Abstract
SHP (short heterodimer partner) is an orphan nuclear receptor that plays an important role in regulating glucose and lipid metabolism. A variety of transcription factors are known to regulate transcription of the PEPCK (phosphoenolpyruvate carboxykinase) gene, which encodes a rate-determining enzyme in hepatic gluconeogenesis. Previous reports identified glucocorticoid receptor and Foxo1 as novel downstream targets regulating SHP inhibition [Borgius, Steffensen, Gustafsson and Treuter (2002) J. Biol. Chem. 277, 49761–49796; Yamagata, Daitoku, Shimamoto, Matsuzaki, Hirota, Ishida and Fukamizu (2004) J. Biol. Chem. 279, 23158–23165]. In the present paper, we show a new molecular mechanism of SHP-mediated inhibition of PEPCK transcription. We also show that the CRE1 (cAMP regulatory element 1; −99 to −76 bp relative to the transcription start site) of the PEPCK promoter is also required for the inhibitory regulation by SHP. SHP repressed C/EBPα (CCAAT/enhancer-binding protein α)-driven transcription of PEPCK through direct interaction with C/EBPα protein both in vitro and in vivo. The formation of an active transcriptional complex of C/EBPα and its binding to DNA was inhibited by SHP, resulting in the inhibition of PEPCK gene transcription. Taken together, these results suggest that SHP might regulate a level of hepatic gluconeogenesis driven by C/EBPα activation.
Keywords: CCAAT/enhancer-binding protein α (C/EBPα), phosphoenolpyruvate carboxykinase (PEPCK), protein–protein interactions, short heterodimer partner (SHP), transcriptional repression
Abbreviations: AF-2, activation function-2; ATF-2, activating transcription factor 2; bZIP, basic region-leucine zipper; C/EBPα, CCAAT/enhancer-binding protein α; CRE, cAMP regulatory element; CREB, CRE binding protein; DBD, DNA-binding domain; DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility shift analysis; ER, oestrogen receptor; FBS, fetal bovine serum; GR, glucocorticoid receptor; GST, glutathione S-transferase; HA, haemagglutinin; HNF-3, hepatocyte nuclear factor 3; LBD, ligand binding domain; NF1, nuclear factor 1; NR, nuclear receptor; PEPCK, phosphoenolpyruvate carboxykinase; RAR, retinoic acid receptor; RXR, retinoid X receptor; SHP, short heterodimer partner; TBST, Tris buffered saline containing 0.05% Tween 20; TR, thyroid hormone receptor
INTRODUCTION
PEPCK (phosphoenolpyruvate carboxykinase) is an enzyme that catalyses a regulatory step in gluconeogenesis. It is expressed primarily in liver, kidney, small intestine and adipose tissue, where its synthesis is regulated under multihormonal control at the level of transcription initiation [1]. The PEPCK promoter integrates cues arising from diverse signalling pathways. PEPCK mRNA is induced by glucocorticoids, thyroid hormone or glucagon [2], whereas insulin causes a repression of the promoter activity in a dominant manner [3–5]. The PEPCK promoter fragment encompassing the region −460 to +73 bp relative to the transcription start site was demonstrated to be sufficient for hormonal regulation in the liver, and many of the transcription factors that bind elements in this region have been identified [6,7]. Proteins that have been shown to bind to, and impact on the regulation of, the PEPCK promoter include CREB (cAMP responsive element binding protein 1), C/EBPα (CCAAT/enhancer-binding protein α), C/EBPβ, ATF-2 (activating transcription factor 2), NF1 (nuclear factor 1), HNF-3 (hepatocyte nuclear factor 3), GR (glucocorticoid receptor), TR (thyroid hormone receptor), RAR (retinoic acid receptor), RXR (retinoid X receptor), HIF-1 (hypoxia-inducible factor 1), calcium-sensing receptor and ERRα (oestrogen-related receptor α) [8–14]. The energy balance state of the cell can affect signals regulating PEPCK gene activation, such as CREB, C/EBPα and C/EBPβ, whereas ATF-2 mediates the stress response signals and hepatic viral protein function that regulates PEPCK expression [15,16].
SHP (small heterodimer partner) is an orphan NR (nuclear receptor) that is specifically expressed in the liver and a limited number of other tissues, and its activities are in some ways opposite to those of RXR [16]. Based on its ability to interact with a variety of NRs, distinct features distinguish SHP from RXR, which is the only known common heterodimerization receptor [17,18]. First, SHP, unlike RXR, interacts with ERs (oestrogen receptors) and agonistic ligands enhance, whereas antagonistic ligands inhibit, these interactions (for a discussion see [19]). Secondly, the C-terminus of SHP, which includes the putative dimerization helix, is dispensable for interactions with C/EBPα and a central LBD (ligand binding domain) region apparently forms the SHP-specific domain for interaction with receptors.
SHP has been suggested to play a generally very negative role in NR signalling. SHP lacks a DBD (DNA-binding domain) and therefore the addition of SHP inhibited DNA binding by NRs with which it interacted in vitro as expected. In mammalian cell cotransfections, SHP repressed transcriptional activation by the same NRs. In transient transfections, SHP inhibits transcriptional activation of its receptor targets and the inhibitory effect of SHP may be potentiated further due to the presence of an intrinsic transcriptional repression domain. SHP has been shown in vitro to inhibit binding of RAR–RXR heterodimers to DNA response elements, suggesting that competitive dimerization may result in novel SHP heterodimers that are unable to bind DNA. Based on earlier studies on SHP and ER [19], it was proposed to be a novel inhibitory mechanism for SHP.
