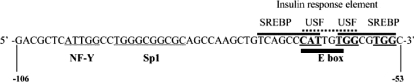Abstract
Dietary PUFAs (polyunsaturated fatty acids) co-ordinately suppress transcription of a group of hepatic genes encoding glycolytic and lipogenic enzymes. Suppression of Fasn (fatty acid synthase) transcription involves two PUFA-responsive regions, but the majority of PUFA sensitivity maps to a region within the proximal promoter containing binding sites for NF-Y (nuclear factor-Y), Sp1 (stimulatory protein 1), SREBP (sterol-regulatory-elementbinding protein), and USF (upstream stimulatory factor). Promoter activation assays indicate that altered NF-Y is the key component in regulation of Fasn promoter activity by PUFA. Using electrophoretic mobility-shift assay and chromatin immunoprecipitation analysis, we demonstrate for the first time that PUFAs decrease in vivo binding of NF-Y and SREBP-1c to the proximal promoter of the hepatic Fasn gene and the promoters of three additional genes, spot 14, stearoyl-CoA desaturase and farnesyl diphosphate synthase that are also down-regulated by PUFA. The comparable 50% decrease in NF-Y and SREBP-1c binding to the promoters of the respective PUFA-sensitive genes occurred despite no change in nuclear NF-Y content and a 4-fold decrease in SREBP-1c. Together, these findings support a mechanism whereby PUFA reciprocally regulates the binding of NF-Y and SREBP-1c to a subset of genes which share similar contiguous arrangements of sterol regulatory elements and NF-Y response elements within their promoters. PUFA-dependent regulation of SREBP-1c and NF-Y binding to this unique configuration of response elements may represent a nutrient-sensitive motif through which PUFA selectively and co-ordinately targets subsets of hepatic genes involved in lipid metabolism.
Keywords: fatty acid synthase (FASN), nuclear factor-Y (NF-Y), stimulatory protein 1 (Sp1), sterol-regulatory-element binding-protein (SREBP), polyunsaturated fatty acids (PUFA), liver
Abbreviations: AMPK, AMP-activated protein kinase; cdk2, cyclin-dependent kinase 2; ChIP, chromatin immunoprecipitation; CPT1α, carnitine palmitoyl transferase 1α; EMSA, electrophoretic mobility-shift assay; FASN, fatty acid synthase; FDPS, farnesyl diphosphate synthase; FF, fat free; FO, FF diet supplemented with 10% fish oil; NF-Y, nuclear factor-Y; PUFA, polyunsaturated fatty acid; RR, response region; S14, spot 14; SCD1, stearoyl-CoA desaturase 1; Sp1, stimulatory protein 1; SRE, sterol regulatory element; SREBP, sterol-regulatory-element-binding protein; USF, upstream stimulatory factor
INTRODUCTION
Dietary n-6 and n-3 PUFAs (polyunsaturated fatty acid), but not mono-unsaturated or saturated fatty acids, co-ordinately suppress the transcription of several hepatic genes encoding glycolytic and lipogenic enzymes [e.g. FASN (fatty acid synthase); acetyl-CoA carboxylase, SCD1 (stearoyl-CoA desaturase 1) and pyruvate kinase] [1–6]. This suppressive effect of PUFA is associated with a significant reduction in the rate of hepatic malonyl-CoA production and fatty acid biosynthesis [1]. The decrease in malonyl-CoA facilitates fatty acid oxidation by releasing carnitine palmitoyltransferase from inhibition by malonyl-CoA [7]. In addition, PUFAs activate peroxisome proliferator-activated receptor α and thereby increase the expression of genes encoding enzymes of fatty acid oxidation (e.g. carnitine palmitoyltransferase and acyl-CoA oxidase) [5,8,9]. The net result is a diversion of fatty acids away from triacylglycerol (triglyceride) synthesis and secretion, and towards fatty acid oxidation and ketogenesis.
PUFAs suppress the expression of some hepatic lipogenic genes by decreasing the nuclear content of the potent lipogenic transcription factor SREBP-1 (sterol-regulatory-element-binding protein 1) [10–13]. Dietary PUFAs regulate the nuclear level of mature SREBP-1 in two ways. First, PUFAs inhibit the proteolytic release of SREBP-1 from a precursor pool anchored to the endoplasmic reticulum membrane [11,14]; and secondly, PUFAs decrease the pool size of precursor, membrane anchored SREPB-1 [5,10,11,13]. The inhibition of proteolysis by dietary PUFAs is rapid (<3 h), and occurs without a reduction in the amount of membrane precursor [11]. In contrast, the decrease in amount of precursor SREBP-1 resulting from PUFA ingestion is a long-term adaptive response (>2 days) that reflects a reduction in SREBP-1 mRNA caused by accelerated transcript decay [11].
The down-regulation of expression and nuclear content of hepatic SREBP-1 by dietary PUFAs is correlated with an inhibition of FAS gene expression, suggesting that PUFAs inhibit hepatic Fasn gene transcription by reducing nuclear SREPB-1 [12]. However, we have reported previously [15] that PUFA suppression of Fasn transcription is a complex event that involves two response regions (PUFA-RRFASN): a distal PUFA-RR that accounts for 35% of the PUFA regulation of the Fasn promoter activity, and a proximal PUFA-RR that accounts for 65% of the PUFA regulation of Fasn promoter activity. Notable recognition sequences located in the proximal PUFA-RR include binding sites for SREBP-1, Sp1 (stimulatory protein 1) and NF-Y (nuclear factor-Y). Previous evidence indicates that SREBP-1 is a weak activator of transcription and it functions efficiently only when activated by co-activating transcription factors such as NF-Y and Sp1 [16–22]. Such is the case for the rat and human Fasn genes, where optimal stimulation of the Fasn promoter activity by SREBP-1c, as well as sterol inhibition of the Fasn promoter activity requires an SREBP-1 interaction with NF-Y and/or Sp1 [15,20,23]. However, these observations have not linked in vivo reduction in Fasn promoter activity to lower nuclear content of SREBP-1 by PUFAs. This led us to hypothesize that the inhibition of hepatic Fasn gene transcription involves a PUFA-mediated reduction in NF-Y binding to the promoter. Using EMSA (electrophoretic mobility-shift assay) and ChIP (chromatin immunoprecipitation), we demonstrate for the first time that PUFA produce a comparable 2-fold decrease of in vivo binding of NF-Y and SREBP-1c to the Fasn proximal promoter and to the promoters of three additional PUFA-sensitive genes. The common feature of all four genes is that they have similar contiguous arrangements of SRE (sterol regulatory element) and NF-Y sites within their promoters. Our findings support a mechanism whereby the PUFA-dependent decrease in nuclear SREBP-1c protein regulates the binding of NF-Y to adjacent NF-Y response elements within the promoters of PUFA-response genes.
EXPERIMENTAL
Primary hepatocyte culture and transfection
Male Sprague–Dawley rats (weighing between 150 and 200 g) were fed a high carbohydrate FF (fat-free) diet for 3–5 days and subsequently fasted for 24 h prior to hepatocyte isolation using collagenase perfusion [24,25]. Briefly, hepatocytes were plated onto 6-well plates that were coated with rat-tail collagen. Cells were allowed to attach to the plates in Waymouth MB 752/1 medium (Invitrogen) supplemented with 0.4 mM alanine, 0.5 mM serine, 26 mM sodium bicarbonate and 2.5% (v/v) foetal calf serum. After a 6 h attachment period, the hepatocytes were transfected with the various pFASN.reporter constructs described in the legends to the Figures. Transfection was conducted using Lipofectin (Invitrogen) in serum-free medium. Following a 12 h transfection period, the medium was changed to one containing 1 μM insulin and dexamethasone, 1 mM α-tocopherol and 200 mM albumin-bound C18:1, n−9, C20:3, n−9, C20:4, n−6, or C20:5, n−6. Fatty acids were purchased as >95% pure from Nu-Chek Prep. The fatty acid to albumin ratio was 4:1. Cells not treated with fatty acids were maintained in a medium containing an amount of fatty acid-free albumin equivalent to that added with the fatty acids. The impact of fatty acids on cell viability was assessed by Trypan Blue exclusion (>95% exclusion) and by measuring the leakage of lactate dehydrogenase in to the media. Luciferase reporter activity was determined in total cell lysates (n=3–6 plates per point) prepared 48 h after transfection [24]. Each construct was evaluated in 3–7 different hepatocyte preparations. Reporter activity is expressed as units of activity per μg of protein. Preparation of the 5′-pFASN.LUC reporter vectors was described previously [15,26,27]. The −150 to −43 bp region (relative to the transcription start site) of the rat Fasn promoter that contains a classical sterol response element (−150), NF-Y (−99 to −93 bp) and Sp1 (−90 to −81 bp) recognition sites, and the SREBP-1/USF (upstream stimulatory factor) sites of the insulin response element (−72 to −53 bp) was fused to a generic TATA box and was placed upstream of the luciferase reporter gene in the pGL2 luciferase reporter vector (Promega) [18]. Using the −150 to −43 bp region as a template for PCR, mutations were generated in the Sp1, NF-Y and insulin response element (Table 1) [20].
