Abstract
In the absence of Spo13, budding yeast cells complete a single meiotic division during which sister chromatids often separate. We investigated the function of Spo13 by following chromosomes tagged with green fluorescent protein. The occurrence of a single division in spo13Δ homozygous diploids depends on the spindle checkpoint. Eliminating the checkpoint accelerates meiosis I in spo13Δ cells and allows them to undergo two divisions in which sister chromatids often separate in meiosis I and segregate randomly in meiosis II. Overexpression of Spo13 and the meiosis-specific cohesin Rec8 in mitotic cells prevents separation of sister chromatids despite destruction of Pds1 and activation of Esp1. This phenotype depends on the combined overexpression of both proteins and mimics one aspect of meiosis I chromosome behavior. Overexpressing the mitotic cohesin, Scc1/Mcd1, does not substitute for Rec8, suggesting that the combined actions of Spo13 and Rec8 are important for preventing sister centromere separation in meiosis I.
Keywords: Centromere cohesion, meiosis I, homolog segregation, Spo13, Rec8, yeast
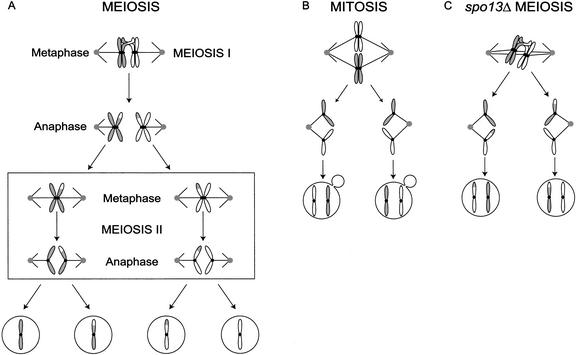
The meiotic pattern of chromosome segregation is created by differential regulation of the linkage between sister chromatid arms and centromeres (Fig. 1A,B). In meiosis I, homologous chromosomes are attached to one another by chiasmata, which are stabilized by sister chromatid cohesion. At anaphase I, homologs segregate away from each other when cohesion is lost between sister chromatid arms, but sister centromeres remain attached to each other because cohesion is maintained here (Lee and Orr-Weaver 2001; Nasmyth 2001). Kinetochore structure is critical in meiosis I, because both sister kinetochores must attach to the same spindle pole.
Figure 1.
Diagram of wild-type meiosis and mitosis, and meiosis in spo13Δ. (A) In meiosis I, homologous chromosomes are held together by chiasmata and segregate away from one another when cohesion is released from sister chromatid arms. In meiosis II, sister chromatids segregate away from one another when cohesion is released from sister centromeres. (B) Mitosis is a single division in which sister chromatids segregate from each other and cohesion is lost simultaneously at the arms and centromeres. (C) In spo13Δ mutants, a single meiotic division occurs in which sister chromatids segregate from each other as cohesion is lost simultaneously from arms and centromeres.
In both meiosis and mitosis, a multisubunit complex called cohesin mediates sister chromatid cohesion (Nasmyth 2001). The mitotic and meiotic complexes differ by at least one subunit, as the mitotic Scc1/Mcd1 protein (Guacci et al. 1997; Michaelis et al. 1997) is replaced by Rec8 in meiosis, which localizes to both sister chromatid arms and centromeres in meiosis I (Klein et al. 1999; Watanabe and Nurse 1999). At anaphase of meiosis I, Rec8 disappears from chromosome arms and homologs disjoin (reductional segregation) (Buonomo et al. 2000), but Rec8 remains bound near sister centromeres, maintaining the linkage between sister centromeres. At anaphase of meiosis II, centromeric Rec8 is removed and sister centromeres segregate from each other (equational segregation) (Klein et al. 1999; Watanabe and Nurse 1999). Replacing meiotic cohesin with its mitotic counterpart deranges meiosis I. Although sister centromeres are linked in metaphase I, Scc1/Mcd1 cannot be protected at sister centromeres and sister centromeres separate from each other as cells enter anaphase I (Toth et al. 2000).
The cell cycle machinery regulates cohesin removal from chromosomes. The anaphase promoting complex (APC), a multisubunit ubiquitin ligase (King et al. 1995; Sudakin et al. 1995), targets the anaphase inhibitor Pds1 (securin) for destruction by the proteasome (Cohen-Fix et al. 1996). Destruction of Pds1 allows its binding partner, separase (Esp1) to cleave Rec8 in meiosis or Scc1/Mcd1 in mitosis, thus destroying the linkage between sister chromatids (Ciosk et al. 1998; Buonomo et al. 2000). Despite an active separase, centromeric Rec8 is protected during meiosis I until meiosis II when separase is reactivated and removes Rec8 at the onset of anaphase I.
Spo13 is a candidate regulator of centromeric cohesion (Klapholz and Esposito 1980b; McCarroll and Esposito 1994). Mutations in SPO12 and SPO13 were identified in a natural yeast isolate that undergoes a single meiotic division, often separates sister centromeres, and produces two viable diploid spores (Klapholz and Esposito 1980a,b). In addition, spo13Δ mutants have much less Rec8 on centromeres during anaphase I, suggesting that Spo13 may regulate Rec8 destruction during anaphase of meiosis I (Klein et al. 1999). These data suggest that the spo13Δ mutant has three defects (Fig. 1C) as follows: Only one meiotic division occurs, sister kinetochores often attach to opposite poles, and sister centromeres often separate from each other in the single division (Klapholz and Esposito 1980; Sharon and Simchen 1990). We have studied these three defects in the spo13Δ mutant. We show that the spindle checkpoint delays anaphase of meiosis I in spo13Δ cells, causing the single division meiosis. To test the hypothesis that Spo13 protects centromeric Rec8 from cleavage by separase, we overexpressed both Rec8 and Spo13 in mitotic cells. In these cells, sister centromeres still attach to opposite spindle poles, but they fail to separate from each other at anaphase, mimicking one aspect of meiosis I chromosome behavior.
Results
Chromosome segregation in spo13Δ
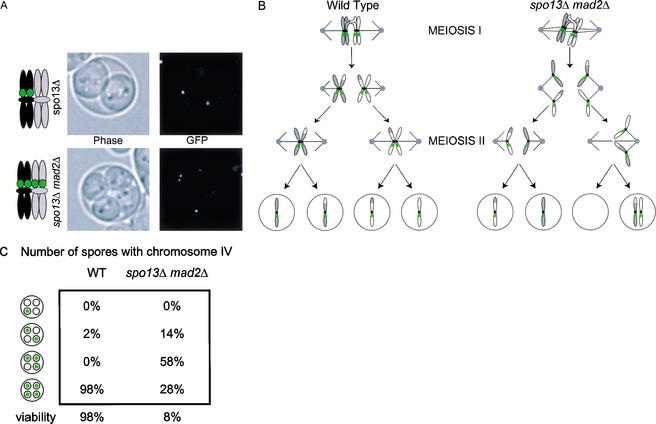
We began by studying chromosome behavior in the single meiotic division that occurs in cells lacking Spo13. Diploids homozygous for the spo13Δ mutation undergo a single meiotic division, forming dyads. Chromosome segregation is mixed, as sister chromatids segregate away from one another in some cells but go to the same pole in others. Previous studies used genetic analysis to follow chromosome segregation in the viable products of meiosis in spo13Δ cells (Klapholz and Esposito 1980b; Hugerat and Simchen 1993). We used a cytological marker that could be scored in both viable and inviable spores; the binding of a GFP–Lac repressor fusion to a repeated array of Lac operators, which produces a microscopically visible, fluorescent dot (Straight et al. 1996). The LacO array was integrated at trp1, 12 kb from the centromere of chromosome IV. In spo13Δ homozygous diploids, only one copy of chromosome IV was marked with the LacO array, therefore, a single GFP dot in each spore of the dyad shows that sister centromeres have segregated away from one another (e.g., see Fig. 4a, below). We tested the segregation of three chromosomes in spo13Δ mutants, as differences in segregation have been reported for different chromosomes (Klapholz and Esposito 1980b; Hugerat and Simchen 1993). In spo13Δ cells, the sister centromeres of chromosome III separate in 55% of divisions, those of chromosome IV separate in 80% of divisions, and those of chromosome VIII separate in 62% of divisions (Table 1). By genetic analysis, Hugerat and colleagues found that the spo13Δ mutant separated sister centromeres of chromosome IV in 66% of divisions and chromosome III in 38% of divisions (Hugerat and Simchen 1993). These results are qualitatively consistent with our findings. Eliminating recombination in spo13Δ cells with the spo11Δ mutation causes all sister chromatids to separate in the first meiotic division (data not shown; Klapholz et al. 1985). Our results confirm the earlier genetic analysis, which could only report on chromosome segregation in meioses that produced two viable spores.