C/EBPα was purified as an active molecule that bound to consensus enhancer elements and to the CCAAT box motif. C/EBPα protein is a member of the bZIP (basic region-leucine zipper) class of transcription factors [20,21]. Leucine zippers accommodate both homo- and hetero-dimerization. The first C/EBP protein characterized, C/EBPα, is one of at least five gene products comprising the C/EBP gene family [22]. When not bound to DNA, the subunits of bZIP dimers are in a rapid monomer:dimer equilibrium such that the lifetime of dimers is estimated in seconds [23]. The ready dissociation of bZIP dimers in vitro suggests that heterodimers with potentially unique regulatory properties may form in vivo.
C/EBPα activates transcription of several liver and fat cell-specific genes [23]. Support for the notion that C/EBPα plays a central role in regulating energy homoeostasis [24] was provided by targeted gene disruption. Homozygous C/EBPα knockout mice are born with apparently normal blood glucose levels, but become severely hypoglycemic within minutes. These animals exhibit glycogen storage defects and morphological anomalies in fat and liver tissues. Although these defects are consistent with a role for C/EBPα in energy homoeostasis, expression of several putative C/EBPα target genes was normal [25]. Thus, it has been difficult to identify genes that are C/EBPα targets in vivo.
Since it is well known that SHP regulates glucose and lipid metabolism, we aimed to examine the effect of SHP on PEPCK gene expression. Consistent with other results, SHP inhibited transcriptional activation of the PEPCK promoter [1,2]. However, in the present paper, we show a different molecular mechanism by which SHP affects PEPCK expression through inhibition of C/EBPα-dependent transcriptional activation. These results also showed that the cross-talk of different kinds of transcription factor families, SHP and C/EBPα, functionally affect each other in the regulation of PEPCK gene expression.
EXPERIMENTAL
Plasmids and cloning
The SHP gene and promoter–reporter plasmids were provided by Dr Heung Sik Choi (Hormone Research Centre, Chonnam University, Kwangju, Korea). A full-length C/EBPα cDNA was generated by reverse transcription followed by PCR and verified by sequencing. The SHP and C/EBPα genes were subcloned into pCMX1 as BamHI and BamHI/KpnI fragments respectively. pCMX1 was a gift from Jon Shuman (Laboratory of Immunogenetics, National Institute of Allergy and Infectious Disease, National Institutes of Health, Rockville, MD, U.S.A.) and was used in the in vitro transcription/translation assay. In vitro translation products were verified by [35S]Met incorporation and SDS/PAGE analysis. HA (haemagglutinin)-tagged SHP and C/EBPα constructs were generated by subcloning into the pcDNA3-HA plasmid. The reporter plasmids PEPCK-275-luc and PEPCK-543-luc were constructed by PCR amplification of rat genomic DNA encompassing positions −275 or −543 to +73 bp of the PEPCK promoter. The reporter plasmid of PEPCK-CRE1 (cAMP regulatory element 1) was prepared with two tandem repeats of the PEPCK CRE1 (−99 to −76 bp relative to the transcription start site). The CRE1-mutant reporter, PEPCK-275m-luc, was prepared by standard mutagenesis by replacing (−99) 5′-CCGGCCCCTTACGTCAGAGGCG-3′ (−76) with 5′-CCGGCCCCTTTTTTCAGAGGCG-3′ (the mutated positions are in bold).
Transient transfection and luciferase assays
HepG2 hepatoma cells were maintained in DMEM (Dulbecco's modified Eagle's medium) containing 5 mM glucose, supplemented with 10% (v/v) FBS (fetal bovine serum) at 37 °C, in a 5% CO2/95% air atmosphere.
HepG2 cells were transfected using the standard calcium phosphate method [18]. Cells were incubated with DNA precipitates for 16 h, washed and maintained in complete medium for 48 h prior to harvesting. Relative luciferase and β-galactosidase activities were determined as described previously [19]. Basal promoter activity is reported as the activity observed after transfection of the reporter plus an appropriate amount of empty expression vector. In all cases, transfection results are presented as means±S.E.M. for three independent experiments.
Preparation of antisera and co-immunoprecipitation analysis
The human SHP cDNA was amplified by PCR and subcloned in between the EcoRI and BamHI sites of the pGEX4T vector (Amersham Biosciences) for protein expression in bacteria. GST–SHP was affinity purified and the antigen was excised from an SDS/10% (v/v) PAGE gel. This protein was used to prepare antiserum. Antisera were raised according to standard protocols [26] and appropriate reactivity was verified against recombinant GST–SHP. Freshly prepared rat liver nuclear extract was pre-cleared with an irrelevant antiserum and protein A–agarose beads (Life Technologies). After centrifugation at 13400 g for 20 min, supernatants were separated and adjusted to 50 mM Tris/HCl (pH 8.0), 15% (w/v) glycerol, 0.25 M NaCl, 1 mM MgCl2, 0.1 mM EDTA and 1% (v/v) Nonidet P-40. C/EBPα antibody (3 μl; Santa Cruz Biotechnology) was added to the supernatants and incubated overnight at 4 °C with constant rocking. Precipitates were collected using protein A–agarose beads and washed five times with 500 μl of 50 mM Tris/HCl (pH 8.0), 150 mM NaCl and 1% (v/v) Nonidet P-40. After SDS/PAGE and electroblotting, proteins were detected with anti-SHP antibodies (see below).
Western blotting
Cells were harvested in ice-cold lysis buffer [150 mM NaCl, 1% (v/v) Nonidet P-40 and 50 mM Tris/HCl, pH8.0] containing PMSF at 4 °C. The protein content of cell lysates was determined with Bradford reagent (Bio-Rad Laboratories) using BSA as the standard. After heating to 100 °C for 10 min in Laemmli sample buffer, the samples were separated by SDS/PAGE. The resolved gels were either stained with Coomassie Blue or transferred to PVDF (Immobilon-P) membranes (Millipore). For Western blotting, the membrane was blocked with 5% (w/v) non-fat dried milk in TBST (Tris-buffered saline containing 0.05% Tween 20) for 1 h at room temperature (25 °C) and then probed with anti-HA antibody for 1 h at room temperature. After washing three times with ice-cold TBST, the blotted membranes were incubated with horseradish-peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). After washing three times with ice-cold TBST, the protein bands were visualized by ECL® (Amersham Biosciences) according to the manufacturer's instructions.
