Abstract
RNA–protein interactions are central to many aspects of cellular metabolism, cell differentiation, and development as well as the replication of infectious pathogens. We have devised a versatile, broadly applicable in vivo system for the analysis of RNA–protein interactions in yeast. TRAP (translational repression assay procedure) is based on the translational repression of a reporter mRNA encoding green fluorescent protein by an RNA-binding protein for which a cognate binding site has been introduced into the 5′ untranslated region. Because protein binding to the 5′ untranslated region can sterically inhibit ribosome association, expression of the cognate binding protein causes significant reduction in the levels of green fluorescent protein fluorescence. By using RNA–protein interactions with affinities in the micromolar to nanomolar range, we demonstrate the specificity of TRAP as well as its ability to recover the cDNA encoding a specific RNA-binding protein, which has been diluted 500,000-fold with unrelated cDNAs, by using fluorescence-activated cell sorting. We suggest that TRAP offers a strategy to clone RNA-binding proteins for which little else than the binding site is known, to delineate RNA sequence requirements for protein binding as well as the protein domains required for RNA binding, and to study effectors of RNA–protein interactions in vivo.
RNA–protein interactions govern a plethora of biological processes and programs as diverse as early development, sexual differentiation, human intelligence, and viral replication (1). In many cases, RNA-binding proteins exert their functions in cells or tissues that are not readily amenable to biochemical analysis, such as specific areas of the brain or in the germ line. The paucity of biochemical material imposes cumbersome limitations to the identification and cloning of biologically important RNA-binding proteins operating in such systems, particularly when confounded by a lack of possible genetic approaches.
To circumvent such limitations, alternative strategies to identify and study RNA–protein interactions have been devised, based on phage display (2), transcription termination (3), or translation (4) in Escherichia coli, or on transcription initiation in yeast (5, 6). We have developed an approach to study RNA–protein interactions in the cytoplasm of the unicellular eukaryote Saccharomyces cerevisiae. This approach is based on the realization that protein binding to specific sites near the 5′ end of an eukaryotic mRNA causes its translation to be repressed both in mammalian cells and in yeast (7–9). Because of its steric nature, translational repression appears not to be restricted by the physiological function of the RNA-binding proteins. We will refer to this approach as TRAP (for translational repression assay procedure). By using three different well-characterized RNA–protein interactions, the binding of the spliceosomal protein U1A to loop 2 of U1 snRNA (10), the binding of iron regulatory protein (IRP)-1 to iron-responsive elements (IREs) (11), and the the binding of bacteriophage MS2 coat protein to the MS2 replicase mRNA (12, 13), we demonstrate that TRAP allows the rapid and specific identification of RNA–protein interactions in vivo and cloning of cDNAs encoding a specific, cognate RNA-binding protein.
MATERIALS AND METHODS
Plasmid Construction.
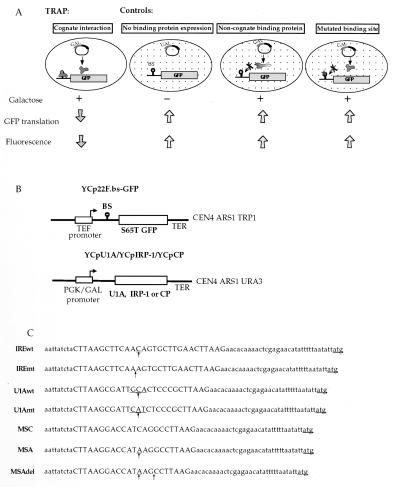
Green fluorescent protein (GFP) indicator plasmids YCp22F.bs-GFP are driven by the TEF1 (translation elongation factor) promoter and contain the TRP1 selection marker. For the construction, the corresponding YCp22FL plasmids (8, 14) were digested with NdeI, the NdeI cohesive ends were filled in by using the Klenow fragment of E. coli DNA polymerase I, before an additional digestion with XbaI and gel purification using QIAquick columns (Qiagen). The Ser-65->Thr mutant GFP coding sequence was amplified by PCR (Boehringer Mannheim PCR Master kit) with primers 5′-TTTTTAATTATGAGCAAAGGAGAAGAAGAACTTTTC-3′ and 5′-TTTTTTCTAGATTATTTGTATAGTTCATCCATG-3′, digested with XbaI, and then introduced into the NdeI blunted-XbaI plasmids to create plasmids YCp22F.IREwt-GFP, YCp22F.IREmt-GFP, YCp22F.U1Awt-GFP, YCp22F.U1Amt-GFP, YCp22F.MSC-GFP, YCp22F.MSA-GFP, and YCp22F.MSAdel-GFP.