Table 1. Identification of regulatory elements localized in the Fasn gene and consensus oligonucleotides used for the EMSAs.
The mutations are underlined and the consensus binding sites are in bold.
EMSA
Male Sprague–Dawley rats (weighing 150–200 g) were trained to consume their daily food during one 3 h period. The rats were then fed an FF diet or an FO diet (fat free diet supplemented with 10% fish oil) for 10 days prior to nuclear protein extraction [28]. Nuclear proteins were prepared immediately following the meal from rats fed a high glucose FF diet or the FO diet [29]. The oligonucleotide sequences of the Fasn gene used in the EMSAs for the candidate NF-Y and Sp1 sites are presented in Table 1. The mutation sites were identical to those introduced for the promoter–reporter assays described above. Double-stranded oligonucleotides were end-labelled with γ[32P]ATP (PerkinElmer Life Sciences) and T4 DNA polynucleotide kinase (20000–50000 c.p.m per reaction) and incubated with nuclear protein extracts (5 μg/reaction) in a reaction buffer containing 10 mM Hepes (pH 7.9) 75 mM KCl, 1 mM EDTA, 5 mM dithiothreitol, 5 mM MgCl2, 10% (v/v) glycerol and 1 μg poly(dI-dC)·(dI-dC). Protein concentrations were determined using the bicinochoninic acid protein assay (Pierce). For competition reactions, 100-fold molar excess of unlabelled nucleotide, as indicated, was added to the reaction mixture. After 30 min on ice, the reaction components were loaded on a 5% (v/v) non-denaturing polyacrylamide gel and electrophoresed at 200 V for approximately 2 h in 1x TGE buffer (25 mM Tris/HCl, 190 mM glycine and 1 mM EDTA, pH 8.5). Super-shift analyses for NF-Y and Sp1 were conducted using antibodies against Sp1 (sc-59; Santa Cruz Biotechnology) and NF-YA (sc-7712; Santa Cruz Biotechnology) coupled for 8–12 h.
ChIP assays
Rats trained to consume their daily food during one 3 h period were fed an FF diet or an FO diet for 7 days as described above. Cross-linked chromatin was prepared using the methodology described by Latasa et al. [30]. ChIP was performed as described in [31] with some modifications. Chromatin samples at A260=4 were diluted to a final volume of 1000 μl, incubated with 100 μl of blocked protein G beads (Roche) for 2 h on a rotating wheel at 4 °C, and the beads were collected by centrifugation at 15000 g for 1 min. The beads were incubated with 500 μg of sheared salmon sperm DNA (Invitrogen) and BSA (Roche) for 30 min at 4 °C to block non-specific binding. The supernatant was transferred to clean tubes and incubated with 5 μg of NF-YA antibody (sc-7712), Sp1 antibody (sc-59), SREBP-1 antibody (sc-8984; Santa Cruz Biotechnology), USF antibody (sc-229; Santa Cruz Biotechnology), goat IgG (Santa Cruz Biotechnology), or rabbit IgG (Santa Cruz Biotechnology) overnight at 4 °C on a rotating wheel. The samples were centrifuged for 10 min at 15000 g then the supernatant was transferred to a new tube and incubated with 100 μl of blocked protein G beads for 90 min at 4 °C on a rotating wheel. The samples were washed extensively, eluted, the crosslinks reversed and treated with Proteinase K (Roche) as described [31]. The samples were extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, by vol.) (Amresco) and precipitated by 40 μg glycogen (Roche) and 2.4 vol. of ethanol at −20 °C overnight. Samples were washed once with 70% ethanol and resuspended in 40 μl of water. Primers were designed to amplify a 152 bp fragment (−153 to −2 bp) of the proximal promoter region in the rat Fasn gene. Primer sequences were: 5′-GGCATCACCCCACCGACG-3′ (forward) and 5′-GCTCCCTCTAGGCCGCGC-3′ (reverse). Additional controls included the proximal promoters of the CPT1α (carnitine palmitoyltransferase 1α), S14 (spot 14), Scd1 and Fdps (farnesyl diphosphate synthase) genes. The primers used were as follows: for CPT1α (−187 to +100 bp), forward, 5′-AGGTCTGTAGTTCACAAGCTC-3′; reverse, 5′-CCACGCTCCCAAAGGTCATAC-3′; for S14 (−190 to −33), forward, 5′-ACCTGAAGTGACAAGCAGAAGC-3′; reverse, 5′-TGAGCAGACAGCAGATTGACAG-3′; for SCD1 (−505 to −244), forward, 5′-AAAGTCATCCATCAGCCCAC-3′; reverse, 5′-TAGATTCCAGAGTAGACCTC-3′; for FDPS (−397 to −154), forward, 5′-CTTGAAGTGCTACAAACAGG-3′; reverse, 5′-ATAATACTACGACTCCCAGC-3′. Owing to the high GC content of the Fasn proximal promoter, the Fasn PCR reactions were performed with 4 μl of immunoprecipitate, 25 μM of each primer, 25 μl of FailSafe™ PCR 2× PreMix G and 2.5 units Taq polymerase (Epicentre) in a total volume of 50 μl. PCR conditions were: one cycle of 96 °C for 2 min; 30 cycles of 96 °C for 1 min, 55 °C for 1 min and 72 °C for 45 s; and one cycle of 72 °C for 1 min. The PCR for S14, Scd1, Fdps and Cpt-1α were performed with 2 μl of immunoprecipitate, in a final volume of 25 μl, 10 μM of each primer, and 1× EX Taq RT-PCR mix (TaKaRa) in a Smart Cycler instrument (Cepheid). Samples were incubated for an initial denaturation at 96 °C for 30 s, followed by 30 cycles of 95 °C for 3 s, 60 °C for 10 s, and 72 °C for 15 s. PCR products were separated on a 1.5% agarose gel and analysed by ethidium bromide staining. The stained gels were visualized and the PCR products from three to six individual experiments were quantified using a Versadoc imaging system (Bio-Rad Laboratories). The numbers are derived from the average intensity of the PCR products from the liver samples of the rats fed the FO diet compared with the average intensity of those fed the FF diet, which was set at 100%.
Immunoblot analysis
Nuclear extracts (25 μg) prepared from livers of rats fed the FF and FO diets for 7 days (as described above) were diluted in 6× Laemmli loading buffer [375 mM Tris/HCl (pH 6.8), 10% (w/v) SDS, 30% (v/v) glycerol, 0.012% Bromphenol Blue and 30% (v/v) 2-mercaptoethanol], boiled for 5 min, resolved by SDS/PAGE [10% (v/v) polyacrylamide gels], and electrotransferred onto PVDF membranes. Membranes were probed with antibodies directed against NF-YA (sc-7712), NF-YB (sc-7711; Santa Cruz Biotechnology), NF-YC (sc-7714; Santa Cruz Biotechnology), SREBP-1 (sc-13551; Santa Cruz Biotechnology) and Sp1 (sc-59) and visualized by standard ECL procedure. Quantification of immunoblot signals was performed by densitometry using the Versadoc system (Bio-Rad Laboratories).
Co-immunoprecipitation and immunoblot analysis
Nuclear extracts were prepared from livers of rats fed the FF and FO diets for 7 days as described above. Samples (150 μg) of nuclear proteins from each rat were pre-cleared with protein-G for 2 h, and then incubated with 5 μg of NF-YA (sc-7712), Sp1 (sc-59), SREBP-1 (sc-8984), goat pre-immune IgG, or rabbit pre-immune IgG overnight. The samples were centrifuged for 1 min at 4000 g at 4 °C and the supernatants were removed and retained to evaluate the immunoprecipitation efficiency of each antibody. The remaining pellets were washed five times with cold RIPA buffer [50 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1% (v/v) Nonidet P40, 0.25% sodium deoxycholate and 1 mM EDTA] at 10 min intervals, denatured and resolved by SDS/PAGE [10% (v/v) gels] along with the supernatants from the corresponding samples. Gels were electrophorectically transferred on to PVDF membranes, which were probed with anti-NF-YA (sc-17753; Santa Cruz Biotechnology), anti-Sp1 (sc-59), anti-SREBP1 (sc-13551) and anti-phosphoserine (Zymed 61-8100) antibodies. The detected bands were visualized and quantified as described above.