Figure 4.
Two meiotic divisions occur in spo13Δ mad2Δ homozygous diploids. (A) GFP-tagged chromosomes were observed in spores by fluorescence microscopy. (Top panel) Pictures of sister chromatid separation in a dyad formed by spo13Δ homozygous diploids (MAS 314). One copy of chromosome IV is marked with a LacO array located 12 kb from CENIV. (Bottom panel) Pictures of random chromosome segregation in a tetrad formed by spo13Δ mad2Δ homozygous diploid (MAS 356). Both copies of chromosome IV are marked with GFP. (B) Cartoon of chromosome segregation in wild-type meiosis (left) and one of several possible patterns of chromosome segregation in spo13Δ mad2Δ homozygous diploids. Sister chromatids are shown separating to opposite poles at anaphase of meiosis I and then segregating at random in the second meiotic division. (C) Chromosome segregation and viability of wild-type and spo13Δ mad2Δ homozygous diploids. Cells were treated as in A. Viability was determined by picking and germinating spores. Chromosome segregation is close to random in spo13Δ mad2Δ homozygous diploids and 92% of spores are inviable.
Table 1.
Meiosis I sister chromatid separation in spo13Δ cells
| Strain
|
Chromosome
|
% Sister separation in Meiosis I: GFP tag
|
|---|---|---|
| Wild type | IV | 0% |
| spo13Δ/spo13Δ | IV | 80% |
| spo13Δ/spo13Δ | VIII | 62% |
| spo13Δ/spo13Δ | III | 62% |
Sister chromatid separation was followed by marking one of the two homologs with a LacO array and determining the number of spores in the dyads that contained a fluorescent dot caused by the binding of the GFP–LacI fusion to the operator. For wild-type cells, the extent of sister separation in meiosis I was inferred from the segregation of homologs in tetrads. All LacO arrays were located less than 22 kb from their respective centromeres, making recombination between the centromere and the LacO array very unlikely. For each strain, at least 200 meioses were scored.
Bipolar attachment in spo13Δ
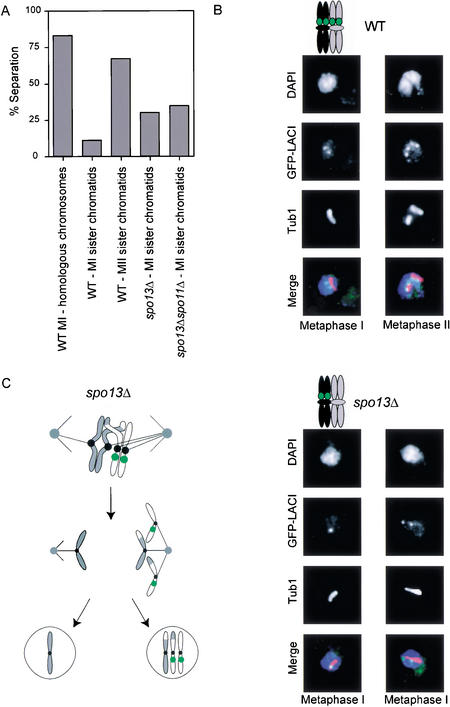
Because chromosome segregation is variable in spo13Δ cells, we wondered whether chromosome attachment to the spindle was normal. Sister chromatids that are attached to opposite spindle poles are bi-oriented, whereas those attached to only one pole are mono-oriented. In mitotic metaphase, cells of animals (Skibbens et al. 1993) and yeast (Goshima and Yanagida 2000), the pulling force on bi-oriented chromosomes produces a visible separation or stretch between sister centromeres, but no separation between sister chromatid arms. We monitored bi-orientation by following the behavior of lactose operator arrays integrated very near the centromere of chromosome VIII. In meiosis, we expect centromere behavior to differ between meiosis I and meiosis II. In metaphase I, the centromeres of homologous chromosomes should be bi-oriented, whereas sister centromeres should be mono-oriented. In meiosis II, sister centromeres should become bi-oriented, as they do in mitosis. Our results confirm these predictions. Wild-type cells with marked centromeres were sporulated, fixed, and indirect immunofluorescence was performed against tubulin, to identify metaphase spindles and GFP to monitor the LacO arrays near centromeres. In wild-type cells, homologous centromeres were stretched apart from each other, and thus bi-oriented in 83% of cells with metaphase I spindles. Sister centromeres were stretched in 11% of metaphase I cells and in 70% of cells with metaphase II spindles (Fig. 2A,B).
Figure 2.
spo13Δ and spo13Δ spo11Δ mutants separate sister centromeres during meiosis. (A) Wild-type (MAS 651), spo13Δ (MAS 875), and spo13Δ spo11Δ (MAS 676) homozygous diploids were sporulated and fixed for indirect immunofluorescence against α tubulin and GFP–LacI binding to a Lac operator array located 2.1 kb from CEN VIII. The percentage of cells with separated homologous centromeres was quantified for wild-type cells in metaphase I. The percentage of cells with separated sister centromeres was quantified for wild-type cells in metaphase of meiosis II. The percentage cells with separated sister centromeres of at least 100 cells in meiosis I was quantified for spo13Δ and spo13Δ spo11Δ. Centromeres were considered separated if two LacI signals could be resolved. (B) Pictures of separated wild-type homologous centromeres in metaphase of meiosis I (left panel, top) and separated sister centromeres in metaphase of meiosis II (right panel, top). Pictures of spo13Δ metaphase I sister centromeres that fail to separate (left panel, bottom) and those that separate (right panel, bottom). (C) Cartoon of spo13Δ mutants illustrating bi-orientation of homologs and sister chromatid separation near the centromere.