Plasmids YCpIRP-1 [described as YCpIRF by Oliveira et al. (14)], YCpU1A (8), and YCpCP (8) express IRP-1, U1A, and MS2 coat protein, respectively, under the control of PGK/GAL fusion promoter and contain URA3 as a selection marker.
Yeast Culture Conditions and Preparation of Protein Extracts.
The S. cerevisiae haploid strain RS453 (ade 2–1, trp 1–1, leu 2–3, his 3–11, ura 3–52, lys+, can 1–100) was grown at 30°C in YPD medium (1% peptone/1% yeast extract/2% glucose) or YPD-agar plates. The yeast selective defined media contained 0.67% (wt/vol) yeast nitrogen base without amino acids, 2% glucose or 2% galactose, amino acids and nucleotides as follows: adenine 20 mg/liter, arginine 20 mg/liter, histidine 20 mg/liter, leucine 60 mg/liter, tryptophan 20 mg/liter, and uracil 20 mg/liter. Solid defined media preparation followed exactly the protocol above with the addition of 2% agar.
Yeast transformation was performed by the lithium acetate method (15). For the mixed plasmid transformation, YCpIRP-1 and YCpU1A were mixed in 1:500,000 ratio (DNA concentration was estimated by measuring the absorption at 260 nm and by ethidium bromide staining). The mixed plasmids were used to transform RS453 cells carrying plasmid YCp22F.IREwt-GFP. Transformants were selected on plates lacking tryptophan and uracil. Approximately 3 × 106 transformants were pooled in liquid selective medium and used for sorting the cells expressing IRP-1, as described below.
For induction of binding protein expression, cultures were grown overnight in selective defined media containing 2% galactose. Yeast protein extracts were prepared as follows: yeast cultures of 10–20 ml were harvested by centrifugation. The pellet was washed once with 10 ml of sterile distilled water and resuspended in 1 ml of yeast lysis buffer (100 mM NaCl/50 mM Tris⋅HCl, pH 7.4). The cell suspension was briefly centrifuged again and resuspended in 150 μl of lysis buffer containing 1 mM phenylmethylsulfonyl fluoride, 10 mg/ml leupeptin, and 1 mM DTT. Cells were disrupted with glass beads (0.45–0.50 mm) at 4°C by three cycles of 1-min vortexing and 1-min incubation on ice, followed by centrifugation for 2 min at 6,000 g in a microfuge. The supernatant was centrifuged again for 5 min at 1,500 g, transferred into a fresh tube and stored at −80°C. Protein concentration of the extracts was determined by using the Bio-Rad protein assay.
Gel Retardation Assays.
32P-labeled IRE and U1snRNA loop 2 probes (specific activity ≈2.9 × 107 cpm/mg RNA) were synthesized in vitro from XbaI-linearized plasmids IREwt-CAT (16) and U1Awt-CAT (7) with T7 RNA polymerase (Stratagene). Aliquots (10–40 mg each) of yeast extracts were incubated for 20 min at room temperature for IRP-1 or ice for U1A with 1 ng of probe in 10 ml of lysis buffer. Subsequently, samples were incubated for 10 min with heparin (0.5 mg/ml) and analyzed by nondenaturing gel electrophoresis (17).
Fluorescence-Activated Cell Analysis and Sorting.
A FACScan (Becton Dickinson) flow cytometer was used to analyze the fluorescence levels of cells. The FACScan uses an argon ion laser fixed at 488 nm for excitation and a 530-nm bandpass filter for collection of the emitted fluorescence. Measurements were made of the cell size, internal complexity, and relative fluorescence intensity of single cells. Debris were excluded from the analysis by gating on the forward scatter versus side scatter parameters.