RESULTS
Role of NF-Y and Sp1 sites in maintenance of Fasn proximal promoter activation
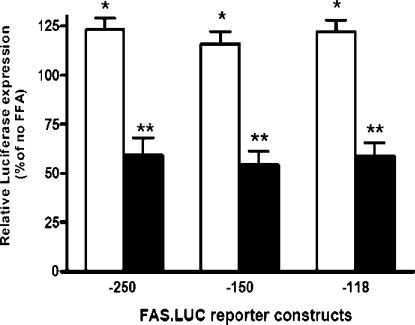
We have demonstrated previously [15] that the Fasn gene contains two PUFA response regions and the proximal PUFA-RRFASN accounted for the majority of the PUFA response. A better understanding of the key cis- and trans-regulatory elements within this region is essential to further understand how PUFAs suppress Fasn gene expression. The −265 to +65 bp region of the Fasn gene contains several PUFA-sensitive sites, including consensus SREs (−150 to −142 bp, −71 to −64 bp and −61 to −53 bp), an insulin response element (−71 to −53 bp), and recognition sites for NF-Y (−99 to −93 bp) and Sp1 (−90 to −81 bp) [17,22,32]. Figure 1 shows that deletion of the SRE between −150 and −118 bp neither reduced promoter activity nor diminished the inhibitory action of PUFA C20:4, n−6. Our previous work [15] showed that the majority of PUFA sensitivity was retained when the SRE at −61 to −53 bp was mutated, implicating the SRE at −71 to −64 bp as the essential PUFA-sensitive SREBP-1c binding site.
Figure 1. PUFA suppression of Fasn gene transcription does not involve −150 SRE.
Rat hepatocytes in primary culture were transfected with luciferase (LUC) reporter constructs containing Fasn 5′-flanking sequences −250 to +65 bp, −150 to −53 bp, or −118 to −53 bp. Cells were treated with 200 μM albumin-bound C18:1(n−9) (open bars) or C20:4(n−6) (closed bars). Reporter activity is expressed relative to cells treated with albumin alone as 100% (no FFA). Luciferase activity in the absence of fatty acid was 4905, 2240, and 1855 relative luciferase units/μg protein for the −250, −150, and −118 constructs respectively (n=6 plates per treatment). *P<0.05 for C18:1(n−9) induction; **P<0.01 for C20:4(n−6) suppression. FFA, free fatty acids.
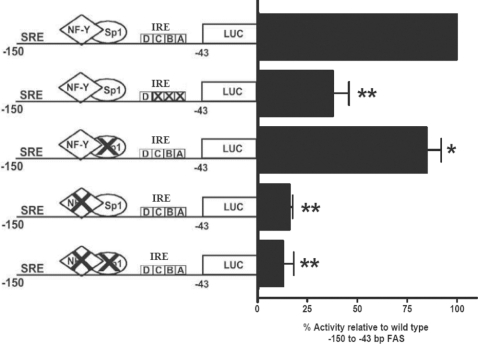
Our previous work [15] also established that the NF-Y (−99 to −93 bp) site within this region is essential for PUFA sensitivity, whereas the Sp1 (−90 to −81 bp) site appears to play a less crucial role. To examine the significance of these sites in maintenance of promoter activation, basal promoter activity was measured in constructs after mutation of each element in primary rat hepatocytes. Figure 2 shows that mutation of the Sp1 site had only a modest effect on basal activity, while mutation of the NF-Y site reduced promoter activity by 89% relative to the control. Mutation of both NF-Y and Sp1 sites had no greater effect than mutating the NF-Y site alone. Together, these results show that maintenance of basal transcriptional activity of the Fasn promoter is dependent upon binding of NF-Y to the promoter.
Figure 2. Mutations in the insulin response element or NF-Y site reduce Fasn promoter activity.
Rat hepatocytes were transfected with a construct that contained the luciferase (LUC) reporter gene driven by the Fasn gene region of −150 to −43 bp fused to a generic TATA box [18]. The region contains a classical SRE, an insulin response element (IRE) at −72 to −53 bp, an Sp1 site (−91 to −81 bp), and an NF-Y site (−99 to −93 bp). Mutations (X) were introduced into the respective sites (see Table 1) as described in the Experimental section. Results are expressed relative to the wild-type sequence, and are means±S.E.M. for two or three independent hepatocyte preparations with 4–6 replications per construct within a cell preparation. *P<0.05 for the effect of the Sp1 mutation; **P<0.01 for effect of the IRE, NF-Y and NF-Y/Sp1 mutations.
Introducing mutations that interfered with the USF and SREBP-1c binding sites (i.e. mutant ABC) of the insulin response element (−71 to −53 bp) reduced basal promoter activity by 65% (Figure 2). Nevertheless, as we established previously [15], the mutated promoter retained 75–80% of its sensitivity to suppression by PUFA. The role of the SRE at −71 to −64 bp was not completely examined, since the mutant ABC leaves a partial E-box triplet. However, the SRE sites at −150 bp and −61 to −53 bp have minimal importance for PUFA responsiveness in the Fasn promoter (Figure 1 and [15]). Together, these observations suggest that PUFA inhibition of Fasn gene transcription involves a mechanism more complex than simply the ability of PUFA to suppress the nuclear content of SREPB-1c [5].
Dietary PUFA decreases the DNA binding activity of hepatic NF-Y and Sp1
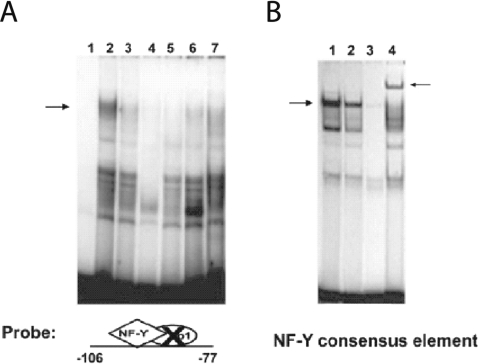
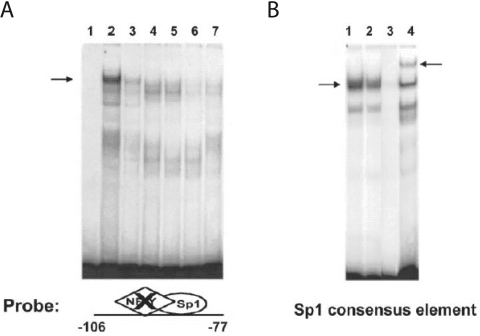
To assess the significance of diet-induced changes in binding of either Sp1 or NF-Y to the PUFA-sensitive region of the Fasn promoter, two complementary approaches (EMSA and ChIP assays) were used. First, we constructed two DNA fragments for evaluation in EMSA assays: (1) the −106/−77Sp1mt fragment prevents Sp1 binding and was used to evaluate NF-Y binding, and (2) the −106/−77NF−Ymt fragment prevents NF-Y binding and was used to evaluate Sp1 binding. The effects of dietary PUFAs on DNA binding activity were evaluated using these fragments in EMSAs with hepatic nuclear protein extracts from meal-trained rats fed with either the high carbohydrate FF diet or the FO diet. EMSAs using the −106/−77Sp1mt fragment revealed that NF-Y binding activity in hepatic nuclear protein extracts from PUFA-fed (FO) rats was markedly lower than that found in nuclear extracts from rats fed the high carbohydrate FF diet (Figure 3A, lanes 2 and 3). PUFA was also found to reduce NF-Y binding to an oligonucleotide containing a consensus NF-Y binding sequence (Figure 3B, lane 2). The anti-(NF-YA) antibody inhibited NF-Y binding to the −106/−77Sp1mt fragment (Figure 3A, lane 7) and anti-(NF-YA) super-shifted the consensus NF-Y oliognucleotide (Figure 3B, lane 4). The specificity of NF-Y binding to the −99 to −93 bp site was further confirmed by showing that excess unlabelled −106/−77Sp1mt, unlabelled consensus NF-Y oligonucleotide and unlabelled wild type −106 to −77 all effectively competed with −106/−77Sp1mt for NF-Y binding (Figure 3A, lanes 4–6).
Figure 3. Dietary PUFAs impair NF-Y DNA binding activity.
Hepatic nuclear protein extracts (5 μg per reaction) were prepared from rats fed an FF diet or the FO diet. (A) EMSAs were conducted using the Fasn promoter region of −106 to −77 bp in which the candidate Sp1 site was mutated (−106/−77Sp1mt) (see Table 1). Lane 1 is free probe. Lanes 2 and 3 show NF-Y binding activity in extracts from rats fed the FF diet or the FO diet respectively. Specificity of NF-Y binding to the −106/−77Sp1mt fragment (A) was determined using nuclear proteins from rats fed the FF diet incubated with a 100-fold molar excess of unlabelled −106/−77Sp1mt (lane 4), NF-Y consensus sequence (lane 5), wild-type −106 to −77 bp Fasn sequence (lane 6), or the anti-(NF-YA) antibody (lane 7). (B) EMSAs were conducted using an oligonucleotide containing a consensus NF-Y binding sequence (see Table 1) and nuclear protein extracts from rats fed the FF diet (lane 1) or the FO diet (lane 2). Specificity of NF-Y binding to the consensus sequence was determined using nuclear proteins from rats fed the FF diet incubated with a 100-fold molar excess of unlabelled consensus NF-Y site (lane 3) or the anti-(NF-YA) antibody (lane 4). The left-hand arrows indicate the NF-Y binding complex, and the right-hand arrow indicates the super-shift resulting from treatment with anti-(NF-YA) antibody. Reduced NF-Y binding by dietary FO was demonstrated in four rats with a mean decrease in binding activity of 50±7% (P<0.05) for the −106/−77Sp1mt and consensus NF-Y sequences.
