociation of the inhibitory sequence from the kinase domain. MSK1 is additionally phosphorylated on a further four sites by an unidentified kinase (green circles), and the function of these phosphorylations is unclear. The activated C-terminal kinase domain then activates the N-terminal kinase domain via the phosphorylation of three sites, Ser212, Ser376 and Ser381 (yellow circles; C). Once activated, the N-terminal kinase domain is able to phosphorylate substrates, and additionally phosphorylates three sites, Ser750, Ser752 and Ser758 (blue circles; D) at the C-terminus of MSK1.
In total, 13 phosphorylation sites have now been identified in MSK1, of which at least six have significant roles in MSK1 activation. The role of the remaining sites is unclear, but may influence other processes such as localization or protein–protein interactions in vivo. MSK1 is sequentially activated through phosphorylation by ERK1/2 or p38 to activate the C-terminal kinase domain, which then activates the N-terminal kinase domain; however, the present study suggests that this may also be influenced by phosphorylation of a C-terminal autoinhibitory domain. To fully understand the mechanism by which phosphorylation regulates MSK1 will, however, require three-dimensional structural studies on the phosphorylated and dephosphorylated enzymes.
Acknowledgments
We thank the antibody purification teams (Division of Signal Transduction Therapy, University of Dundee) co-ordinated by Hilary McLauchlan and James Hastie for the phospho-Thr700 MSK1 antibody and the Sequencing Service (School of Life Sciences, University of Dundee) for DNA sequencing. This research was supported by the U.K. Medical Research Council, AstraZeneca, Boehringer–Ingelheim, GlaxoSmithKline, Merck and Co., Merck KGaA and Pfizer. C.E.M. was the recipient of a Royal Society Ph.D. studentship.
References
- 1.Arthur J. S. C., Fong A. L., Dwyer J. M., Davare M., Reese E., Obrietan K., Impey S. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J. Neurosci. 2004;24:4324–4332. doi: 10.1523/JNEUROSCI.5227-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes J. P., Staton P. C., Wilkinson M. G., Strijbos P. J., Skaper S. D., Arthur J. S., Reith A. D. Mitogen and stress response kinase-1 (MSK1) mediates excitotoxic induced death of hippocampal neurones. J. Neurochem. 2003;86:25–32. doi: 10.1046/j.1471-4159.2003.01830.x. [DOI] [PubMed] [Google Scholar]
- 3.Arthur J. S., Cohen P. MSK1 is required for CREB phosphorylation in response to mitogens in mouse embryonic stem cells. FEBS Lett. 2000;482:44–48. doi: 10.1016/s0014-5793(00)02031-7. [DOI] [PubMed] [Google Scholar]
- 4.Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 2002;22:2871–2881. doi: 10.1128/MCB.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C. W., Nam J. S., Park Y. K., Choi H. K., Lee J. H., Kim N. H., Cho J., Song D. K., Suh H. W., Lee J., et al. Lysophosphatidic acid stimulates CREB through mitogen- and stress-activated protein kinase-1. Biochem. Biophys. Res. Commun. 2003;305:455–461. doi: 10.1016/s0006-291x(03)00790-3. [DOI] [PubMed] [Google Scholar]
- 6.Gustin J. A., Pincheira R., Mayo L. D., Ozes O. N., Kessler K. M., Baerwald M. R., Korgaonkar C. K., Donner D. B. Tumor necrosis factor activates CRE-binding protein through a p38 MAPK/MSK1 signaling pathway in endothelial cells. Am. J. Physiol. Cell Physiol. 2004;286:C547–C555. doi: 10.1152/ajpcell.00332.2002. [DOI] [PubMed] [Google Scholar]
- 7.Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M. J., Mahadevan L. C. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie J. R. MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved. Science STKE 2003. 2003:PE33. doi: 10.1126/stke.2003.195.pe33. [DOI] [PubMed] [Google Scholar]
- 10.Strelkov I. S., Davie J. R. Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res. 2002;62:75–78. [PubMed] [Google Scholar]
- 11.Schuck S., Soloaga A., Schratt G., Arthur J. S., Nordheim A. The kinase MSK1 is required for induction of c-fos by lysophosphatidic acid in mouse embryonic stem cells. BMC Mol. Biol. 2003;4:6. doi: 10.1186/1471-2199-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song K. S., Lee W. J., Chung K. C., Koo J. S., Yang E. J., Choi J. Y., Yoon J. H. Interleukin-1 beta and tumor necrosis factor-α induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases–MSK1–CREB activation in human airway epithelial cells. J. Biol. Chem. 2003;278:23243–23250. doi: 10.1074/jbc.M300096200. [DOI] [PubMed] [Google Scholar]
- 13.Darragh J., Soloaga A., Beardmore V. A., Wingate A., Wiggin G. R., Peggie M., Arthur J. S. MSKs are required for the transcription of the nuclear orphan receptors Nur77, nurr1 and nor1 downstream of MAP kinase signalling. Biochem. J. 2005;390:749–759. doi: 10.1042/BJ20050196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deak M., Clifton A. D., Lucocq L. M., Alessi D. R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.New L., Zhao M., Li Y., Bassett W. W., Feng Y., Ludwig S., Padova F. D., Gram H., Han J. Cloning and characterization of RLPK, a novel RSK-related protein kinase. J. Biol. Chem. 1999;274:1026–1032. doi: 10.1074/jbc.274.2.1026. [DOI] [PubMed] [Google Scholar]