For sorting, the cells were analyzed and isolated on a FACS Vantage (Becton Dickinson) cell sorter. Excitation was at 488 nm, and the emitted fluorescence was collected with a 530-nm bandpass filter. Sorting gates were set by using forward scatter width versus side scatter height signals, to exclude debris and clumps, and forward scatter height versus fluorescence height signal, to sort cells that fell within a certain fluorescence intensity.
RESULTS
The Principle of TRAP.
Fig. 1A outlines the functional principle of TRAP: RS453 cells (genotype ade 2–1, trp 1–1, leu 2–3, his 3–11, ura 3–52, lys+, can 1–100) are transformed with plasmids for the expression of the RNA-binding protein (or a cDNA expression library) from a galactose-inducible promoter (that is repressed in the presence of glucose), and the GFP indicator (18) bearing the protein binding site in the 5′ untranslated region (UTR) of the encoded mRNA from a constitutively active promoter. Both plasmids are centromeric and maintained at 1–2 copies per cell. In the presence of glucose, GFP is expressed and yields high fluorescence levels. Replacement of glucose by galactose in the growth medium activates the PGK/GAL promoter, and the expression of the RNA-binding protein is induced. As a consequence, GFP expression is translationally inhibited, and the fluorescence level of the cells bearing the cDNA expression vector for the cognate binding protein is diminished. This effect can be quantitated by flow cytometry, cells displaying different GFP levels can be separated by fluorescence activated cell sorting (FACS), and viable cells recovered for additional analysis or cycles of sorting.
Figure 1.
Schematic representation of TRAP. (A) The principle of TRAP. RS453 cells are transformed with the GFP indicator plasmid that carries within the GFP 5′UTR the binding site of the protein under study and the plasmid for expression of the RNA-binding protein (or a cDNA library) under the control of a galactose-inducible promoter. TRAP: In galactose medium, expression of the RNA-binding protein is induced. The RNA–protein interaction at the 5′ end of the GFP mRNA represses its translation and the fluorescence of the cell is diminished (cognate interaction). Controls: GFP mRNA translation yields high fluorescence levels when expression of the RNA-binding protein is repressed in glucose medium (no binding protein expression), or when the expressed protein cannot interact with the GFP mRNA (noncognate binding protein or mutated binding site). (B) Description of the plasmids used. The GFP reporter plasmids YCp22F.BS-GFP contain the GFP S65T mutant ORF. The IRE and U1 snRNA loop 2 wild type and mutated, as well as the MSC, MSA, and MSAdel binding sites (BS) are cloned into the AflII site. The binding sites are located 9 nucleotides downstream from the transcription start site and 32 nucleotides upstream from the GFP translation initiation codon. The plasmids harbor a TRP1 selection marker. YCpU1A, YCpIRP-1, and YCpCP are used for the galactose-inducible expression of U1A, IRP-1, and MS2 coat protein (CP), respectively. They contain a URA3 selection marker. (C) Sequences and nomenclature of the GFP reporter plasmids. Binding site insertions are represented by capital letters. Differences between wild-type and mutated binding sites are indicated by arrows.
Specificity of TRAP.
The specificity of TRAP was challenged with three well-characterized RNA–protein interactions: IRE/IRP-1, U1 snRNA loop 2/U1A, and the binding of bacteriophage MS2 coat protein to the MS2 replicase mRNA. The coding sequence for the S65T mutant GFP, which displays improved excitation/emission characteristics for fluorometry and FACS (19, 20), was subcloned into reporter gene plasmids to generate YCp22F.IREwt-GFP, YCp22F.IREmt-GFP, YCp22F.U1Awt-GFP, and YCp22F.U1Amt-GFP (Fig. 1 B and C). Yeast cells were cotransformed with one of these GFP reporter plasmids and either YCpIRP-1 or YCpU1A (Fig. 1B). Double transformants were selected on plates lacking tryptophan and uracil, grown in selective liquid medium containing either 2% glucose or 2% galactose for 14 hr to a density of OD600 0.5–1, and GFP fluorescence was analyzed by flow cytometry (Fig. 2). Cells grown in glucose and expressing GFP from YCp22F.bs-GFP plasmids display high fluorescence compared with control cells lacking a GFP reporter plasmid (Fig. 2 a and d). Induction of the cognate RNA-binding proteins in galactose-containing media significantly decreases the mean fluorescence, manifested by a leftward shift of the fluorescence distribution curve toward lower values (Fig. 2 b and e) and the repression of the mean fluorescence of cells (Table 1). This reduction specifically requires the presence of the wild-type binding site and the cognate binding protein and is not seen with either the mutant binding sites (Fig. 2 c and f) or the noncognate binding proteins (Fig. 2 a and d).