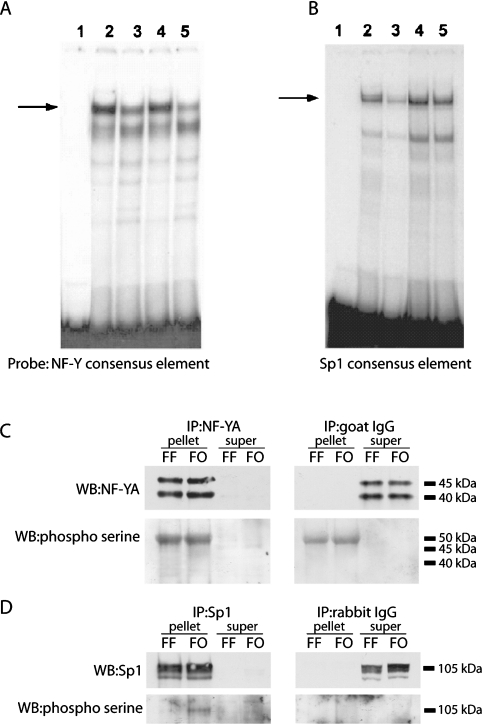
The Sp1 site at the −91 to −81 bp region is also important for optimal rates of Fasn gene transcription (Figure 2). Thus, the impact of dietary PUFA on its DNA binding activity was examined using the −106 to −77 bp region in which the NF-Y binding site was mutated at the −92 bp position (i.e. −106/−77NF−Ymt). EMSAs with the −106/−77NF−Ymt fragment revealed that Sp1 DNA binding activity in hepatic nuclear protein extracts from PUFA-fed rats was significantly lower (P<0.05) than in extracts from rats fed a high carbohydrate FF diet (Figure 4A, lanes 2 and 3). FO ingestion also reduced Sp1 binding to an oligonucleotide containing a consensus Sp1 binding sequence (Figure 4B, lanes 1 and 2), and anti-Sp1 antibody super-shifted the binding to the consensus Sp1 oligonucleotide (Figure 4B, lane 4). Furthermore, excess unlabelled −106/−77NF−Ymt, unlabelled consensus Sp1 oligonucleotide and unlabelled wild type −106 to −77 bp all effectively prevented Sp1 binding to the −106/−77NF−Ymt (Figure 4A, lanes 4–6). Taken together, DNA binding activity of NF-Y and Sp1 to probes corresponding to their sites within the Fasn promoter was reduced in nuclear extracts from livers of PUFA-fed rats.
Figure 4. Dietary PUFAs impair Sp1 DNA binding activity.
Hepatic nuclear protein extracts (5 μg per reaction) were prepared from rats fed an FF diet or the FO diet. (A) EMSAs were conducted using the Fasn promoter region of −106 to −77 bp in which the NF-Y site was mutated (−106/−77NF−Ymt) (see Table 1). Lane 1 is free probe. Lanes 2 and 3 show Sp1 binding activity for extracts from rats fed the FF or the FO diet. Specificity of Sp1 binding to the −106/−77NF−Ymt fragment (A) was determined using nuclear proteins from rats fed the FF diet incubated with a 100-fold molar excess of unlabelled −106/−77NF−Ymt, (lane 4), the Sp1 consensus sequence (lane 5), the wild-type −106 to −77 bp Fasn sequence (lane 6) or the anti-Sp1 antibody (lane 7). (B) EMSAs were conducted using an oligonucleotide containing a consensus Sp1 binding sequence (see Table 1) and nuclear protein extracts from rats fed the FF diet (lane 1) or the FO diet (lane 2). Specificity of Sp1 binding to the consensus sequence was determined using nuclear proteins from rats fed the FF diet incubated with a 100-fold molar excess of unlabelled consensus Sp1 site (lane 3) or the anti-Sp1 antibody (lane 4). The left-hand arrows designate the Sp1 binding complex, and the right-hand arrow designates the super-shift resulting from treatment with anti-Sp1 antibody. Reduced Sp1 binding by dietary FO was demonstrated in seven rats with a mean decrease in binding activity of 52±4% (P<0.05) for the −106/−77NF−Ymt and consensus Sp1 sequences.
To test whether the reduction in NF-Y and Sp1 DNA binding activity associated with the ingestion of PUFA involved a phosphorylation/dephosphorylation mechanism, nuclear extracts from each dietary regimen were incubated with protein phosphatase 1A or alkaline phosphatase prior to the assay. Neither protein phosphatase 1A (Figure 5A) nor alkaline phosphatase (results not shown) enhanced binding of NF-Y to the consensus NF-Y oligonucleotide in nuclear extracts from either group. In contrast, the decrease in Sp1 DNA binding activity observed in nuclear extracts from rats fed the FO diet was completely reversed by pre-incubating the extracts with protein phosphatase 1A (Figure 5B, lane 5). Sp1 binding activity was also increased by protein phosphatase 1A in nuclear extracts from rats fed the FF diet (Figure 5B, lane 4), suggestive of inhibitory regulation in extracts from control rats that is further accentuated by the FO diet. To further explore post-translational regulation of Sp1 and NF-Y, nuclear extracts from each dietary group were immunoprecipitated with anti-Sp1 or anti-NF-Y antibodies, and the pellets were probed with an anti-phosphoserine antibody. The efficiency of immunprecipitation was near 100% for both transcription factors (Figures 5C and 5D), but interpretation of Figure 5(C) is complicated by cross-reactivity of the phosphoserine antibody with the goat anti-(NF-Y) IgG used for immunprecipitation. The band at ∼50 kDa that migrates near the same size as the larger NF-YA isoform is also observed when the immunoprecipitation reaction is conducted with goat IgG rather than the NF-Y antibody (Figure 5C). However, the data provides no evidence that the lower molecular mass isoform of NF-Y is phosphorylated in extracts from either the FF or FO fed rats. In contrast, a faint but detectable band that co-migrates with Sp1 was detected by the phosphoserine antibody in the FF nuclear extracts and its intensity was significantly increased in extracts from FO fed rats (Figure 5D). Viewed together, the data support the hypothesis that Sp1 binding activity and phosphorylation state can be modulated by a protein phosphatase 1A-sensitive site. In contrast, the mechanism responsible for reduced DNA binding activity of NF-Y remains unclear.
Figure 5. Post-translational regulation of Sp1 and NF-Y DNA binding activity and phosphorylation state by dietary FO.
Hepatic nuclear protein extracts were prepared from rats fed an FF diet or the FO diet. The extracts were treated with phosphatase 1a (0.1 units per μg of protein) for 60 min. NF-Y (A) and Sp1 (B) DNA binding activity was determined by EMSA. Lane 1 in both (A) and (B) is free probe. Lanes 2 show the NF-Y (A) and Sp1 (B) binding activity for the FF diet, and lanes 3 show the NF-Y (A) and Sp1 (B) binding activity for the FO diet. Lanes 4 (FF) and 5 (FO) show the change in NF-Y (A) and Sp1 (B) binding activity following phosphatase 1a treatment. Extracts prepared from rats fed the FO diet (n=4) displayed 50±7% and 45±7% less NF-Y binding activity before and after phosphatase 1a treatment respectively. Sp1 binding activity in the same nuclear protein extracts was reduced 52±5% in the absence of phosphatase, but only 7±1% (P<0.05) after phosphatase treatment. (C) Immunoprecipitation of nuclear extracts from livers of rats fed either the FF or FO diet were performed using the NF-YA antibody to evaluate effects of fish oil on serine phosphorylation of NF-YA. In conjunction with immunoprecipitation using an antibody specific for NF-YA (left-hand panels), we also performed immunoprecipitations using equivalent amounts of normal goat IgG (the source of the NF-YA immunoprecipitation antibody; right-hand panels). The pellets and supernatants from each immunoprecipitation were retained and blotted for NF-YA (top panels) and phosphoserine (bottom panels). (D) Western blots of Sp1 (top panels) and phosphoserine (bottom panels) on pellets and supernatants from representative samples of each group immunoprecipitated with Sp1 antibody (left-hand panels) or equivalent amounts of normal rabbit IgG (the source of the Sp1 immunoprecipitation antibody; right-hand panels). IP, immunoprecipitation; WB, Western blot.