Yeast two-hybrid interaction assay
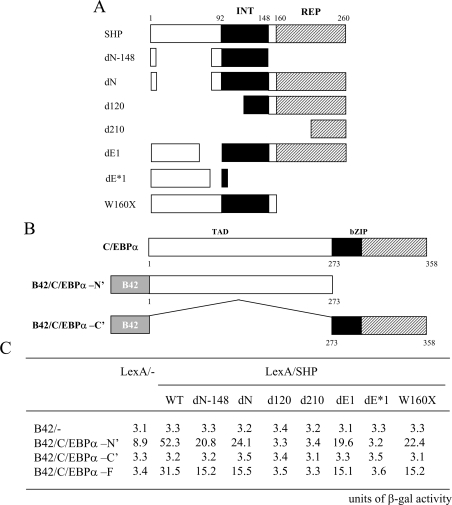
Yeast two-hybrid interaction assays were performed as described previously [18]. Briefly, full-length or deletions of murine SHP were fused to LexA (see Figure 4A and B) [27] and co-transformed with B42 C/EBPα full-length or deletion constructs into Saccharomyces cerevisae EGY48 strain containing the β-galactosidase reporter plasmid 8H18-34. The transformants were selected on plates with appropriate selection markers. The β-galactosidase assay was carried out on the plates using the method described previously [17].
Figure 4. Interaction region of SHP with C/EBPα in yeast two-hybrid assay.
An illustration of the domain structure and deletion constructs of SHP (A) and C/EBPα (B). INT, interaction domain of SHP; REP, repression domain of SHP; TAD, transcriptional activation and regulatory domain of C/EBPα; bZIP, basic and leucine zipper domain of C/EBPα. (C) The indicated B42 and LexA plasmids were transformed into EGY48 yeast cells containing the appropriate β-galactosidase reporter gene. At least three separate transformants from each transformation were transferred to indicator plates containing 5-bromo-4-chloroindol-3-yl β-D-galactopyranoside (X-Gal) and reproducible results were obtained using colonies from a separate transformation. The number indicates the β-galactosidase activity, measured using the β-galactosidase enzyme assay (Promega).
Mammalian two-hybrid assay
Cells were seeded in DMEM supplemented with 10% FBS and 1% (w/v) antibiotics, and co-transfected with expression vectors encoding Gal4–DBD fusions [pCMX-Gal4-N-C/EBPα (amino acids 1–272), pCMX-Gal4-C-C/EBPα (amino acids 272–358) or pCMX-Gal4-F-C/EBPα (amino acids 1–358)] and VP16–activation domain fusions (pCMX-VP16, pCMX-VP16-SHP, pCMX-VP16-C/EBPα and pCMX-VP16-ATF-2) as well as the Gal4-tk-luc reporter plasmid [19], which has been described previously. After 48 h, the cells were harvested and the luciferase activity was measured. The results were normalized to the β-galactosidase expression levels. The results represent the means for at least three independent experiments.
EMSA (electrophoretic mobility shift analysis)
Nuclear extracts were prepared from HepG2 cells at the indicated times in the following buffer [50 mM Tris/HCl, pH 8.0, 15% (v/v) glycerol, 0.25 M NaCl, 1 mM MgCl2, 0.1 mM EDTA and 1% (v/v) Nonidet P40] as described previously [26]. Approx. 10 μg nuclear extract proteins were incubated with a double-stranded oligonucleotide encoding the PEPCK CRE1 sequence (promoter positions −99 to −76 bp) 5′-CCGGCCCCTTACGTCAGAGGCG-3′ as the probe [26,28]. Binding reactions were carried out without probe and incubated for 5 min on ice, followed by 5 min at room temperature. The probe (approx. 50000 cpm) was added and the samples were incubated at room temperature for a further 30 min. Samples were separated in 4% (w/v) acrylamide, 0.5×TBE [0.045 M Tris, 0.045 M boric acid, 1.0 mM EDTA (pH 8.0)] gels run at 200 V constant voltage [19].
GST-pull down assay
Equal amounts (approximately 1 μg) of GST-fusions to the full-length and deletion mutant series of C/EBPα, and the full-length human SHP were induced and purified from E. coli BL-21 strain. Full-length SHP and C/EBPα were in vitro translated in the presence of [35S]methionine. GST-fusion proteins were immobilized on glutathione–Sepharose CL-4B beads (Amersham Pharmacia Biotech) in GST-pull down reaction buffer [25 mM Hepes, pH 7.6, 20% (v/v) glycerol, 120 mM NaCl, 0.2 mM EDTA, 1 mM DTT, 0.1% Triton X-100 and 0.1% BSA] and then incubated with radiolabelled [35S]SHP or [35S]C/EBPα for 4 h at 4 °C. After washing three times with PBS containing 0.1 % Tween 20, the bound proteins were eluted with 10 mM GSH in 50 mM Tris/HCl (pH 8.0) and 120 mM NaCl, and boiled with an equal volume of 2×Laemmli sample buffer at 100 °C for 3 min prior to electrophoresis. After electrophoresis, the gel was dried and analysed with a Molecular Imager Fx (Bio-Rad Laboratories).