Figure 2.
Fluorescence of GFP-expressing cells is specifically reduced by cognate RNA–protein interactions. Cells cotransformed with a GFP indicator plasmid and either plasmid YCpIRP-1 or YCpU1A were grown in glucose or galactose containing medium. The fluorescence of cells grown in glucose (green curves) or galactose (red curves) media was analyzed by flow cytometry. Control cells not carrying a GFP plasmid are represented by the black curves in a and d. Fluorescence drops when expression of the cognate binding protein (b and e) is induced by the addition of galactose. Expression of a noncognate binding protein (a and d) does not affect fluorescence levels. No change of fluorescence is observed with a mutated binding site (c and f) present within the 5′UTR of GFP mRNA. The fluorescence intensity is plotted against counts (number of cells analyzed per channel of fluorescence).
Table 1.
Quantitative analysis of the repression of GFP fluorescence by RNA–protein interactions with different binding affinities
| Interaction | Approx. affinity (references)* | GFP repression† |
|---|---|---|
| IRE/IRP-1 | 1.0–10 nM (21–23) | ∼60–70% |
| U1 loop 2/U1A | 0.02–80 nM (24–26) | ∼40% |
| MSC/MS2 coat protein | 0.02–0.1 nM (12, 13) | ∼70% |
| MSA/MS2 coat protein | 0.1–1 mM (13) | ∼50% |
Binding affinities for the different RNA–protein interactions as reported.
GFP fluorescence of cells grown in glucose media was defined as 100%. The remaining mean fluorescence of cells after growth in galactose media was expressed as a percentage of the above, and the repression level calculated by substraction from 100%.
Whereas the IRE/IRP-1 and the U1 snRNA loop 2/U1A interactions display binding affinities in the low nanomolar range (refs. 21–26, Table 1), many specific and physiologically relevant RNA–protein interactions display lower binding affinities. To evaluate the utility of TRAP to study such lower binding affinity interactions by using a well-characterized example, defined binding sites for the MS2 coat protein (MS2 CP) in the subnanomolar [MSC, (12, 13)] and low micromolar [MSA, 13)] range were introduced into the GFP reporter plasmid and compared with a negative control (plasmid YC22F.MSAdel-GFP, Fig. 1 B and C). Yeast cells were cotransformed with one of the three GFP reporter plasmids and the MS2 coat protein expression plasmid YCpCP (Fig. 1B). Induction of MS2 CP expression in galactose-containing media resulted in specific repression of GFP expression (Table 1), whereas the MSAdel construct was unaffected (data not shown). Importantly, the lower affinity MSA/MS2 CP interaction caused a 50% repression of GFP fluorescence, which even exceeded the effect caused by the higher affinity U1 loop 2/U1A interaction (see Discussion). We conclude that TRAP allows study of specific RNA–protein interactions with affinities spanning a broad physiological range. Interestingly, for a given RNA-binding protein the degree of repression appears to reflect its binding affinity although different RNA–protein interactions cannot be quantitatively compared, presumably because additional parameters affect the degree of repression (see Discussion).
Isolation of Cells Expressing Cognate RNA-Binding Proteins by FACS.
The specific reduction of GFP fluorescence caused by the interaction of cognate RNA-binding proteins with their wild-type binding sites introduced into the 5′ UTR of GFP reporter mRNAs (Fig. 2) suggests the possibility to recover cells expressing the cognate RNA binding protein from a background pool of cells expressing control proteins by FACS. Cells cotransformed with plasmid YCp22F.IREwt-GFP and either YCpIRP-1 or YCpU1A were mixed at a ratio of 1:10,000. The mixed cell suspension was grown in galactose-containing medium to induce the expression of the RNA-binding proteins and sorted by FACS according to their GFP fluorescence. Cells displaying low fluorescence levels were regrown in galactose medium and sorted for a second time.