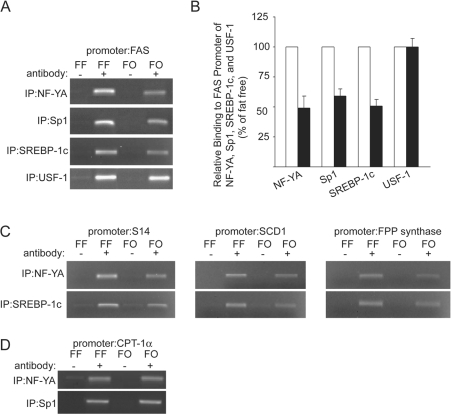
To test whether NF-YA, Sp1, SREBP-1 and USF binding to the proximal promoter of the endogenous Fasn gene was modulated by PUFA in vivo, we isolated chromatin from livers of rats fed with either an FO diet or a high carbohydrate FF diet. After immunoprecipitation with non-specific IgGs or with antibodies against NF-YA, Sp1, SREBP-1 and USF, initial PCRs were conducted with serial dilutions of input material from each immunoprecipitation to establish the appropriate cycling conditions to accurately compare template content across treatments (results not shown). Using an equal amount of chromatin from each input, Figure 6(A) shows that the 152 bp fragment of the proximal Fasn promoter was not amplified when non-specific IgG was the immunoprecipitation antibody. Consistent with the EMSA data presented in Figures 3 and 4, the relative binding of NF-YA and Sp1 to the proximal promoter was decreased by 40–50% in chromatin extracted from livers of rats receiving PUFA in their diets (Figures 6A and 6B). To expand our insight into the effect of PUFA on the regulation of the proximal Fasn promoter, we also performed ChIP analysis with an antibody specific for SREBP-1c. Similar to that of NF-YA and Sp1, PUFA supplementation also reduced the binding of SREBP-1c to the Fasn proximal promoter by approximately 50% (Figures 6A and 6B), suggestive of a co-ordinated mechanism responsible for the decreased binding of these factors. In contrast, equivalent amounts of the promoter fragment were pulled down by USF-1 antibody regardless of diet (Figures 6A and 6B). This data indicates that USF-1 binding to the Fasn proximal promoter was unaffected by the FO diet.
Figure 6. Dietary PUFA decrease NF-Y, Sp1 and SREBP-1c binding to the promoters of several PUFA-sensitive genes.
(A) ChIP analysis of the Fasn promoter was conducted using livers from rats fed with the FF or FO diet. Chromatin fragments imunoprecipitated with anti-NF-Y, anti-Sp1, anti-SREBP-1c and anti-USF-1 antibodies (+) were amplified by PCR with primers spanning the proximal PUFA-sensitive region of the rat Fasn gene promoter (−153 to −2 bp). Immunoprecipitation with normal goat IgG (the source of the NF-YA immunoprecipitation antibody) and normal rabbit IgG (the source of the Sp1, SREBP-1c and USF-1 immunoprecipitation antibodies) were used as negative controls (−). The results shown are representative of three to six individual experiments. (B) The results of the experiments displayed in (A) were quantified by measuring the density of the PCR products separated on agarose gels. The numbers are derived from the average density of the PCR products from the liver samples from rats fed the FO diet (closed bars) compared with the average intensity of those from rats fed an FF diet (open bars), which are set at 100%. (C) Chromatin immunoprecipitated by the NF-YA and SREBP-1 antibodies was amplified using primers spanning the PUFA-sensitive proximal promoter regions of S14 (left-hand panel), Scd1 (middle panel), and Fdps (FPP synthase; right-hand panel). The PCR products are representative of three to six individual experiments. (D) Chromatin immunoprecipitated by the NF-YA and Sp1 antibodies was amplified using primers spanning the NF-Y and Sp1 sites of the Cpt-1α promoter. The PCR products are representative of three to six individual experiments.
To assess the specificity of the PUFA-dependent decrease in binding of NF-Y and SREBP-1c to the Fasn promoter, their binding to the promoters of additional genes regulated by PUFA was evaluated. Like Fasn, gene transcription for S14, scd1 and Fdps is repressed by PUFA [33–35]. In addition, the proximal promoters of these genes contain adjacent sites for both SREBP-1c and NF-Y [19,34,36]. Although the requirement for NF-Y and SREBP-1c in the PUFA-induced repression of these genes has not been examined, it has been shown that a functional interaction between NF-Y and SREBP-1c is required for up-regulation of Scd1 [36] and Fdps [19,37] by sterol as well as the T3-mediated transactivation of the S14 gene [38]. Figure 6(C) shows that dietary PUFA produced an average 50% decrease in NF-Y binding to the promoters of S14 (−51.3±4.2%; n=4), Scd1 (−49.7±5.6%; n=4) and Fdps (−53.7±5.9%; n=4) compared with the samples from rats fed the FF diet. As with Fasn, the binding of SREBP-1c to the promoters of S14 (−44±1.9%; n=4), Scd1 (−57.7±8.2%; n=4) and Fdps (−53.7±5.9%; n=4) was decreased by 50% in samples from rats fed the FO diet (Figure 6C). These results argue that PUFA produces a co-ordinated decrease in NF-Y and SREBP-1c binding to the promoters of these PUFA-sensitive genes. In contrast with Fasn, S14, Scd1 and Fdps, Cpt-1α is not down-regulated by dietary PUFA [28]. However, its proximal promoter is dependent upon NF-Y to drive basal expression, making it very sensitive to changes in NF-Y binding [39]. Figure 6(D) shows that PUFA had no effect on binding of NF-Y to its site in the Cpt-1α promoter. Real time PCR of liver RNA from the same samples revealed that PUFA had no effect on Cpt-1α mRNA (Cpt-1α/cyclophilin mRNA ratio; FF=0.29±0.02 compared with FO=0.27±0.02). These findings support the conclusion that PUFA does not produce a generalized decrease in NF-Y binding to promoters of genes with this regulatory element.
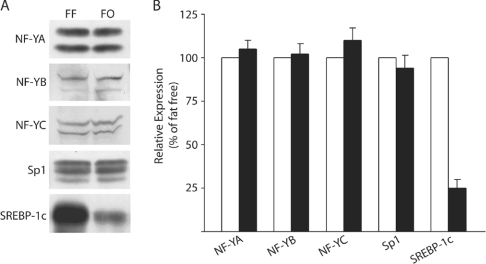
Mechanisms of PUFA-dependent effects on SREBP-1c, NF-Y and Sp1 binding to the Fasn proximal promoter
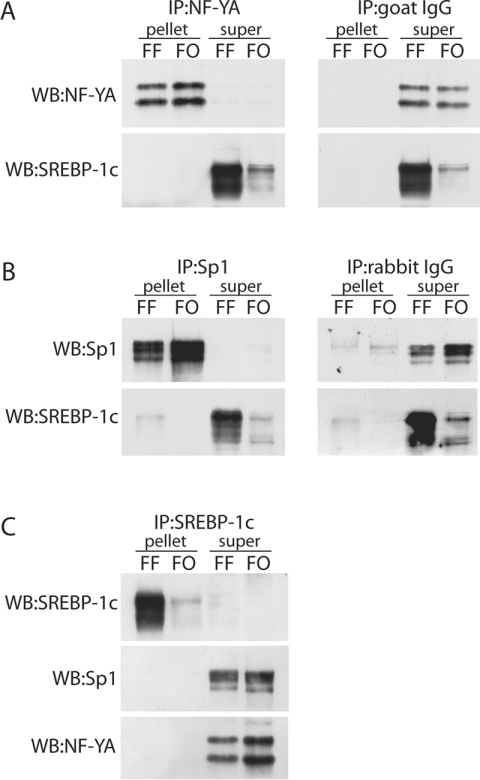
To explore the mechanism of PUFA-dependent down-regulation of in vivo Fasn promoter activity, the expression and physical interaction of the transcription factors which bind to the PUFA-sensitive region of the Fasn promoter were examined. Immunoblot analysis of NF-YA, NF-YB, NF-YC, Sp1 and SREBP-1c in nuclear extracts shows that FO did not alter the nuclear content of Sp1 or any of the NF-Y isoforms (Figures 7A and 7B). However, as reported previously [5] and expected, FO produced a 4-fold decrease in SREBP1-c expression (Figures 7A and 7B). Therefore, the 2-fold decrease in NF-Y binding to PUFA-sensitive gene promoters with adjacent SRE sites suggested the interesting possibility that a physical interaction between SREBP-1c and NF-Y could form a complex that regulates delivery of transcription factors to the promoter. If such a physical interaction occurred, it could be argued that the PUFA-dependent decrease in SREBP-1c nuclear protein would directly result in the reduced binding of NF-Y and Sp1. To test the hypothesis that this type of transcriptional complex formed through protein–protein interactions, co-immunoprecipitation of SREBP-1c with NF-Y (or Sp1) was performed using antibodies specific for NF-YA, Sp1 and SREBP-1, or equivalent amounts of either normal goat IgG (the source of the NF-YA immunoprecipitation antibody) or normal rabbit IgG (the source of Sp1 and SREBP-1c immunoprecipitation antibodys) as negative controls. Western blotting of the pellets and supernatant after immunoprecipitation with NF-YA antibody shows that equivalent amounts of NF-YA were present in pellets from each treatment group (Figure 8A). No NF-YA was detected in the supernatant of either sample, indicating that the immunoprecipitation was highly efficient (Figure 8A). When the NF-YA immunoprecipitation was immunblotted for SREBP-1c, we detected no SREBP-1c in either pellet. In contrast, essentially all SREBP-1c was found in the supernatants and at ratios that reflected the FO-mediated reduction in total SREBP-1c (compare Figure 7A with Figure 8A). The negative controls presented in Figure 8(A) show that no NF-YA or SREBP-1c was immunoprecipitated with non-specific goat IgG. Collectively, these findings suggest that NFY-A and SREBP-1c do not form a complex that can be detected using a standard co-immunoprecipitation technique. A similar conclusion was reached when the immunoprecipitation was conducted with anti-Sp1 antibody, in the sense that equivalent amounts of Sp1 were found in the pellets from each treatment group (Figure 8B) and essentially no Sp1 was detected in the supernatants (Figure 8B). When