We next asked whether there is a defect in bi-orientation of sister centromeres in spo13Δ and spo13Δ spo11Δ homozygous diploids (Fig. 2A,B). In spo13Δ cells, sister centromeres were stretched in 32% of cells, somewhat lower than the 62% of sister chromatids that ultimately separate in anaphase. In spo11Δ spo13Δ double mutants, sister centromeres were stretched in 35% of cells with metaphase I spindles, even though all cells are destined to separate their sister chromatids in anaphase I. Thus, metaphase sister centromere separation in spo13Δ cells is greater than that seen in wild-type cells in meiosis I, but less than that observed for wild-type cells in meiosis II. These results suggest that there are two defects in the interactions between kinetochores and microtubules in cells lacking Spo13 (Fig. 2C). The first defect, predicted from observations of chromosome behavior, is that sister centromeres are more likely to attach to opposite poles in cells lacking Spo13. The second defect is that sister centromeres are less likely to be visibly bi-oriented in cells lacking Spo13 in meiosis I than they are in wild type in meiosis II. This observation suggests that the absence of Spo13 reduces the force acting on sister kinetochores, their probability of binding to microtubules, or both.
The spindle checkpoint delays metaphase I in spo13Δ
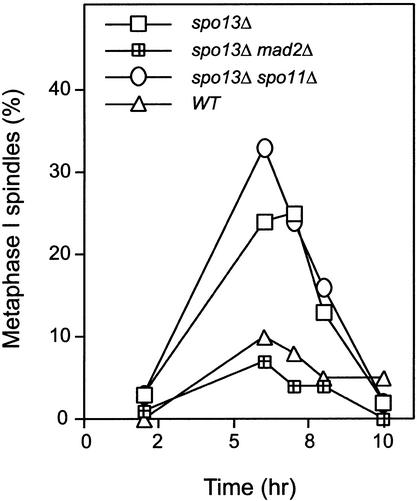
We asked why spo13Δ cells perform only one meiotic division. One possibility is that their first meiotic division is slow and there is no time for a second division before spore formation. We followed meiotic progression in spo13Δ diploids, assessing entry into and exit from meiosis I by the formation and disappearance of metaphase I spindles. Once recombination is complete, short spindles form (Padmore et al. 1991) and are maintained, whereas chromosomes attach to the spindle (prometaphase) and prepare for division (metaphase). Short spindles disappear as cells enter anaphase. Because the synchrony of meiotic divisions is poor, we calculated the duration of meiosis I by integrating the fraction of cells that had short spindles over time. Throughout meiosis I, the fraction of spo13Δ cells with metaphase I spindles was higher than wild type (Fig. 3), and we calculate the average duration of meiosis I to be 2.3 times longer in spo13Δ cells.
Figure 3.
The spindle checkpoint delays spo13Δ mutants in metaphase of meiosis I. (▵) Wild type (MAS118 × MAS119); (□) spo13Δ (MAS278 × MAS279); (○) spo13Δ spo11Δ (MAS 659 × MAS 660); and (│) spo13Δ mad2Δ (MAS442 × MAS443) homozygous diploids were sporulated and fixed for indirect immunofluorescence against α tubulin. The percentage of cells with metaphase spindles was quantified by fluorescence microscopy and graphed against time during sporulation. At least 100 cells were counted for each time point and the experiment was repeated three times with similar results. The spindle checkpoint delays spo13Δ and spo13Δ spo11Δ during metaphase I.
Because the spindle checkpoint is known to arrest cells in prometaphase, we asked whether the metaphase I delay in spo13Δ could be eliminated by removing the checkpoint protein Mad2. In the spo13Δ mad2Δ double mutant, cells proceed through metaphase I at the same rate as in wild type, showing that spo13Δ mutants are delayed by the spindle checkpoint in prometaphase of meiosis I (Fig. 3).
What defect does the spindle checkpoint sense in spo13Δ cells? One possibility is that the mixed homolog and sister segregation in the single meiosis of spo13Δ mutants leaves some kinetochores unattached to microtubules, and these kinetochores trigger the spindle checkpoint. In the absence of recombination and Spo13, sister chromosomes always segregate apart (Klapholz et al. 1985), suggesting that all sister kinetochores attach to the spindle. We tested whether spo13Δ spo11Δ homozygous diploids delay in metaphase I, and find that these cells proceed through metaphase at the same slow rate as spo13Δ cells (Fig. 3), suggesting that mixed homolog and sister chromatid segregation does not trigger the spindle checkpoint. Another possible cause of the spindle checkpoint delay is that microtubule-kinetochore attachments are defective. As shown in Figure 2A, both spo13Δ mutants and spo13Δ spo11Δ double mutants decrease sister centromere stretching as compared with wild-type homologous centromere stretching in metaphase I or wild-type sister centromere stretching in meiosis II. These observations are consistent with the idea that the checkpoint senses either a lack of attachment or tension at kinetochores formed in the absence of Spo13.
Eliminating the spindle checkpoint in spo13Δ allows two divisions
To our surprise, spo13Δ mad2Δ double mutants complete two meiotic divisions and form four spores (Table 2; Fig. 4A). Similar results were obtained when spo13Δ was combined three other checkpoint mutants, mad1Δ, mad3Δ (Li and Murray 1991), and a dominant mutation in CDC20 (Hwang et al. 1998), the target of the spindle checkpoint (data not shown). We suggest that the spindle checkpoint delay causes a single division meiosis in spo13Δ mutants. In spo13Δ mutants, metaphase I is extended to twice its normal length (Fig. 3). As a result of this delay, we suggest that spore formation begins before a second division can occur, effectively cutting off a second division. Our results suggest that although spo13Δ mutants are mechanically capable of two divisions, the spindle checkpoint normally limits them to one.
Table 2.
Formation of tetrads in spo13Δ cells lacking the spindle checkpoint
| Strain
|
% Dyads
|
% Tetrads
|
|---|---|---|
| Wild type | 3 | 97 |
| mad2Δ | 28 | 72 |
| spo13Δ | 100 | 0 |
| spo13Δmad2Δ | 36 | 64 |
Diploid cells were sporulated for 2 d in 2% potassium acetate and the frequency of dyad or tetrad formation was determined by light microscopy. At least 200 asci were counted for each strain.
As spo12Δ mutants have a similar phenotype to spo13Δ mutants, we wondered whether the single division in spo12Δ could also be suppressed by eliminating the spindle checkpoint. We find that spo12Δ mad2Δ double mutants form dyads. In addition, the spo13Δ spo12Δ mad2Δ triple mutant forms dyads (data not shown). What does this analysis suggest about the function of Spo12? It has been proposed that Spo12 is important for completion of two meiotic divisions (Grether and Herskowitz 1999), and our observations are consistent with that conclusion. Our analysis suggests that there are two classes of mutations that cause a single division meiosis, those that prevent events that occur after anaphase such as spo12Δ and those that delay in metaphase of meiosis I, such as spo13Δ.
Undergoing two meiotic divisions is catastrophic for spo13Δ mad2Δ double mutants. Spores formed by spo13Δ mad2Δ double mutants have low viability because they are aneuploid. When sister centromeres separate from each other in meiosis I, as often occurs in the absence of Spo13, the unlinked sisters segregate randomly in meiosis II (Fig. 4B,C). If all of the sister centromeres separated in meiosis I, a strain with two LacO marked homologs of chromosome IV should give a 1:2:1 ratio of tetrads with 2, 3, or 4 GFP-labeled spores (Fig. 4C). We observe a ratio of 0.5:2:1, which, although broadly similar to the expectation, is significantly different from it (p = 0.0015 by the χ2 test) as it is from every other detailed model we have tested. Nevertheless, there is evidence for a high rate of random segregation in meiosis II in spo13Δ mad2Δ double mutants, and the most common outcome of chromosome segregation is the presence of the marked chromosome (IV) in three of four tetrads (Fig. 4A,C).