RESULTS AND DISCUSSION
SHP inhibits transcriptional activation of the PEPCK promoter
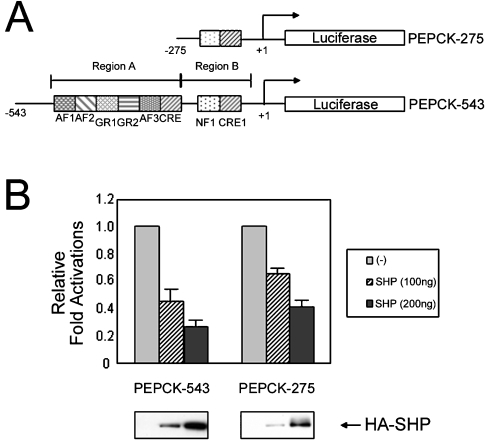
To address the effect of SHP on the regulation of PEPCK promoter activity, HepG2 cells were transiently co-transfected with an expression vector encoding SHP and either PEPCK-275-luc or PEPCK-543-luc, two PEPCK promoter–reporter plasmids (Figure 1A). Region A (−543 to −276 bp) of the PEPCK promoter is shown to contain many NR responsive elements, including GR and RXR, which are known to interact with SHP [29]. Since SHP is known to inhibit GR- and RXR-mediated transcriptional activation by direct protein interaction, it was suggested that the transcription of PEPCK might be decreased by SHP expression in a region A-dependent manner. Interestingly, transfection with SHP expression plasmid decreased luciferase activity driven by both the PEPCK-275-luc and the PEPCK-543-luc promoter–reporter constructs (Figure 1B). Region B (−275 to +73 bp) of the PEPCK promoter contains a CRE1 located at −99 to −76 bp relative to the transcription start site and is immediately adjacent to an NF1-binding site; however, this region lacks an NR responsive element. SHP expression repressed base-line luciferase activity driven by the PEPCK-275-luc and PEPCK-543-luc promoter–reporter constructs to 25% and 40% respectively. Thus, this result indicates that region B of the PEPCK promoter also contains an SHP-responsive element that mediates the SHP-dependent transcriptional repression of the PEPCK promoter.
Figure 1. SHP represses transcriptional activation of the PEPCK promoter.
(A) Schematic representation of two luciferase reporter constructs of the PEPCK promoter are shown. Region A contains the glucocorticoid response unit composed of two glucocorticoid regulatory elements (GR1 and 2), three accessory factor-binding sites (AF1–3) and a CRE. This region provides several factor-binding sites for RAR, RXR, GR, TR, C/EBP, and HNF-3 [11,15–17]. Region B contains CRE1 (at −99 to −76 bp relative to the transcription start site) and is immediately adjacent to a nuclear factor 1-binding site (NF1). (B) SHP decreases the transcriptional activity of the PEPCK promoter. HepG2 cells were transiently transfected with reporter constructs PEPCK-275 or PEPCK-543 along with the indicated expression plasmids. The cells were incubated for 48 h after transfection, then harvested and the luciferase activity measured. All the transfection results were normalized to β-galactosidase activity, and the results represent the means±S.E.M. for three independent experiments, with fold induction over the level observed with the reporter alone.
C/EBPα is associated with SHP-mediated repression of PEPCK transcription
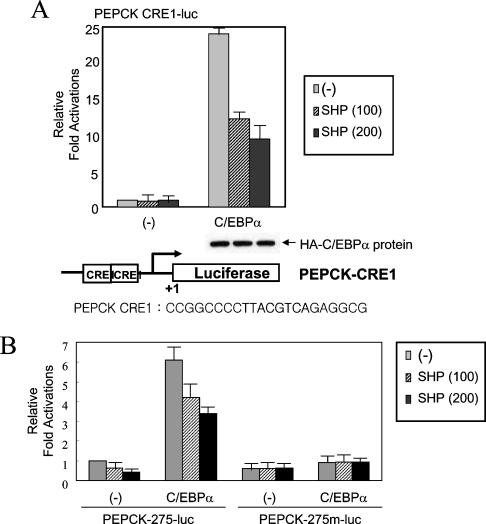
Since the CRE1 region of the PEPCK promoter is known to be responsive to a variety of extracellular stimuli including cAMP and insulin [3,9–11,29], we next investigated the importance of the CRE1 region in SHP-mediated PEPCK transcriptional repression. Previous reports have shown that C/EBPα and liver-enriched transcription factor proteins bind to the CRE1 region of the PEPCK promoter [16,26,28]. Thus, it was examined whether overexpression of C/EBPα affected the SHP-induced transcriptional repression of the PEPCK promoter. Co-expression of C/EBPα with increasing amounts of SHP protein exerted an inhibitory effect on the luciferase reporter activity of the PEPCK-CRE1 construct by C/EBPα in a dose-dependent manner (Figure 2A). Equal expression of C/EBPα (HA-tagged form of C/EBPα) in the transfected cells was confirmed by Western blotting using an antibody against the HA-tag, indicating that SHP expression did not affect C/EBPα expression levels in the cells.
Figure 2. SHP inhibits C/EBPα-mediated transcription activation of the PEPCK promoter.
(A) An expression vector for C/EBPα and increasing amounts of SHP were co-transfected into HepG2 cells with the reporter construct PEPCK-CRE1. The expression level of C/EBPα (HA-tagged) was confirmed by Western blotting using an antibody against the HA-tag. (B) The inhibitory function of SHP on C/EBPα transcription activation is dependent on the CRE1 sequence of the PEPCK promoter. The same transient transfection assay as shown in (A) was performed using wild-type PEPCK-275-luc and mutant PEPCK-275m-luc reporter constructs instead of PEPCK-CRE1. The mutated sequence of the CRE1 of PEPCK-275m-luc was described in the Experimental section. The cells were incubated for 48 h after transfection, then harvested and the luciferase activity measured. All the transfection results were normalized to β-galactosidase activity, and the results represent the means±S.E.M for four independent experiments, with fold induction over the level observed with the reporter alone.