the Sp1 immunoprecipitation was immunblotted for SREBP-1c, we detected a small amount of SREBP-1c in the pellet from the rats fed the FF diet, while the vast majority was found in the supernatants (Figure 8B). However, an equivalent amount of SREBP-1c was also observed in the pellet from the immunoprecipitation using normal rabbit IgG as a negative control (Figure 8B). This finding indicates that the SREBP-1c observed in the Sp1 immunoprecipitation is probably due to non-specific binding of SREBP-1c to the IgG. Therefore, we are unable to conclude that Sp1 and SREBP-1c can be co-immunoprecipitated from either treatment group. Finally, we immunoprecipitated SREBP-1c and performed Western blots for SREBP-1c, NF-YA and Sp1. The immunoprecipitation of SREBP-1c was highly efficient as essentially all SREBP-1c was found in the pellet with none in the supernatant. However, when the pellets and supernatants were probed for NF-Y or Sp1 (Figure 8C) neither protein could be detected in the pellets because proteins were found exclusively in the supernatants. These findings provide no evidence for a physical association between SREBP-1c and NF-Y or Sp1, and argue collectively that the three proteins do not form a transcriptional complex prior to binding to their requisite sites within each promoter.
Figure 7. Dietary PUFA decreases nuclear content of SREBP-1c protein, but does not affect NF-YA and Sp1 expression.
Protein expression for SREBP1, NF-YA, NF-YB, NF-YC and Sp1 in the nuclear extract of each sample was determined by Western blotting. (A) Blots of representative samples from rats fed the FF or the FO diet. (B) The results expressed as relative integrated units for the FF (open bars) or FO (closed bars) diets and represent the means±S.E.M. for four rats per treatment.
Figure 8. Physical interactions between SREBP-1c, NF-Y and Sp1 are not required for PUFA-dependent decrease in Fasn transcription.
Immunoprecipitation (IP) of nuclear extracts from livers of rats on each diet were conducted with NF-Y, Sp1 and SREBP-1 antibodies to evaluate the effects of FO on protein–protein interactions involving SREBP-1c, NF-Y and Sp1. In conjunction with immunoprecipitation using antibodies specific for NF-YA, Sp1 and SREBP-1, we also performed immunoprecipitations using equivalent amounts of either normal goat IgG (the source of the NF-YA IP antibody) and normal rabbit IgG (the source of the Sp1 and SREBP-1c IP antibodies) to control for the formation of non-specific complexes. The supernatants and pellets from each immunoprecipitation were retained and Western blotted (WB) for NF-YA, Sp1 and SREBP-1c, and relative band intensity was analysed on the Versadoc system (Bio-Rad Laboratories). (A) Western blots of NF-YA (top panel) and SREBP-1c (bottom panel) on pellets and supernatants from representative samples of each group immunoprecipitated with the NF-YA antibody. (B) Western blots of SP1 (top panel) and SREBP-1c (bottom panel) on pellets and supernatants from representative samples of each group immunoprecipitated with the Sp1 antibody. (C) Western blots of SREBP-1c (top panel), Sp1 (middle panel) and NF-YA (bottom panel) on pellets and supernatants from representative samples of each group immunoprecipitated with the SREBP-1 antibody.
DISCUSSION
The proximal PUFA-RRFASN accounts for 65% of the PUFA-dependent inhibition of Fasn gene transcription [15]. This region contains DNA recognition sites for SREBP-1 (−150 to −142, −71 to −66 bp and −59 −52 bp), USF (−65 to −60 bp), Sp1 (−91 to −81 bp) and NF-Y (−99 to −93 bp). Overexpression of mature SREBP-1 in mice increases lipogenic gene expression and promotes the development of fatty livers [40,41]. Moreover, PUFA inhibition of lipogenic gene transcription is paralleled by a 65–85% reduction in the nuclear content of hepatic SREBP-1 [5]. The close association between the nuclear content of SREBP-1 and the transcription of lipogenic genes suggests that PUFAs suppress lipogenic gene transcription by lowering the nuclear concentration of SREBP-1 [5,13,14]. However, we demonstrated that mutation of the −59 to −52 bp SREBP-1 site plus the −65 to −60 bp USF sites of the insulin response element eliminate only 25% of the PUFA-dependent inhibition of Fasn promoter activity [15]. In addition, we show in the present work (Figure 2) that the SRE at −150 to −142 bp, shown previously [42] to mediate the in vivo fasting/re-feeding regulation of the Fasn promoter, does not participate in PUFA regulation of the promoter (Figure 2). These observations indicate that PUFA inhibition of Fasn gene transcription is more complex than a simple down-regulation of SREBP-1.
By themselves SREBP-1a and -1c possess low trans-activating efficacy, but their transcriptional effects on lipogenic genes are amplified by interaction with additional transcription factors [16–22]. SREBP-1a, the predominant isoform in cultured cells, interacts synergistically with NF-Y to activate Fasn promoter activity [20]. In contrast, SREBP-1c (the prevalent hepatic in vivo isoform) activates Fasn promoter activity by interacting with both NF-Y and Sp1 [18,23]. Mutating the NF-Y recognition site (−99 to −93 bp) eliminates activation of the Fasn promoter by glucose and insulin [20], and we have shown previously [15] that the −99 to −93 bp mutation eliminated 50% of the PUFA inhibition of Fasn promoter activity, whereas mutation of the Sp1 site had only a minimal effect. Viewed alongside current findings showing that basal promoter activity is also dependent upon NF-Y binding to this site, the collective interpretation is that modulation of NF-Y binding to this response element is a key common mechanism for regulating transcriptional activity of the Fasn promoter.
The specificity and roles for NF-Y and Sp1 in PUFA regulation of the Fasn promoter is supported by in vitro EMSA data showing that hepatic nuclear protein extracts prepared from rats fed a diet rich in PUFA displayed 50% less NF-Y binding to the Fasn promoter (−99 to −93 bp fragment) and to an oligonucleotide containing a consensus NF-Y element (Figure 3). Sp1 binding to its recognition site at −91 to −81 bp of the Fasn promoter and to a consensus Sp1 oligonucleotide was also reduced by the FO diet (Figure 4). Given the absence of PUFA-dependent changes in nuclear content of NF-Y and Sp1 (Figure 7), these findings raise the interesting possibility that the FO diet altered NF-Y and Sp1 DNA binding activity through kinase-dependent post-translational mechanisms [36,43]. This prediction was supported for Sp1, where pre-incubation with protein phosphatase 1 fully restored the reduced Sp1 DNA binding activity in nuclear extracts from the PUFA-fed rats (Figure 5B). In addition, PUFA increased phosphoserine content of immunoprecipitated Sp1 in nuclear extracts from rats fed the FO diet compared with the FF diet (Figure 5D). These findings support the concept that dietary PUFA post-translationally regulates Sp1 through a phosphatase 1A-dependent site and suggests that Sp1 is a target for nutritionally sensitive signalling inputs which reciprocally modulate its binding activity. For example, previous work has shown that glucose regulates transcription of acetyl-CoA carboxylase by regulation of the phosphorylation state of Sp1 [44]. The kinase(s) responsible for phosphorylating Sp1 and translating PUFA- and glucose-dependent effects into changes in Sp1 binding activity are yet to be identified. AMPK (AMP-activated protein kinase) represents an attractive candidate, based on our recent report [28] showing that hepatic AMPK activity is increased by dietary PUFA. In contrast with Sp1, the data provide no support for the hypothesis that NF-Y binding activity is regulated by PUFA through protein phosphatase 1A (Figure 5A) or alkaline phosphatase-dependent sites. Moreover, we could find no evidence that PUFA increased the phosphoserine content of immunoprecipitated NF-Y (Figure 5C). Previous evidence indicates that NF-Y can be phosphorylated by cdk2 (cyclin-dependent kinase 2) [45], but it seems unlikely that dietary PUFA is influencing NF-Y binding activity through cdk2.