Spo13 and Rec8 prevent separation of mitotic sister chromatids
The behavior of centromeres differs between anaphase of meiosis I and mitosis. During anaphase of meiosis I, the linkage between sister centromeres is maintained, whereas it dissolves before anaphase in mitotic cells. Cells lacking either Spo13 or Rec8 separate their sister centromeres in meiosis I and spo13Δ mutants fail to protect Rec8 at centromeres (Klein et al. 1999). On the basis of these observations, we reasoned that the presence of Spo13 and Rec8 might be sufficient to prevent sister centromere separation. This hypothesis predicts that expressing Spo13 and Rec8 in mitotic cells will inhibit sister chromatid separation during anaphase.
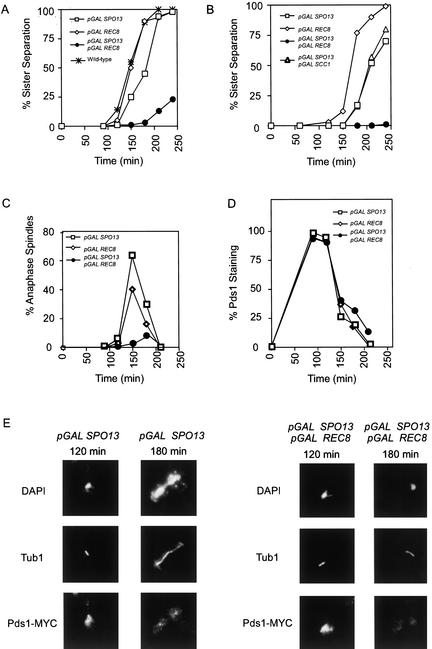
We tested this idea by expressing SPO13 and REC8 genes in mitotic cells under the control of the galactose-inducible promoter and monitoring both biochemical and cytological aspects of anaphase. Our biochemical marker was the destruction of Pds1, which requires activation of the anaphase-promoting complex (APC) and occurs at the time of anaphase initiation (Cohen-Fix et al. 1996). The cytological markers were the separation of sister chromatids and the elongation of the spindle. Cells were arrested in G1 with α factor, released into galactose-containing medium and sister chromatid separation of GFP-tagged chromosomes and spindle elongation were monitored by fluorescence microscopy (Fig. 5A,C). Overexpressing Rec8 had no effect on sister chromatid separation (Fig. 5A). Overexpressing Spo13 resulted in a small delay in sister chromatid separation without an associated delay in Pds1 destruction (Fig. 5A,D), suggesting that Spo13 has a minor effect on the removal of mitotic cohesin. It was reported previously that overexpression of SPO13 arrests cells in metaphase (McCarroll and Esposito 1994). We find that integration of a galactose-inducible version of SPO13 gene gives transformants with one of three phenotypes, a metaphase arrest similar in phenotype to that reported (McCarroll and Esposito 1994) and two novel phenotypes, a telophase arrest and slow growth (data not shown). All experiments reported here use a galactose-inducible SPO13 that arrests in telophase on galactose-containing medium.
Figure 5.
Sister chromatids do not separate, but Pds1 is destroyed in strains overexpressing Spo13 and a meiotic cohesin. (A) GFP-tagged chromosomes were observed throughout the cell cycle by fluorescence microscopy in cells overexpressing various combinations of Spo13, a meiotic cohesin subunit (Rec8), and its mitotic counterpart (Scc1/Mcd1), as indicated. Sister chromatid separation was quantified and the percentage of cells with separated sister chromatids is graphed against time after release from a G1 arrest. For each timecourse, at least 100 cells were counted at every timepoint and the experiments were repeated at least twice. Strains: (□) pGAL–SPO13 (MAS 774); (⋄) pGAL–REC8 (MAS 786); (●) pGAL–REC8 and pGAL–SPO13 (MAS 775); or (✠) wild type (SBY 214). (B) Sister chromatid separation assessed as in A. Strains: (□) pGAL–SPO13 (MAS 774); (⋄) pGAL–REC8 (MAS 786); (●) pGAL–REC8 and pGAL–SPO13 (MAS 775); (▵) pGAL–SCC1 pGAL–SPO13 (MAS 874). (C) Indirect immunofluorescence microscopy was performed against α tubulin and percentage of cells with anaphase spindles were quantified and graphed against time after release from a G1 arrest into galactose-containing medium. Strains: (□) pGAL–SPO13 (MAS 774); (⋄) pGAL–REC8 (MAS 786); (●) pGAL–REC8 and pGAL–SPO13 (MAS 775). (D) Pds1 destruction was quantified and graphed against time after release from a G1 arrest into galactose-containing medium. PDS1–MYC was detected by indirect immunofluorescence throughout the cell cycle in the indicated strains: (□) pGAL–SPO13 (MAS 774); (⋄) pGALREC8 (MAS785); (●) pGAL–SPO13 and pGAL–REC8 (MAS 775). Punctate staining is GFP–LacI. (E) Images from the experiment quantified in D, T = 120 min (left panels), T = 180 min (right panels). DAPI staining shown at top panels, anti-α tubulin staining (middle panels), and anti-MYC staining (bottom panels). Sister chromatids failed to separate and the spindle did not elongate in cells expressing pGAL–SPO13 and pGAL–REC8 despite the destruction of Pds1.
Because overexpression of Spo13 has some effect on sister chromatid separation (Fig. 5A), we asked whether overexpressing both Spo13 and Scc1/Mcd1 prevents separation of sister chromatids. Overexpression of both Spo13 and Scc1/Mcd1 had no effect on sister chromatid separation beyond the slight delay that is seen in cells that only overexpress Spo13 (Fig. 5B). These results indicate that Scc1/Mcd1 is a poor substrate for protection by Spo13.
Overexpressing both Spo13 and Rec8 prevented sister chromatid separation and spindle elongation (Fig. 5A,C). This phenotype could reflect a metaphase arrest caused by the spindle checkpoint, inactivation of the APC, or a physical block to chromosome separation. We excluded a cell cycle arrest by showing that cells that overexpressed both Rec8 and Spo13 destroyed Pds1 with the same time course as cells that expressed only one of the two proteins (Fig. 5D,E). Pds1 was destroyed in the absence of spindle elongation in cells expressing both Spo13 and Rec8 (Fig. 5E). Sister chromatids did not separate when the spindle checkpoint was removed (data not shown). Thus, we suggest that mitotically expressing Spo13 and Rec8 mimics meiosis I chromosome behavior, because the cell cycle continues, but sister chromatids stay linked to each other.
The phenotype we observe is similar to normal chromosome behavior in meiosis I, in which centromeric cohesin is retained at anaphase, promoting sister chromatid attachment. However, our mimicry of meiosis I kinetochore structure is not complete. If it were, the two sister kinetochores should mono-orient, leading to spindle elongation in the absence of sister chromatid separation. Instead, we see a short spindle, suggesting that the sister kinetochores bi-orient. Coexpression of Mam1, a component of the meiosis I kinetochore required to unite the two sister kinetochores (Toth et al. 2000), failed to confer this meiotic character on the mitotic kinetochores (data not shown).