To further determine whether the PEPCK CRE1 is responsible for this SHP-induced transcriptional repression in region B, a PEPCK-275 mutant promoter containing a mutated CRE1 (PEPCK-275m-luc; described in the Experimental section) was used in the absence or presence of C/EBPα overexpression. It was confirmed that the CRE1-mutated PEPCK promoter was not responsive to the increasing expression of SHP (Figure 2B). These findings provide the first clue that SHP mediated inhibition of PEPCK is dependent on C/EBPα, one of the bZIP family proteins, in addition to NRs.
SHP interacts physically with C/EBPα
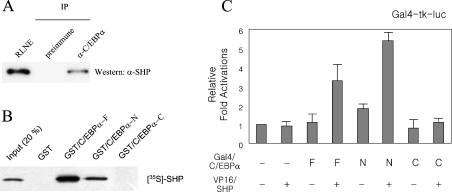
Two previous findings [18,26], which showed that SHP and C/EBPα proteins were highly expressed in liver, and that SHP functionally inhibited the C/EBPα-mediated PEPCK gene transcription, led us to investigate whether the two transcription factors interacted mutually in hepatic cells. To address the possibility, freshly prepared rat liver nuclear extracts were subjected to immunoprecipitation with affinity purified antibody against C/EBPα. C/EBPα was immunoprecipitated, gel-fractionated and the co-precipitation of SHP was analysed by Western blotting using an anti-SHP antibody. As shown in Figure 3(A), endogenous SHP was detected as a co-precipitant in C/EBPα immunoprecipitates. A parallel immunoprecipitation experiment using pre-immune serum failed to show SHP immunoreactivity. These results support the observation that C/EBPα is able to associate with SHP in hepatic cells.
Figure 3. SHP interacts with C/EBPα.
(A) SHP co-precipitates with C/EBPα from rat liver nuclear extracts. SHP and C/EBPα were co-immunoprecipitated (IP) from rat liver nuclear extract by incubation with anti-C/EBPα IgG. Antibody complexes were captured on protein A–Sepharose. The beads were washed three times with binding buffer and eluted into Laemmli sample buffer. Proteins were resolved by SDS/PAGE (10% gels), electroblotted and detected by Western blotting by using an anti-SHP antibody. Lane 1, rat liver nuclear extract; lane 2, co-immunoprecipitates from samples using preimmune serum; lane 3, co-immunoprecipitates from samples treated with anti-C/EBPα IgG. (B) The N-terminal domain of C/EBPα interacts with SHP in vitro. GST fusions to full-length (C/EBPα-F), N-terminal domain (C/EBPα-N) and C-terminal domain (C/EBPα-C) of C/EBPα were purified from E. coli. In vitro translated SHP protein in the presence of [35S]-methionine were incubated with GST or GST-fusion proteins immobilized on glutathione–resin. The bound proteins were eluted by GSH and resolved by SDS/PAGE. (C) SHP associates with the N-terminal transcriptional activation domain of C/EBPα in cells. The mammalian expression plasmids encoding VP16–SHP, and GAL4–full length C/EBPα (F), Gal4–N-terminal (N) and Gal4–C-terminal (C) domains were transfected into HepG2 cells as indicated. The cells were incubated for 48 h after transfection, then harvested and the luciferase activity measured. The results represent the means for three independent experiments, with fold induction over the level observed with the reporter alone.
To identify whether SHP protein interacts directly with C/EBPα and to delineate the interaction region of C/EBPα with SHP, we used GST pull-down assays. GST-fusion proteins encoding the full-length (C/EBPα-F, amino acids 1–358), N-terminal domain (C/EBPα-N, amino acids 1–273) and C-terminal domain (C/EBPα-C, amino acids 273–358) of C/EBPα were produced in bacteria, immobilized on glutathione–Sepharose beads and incubated with in vitro translated 35S-labelled SHP. Consistent with the co-immunoprecipitation result described in Figure 3(A), the full-length C/EBPα protein interacted with SHP in vitro (Figure 3B). The N-terminal transcriptional activation domain of C/EBPα interacted with SHP, but its C-terminal bZIP domain did not (Figure 3B). This result indicates that SHP might inhibit the C/EBPα function by masking its transcriptional activation domain. To further confirm the association of SHP with the transcriptional activation domain of C/EBPα in vivo, we utilized the mammalian two-hybrid assay. Co-expression of the full length and the N-terminal domain of C/EBPα fused to the Gal4-DBD with VP16–SHP enhanced the promoter activity of the Gal4-tk-luc reporter plasmid, whereas the Gal4–C/EBPα C-terminal domain was inert (Figure 3C). These results suggest that the transcriptional activation domain of C/EBPα interacts with SHP in vivo.
Next, to determine which SHP region was required for the interaction with C/EBPα, a series of LexA-fused deletion constructs of SHP were used as outlined in Figure 4. Yeast two-hybrid interactions revealed that the deletion constructs containing the entire interaction domain (amino acids 92–148), such as dN–148, dN, dE1, and W160stop, were sufficient for the interaction with the N-terminal region of C/EBPα, whereas other regions of SHP failed to interact with C/EBPα and thus did not increase the β-galactosidase reporter activity (Figure 4). We conclude that the central domain of SHP interacts directly with the N-terminal transcription activation domain of C/EBPα.