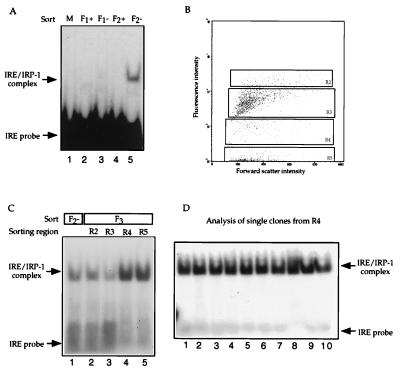
The enrichment for cells carrying plasmid YCpIRP-1 was tested in a gel retardation assay for IRE-binding activity by using extracts prepared from sorted cells (Fig. 3A). After two rounds of sorting (F2), the extracts from galactose-induced cultures of cells selected for low fluorescence levels (Fig. 3A, F2−, lane 5) display IRE-binding activity comparable to that of an extract of IRP-1 expressing cells and U1A expressing cells mixed at a ratio of 1:100 (not shown). No binding activity was detectable in the initial 1:10,000 starting population (Fig. 3A, M, lane 1), in extracts from cells selected for high fluorescence levels (Fig. 3A, +, lanes 2 and 4), or sorted only once (Fig. 3A, F1−, lane 3). The F2− cells subsequently were regrown and sorted again (F3) into four different pools of descending fluorescence (R2-R5, Fig. 3B). R2-R5 extracts were compared in a gel retardation assay to the F2 extracts (Fig. 3C). A substantial further enrichment in IRE-binding activity is apparent in cell populations exhibiting low fluorescence, R4 (Fig. 3C, lane 4) and R5 (Fig. 3C, lane 5). Cells from culture R4 were plated on selective medium, single colonies picked and extracts made from cultures of these clones tested for IRP-1 expression (Fig. 3D). Ten of 10 clones tested from R4 expressed IRP-1 (Fig. 3D). Thus, starting from 1:10,000 (0.01%) cells carrying plasmid YCpIRP-1, the IRP-1 expressing cells represented 100% of the cells analyzed from F3/R4.
Figure 3.
IRP-1 expressing cells can be recovered from a mixture with U1A expressing cells by TRAP. (A) Cells cotransformed with plasmid YCp22F-IREwt.GFP and either YCpIRP-1 or YCpU1A were mixed in a ratio of 1:104 and grown overnight in galactose-containing medium (M). Cells displaying high (F1+) or low fluorescence levels (F1−) were separated by FACS. Low fluorescent cells were regrown and sorted for a second time (F2+ and F2−). Total cell extracts were incubated with 1 ng of IRE probe and analyzed by native PAGE. (B) Dot plot profile of cells from F2−. Cells were analyzed by flow cytometry, plotted according to forward scatter (size) vs. fluorescence intensity and sorted for a third time (F3) to regions R2-R5 according to their fluorescence. (C) Extracts from cells sorted twice (F2−, lane 1) and three times (F3, lanes 2–5) were incubated with 1 ng of IRE probe and analyzed by native PAGE. (D) Extracts from single cell clones of culture R4 were analyzed in a gel retardation assay (lanes 1–10).
We next performed an analogous experiment for the U1A-encoding plasmid with YCp22F.U1Awt-GFP, by using a 10,000-fold excess of YCpIRP-1 as a background (Fig. 4 A–D). After two rounds of initial sorting (F1, F2, Fig. 4A), the F2− cells were regrown and sorted into regions (F3, R2-R5, Fig. 4B). As before, the strongest enrichment was observed in R4, but there was substantially less U1A activity in R3 and R5 (Fig. 4C). Single colony analysis of F3/R4 revealed successful cloning of YCpU1A in eight of 10 samples tested (Fig. 4D). Taken together, the results obtained for IRP-1 and U1A underscore the potential of TRAP to be used for the identification and recovery of cells expressing an RNA-binding protein from a population of cells expressing unrelated proteins.
Figure 4.