Spo13 protects only meiotic cohesin
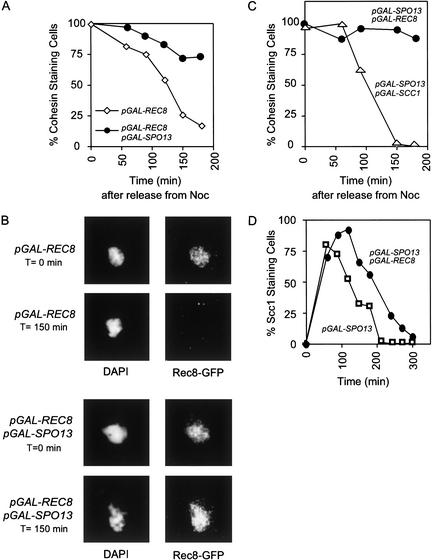
Coexpression of Spo13 and Rec8 in mitotic cells creates a physical block to sister chromatid separation. We asked whether Spo13 prevents removal of Rec8 from chromosomes by making a protein fusion between GFP and Rec8 and analyzing localization of Rec8 on chromosome spreads in the presence or absence of Spo13. Cells were synchronized in G1 and released into galactose-containing medium plus nocadazole to induce a metaphase arrest. After 3 h, cells were washed free of nocodazole and galactose, and placed in glucose-containing medium. This manipulation activates the APC and represses the transcription of Rec8 and Spo13. In cells lacking Spo13, Rec8–GFP is bound to chromosomes in nocodazole-arrested cells and is removed from chromosomes after release from this arrest (Fig. 6A,B). In cells expressing both Spo13 and Rec8, Rec8–GFP remained bound to chromosome spreads even after cells are released from nocodazole. Does Spo13 protect all cohesin complexes or just those containing Rec8? The same experiment was performed with cells expressing galactose-inducible SCC1/MDC1 (mitotic cohesin) and SPO13 (Fig. 6C). We find that Spo13 does not protect mitotic cohesin. These results suggest that Spo13 is important for protection of the meiotic form of centromeric cohesin at anaphase I.
Figure 6.
Rec8 is protected on chromosomes in the presence of Spo13. (A) A Rec8–GFP fusion was detected by indirect immunofluorescence at the indicated times after release from nocodazole arrest in strains expressing either (⋄) pGAL–REC8–GFP (MAS 850) or (●) pGAL–REC8–GFP pGAL–SPO13 (MAS 866). Rec8–GFP destruction was quantified by fluorescence microscopy and graphed against time after release from nocodazole arrest. (B) Images from the experiment quantified in A, at T = 0 min (top panels), T = 150 (bottom panels). DAPI staining is shown in left panels, anti-GFP staining is shown in right panels. Rec8–GFP is protected on chromosomes in cells expressing Spo13. (C) Cohesin staining was detected by indirect immunofluorescence against SCC1–HA3 (Uhlmann and Nasmyth 1998) after release from nocodazole arrest into glucose-containing medium. Strains expressed either (●) pGAL–REC8–GFP pGAL–SPO13 (MAS 866) or (▵) pGAL–SCC1–HA3 (Uhlmann and Nasmyth 1998), pGAL–SPO13 (MAS 874). (D) Indirect immunofluorescence was performed on Scc1/Mcd1–3XHA expressed from its own promoter in strains expressing either (□) pGAL–SPO13 (MAS 823) or (●) pGAL–SPO13 pGAL–REC8 (MAS 821). Staining of endogenous Scc1/Mcd1–3XHA is graphed against time after release from a G1 arrest. Scc1/Mcd1–3XHA is removed from chromosomes in the presence of Spo13 and Rec8.
We asked whether the simultaneous overexpression of Spo13 and Rec8 leads to protection of mitotic cohesin. Mitotic cohesin was epitope tagged (Scc1/Mcd1–3XHA) and expressed from the gene's normal promoter. Removal of Scc1/Mcd1–3XHA was assayed on chromosome spreads in strains that overexpressed Spo13 alone or the combination of Spo13 and Rec8 (Fig. 6D). In cells expressing Spo13, Scc1/Mcd1–3XHA was completely removed from chromatin after 210 min, reflecting the brief delay in chromosome segregation in these strains (Fig. 5A). In cells expressing Spo13 and Rec8, Scc1/Mcd1–3XHA staining disappeared from spreads after 270 min. Although separase cleavage of Scc1/Mcd1 is delayed, unlike Rec8, Scc1/Mcd1 is removed from chromosomes, showing that Spo13 protects meiotic cohesin from active separin protease.
Discussion
We investigated the role of Spo13 in meiosis. We find that the single division in spo13Δ mutants is caused by a delay induced by the spindle checkpoint. We show that kinetochore structure is disrupted in the spo13Δ mutant as sister kinetochores are improperly activated at meiosis I, and suggest that this defect is sensed by the spindle checkpoint. Finally, we show that expression of Spo13 and Rec8 in mitotic cells prevents sister chromatid separation, suggesting that these two meiosis-specific components combine to induce one aspect of meiotic centromere behavior.
Meiosis I centromere behavior in spo13Δ mutants
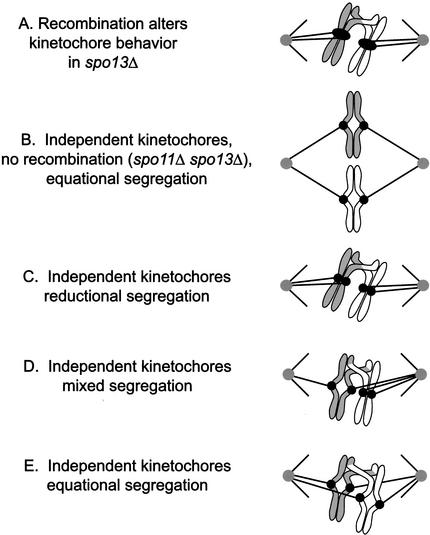
We began our investigation of Spo13 by re-examining chromosome behavior in spo13Δ cells. In spo13Δ mutants, both homologs and sister chromatids can segregate away from one another in the single meiotic division (Klapholz and Esposito 1980b; Hugerat and Simchen 1993). This pattern of mixed segregation is influenced by recombination (Rutkowski and Esposito 2000), and eliminating recombination results in 100% sister separation (Klapholz et al. 1985). Recombination could influence chromosome segregation in spo13Δ mutants in two ways. In the first scheme, recombination influences kinetochore behavior in spo13Δ mutants in some cases, causing the two sister kinetochores to act as a single functional unit and go to the same pole in anaphase (Fig. 7A). In the second scheme, all four sister kinetochores are activated in the absence of Spo13 (Fig. 7B–E). Only kinetochores that are under tension have a stable linkage to a pole of the spindle (Nicklas and Koch 1969). If recombination does not occur (spo13Δ spo11Δ), then tension can only be generated when sister kinetochores attach to opposite poles (Fig. 7B). However, if recombination links homologous chromosomes, tension can be generated if a pair of sister kinetochores attach to the same pole and at least one of the other two kinetochores attaches to the opposite pole (Fig. 7D). Thus, in spo13Δ cells, a pair of sister kinetochores can be segregated to the same pole despite the complete absence of meiosis I kinetochore structure. Sister centromeres segregate from each other if they attach to opposite poles (Fig. 7E). Finally, homologous pairs can also undergo mixed homolog and sister segregation, as has been observed (Hugerat and Simchen 1993; M. Shonn, unpubl.; Fig. 7D).
Figure 7.