SHP inhibits C/EBPα–DNA complex formation
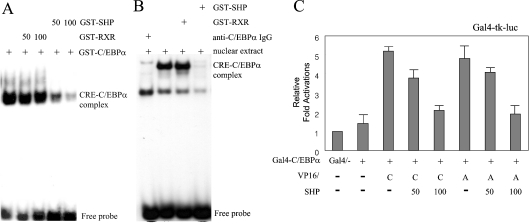
As expected from its lack of a DBD, SHP inhibited in vitro DNA binding by NRs with which it interacted and repressed the transcriptional activation of their target genes [16,17,27]. These findings, combined with the results presented in Figures 2 and 3, prompted us to investigate whether SHP inhibited DNA-binding of C/EBPα to the PEPCK CRE1 sequence. In order to investigate this, we prepared GST-fusion proteins for full-length C/EBPα and SHP, and then applied to an EMSA using the 32P-labelled CRE1 region from the PEPCK promoter as the probe. GST–RXR was used as a negative control for molecular interaction with C/EBPα. As shown in Figure 5(A), while GST–RXR did not inhibit DNA-binding activity of C/EBPα, increasing amounts of SHP protein blocked the formation of the C/EBPα–CRE1 complex in a dose-dependent manner. This result suggests that SHP inhibits the formation of functional C/EBPα homodimers by competitive interaction, thereby leading to a squelching of the functional C/EBPα proteins that are required for specific DNA binding. It would be of interest to provide a comparison of the retarded band obtained in this way with the EMSA pattern seen with liver cell nuclear extract. For this, we prepared rat liver nuclear extract and performed a similar experiment to that shown in Figure 5(A). The band that contained the specific protein–CRE complex disappeared by treatment with anti-C/EBPα IgG, indicating the presence of an endogenous C/EBPα–CRE complex. Consistent with the result of Figure 5(A), addition of GST–SHP also inhibited the formation of the C/EBPα–CRE complex (Figure 5B).
Figure 5. SHP inhibits the DNA binding and homo/heterodimerization of C/EBPα.
(A) SHP blocks C/EBPα–DNA complex formation. A double-stranded oligonucleotide probe containing the CRE1 site of the PEPCK promoter was used in EMSAs. Rat liver nuclear extracts (20 ng) were incubated as indicated, followed by the addition of the probe. Mobility shifts were demonstrated by incubation of the nuclear extract proteins with 100 ng of GST–RXR or SHP proteins. The specificty of the C/EBPα–CRE complex was confirmed by incubating the nuclear extracts with anti-C/EBPα IgG. (B) Bacterially expressed and purified GST–C/EBPα, GST–RXR and GST–SHP were incubated as indicated, followed by the addition of a DNA probe. Mobility shifts were demonstrated by incubation of 50 ng of GST–C/EBPα with increasing amounts of GST–RXR or SHP proteins (50 and 100 ng). (C) SHP inhibits the homo- and hetero-dimerization of C/EBPα. The mammalian expression plasmids encoding VP16–C/EBPα or –ATF-2 and GAL4–C/EBPα were transiently transfected with the Gal4-tk-luc reporter plasmid into HepG2 cells as indicated. The cells were incubated for 48 h after transfection, then harvested and the luciferase activity measured. The results represent the means for three independent experiments, with fold induction over the level observed with the reporter alone. C, C/EBPα; A, ATF-2.
To examine further whether SHP prevents the formation of a transcriptionally active C/EBPα homodimer or a heterodimer between C/EBPα and another bZIP protein, ATF-2, we performed an in vivo protein interaction assay using the mammalian two-hybrid system, with Gal4–C/EBPα as the bait protein and either VP16–C/EBPα, as a homodimerization partner, or VP16-ATF-2, as a heterodimerization partner, in the absence or presence of ectopic SHP expression. SHP expression inhibited the dimerization of C/EBPα with both C/EBPα and ATF-2 in a dose-dependent manner (Figure 5C). Taken together, these results indicate that direct interaction of SHP and C/EBPα inhibits the formation of an active transcriptional complex and therefore inhibits the DNA-binding and transcription activation activities of C/EBPα.
SHP prevents formation of a functional complex of C/EBPα and RFC140
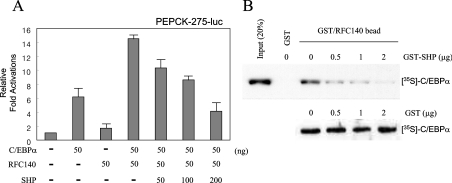
Previous studies have shown that SHP itself carries a novel autonomous repression domain in its C-terminal region [27]. The existence of this domain suggested that SHP could inhibit transcription if it was able to interact with DNA-bound receptor targets. Support for this alternative possibility was provided previously by results demonstrating that SHP is an effective inhibitor of ER transcription activation, even though it does not block binding of ER to oestrogen response elements [17]. The results demonstrated that SHP and AF-2 (activation function-2) co-activators such as TIF2 (transcriptional intermediary factor 2) compete directly for binding to ERs, suggesting either that SHP and AF-2 co-activators contact a common surface or, alternatively, that binding of SHP to the LBD induces conformational changes that cause the dissociation of AF-2 co-activators. Since we previously identified that RFC140 increased C/EBPα-mediated transcription activation via direct protein interaction as a co-activator [15], it was addressed whether SHP expression in cultured cells affected the co-operative transcription activation of PEPCK by C/EBPα and RFC140. HepG2 cells were transiently transfected with expression plasmids encoding C/EBPα and RFC140 along with a promoter–reporter plasmid in the absence or presence of increasing amounts of SHP as indicated. Overexpression of SHP inhibited the synergistic transcription activation by RFC140 and C/EBPα in a dose-dependent manner (Figure 6A).
Figure 6. SHP inhibits the functional complex of C/EBPα and RFC140.