U1A expressing cells can be recovered from a mixture with IRP-1 expressing cells by TRAP. (A) Cells cotransformed with plasmid YCp22F-U1Awt.GFP and either YCpU1A or YCpIRP-1 were mixed in 1:104 ratio and grown overnight in galactose-containing medium (M). Cells displaying low fluorescence levels (F1−) were isolated by FACS, regrown, and sorted for a second time (F2−). Total cell extracts were incubated with 1 ng of U1A probe and analyzed by native PAGE. (B) Dot plot profile of cells from F2−. Cells were analyzed by flow cytometry, plotted according to forward scatter (size) vs. fluorescence intensity and sorted for a third time (F3) into regions R2-R4 according to their fluorescence. (C) Extracts from cells sorted twice (F2−, lane 1) and three times (F3, lanes 2–5) were incubated with 1 ng of U1A probe and analyzed by native PAGE. (D) Extracts from single cell clones of culture R4 were analyzed in a gel retardation assay (lanes 1–10).
Cloning by TRAP.
To further challenge the utility of TRAP for cloning, we attempted to reclone the IRP-1 cDNA. To mimic the cloning process with a precisely preset required enrichment factor, we mixed the plasmids YCpIRP-1 and YCpU1A in a 1:500,000 ratio and transformed RS453 cells carrying YCp22F.IREwt-GFP. Double transformants were selected on plates lacking tryptophan and uracil, ≈3 × 106 transformants were pooled, expanded in selective medium containing galactose to induce expression of the binding proteins, and sorted by FACS. After two rounds of sorting, cells from culture F2− showed IRE-binding activity (Fig. 5A, lane 3). F2− cells were further sorted into regions of cells with different fluorescence (Fig. 5B), and the greatest further enrichment was again found in R4 (Fig. 5A, lane 6). When single colonies from F3/R4 were analyzed, 1 of 20 expressed IRP-1 (not shown). Thus, TRAP allowed successful recloning of the cDNA encoding a cognate RNA-binding protein from a 500,000-fold background.
Figure 5.
Cloning of IRP-1 by TRAP. (A) Recovery of IRP-1 expressing cells from a transformation with mixed plasmid DNA. RS453 cells carrying plasmid YCpIREwt-GFP were transformed with a DNA mixture containing plasmids YCpIRP-1 and YCpU1A in a ratio of 1:500,000. Transformants were pooled and grown in galactose medium. Cells with low fluorescence were sequentially sorted twice. Total extracts were analyzed in a gel retardation assay with 1 ng of IRE probe (lanes 1–3). The extract of cells sorted twice shows IRE binding activity (lane 3). IRE binding activity is below detection limit in the extract from pooled transformants (lane 1), or cells sorted once (lane 2). Cells from F2− were sorted for a third time (F3) into populations R2-R5. The IRE binding activity of extracts from cultures R2-R5 was analyzed (lanes 4–7). The highest activity is present in extract R4. (B) Fluorescence analysis and sorting of cells from F2− into regions R2-R4 according to their fluorescence.
DISCUSSION
The observation that RNA–binding proteins can sterically repress translation from binding sites introduced close to the 5′ end of a mRNA has paved the way for the development of TRAP. The physiological principle (7, 8) appears to apply to organisms as diverse as mammals and yeast, and not to be limited by the origin or physiological functions of the RNA-binding proteins under study. Our demonstration that a cDNA can be recloned from a 0.5 million-fold background (Fig. 5) indicates the potential of TRAP for the cloning of RNA-binding proteins for which little other than the binding site needs to be known. Because we have not attempted recloning from a higher than 0.5 million-fold excess of unrelated plasmids, we have no reason to expect this figure to reflect the upper limit.