On the surface, the EMSA findings present a caveat in the sense that they predict PUFA would produce a generalized decrease in NF-Y binding activity among the ∼30% of genes estimated to have NF-Y sites within their promoters. Given the inherent limitations of EMSA [46] and the fact that PUFA are known to affect the expression of only a modest subset of hepatic genes, we employed ChIP to provide an unbiased in vivo measure of NF-Y binding to the Fasn promoter and PUFA-insensitive genes. ChIP analysis revealed that, relative to the FF diet, the FO diet decreased NF-Y binding to the PUFA-sensitive region within the Fasn promoter by ∼2-fold (Figure 6A). In contrast, the Cpt-1α promoter is sensitive to changes in NF-Y binding, but PUFA neither altered mRNA expression nor affected NF-Y binding to the Cpt-1α promoter (Figure 6D). The key difference is that the NF-Y site in the Cpt-1α proximal promoter is not adjacent to an SRE, whereas in the Fasn proximal promoter the NF-Y and Sp1 sites are flanked by an SRE. These data suggest the interesting possibility that PUFA selectively regulates NF-Y and SREBP-1c binding to specific motifs within promoters that have this tandem arrangement of response elements. To test this hypothesis, we examined SREBP-1c and NF-Y binding to promoters of three additional PUFA-sensitive genes (S14, Scd1, Fdps) with similar contiguous arrangement of these sites within their promoters. In all cases, PUFA produced a similar 2-fold decrease in SREBP-1c and NF-Y binding to the respective promoters, compared with the FF diet. Although the underlying cause for this uniform decrease in transcription factor binding remains unclear, these findings led us to hypothesize that PUFA suppression of subsets of hepatic genes is co-ordinated through a mechanism that links reduction in nuclear content of SREBP-1c to reduction of NF-Y binding activity. The functional outcome of this co-ordinated suppression of hepatic lipogenesis could ultimately explain the decreased triacylglycerol production observed with diets rich in fish oil.
The central question that evolves from these studies is the mechanism linking reduced SREBP-1c expression to reduced NF-Y binding to adjacent response elements within promoters of PUFA-sensitive genes. One attractive hypothesis is that SREBP-1c co-ordinates assembly of a transcriptional complex containing NF-Y and Sp1, and effectively regulates presentation of NF-Y (and Sp1) to the Fasn proximal promoter. According to this hypothesis, the stoichiometry of proteins present in the transcriptional complex would be regulated by PUFA-dependent repression of SREBP-1c expression, such that PUFA would effectively decrease the presentation of NF-Y to the Fasn promoter. The alternative hypothesis is that the stoichiometry of response element occupancy is governed during or after binding of SREBP-1c and NF-Y to their adjacent sites in the promoter. As in the transcriptional complex model, the PUFA-dependent decrease in SREBP-1c expression would play a key role in regulating the binding of NF-Y to the promoter by decreasing its occupancy on adjacent SREs. Using co-immunoprecipitation to test for protein–protein interactions, we obtained no evidence to support a physical interaction between SREBP-1c and NF-Y (or Sp1). These findings argue against the formation of an SREBP-1c–NF-Y–Sp1 transcriptional complex regulating presentation of NF-Y to the promoter. Our findings are most consistent with a model in which an SRE/NF-Y response element motif regulates the occupancy of the NF-Y site through PUFA-dependent regulation of occupancy of the SRE site by SREBP-1c. An important strength of the ChIP approach is that it provides a measure of the integration of all regulatory inputs into the transcription factor binding process. When applied to nuclear extracts from the livers of FF and FO fed rats, the current findings provide important new insights into the in vivo mechanisms of dietary modulation of gene expression by PUFA. Collectively, these findings support a mechanism whereby PUFA-dependent regulation of SREBP-1c and NF-Y binding to a contiguous arrangement of their response elements within target gene promoters may represent a common mechanism through which dietary PUFA selectively and co-ordinately regulates subsets of hepatic genes involved in lipid metabolism.
Acknowledgments
Supported by the National Institutes of Health (NIH) DK 53872 and DK06156 (T.W.G.), the Universidad Nacional Autonoma de Mexico, Instituto de Investigaciones Biomedicas (M.T.-G.), P30 DK072476 (A.W.A.) and the sponsors of the M.M. Love Chair in Nutritional, Cellular, and Molecular Sciences at The University of Texas (S.D.C.). A.W.A. is supported by NIH Training Grant T32 DK064584-02.
References
- 1.Wilson M. D., Blake W. L., Salati L. M., Clarke S. D. Potency of polyunsaturated and saturated fats as short-term inhibitors of hepatic lipogenesis in rats. J. Nutr. 1990;120:544–552. doi: 10.1093/jn/120.6.544. [DOI] [PubMed] [Google Scholar]
- 2.Jump D. B., Clarke S. D., Thelen A., Liimatta M. Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J. Lipid Res. 1994;35:1076–1084. [PubMed] [Google Scholar]
- 3.Katsurada A., Iritani N., Fukuda H., Matsumura Y., Nishimoto N., Noguchi T., Tanaka T. Effects of nutrients and hormones on transcriptional and post- transcriptional regulation of fatty acid synthase in rat liver. Eur. J. Biochem. 1990;190:427–433. doi: 10.1111/j.1432-1033.1990.tb15592.x. [DOI] [PubMed] [Google Scholar]
- 4.Ntambi J. M. Dietary regulation of stearoyl-CoA desaturase 1 gene expression in mouse liver. J. Biol. Chem. 1992;267:10925–10930. [PubMed] [Google Scholar]
- 5.Xu J., Nakamura M. T., Cho H. P., Clarke S. D. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. A mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J. Biol. Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 6.Liimatta M., Towle H. C., Clarke S., Jump D. B. Dietary polyunsaturated fatty acids interfere with the insulin/glucose activation of L-type pyruvate kinase gene transcription. Mol. Endocrinol. 1994;8:1147–1153. doi: 10.1210/mend.8.9.7838147. [DOI] [PubMed] [Google Scholar]
- 7.McGarry J. D., Brown N. F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 8.Baillie R. A., Takada R., Nakamura M., Clarke S. D. Coordinate induction of peroxisomal acyl-CoA oxidase and UCP-3 by dietary fish oil: a mechanism for decreased body fat deposition. Prostaglandins Leukot. Essent. Fatty Acids. 1999;60:351–356. doi: 10.1016/s0952-3278(99)80011-8. [DOI] [PubMed] [Google Scholar]
- 9.Brandt J. M., Djouadi F., Kelly D. P. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor α. J. Biol. Chem. 1998;273:23786–23792. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- 10.Mater M. K., Thelen A. P., Pan D. A., Jump D. B. Sterol response element-binding protein 1c (SREBP1c) is involved in the polyunsaturated fatty acid suppression of hepatic S14 gene transcription. J. Biol. Chem. 1999;274:32725–32732. doi: 10.1074/jbc.274.46.32725. [DOI] [PubMed] [Google Scholar]
- 11.Xu J., Teran-Garcia M., Park J. H., Nakamura M. T., Clarke S. D. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J. Biol. Chem. 2001;276:9800–9807. doi: 10.1074/jbc.M008973200. [DOI] [PubMed] [Google Scholar]
- 12.Worgall T. S., Sturley S. L., Seo T., Osborne T. F., Deckelbaum R. J. Polyunsaturated fatty acids decrease expression of promoters with sterol regulatory elements by decreasing levels of mature sterol regulatory element-binding protein. J. Biol. Chem. 1998;273:25537–25540. doi: 10.1074/jbc.273.40.25537. [DOI] [PubMed] [Google Scholar]
- 13.Yahagi N., Shimano H., Hasty A. H., Amemiya-Kudo M., Okazaki H., Tamura Y., Iizuka Y., Shionoiri F., Ohashi K., Osuga J., et al. A crucial role of sterol regulatory element-binding protein-1 in the regulation of lipogenic gene expression by polyunsaturated fatty acids. J. Biol. Chem. 1999;274:35840–35844. doi: 10.1074/jbc.274.50.35840. [DOI] [PubMed] [Google Scholar]