- 16.Pierrat B., Correia J. S., Mary J. L., Tomas-Zuber M., Lesslauer W. RSK-B, a novel ribosomal S6 kinase family member, is a CREB kinase under dominant control of p38α mitogen-activated protein kinase (p38αMAPK) J. Biol. Chem. 1998;273:29661–29671. doi: 10.1074/jbc.273.45.29661. [DOI] [PubMed] [Google Scholar]
- 17.Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 18.Frodin M., Antal T. L., Dummler B. A., Jensen C. J., Deak M., Gammeltoft S., Biondi R. M. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 2002;21:5396–5407. doi: 10.1093/emboj/cdf551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frodin M., Jensen C. J., Merienne K., Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams M. R., Arthur J. S., Balendran A., van der Kaay J., Poli V., Cohen P., Alessi D. R. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- 21.Dummler B. A., Hauge C., Silber J., Yntema H. G., Kruse L. S., Kofoed B., Hemmings B. A., Alessi D. R., Frodin M. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J. Biol. Chem. 2005;280:13304–13314. doi: 10.1074/jbc.M408194200. [DOI] [PubMed] [Google Scholar]
- 22.Beardmore V. A., Hinton H. J., Eftychi C., Apostolaki M., Armaka M., Darragh J., McIlrath J., Carr J. M., Armit L. A., Clacher C., Kollias G., Arthur J. S. C. Generation and characterisation of p38b (MAPK11) gene targeted mice. Mol. Cell. Biol. 2005;25:10454–10464. doi: 10.1128/MCB.25.23.10454-10464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caivano M., Cohen P. Role of mitogen-activated protein kinase cascades in mediating lipopolysaccharide-stimulated induction of cyclooxygenase-2 and IL-1β in RAW264 macrophages. J. Immunol. 2000;164:3018–3025. doi: 10.4049/jimmunol.164.6.3018. [DOI] [PubMed] [Google Scholar]
- 24.McCoy C. E., Campbell D. G., Deak M., Bloomberg G. B., Arthur J. S. MSK1 activity is controlled by multiple phosphorylation sites. Biochem. J. 2005;387:507–517. doi: 10.1042/BJ20041501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomas-Zuber M., Mary J. L., Lesslauer W. Control sites of ribosomal S6 kinase B and persistent activation through tumor necrosis factor. J. Biol. Chem. 2000;275:23549–23558. doi: 10.1074/jbc.M002586200. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E., Lane D. Plainview, NY: Cold Spring Harbor Laboratory Press; 1998. Antibodies: A Laboratory Manual. [Google Scholar]
- 27.Unwin R. D., Griffiths J. R., Leverentz M. K., Grallert A., Hagan I. M., Whetton A. D. Multiple reaction monitoring to identify sites of protein phosphorylation with high sensitivity. Mol. Cell. Proteomics. 2005;4:1134–1144. doi: 10.1074/mcp.M500113-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Smith K. J., Carter P. S., Bridges A., Horrocks P., Lewis C., Pettman G., Clarke A., Brown M., Hughes J., Wilkinson M., et al. The structure of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein. Structure. 2004;12:1067–1077. doi: 10.1016/j.str.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 29.Roux P. P., Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomas-Zuber M., Mary J. L., Lamour F., Bur D., Lesslauer W. C-terminal elements control location, activation threshold, and p38 docking of ribosomal S6 kinase B (RSKB) J. Biol. Chem. 2001;276:5892–5899. doi: 10.1074/jbc.M005822200. [DOI] [PubMed] [Google Scholar]
- 31.Waskiewicz A. J., Flynn A., Proud C. G., Cooper J. A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheper G. C., Morrice N. A., Kleijn M., Proud C. G. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 2001;21:743–754. doi: 10.1128/MCB.21.3.743-754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukunaga R., Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ben-Levy R., Leighton I. A., Doza Y. N., Attwood P., Morrice N., Marshall C. J., Cohen P. Identification of novel phosphorylation sites required for activation of MAPKAP kinase-2. EMBO J. 1995;14:5920–5930. doi: 10.1002/j.1460-2075.1995.tb00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engel K., Schultz H., Martin F., Kotlyarov A., Plath K., Hahn M., Heinemann U., Gaestel M. Constitutive activation of mitogen-activated protein kinase-activated protein kinase 2 by mutation of phosphorylation sites and an A-helix motif. J. Biol. Chem. 1995;270:27213–27221. doi: 10.1074/jbc.270.45.27213. [DOI] [PubMed] [Google Scholar]
- 36.Underwood K. W., Parris K. D., Federico E., Mosyak L., Czerwinski R. M., Shane T., Taylor M., Svenson K., Liu Y., Hsiao C. L., et al. Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme. Structure. 2003;11:627–636. doi: 10.1016/s0969-2126(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 37.Meng W., Swenson L. L., Fitzgibbon M. J., Hayakawa K., Ter Haar E., Behrens A. E., Fulghum J. R., Lippke J. A. Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J. Biol. Chem. 2002;277:37401–37405. doi: 10.1074/jbc.C200418200. [DOI] [PubMed] [Google Scholar]
- 38.Yang J., Cron P., Thompson V., Good V. M., Hess D., Hemmings B. A., Barford D. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol. Cell. 2002;9:1227–1240. doi: 10.1016/s1097-2765(02)00550-6. [DOI] [PubMed] [Google Scholar]
- 39.Collins B. J., Deak M., Arthur J. S., Armit L. J., Alessi D. R. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 2003;22:4202–4211. doi: 10.1093/emboj/cdg407. [DOI] [PMC free article] [PubMed] [Google Scholar]