The specific features of TRAP suggest that it complements the repertoire of recently reported strategies for studying RNA-binding proteins (2–6) in several useful ways: it represents a system that combines the use of a eukaryotic host cell with the advantages implicit in assaying cytoplasmic (rather than nuclear) expression and the possibilities afforded by successive rounds of enrichment. TRAP allows the study of RNA-binding proteins in their native form without having to fuse additional protein domains that might interfere with their RNA-binding affinity or specificity. Although TRAP detects the RNA-binding proteins in the cytoplasm, it also was used successfully with the nuclear protein U1A (Figs. 2 and 4), probably because sufficient amounts of newly synthesized, overexpressed U1A remain cytoplasmic to affect the translation of the GFP indicator mRNA. To clone or study RNA binding proteins with more potent nuclear import signals, such signals could be mutated (if known) or the expression library modified by using a vector that directs the expression of fusion proteins with the powerful leucine-rich nuclear export signal of PKI (27). TRAP only requires two components: the RNA-binding protein (or a suitable cDNA expression library) and the indicator mRNA. This simplicity minimizes the potential for inadvertent nonspecific effects on the assay, a consideration that may be particularly relevant when studying pharmacological effectors of RNA-protein interactions. Furthermore, the utilization of FACS permits the rapid processing of large numbers of independent clones and the recovery of living cells after sorting, thus allowing for multiple rounds of sorting while monitoring enrichment. The highest level of specific enrichment for both U1A and IRP-1 was found in the R4 fractions, i.e., cells displaying reduced but not minimal fluorescence. The minimally fluorescent population (R5) is possibly contaminated by cells that may harbor mutations in the GFP cDNA or may have lost the GFP-expressing plasmid, even during culture in selective medium. However, the design of TRAP allows counterselection against nonspecific, constitutive loss of GFP fluorescence, because cells displaying a specific reduction in fluorescence specifically shift to and can be recovered from the high fluorescent pool following incubation in glucose-containing media (data not shown).
What are the current limitations of TRAP? The affinities of the RNA–protein interactions tested have been reported to be approximately 1.0–10 nM for IRE/IRP-1 (21–23), 0.02–80nM for U1 loop 2/U1A (24–26), 0.02–0.1 nM for MSC/MS2 coat protein (12, 13), and 0.1–1.0 μM for MSA/MS2 coat protein (13) interactions. As shown in Table 1 and Fig. 2, the affinities of the interaction of IRP-1 and U1A with the specificity controls YCp22F.IREmt-GFP and YCp22F.U1Amt-GFP, respectively, and of MS2 coat protein for the MSAdel mutant (Fig. 1C and data not shown) are not sufficient to cause diminished GFP fluorescence. The minimally required affinity is currently unknown. It certainly will be subject to case-specific parameters such as the stability of the corresponding RNA-binding protein and the maximal level of its overexpression in yeast, its distribution between different subcellular compartments, or possible complexation with other proteins. Such effects probably account for the lower degree of GFP repression by the high affinity U1 loop 2/U1A interaction compared with the lower affinity MSA/MS2 CP interaction (Table 1). Although TRAP thus appears suitable to study specific RNA–protein interactions in a broad, physiological range, our findings suggest that nonspecific and/or low affinity RNA–protein interactions do not affect the performance of TRAP. This consideration is supported by the lack of translational repression of IREmt, U1Amt, and MSAdel binding site controls as well as the ability to observe a specific interaction with an affinity in the low micromolar range in a cytoplasmic compartment replete with nonspecific RNA binding proteins. Because the three proteins tested here bind to hairpin structures, we currently cannot conclude that proteins binding to less structured RNA motifs will perform equally well. Like the other strategies to detect RNA–protein interactions (2–6), TRAP requires that the protein under investigation be able to bind RNA as a monomer or homopolymer. The specific principle of TRAP also requires the introduction of the RNA-binding region into the 5′ UTR not to interfere with GFP expression per se. Therefore, binding regions that are exceedingly highly structured or harbor inhibitory ORFs may not represent suitable target sequences. Ideally, the binding region should be defined to 100 nucleotides or less, to minimize the probability of introducing inhibitory features into the 5′ UTR, and also to increase the probability of introducing the actual binding site close to the 5′ end of the mRNA, a region that allows maximal translational repression at least in mammalian cells (7, 28, 29). However, a recent analysis of the IRE/IRP-1 interaction in yeast has suggested that 5′ end proximity of the RNA binding site may not be a required feature for translational repression in S. cerevisiae (30).
In summary, TRAP offers a versatile approach for studying RNA–protein interaction pairs in yeast. The principle of TRAP should be adaptable to other eukaryotes, including mammalian cells. In addition to cloning novel RNA-binding proteins, we envisage its utility to demonstrate specific interactions between a protein and an RNA sequence in vivo, to evaluate the impact of mutations of an RNA-binding protein and/or the RNA-binding site on the interaction, and to monitor the effect of pharmacological agents on known RNA–protein interactions.