Centromere behavior in spo13Δ mutants. Model for centromere behavior in spo13Δ mutants. (A) The act of recombination in a spo13Δ cell induces the two sister kinetochores to act as a single functional unit, as they do in wild-type cells in meiosis I. (B) In spo11Δ spo13Δ cells, no recombination occurs. Kinetochores can only come under tension if they attach to opposite poles, explaining why sister centromeres always segregate from each other at meiosis I in these cells. (C–E) Recombination has no direct effect on kinetochore behavior, but connects together four independent kinetochores. (C) Homologs segregate to opposite poles (reductional division) when one set of sister kinetochores attaches to one pole, and the other pair attaches to the opposite pole. (D) A mixed division occurs when one pair of sister kinetochores attaches to opposite poles and the other pair attaches to the same pole. (E) Sister centromeres separate from each other at anaphase (equational segregation) when they attach to opposite poles of the spindle.
The spindle checkpoint is activated in spo13Δ mutants
The spindle checkpoint senses a defect in spo13Δ mutants and delays cells in metaphase of meiosis I. What defect does the checkpoint sense? Our analysis shows that spo13Δ mutants stretch sister centromeres in metaphase I more frequently than wild type in meiosis I and less than wild type in meiosis II (Fig. 2). This decrease in centromere separation can be explained in four ways. (1) Only one of the two sister kinetochores is attached to a microtubule. (2) Sister centromeres are attached to opposite poles, but are subject to forces that are too small to stretch them apart. (3) Sister centromeres are attached to the same pole. In these three cases, kinetochore–microtubule attachment is either defective or fails to establish tension, defects that are sensed by the spindle checkpoint. An alternative possibility is that Rec8 localized near centromeres inhibits centromere stretching. This possibility seems less likely, as sister separation occurs in 70% of wild-type cells during metaphase II, a time when all cohesion is centromere proximal.
Single division meiosis
Why do spo13Δ mutants undergo only a single meiotic division? Previous studies have speculated that spo13Δ mutants either skip meiosis I (Klapholz and Esposito 1980b), or undergo a combination of meiosis I and meiosis II (Hugerat and Simchen 1993). We find that spo13Δ mutants are delayed in meiosis I by the spindle checkpoint and suggest that by delaying meiosis I, spore formation begins before the second division can occur. In support of this model, a delay in meiosis I caused by expressing low levels of a nondegradable version of PDS1 also results in the formation of dyads (Shonn et al. 2000). These observations suggest that there is a certain amount of time allotted for the meiotic divisions and if the divisions are not complete, spores form, packaging chromosomes and effectively ending meiosis.
The spo12Δ mutant (Klapholz and Esposito 1980a) and the clb1Δ, clb3Δ, clb4Δ triple mutant (Dahmann and Futcher 1995) also undergo a single meiotic division and form dyads. Deletion of all three CLB genes is necessary to produce a high frequency of sporulation and segregation of homologous chromosomes (Dahmann and Futcher 1995). Do these mutants form dyads due to spindle checkpoint activation? We find that eliminating the spindle checkpoint in spo12Δ mutants or the triple cyclin deletion mutant does not restore a second meiotic division (data not shown). Spo12 has been implicated in mitotic exit (Grether and Herskowitz 1999; Stegmeier et al. 2002) and the CLB genes encode cyclins, important cell cycle regulators (Dahmann and Futcher 1995). We speculate that there are two classes of mutants that undergo a single meiotic division, those that delay in metaphase of meiosis I (spo13Δ), and those that delay after anaphase of a single meiotic division (spo12Δ, slk19Δ, and the triple clb deletion). Despite this difference, the ultimate reason for the formation of dyads may be the same for both classes, namely a second division fails to occur before the onset of spore formation.
Spo13 protects centromeric Rec8
A long-standing question has been the identity of a molecule that protects sister centromere cohesion in meiosis I. At anaphase I, cohesion is lost from sister chromatid arms but is protected near sister centromeres (Lee and Orr-Weaver 2001; Nasmyth 2001). What molecule(s) protect centromeric Rec8 from cleavage at anaphase I? Spo13 is a candidate because cells lacking Spo13 fail to fully protect Rec8 at sister centromeres in anaphase I (Klein et al. 1999). We have mimicked one aspect of meiotic chromosome behavior in mitotic cells. Expressing two meiotic proteins, Spo13 and Rec8, prevents sister chromatids from separating at mitotic anaphase. Because Spo13 and Rec8 are both required for normal chromosome behavior in meiosis I, we suggest that Spo13 protects a subset of meiotic cohesin during anaphase of meiosis I and metaphase of meiosis II.
Spo13 protects Rec8 from cleavage at the centromere and forces the two kinetochores to act as one. How does it perform both functions? Spo13 could have two independent functions at the centromere, uniting the two kinetochores prior to metaphase I and then protecting Rec8 from cleavage during anaphase I. Alternatively, Spo13 may work through Rec8 to both unite the two sister kinetochores and maintain cohesion in metaphase I. In fission yeast, Rec8 must be present during meiotic S phase to establish meiosis I kinetochore structure (Watanabe et al. 2001). In budding yeast, rec8Δ mutants separate their sister chromatids before anaphase of meiosis I, suggesting that meiosis I kinetochore structure has not been established (Klein et al. 1999).
Building meiosis I chromosomes
How is protection of Rec8 limited to the centromere region in metaphase I? In our experiments, overexpression may obscure localization of Rec8 to centromeres. To achieve localized protection of Rec8, Spo13 may work in concert with additional, unidentified meiotic proteins. Alternatively, meiosis-specific functions of the mitotic proteins Slk19 and Bub1 may be required. Sister centromere cohesion requires Slk19 in budding yeast (Kamieniecki et al. 2000; Zeng and Saunders 2000) and Bub1 in fission (Bernard et al. 2001) and budding (M. Shonn and A. Murray, unpubl.) yeast. Deletion of SLK19 or BUB1 genes in cells expressing Spo13 and Rec8 does not disrupt sister chromatid cohesion (M. Shonn and A. Murray, unpubl.). How is Rec8 removal targeted to sister chromatid arms? When Rec8 is removed from chromosome arms, sister telomeres separate at anaphase I (Buonomo et al. 2000). We monitored telomere separation in mitotic cells expressing both Rec8 and Spo13 and found that telomeres do not separate (M. Shonn and A. Murray, unpubl.). However, telomeres also fail to separate in spo11Δ homozygous diploids, despite removal of Rec8 from chromosome arms (Buonomo et al. 2000), suggesting that telomere separation requires both the formation and the resolution of chiasmata, events that do not occur in spo11Δ or mitotic cells. It is interesting to note that Rec8 contains two separin cleavage sites (Buonomo et al. 2000), both located in regions of the protein poorly conserved with respect to mitotic cohesin. Perhaps this rearrangement of cleavage sites creates conditions favorable for removal of Rec8 along chromosome arms.
In meiosis I, a homolog attaches to a single spindle pole because the sister kinetochores form a single functional unit. This mono-orientation requires Spo13 and the meiosis-specific protein Mam1 (Toth et al. 2000). Despite expressing Mam1 in addition to Spo13 and Rec8 in mitotic cells, we have not been able to prevent sister kinetochores from attaching to opposite poles (M. Shonn and A. Murray, unpubl.). Meiotic proteins or regulation in addition to Mam1 must be needed to unite sister kinetochores.
Centromere cohesion in meiosis I: many solutions to a similar problem?