(A) SHP inhibits the co-activator function of RFC140 in C/EBPα-mediated transcription activation. Cells were co-transfected with constructs encoding C/EBPα and RFC140 and increasing amounts of SHP, along with the PEPCK-275-luc promoter–reporter construct. Results are the means for three independent experiments. (B) SHP inhibits the protein–protein interaction between C/EBPα and RFC140. GST fusions to the C/EBPα-interaction region (residues 151–545) of RFC140 were purified from E. coli. C/EBPα was in vitro translated in the presence of 35S-methionine and then incubated with glutathione-resin immobilized GST–RFC140(151–545) in the presence of increasing amounts of GST–SHP or GST alone. The bound proteins were eluted by GSH and resolved by SDS/PAGE.
To determine further whether the inhibition of transcriptional activation by SHP was due to a competitive interaction between SHP and RFC140 for C/EBPα, we performed a GST-pull down assay using GST or GST–RFC140 resin and 35S-labelled in vitro translated C/EBPα protein with increasing amounts of SHP proteins. As shown in Figure 6(B), increasing amounts of SHP gradually inhibited the interaction between C/EBPα and RFC140 in vitro. Taken together with the results presented in Figure 5, this suggests that a direct molecular interaction of SHP with C/EBPα inhibited the formation of an active transcriptional complex of C/EBPα with other transcription factors and co-activators, thereby preventing its specific promoter–DNA binding and transcription activation of PEPCK.
The transcription factor complex on the PEPCK promoter is a typical enhancersome, comprised of a series of cis-elements, including binding sites for bZIP proteins and NRs. The present paper shows that crosstalk of different kinds of transcription factor families, bZIP proteins and NRs functionally affect each other to regulate target gene expression.
Borgius et al. [30] showed that PEPCK promoter activity was perturbed by SHP through the inhibition of glucocorticoid signalling. In addition, Yamagata et al. [29] reported that bile acids inhibit the expression of gluconeogenic genes, including glucose-6-phosphatase, phosphoenolpyruvate carboxykinase and fructose 1,6-bis-phosphatase in an SHP-dependent manner. SHP affected transcription activation by the orphan receptor HNF-4 [29]. SHP interacted with the same HNF-4 surface that is recognized by transcriptional co-activators and competed with them for binding in vivo. SHP also appeared to regulate the transcription activity of NF-κB (nuclear factor κB) in oxidized low-density lipoprotein-treated resting macrophage cells. The results of the present paper add C/EBPα to the list of SHP targets. Cross-talk between transcription factors of distinct families is an important phenomenon in regulating gene transcription and has recently become the subject of intensive investigation. The potential importance of the functional interaction between SHP and C/EBPα is supported by the enrichment of their expression in liver cells. This is reinforced by the fact that only a minor perturbation of C/EBPα activity results in the deregulation of energy metabolism, suggesting that the modulation of C/EBPα activity by SHP could have important metabolic effects. Since SHP repressed PEPCK transcription, it might lead to decreased hepatic glucose production.
Acknowledgments
This work was supported by the Korean Ministry of Health and Welfare, Republic of Korea (A050404 and 02-PJ1-PG3-20908-0009).
References
- 1.Hanson R. W., Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 2.Loose D. S., Cameron D. K., Short H. P., Hanson R. W. Thyroid hormone regulates transcription of the gene for cytosolic phosphoenolpyruvate carboxykinase (GTP) in rat liver. Biochemistry. 1995;24:4509–4512. doi: 10.1021/bi00338a004. [DOI] [PubMed] [Google Scholar]
- 3.Kioussis D., Reshef L., Cohen H., Tilghman S. M., Iynedjian P. B., Ballard F. J., Hanson R. W. Alterations in translatable messenger RNA coding for phosphoenolpyruvate carboxykinase (GTP) in rat liver cytosol during deinduction. J. Biol. Chem. 1978;253:4327–4332. [PubMed] [Google Scholar]
- 4.Forest C. D., O'Brien R. M., Lucas P. C., Magnuson M. A., Granner D. K. Regulation of phosphoenolpyruvate carboxykinase gene expression by insulin. use of the stable transfection approach to locate an insulin responsive sequence. Mol. Endocrinol. 1990;4:1302–1310. doi: 10.1210/mend-4-9-1302. [DOI] [PubMed] [Google Scholar]
- 5.Scott D. K., O'Doherty R. M., Stafford J. M., Newgard C. B., Granner D. K. The repression of hormone-activated PEPCK gene expression by glucose is insulin-independent but requires glucose metabolism. J. Biol. Chem. 1998;273:24145–24151. doi: 10.1074/jbc.273.37.24145. [DOI] [PubMed] [Google Scholar]
- 6.McGrane M. M., Yun J. S., Moorman A. F., Lamers W. H., Hendrick G. K., Arafah B. M., Park E. A., Wagner T. E., Hanson R. W. Metabolic effects of developmental, tissue-, and cell-specific expression of a chimeric phosphoenolpyruvate carboxykinase (GTP)/bovine growth hormone gene in transgenic mice. J. Biol. Chem. 1990;265:22371–22379. [PubMed] [Google Scholar]