Acknowledgments
We gratefully acknowledge Graham Smith for his help with some initial cell sorting experiments and Kostas Pantopoulos for supplying IRE probe. We thank Bertrand Seraphin and George Simos for helpful discussions and suggestions, and Anne Ephrussi, Juan Valcarcel, and the members of the Hentze group for comments on the manuscript. M.W.H. acknowledges support from the European Commission Biotechnology Program (BIO-CT95-0045). E.P. was the recipient of a Human Capital and Mobility fellowship from the European Commission.
ABBREVIATIONS
- TRAP
translational repression assay procedure
- GFP
green fluorescent protein
- UTR
untranslated region
- FACS
fluorescence activated cell sorting
- IRP
iron regulatory protein
- IRE
iron responsive element
- CP
coat protein
References
- 1.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 2.Laird-Offringa I A, Belasco J G. Proc Natl Acad Sci USA. 1995;92:11859–11863. doi: 10.1073/pnas.92.25.11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada K, Martin S S, Frankel A D. Nature (London) 1996;380:175–179. doi: 10.1038/380175a0. [DOI] [PubMed] [Google Scholar]
- 4.Jain C, Belasco J G. Cell. 1996;87:115–125. doi: 10.1016/s0092-8674(00)81328-8. [DOI] [PubMed] [Google Scholar]
- 5.SenGupta D J, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Putz U, Skehel P, Kuhl D. Nucleic Acids Res. 1996;24:4838–4840. doi: 10.1093/nar/24.23.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stripecke R, Hentze M W. Nucleic Acids Res. 1992;20:5555–5564. doi: 10.1093/nar/20.21.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stripecke R, Oliveira C C, McCarthy J E G, Hentze M W. Mol Cell Biol. 1994;14:5898–5909. doi: 10.1128/mcb.14.9.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray N K, Hentze M W. EMBO J. 1994;13:3882–3891. doi: 10.1002/j.1460-2075.1994.tb06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scherly D, Boelens W, van Venrooij W J, Dathan N A, Hamm J, Mattaj I W. EMBO J. 1989;8:4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentze M W, Kühn L C. Proc Natl Acad Sci USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowary P T, Uhlenbeck O C. Nucleic Acids Res. 1987;15:10483–10493. doi: 10.1093/nar/15.24.10483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witherell G W, Gott J M, Uhlenbeck O C. Progr Nucleic Acids Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 14.Oliveira C C, Goossen B, Zanchin N I T, McCarthy J E G, Hentze M W, Stripecke R. Nucleic Acids Res. 1993;21:5316–5322. doi: 10.1093/nar/21.23.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray N K, Quick S, Goossen B, Constable A, Hirling H, Kühn L C, Hentze M W. Eur J Biochem. 1993;218:657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- 17.Leibold E A, Munro H N. Proc Natl Acad Sci USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 19.Heim R, Cubitt A B, Tsien R Y. Nature (London) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 20.Cubit A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 21.Leibold E A, Laudano A, Yu Y. Nucleic Acids Res. 1990;18:1819–1824. doi: 10.1093/nar/18.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettany A J E, Eisenstein R S, Munro H N. J Biol Chem. 1992;267:16531–16537. [PubMed] [Google Scholar]
- 23.Henderson B R, Menotti E, Bonnard C, Kühn L C. J Biol Chem. 1994;269:17481–17489. [PubMed] [Google Scholar]
- 24.Lutz-Freyermuth C, Query C C, Keene J D. Proc Natl Acad Sci USA. 1990;87:6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jessen T-H, Oubridge C, Hiang Teo C, Pritchard C, Nagai K. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stump W T, Hall B K. RNA. 1995;1:55–63. [PMC free article] [PubMed] [Google Scholar]
- 27.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 28.Goossen B, Caughman S W, Harford J B, Klausner R D, Hentze M W. EMBO J. 1990;9:4127–4133. doi: 10.1002/j.1460-2075.1990.tb07635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goossen B, Hentze M W. Mol Cell Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koloteva N, Mueller P P, McCarthy J E G. J Biol Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]