Although Rec8 has been identified in several species (Bhatt et al. 1999; Klein et al. 1999; Watanabe and Nurse 1999; Pasierbek et al. 2001), little is known about the molecular components that protect it during meiosis I. We find that Rec8 and Spo13 are sufficient to prevent sister chromatid separation at anaphase. This observation suggests that evolving meiosis-specific chromosome behavior may have required only two steps. First, the duplication and divergence of a cohesin subunit (an ancestral cohesin giving rise to both Scc1/Mcd1 and Rec8), and second, the evolution or meiosis-specific recruitment of a protein (Spo13) that could protect a subset of the meiosis-specific cohesin until anaphase of meiosis II. Spo13 has so far only been recognized in fungi, suggesting that it has either diverged rapidly or is a fungus-specific solution to a problem faced by most eukaryotes. In Drosophila, the Mei-S332 protein localizes to centromeres and performs a function similar to Spo13, preventing sister chromatid separation until anaphase II (Kerrebrock et al. 1995). Spo13 and Mei-S332 show no homology, although they appear to perform a similar function.
Materials and methods
Microbial techniques and yeast strain construction
Media and genetic and microbial techniques were essentially as described (Sherman et al. 1974; Rose et al. 1990). All cytological experiments began from a G1 arrest. Cells were arrested in medium containing 2% raffinose and 1 μg/mL α-factor at 23°C for 3.5 h, washed three times in water, and resuspended in medium containing 2% raffinose and 2% galactose. After 1.5 h, α-factor was added back to prevent cells from entering the next cell cycle. For experiments requiring a metaphase arrest, 15 μg/mL nocodazole was added to the medium after release from α-factor. Cells were released from nocodazole by washing three times in water, and were then resuspended in dextrose medium containing 1.5 μg/mL α-factor. All experiments reported were repeated a minimum of two times with similar results. In all experiments, at least 100 cells were counted. Stock solutions of inhibitors were 10 mg/mL nocodazole, 10 mg/mL α-factor in DMSO. All stocks were stored at −20°C. Yeast strains are listed in Table 3 and were constructed by use of standard genetic techniques. The DH5-α strain was used for all bacterial manipulations.
Table 3.
Strains used in this study.
| MAS 314 | MATa/MATαpCYC1-GFP12-lacI12::URA3/uru3-1, leu2-3/leu2-3, his3-11/his3-11, LacO::TRP1/trp1-1, ade2-1/ade2-1, can1-100can1-100, spo13::hisG/spo13::hisG |
|---|---|
| MAS 492 | MATa/MATαpCYC1-GFP12-lacI12::URA3/ura3-1, LacO:LEU2/leu2-3, his3-11/his3-11, trp1-1/trp1-1, ade2-1/ade2-1, can1-100/can1-100, spo13::hisG/spo13::hisG |
| MAS 356 | MATa/MATαpCYC1-GFP12-lacI12::URA3/ura3-1, leu2-3/leu2-3, his3-11/his3-11, LacO::TRP1/LacO::TRP1, ade2-1/ade2-1, can1-100/can1-100, spo13::hisG/spo13::hisG, mad2::LEU2/mad2::LEU2 |
| MAS 386 | MATa/MATαpCYC1-GFP12-lacI12::URA3/ura3-1, leu2-3/leu2-3, his3-11/his3-11, LacO::TRP1/LacO::TRP1, ade2-1/ade2-1, can1-100/can1-100 |
| MAS 651 | MATa/MATαpCYC1-GFP12-lacI12::URA3/ura3-1, leu2-3/leu2-3, his3-11/his3-11, trp1-1/trp1-1, ade2-1/ade2-1, can1-100/can1-100, LacO::TRP1(pMAS80)/LacO::TRP1(pMAS80) |
| MAS 676 | MATa/MATαpCYC1-GFP12-lacI12::URA3/ura3-1, leu2-3/leu2-3, his3-11/his3-11, trp1-1/trp1-1, ade2-1/ade2-1, can1-100/can1-100, LacO::TRP1(pMAS80)/-, spo11::HIS3/spo11::HIS3, spo13::URA3/spo13::URA3 |
| MAS 875 | MATa/MATαpCYC1-GFP12-lacI12::URA3/ura3-1, leu2-3/leu2-3, his3-11/his3-11, trp1-1/trp1-1, ade2-1/ade2-1, can1-100/can1-100, LacO::TRP1(pMAS80)/-, spo13::URA3/spo13::URA3 |
| MAS 774 | MATa, pGAL-SPO13::URA3, leu2-3, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, ade2-1, can1-100, bar1Δ, PDS1::PDS1-myc13::LEU2 |
| MAS 775 | MATa, pGAL-SPO13::URA3, leu2-3, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, pGAL-REC8::ADE2, can1-100, bar1Δ, PDS1::PDS1-myc13::LEU2 |
| MAS 786 | MATa, ura3-1, leu2-3, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, pGAL-REC8::ADE2, can1-100, bar1Δ, PDS1::PDS1-myc13::LEU2 |
| MAS 850 | MATa, ura3-1, leu2-3, his3-11, trp1-1, ade2-1, can1-100, bar1Δ, REC::pGAL:::HIS5-REC8-GFP::KAN, NDC10::NDC10-3HA::URA3 |
| MAS 866 | MATa, pGAL-SPO13::URA3, leu2-3, his3-11, trp1-1, ade-2-1, can1-100, bar1::LEU2, REC8::pGAL::HIS5-REC8-GFP::KAN, NDC10::NDC10-3HA::URA3 |
| MAS 821 | MATa, pGAL-SPO13::URA3, leu2-3, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, pGAL-REC8::ADE2, can1-100, bar1Δ, MCD1::MCD1-3HA::URA3 |
| MAS 823 | MATa, pGAL-SPO13::URA, leu2-3, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, can1-100, bar1Δ, MCD1::MCD1-3HA::URA3 |
| MAS 874 | MATa, pGAL-SPO13::URA3, pGAL-SCC1-3HA::LEU2, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, ade2-1, can1-100, bar1Δ |
| SBY447 | MATa, ura3-1, leu2-3, pCUP1-GFP12-lacI12:HIS3, LacO::TRP1, ade2-1, can1-100, bar1Δ, MCD1::MCD1-3HA::URA3 |
| SBY214 | MATa, ura3-1, leu2-3, pCUP1-GFP12-lacI12::HIS3, LacO::TRP1, ade2-1, can1-100, barIΔ |
| Strains in the SK1 background | |
| MAS 118 | MATa ura3-1, leu2::hisG |
| MAS 119 | MATα ura3-1, leu2::hisG |
| MAS 278 | MATa ura3-1, leu2::hisG, spo13::URA3::hisG, trp1::hisG |
| MAS 279 | MATα ura3-1, leu2::hisG, spo13::URA3::hisG, trp1::hisG |
| MAS 442 | MATa ura3-1, leu2::hisG, mad2::URA3, spo13::URA3::hisG |
| MAS 443 | MATα ura3-1, leu2::hisG, mad2::URA3, spo13::URA3::hisG |
| MAS 659 | MATa, ura3-1, leu2::hisG, spo13::URA3::hisG, spo11::URA3, PDS1-MYC13:LEU2 |
| MAS 660 | MATα, ura3-1, leu2::hisG, spo13::URA3::hisG, spo11::URA3, PDS1-MYC13:LEU2 |
All strains are in the W303 background unless otherwise noted.
Plasmid construction and gene tagging
A lactose operator array that marks the centromere of chromosome VIII was constructed. A region of chromosome VIII from coordinates 108515–107836 was PCR amplified from yeast genomic DNA by use of primers Lac80–BAM (5′-GCG/CGG/ATC/CAG/TGT/AAT/ATG/CAC/CT-3′) and Lac80–SAC (5′-GCG/CGA/GCT/CTC/AAT/TAG/GTT/TAT/CTT/C-3′). The resulting PCR product was digested with BamHI and SacI and ligated into the BamHI–SacI sites of pAFS149 (A. Straight and A. Murray, unpubl.), which contains 128 repeats of the lactose operator sequence. To integrate into yeast, plasmid pMAS80 was digested with ClaI.