- 7.Short M. K., Clouthier D. E., Schaefer I. M., Hammer R. E., Magnuson M. A., Beale E. G. Tissue-specific, developmental, hormonal, and dietary regulation of rat phosphoenolpyruvate carboxykinase-human growth hormone fusion genes in transgenic mice. Mol. Cell. Biol. 1992;12:1007–1020. doi: 10.1128/mcb.12.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai E., Stromstedt P. E., Quinn P. G., Carlstedt-Duke J., Gustaffson J. A., Granner D. K. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol. Cell Biol. 1990;10:4712–4719. doi: 10.1128/mcb.10.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiyama T., Scott D. K., Wang J. C., Granner D. K. Structural requirements of the glucocorticoid and retinoic acid response units in the phosphoenolpyruvate carboxykinase gene promoter. Mol. Endocrinol. 1998;12:1487–1498. doi: 10.1210/mend.12.10.0187. [DOI] [PubMed] [Google Scholar]
- 10.Yamada K., Duong D. T., Scott D. K., Wang J. C., Granner D. K. CCAAT/Enhancer-binding Protein β is an accessory factor for the glucocorticoid response from the cAMP response element in the rat phosphoenolpyruvate carboxykinase gene promoter. J. Biol. Chem. 1999;274:5880–5887. doi: 10.1074/jbc.274.9.5880. [DOI] [PubMed] [Google Scholar]
- 11.Liu J. S., Park E. A., Gurney A. L., Roesler W. J., Hanson R. W. Cyclic AMP induction of phosphoenolpyruvate carboxykinase (GTP) gene transcription is mediated by multiple promoter elements. J. Biol. Chem. 1991;266:19095–19102. [PubMed] [Google Scholar]
- 12.Cheong J., Coligan J. E., Shuman J. D. Activating transcription factor-2 regulates phosphoenolpyruvate carboxykinase transcription through a stress-inducible mitogen-activated protein kinase pathway. J. Biol. Chem. 1998;273:22714–22718. doi: 10.1074/jbc.273.35.22714. [DOI] [PubMed] [Google Scholar]
- 13.Choi J. H., Park M. J., Kim K. W., Choi Y. H., Park S. H., An W. G., Yang U. S., Cheong J. Molecular mechanism of hypoxia-mediated hepatic gluconeogenesis by transcriptional regulation. FEBS Lett. 2005;579:2795–2801. doi: 10.1016/j.febslet.2005.03.097. [DOI] [PubMed] [Google Scholar]
- 14.Herzog B., Cardenas J., Hall R. K., Villena J. A., Budge P. J., Giguere V., Granner D. K., Kralli A. Estrogen-related receptor α is a repressor of phosphoenolpyruvate carboxykinase gene transcription. J. Biol. Chem. 2006;281:99–106. doi: 10.1074/jbc.M509276200. [DOI] [PubMed] [Google Scholar]
- 15.Hong S., Park S. J., Kong H. J., Shuman J. D., Cheong J. Functional interaction of bZIP proteins and the large subunit of replication factor C in liver and adipose cells. J. Biol. Chem. 2001;276:28098–28105. doi: 10.1074/jbc.M010912200. [DOI] [PubMed] [Google Scholar]
- 16.Jurado L. A., Song S., Roesler W. J., Park E. A. Conserved amino acids within CCAAT enhancer-binding proteins (C/EBPα and β) regulate phosphoenolpyruvate carboxykinase (PEPCK) gene expression. J. Biol. Chem. 2002;277:27606–27612. doi: 10.1074/jbc.M201429200. [DOI] [PubMed] [Google Scholar]
- 17.Johansson L., Thomsen J. S., Damdimopoulos A. E., Spyrou G., Gustafsson J.Å., Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERα and ERβ. J. Biol. Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 18.Seol W., Choi H. S., Moore D. D. An orphan nuclear hormone receptor that lacks a DNA binding domain and heterodimerizes with other receptors. Science. 1996;272:1336–1339. doi: 10.1126/science.272.5266.1336. [DOI] [PubMed] [Google Scholar]
- 19.Seol W., Hanstein B., Brown M., Moore D. D. Inhibition of estrogen receptor action by the orphan receptor SHP (short heterodimer partner) Mol. Endocrinol. 1998;12:1551–1557. doi: 10.1210/mend.12.10.0184. [DOI] [PubMed] [Google Scholar]
- 20.Williams S. C., Cantwell C. A., Johnson P. F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 21.Crosson S. M., Roesler W. J. Hormonal regulation of the phosphoenolpyruvate carboxykinase gene. Role of specific CCAAT/enhancer-binding protein isoforms. J. Biol. Chem. 2000;275:5804–5809. doi: 10.1074/jbc.275.8.5804. [DOI] [PubMed] [Google Scholar]
- 22.Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987;1:133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- 23.Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 24.Cao Z., Umek R. M., McKnight S. L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 25.MacDougald O. A., Lane M. D. Transcriptional regulation of gene expression during adipocyte differentiation. Annu. Rev. Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 26.Shuman J. D., Cheong J. H., Coligan J. E. ATF-2 and C/EBP α can form a heterodimeric DNA binding complex in vitro: functional implications for transcriptional regulation. J. Biol. Chem. 1997;272:12793–12800. doi: 10.1074/jbc.272.19.12793. [DOI] [PubMed] [Google Scholar]
- 27.Seol W., Chung M., Moore D. D. Novel receptor interaction and repression domains in the orphan receptor SHP. Mol. Cell. Biol. 1997;17:7126–7131. doi: 10.1128/mcb.17.12.7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croniger C., Leahy P., Reshef L., Hanson R. W. C/EBP and the control of phosphoenolpyruvate carboxykinase gene transcription in the liver. J. Biol. Chem. 1998;273:31629–31632. doi: 10.1074/jbc.273.48.31629. [DOI] [PubMed] [Google Scholar]
- 29.Yamagata K., Daitoku H., Shimamoto Y., Matsuzaki H., Hirota K., Ishida J., Fukamizu A. Bile acids regulate gluconeogenic gene expression via small heterodimer partner-mediated repression of hepatocyte nuclear factor 4 and Foxo1. J. Biol. Chem. 2004;279:23158–23165. doi: 10.1074/jbc.M314322200. [DOI] [PubMed] [Google Scholar]
- 30.Borgius L. J., Steffensen K. R., Gustafsson J. A., Treuter E. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J. Biol. Chem. 2002;277:49761–49796. doi: 10.1074/jbc.M205641200. [DOI] [PubMed] [Google Scholar]