- 14.Hannah V. C., Ou J., Luong A., Goldstein J. L., Brown M. S. Unsaturated fatty acids down-regulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 15.Teran-Garcia M., Rufo C., Nakamura M. T., Osborne T. F., Clarke S. D. NF-Y involvement in the polyunsaturated fat inhibition of fatty acid synthase gene transcription. Biochem. Biophys. Res. Commun. 2002;290:1295–1299. doi: 10.1006/bbrc.2002.6341. [DOI] [PubMed] [Google Scholar]
- 16.Bennett M. K., Osborne T. F. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6340–6344. doi: 10.1073/pnas.97.12.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dooley K. A., Millinder S., Osborne T. F. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J. Biol. Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 18.Ericsson J., Edwards P. A. CBP is required for sterol-regulated and sterol regulatory element-binding protein-regulated transcription. J. Biol. Chem. 1998;273:17865–17870. doi: 10.1074/jbc.273.28.17865. [DOI] [PubMed] [Google Scholar]
- 19.Jackson S. M., Ericsson J., Osborne T. F., Edwards P. A. NF-Y has a novel role in sterol-dependent transcription of two cholesterogenic genes. J. Biol. Chem. 1995;270:21445–21448. doi: 10.1074/jbc.270.37.21445. [DOI] [PubMed] [Google Scholar]
- 20.Magana M. M., Koo S. H., Towle H. C., Osborne T. F. Different sterol regulatory element-binding protein-1 isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J. Biol. Chem. 2000;275:4726–4733. doi: 10.1074/jbc.275.7.4726. [DOI] [PubMed] [Google Scholar]
- 21.Naar A. M., Beaurang P. A., Robinson K. M., Oliner J. D., Avizonis D., Scheek S., Zwicker J., Kadonaga J. T., Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roder K., Wolf S. S., Larkin K. J., Schweizer M. Interaction between the two ubiquitously expressed transcription factors NF-Y and Sp1. Gene. 1999;234:61–69. doi: 10.1016/s0378-1119(99)00180-8. [DOI] [PubMed] [Google Scholar]
- 23.Xiong S., Chirala S. S., Wakil S. J. Sterol regulation of human fatty acid synthase promoter I requires nuclear factor-Y- and Sp-1-binding sites. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3948–3953. doi: 10.1073/pnas.040574197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rufo C., Teran-Garcia M., Nakamura M. T., Koo S. H., Towle H. C., Clarke S. D. Involvement of a unique carbohydrate-responsive factor in the glucose regulation of rat liver fatty-acid synthase gene transcription. J. Biol. Chem. 2001;276:21969–21975. doi: 10.1074/jbc.M100461200. [DOI] [PubMed] [Google Scholar]
- 25.Salati L. M., Clarke S. D. Fatty acid inhibition of hormonal induction of acetyl-coenzyme A carboxylase in hepatocyte monolayers. Arch. Biochem. Biophys. 1986;246:82–89. doi: 10.1016/0003-9861(86)90451-0. [DOI] [PubMed] [Google Scholar]
- 26.Rufo C., Gasperikova D., Clarke S. D., Teran-Garcia M., Nakamura M. T. Identification of a novel enhancer sequence in the distal promoter of the rat fatty acid synthase gene. Biochem. Biophys. Res. Commun. 1999;261:400–405. doi: 10.1006/bbrc.1999.1034. [DOI] [PubMed] [Google Scholar]
- 27.Osborne T. F. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 28.Suchankova G., Tekle M., Saha A. K., Ruderman N. B., Clarke S. D., Gettys T. W. Dietary polyunsaturated fatty acids enhance hepatic AMP-activated protein kinase activity in rats. Biochem. Biophys. Res. Commun. 2005;326:851–858. doi: 10.1016/j.bbrc.2004.11.114. [DOI] [PubMed] [Google Scholar]
- 29.Koo S. H., Towle H. C. Glucose regulation of mouse S(14) gene expression in hepatocytes. Involvement of a novel transcription factor complex. J. Biol. Chem. 2000;275:5200–5207. doi: 10.1074/jbc.275.7.5200. [DOI] [PubMed] [Google Scholar]
- 30.Latasa M. J., Moon Y. S., Kim K. H., Sul H. S. Nutritional regulation of the fatty acid synthase promoter in vivo: sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10619–10624. doi: 10.1073/pnas.180306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adamson A. W., Suchankova G., Rufo C., Nakamura M. T., Teran-Garcia M., Clarke S. D., Gettys T. W. Hepatocyte nuclear factor-4α contributes to carbohydrate-induced transcriptional activation of hepatic fatty acid synthase. Biochem. J. 2006;399:285–295. doi: 10.1042/BJ20060659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon Y. S., Latasa M. J., Kim K. H., Wang D., Sul H. S. Two 5′-regions are required for nutritional and insulin regulation of the fatty-acid synthase promoter in transgenic mice. J. Biol. Chem. 2000;275:10121–10127. doi: 10.1074/jbc.275.14.10121. [DOI] [PubMed] [Google Scholar]
- 33.Kim H. J., Takahashi M., Ezaki O. Fish oil feeding decreases mature sterol regulatory element-binding protein 1 (SREBP-1) by down-regulation of SREBP-1c mRNA in mouse liver. A possible mechanism for down-regulation of lipogenic enzyme mrnas. J. Biol. Chem. 1999;274:25892–25898. doi: 10.1074/jbc.274.36.25892. [DOI] [PubMed] [Google Scholar]
- 34.Jump D. B., Clarke S. D., MacDougald O., Thelen A. Polyunsaturated fatty acids inhibit S14 gene transcription in rat liver and cultured hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8454–8458. doi: 10.1073/pnas.90.18.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Jossic-Corcos C., Gonthier C., Zaghini I., Logette E., Shechter I., Bournot P. Hepatic farnesyl diphosphate synthase expression is suppressed by polyunsaturated fatty acids. Biochem. J. 2005;385:787–794. doi: 10.1042/BJ20040933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabor D. E., Kim J. B., Spiegelman B. M., Edwards P. A. Identification of conserved cis-elements and transcription factors required for sterol-regulated transcription of stearoyl-CoA desaturase 1 and 2. J. Biol. Chem. 1999;274:20603–20610. doi: 10.1074/jbc.274.29.20603. [DOI] [PubMed] [Google Scholar]
- 37.Ericsson J., Jackson S. M., Edwards P. A. Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J. Biol. Chem. 1996;271:24359–24364. doi: 10.1074/jbc.271.40.24359. [DOI] [PubMed] [Google Scholar]
- 38.Jump D. B., Thelen A. P., Mater M. K. Functional interaction between sterol regulatory element-binding protein-1c, nuclear factor Y, and 3,5,3′-triiodothyronine nuclear receptors. J. Biol. Chem. 2001;276:34419–34427. doi: 10.1074/jbc.M105471200. [DOI] [PubMed] [Google Scholar]
- 39.Steffen M. L., Harrison W. R., Elder F. F., Cook G. A., Park E. A. Expression of the rat liver carnitine palmitoyltransferase I (CPT-Iα) gene is regulated by Sp1 and nuclear factor Y: chromosomal localization and promoter characterization. Biochem. J. 1999;340:425–432. [PMC free article] [PubMed] [Google Scholar]
- 40.Horton J. D., Bashmakov Y., Shimomura I., Shimano H. Regulation of sterol regulatory element binding proteins in livers of fasted and refed mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5987–5992. doi: 10.1073/pnas.95.11.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimomura I., Shimano H., Korn B. S., Bashmakov Y., Horton J. D. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 42.Latasa M. J., Griffin M. J., Moon Y. S., Kang C., Sul H. S. Occupancy and function of the −150 sterol regulatory element and −65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol. Cell Biol. 2003;23:5896–5907. doi: 10.1128/MCB.23.16.5896-5907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang D., Rhee S. H. Receptor-mediated signaling pathways: potential targets of modulation by dietary fatty acids. Am. J. Clin. Nutr. 1999;70:545–556. doi: 10.1093/ajcn/70.4.545. [DOI] [PubMed] [Google Scholar]
- 44.Daniel S., Kim K. H. Sp1 mediates glucose activation of the acetyl-CoA carboxylase promoter. J. Biol. Chem. 1996;271:1385–1392. doi: 10.1074/jbc.271.3.1385. [DOI] [PubMed] [Google Scholar]
- 45.Yun J., Chae H. D., Choi T. S., Kim E. H., Bang Y. J., Chung J., Choi K. S., Mantovani R., Shin D. Y. Cdk2-dependent phosphorylation of the NF-Y transcription factor and its involvement in the p53-p21 signaling pathway. J. Biol. Chem. 2003;278:36966–36972. doi: 10.1074/jbc.M305178200. [DOI] [PubMed] [Google Scholar]
- 46.Wang D., Sul H. S. Upstream stimulatory factors bind to insulin response sequence of the fatty acid synthase promoter. USF1 is regulated. J. Biol. Chem. 1995;270:28716–28722. doi: 10.1074/jbc.270.48.28716. [DOI] [PubMed] [Google Scholar]