A clone of the SPO13 gene under the control of the galactose-inducible promoter was constructed. The SPO13 gene was PCR amplified from yeast genomic DNA by use of primers SPO13–NOT (5′-GCG/CGC/GCG/CGG/CCG/CAT/TAT/GGC/ACC/CAG/A-3′) and SPO13–BAM (5′-GCG/CGC/GCG/GAT/CCT/TAA/TTA/AGG/GAA/GAC/T-3′). The resulting PCR product was digested with BamHI and NotI and ligated into the BamHI–NotI sites of PDK20 (gift of Doug Kellogg, University of California, Santa Cruz) to create pMAS63. To integrate into yeast at URA3, it was digested with StuI.
A clone of the REC8 gene under the control of the galactose-inducible promoter was similarly constructed. The REC8 gene was PCR amplified from yeast genomic DNA by use of primers REC8–SAC (5′-GCG/CGC/GCG/AGC/TCT/ACG/TAT/GAT/ATC/GC-3′) and REC8–BAM (5′-GCG/CGC/GGA/TCC/ATG/GCA/CCT/CTT/TCG-3′). The resulting PCR product was digested with BamHI and SacI and ligated into the BamHI–SacI sites of pBS163 (gift of Bodo Stern, Harvard University, Cambridge, MA). To integrate into yeast at ADE2, it was digested with StuI.
The chromosomal copy of REC8 was tagged with the GFP and placed under the control of the galactose-inducible promoter by transformation as described (Longtine et al. 1998). GFP was PCR amplified from plasmid pFA6a-GFP(S65T)-kanMX6 by use of primers REC8–TAG–FORW (5′-TAA/CTA/AAG/ATC/TTA/AAC/TGA/GAA/GAG/AGGACG/AAA/TAA/TTG/TAT/ATG/CCG/GTC/GAC/GGA/TCC/CCG/GGT/T-3′) and REC8–TAG–REV (5′TTT/ACA/TTA/TAT/AGT/GTA/CGT/ATG/ATA/TCG/CTA/GCA/TGA/TG/TAG/TGT/TTT/CGA/TGA/ATT/CGA/GCT/CGT/T-3′) that are designed to integrate after the last codon of the REC8gene. The galactose-inducible promoter was PCR amplified from plasmid pFA6a-His3MX6-PGAL1 by use of primers REC8–GAL–FORW (5′-CAA/CTC/TAA/AGC/ATT/TGC/TAT/ATA/TAG/ATT/AAT/ATT/ACA/AAT/ATT/CTG/CA/GAA/TTC/GAG/CTC/GTT/TAA/AC-3′) and REC8–GAL–REV (5′-GTG/AGG/CCC/TTA/TATTTC/TTG/TCA/TCT/TTA/AAG/TTC/AAC/GAA/AGA/GGT/GCC/ATT/TTG/AGA/TCC/GGG/TTT/T-3′) designed to integrate before the start codon of REC8.
Calculation of average duration of meiosis I
Values for the percentage of meiosis I spindles were added for each timepoint and divided by the sum of the wild-type values. Wild type = 1, spo13Δ = 2.39, spo13Δ spo11Δ = 2.79.
Sporulation
Synchronous sporulation was as described for SK1 (Padmore et al. 1991) with modifications (Shonn et al. 2000). Modifications are as follows: haploid SK1 cells were mated overnight on YPD plates and diploids were selected by micromanipulation and grown overnight at 30°C. Diploids were streaked to YPD and grown 2 d at 30°C. Synchronization of sporulating cells was then completed as described for SK1 (Padmore et al. 1991). Chromosome segregation in meiosis was assessed in the W303 background. Diploid cells were grown overnight in YPD, diluted 1:100 into YEP plus 2% potassium acetate for 24 h, then washed two times and released into an equal volume of SPM (2% potassium acetate, 0.1% raffinose).
Microscopy and immunofluorescence
Microscopy on meiotic cells to localize GFP–LacI was performed on living cells using a 60X objective (Nikon Instruments). Immunofluorescence was carried out as described (Rose et al. 1990). Chromosome spreads were performed as described (Michaelis et al. 1997; Biggins et al. 1999).
Acknowledgments
We thank Dean Dawson, Hiro Funabiki, and the members of the Murray lab for their critical reading of the manuscript. This work is supported by grants from the NIH and Human Frontiers Science Project.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL amurray@mcb.harvard.edu; FAX (671) 496-1541.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.975802.
References
- Bernard P, Maure JF, Javerzat JP. Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat Cell Biol. 2001;3:522–526. doi: 10.1038/35074598. [DOI] [PubMed] [Google Scholar]
- Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999;19:463–472. doi: 10.1046/j.1365-313x.1999.00548.x. [DOI] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes & Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Futcher B. Specialization of B-type cyclins for mitosis or meiosis in S. cerevisiae. Genetics. 1995;140:957–963. doi: 10.1093/genetics/140.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Grether ME, Herskowitz I. Genetic and biochemical characterization of the yeast spo12 protein. Mol Biol Cell. 1999;10:3689–3703. doi: 10.1091/mbc.10.11.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugerat Y, Simchen G. Mixed segregation and recombination of chromosomes and YACs during single-division meiosis in spo13 strains of Saccharomyces cerevisiae. Genetics. 1993;135:297–308. doi: 10.1093/genetics/135.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Kamieniecki RJ, Shanks RM, Dawson DS. Slk19p is necessary to prevent separation of sister chromatids in meiosis I. Curr Biol. 2000;10:1182–1190. doi: 10.1016/s0960-9822(00)00723-5. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Moore DP, Wu JS, Orr-Weaver TL. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Klapholz S, Esposito RE. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics. 1980a;96:567–588. doi: 10.1093/genetics/96.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Recombination and chromosome segregation during the single division meiosis in SPO12-1 and SPO13-1 diploids. Genetics. 1980b;96:589–611. doi: 10.1093/genetics/96.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S, Waddell CS, Esposito RE. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Lee JY, Orr-Weaver TL. The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol. 2001;17:753–777. doi: 10.1146/annurev.cellbio.17.1.753. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- McCarroll RM, Esposito RE. SPO13 negatively regulates the progression of mitotic and meiotic nuclear division in Saccharomyces cerevisiae. Genetics. 1994;138:47–60. doi: 10.1093/genetics/138.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome manipulation III. Induced reorientation and the experimental control of segregation in meiosis. J Cell Biol. 1969;43:40–50. [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes & Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Winston F, Heiter P. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1990. [Google Scholar]
- Rutkowski LH, Esposito RE. Recombination can partially substitute for SPO13 in regulating meiosis I in budding yeast. Genetics. 2000;155:1607–1621. doi: 10.1093/genetics/155.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Simchen G. Mixed segregation of chromosomes during single-division meiosis of Saccharomyces cerevisiae. Genetics. 1990;125:475–485. doi: 10.1093/genetics/125.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: A push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hersko J, Luca F, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitination ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Rabitsch KP, Galova M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis i. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 2001;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- Zeng X, Saunders WS. The Saccharomyces cerevisiae centromere protein Slk19p is required for two successive divisions during meiosis. Genetics. 2000;155:577–587. doi: 10.1093/genetics/155.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]