Abstract
Developmental responses to the plant hormone auxin are thought to be mediated by interacting pairs from two protein families: short-lived inhibitory IAA proteins and ARF transcription factors binding to auxin-response elements. monopteros mutants lacking activating ARF5 and the auxin-insensitive mutant bodenlos fail to initiate the root meristem during early embryogenesis. Here we show that the bodenlos phenotype results from an amino-acid exchange in the conserved degradation domain of IAA12. BODENLOS and MONOPTEROS interact in the yeast two-hybrid assay and the two genes are coexpressed in early embryogenesis, suggesting that BODENLOS inhibits MONOPTEROS action in root meristem initiation.
Keywords: Arabidopsis, embryo patterning, BODENLOS, MONOPTEROS, auxin response, root meristem initiation
Plant embryogenesis establishes a multicellular body organization that is elaborated postembryonically by the stem-cell systems of the shoot and root meristems to give the species-specific architecture of the adult plant (Jürgens 2001). The two primary meristems themselves originate from distinct regions along the apical-basal axis of the early embryo. Following the asymmetric division of the zygote, the smaller apical daughter cell generates an eight-celled proembryo consisting of two tiers of four cells each. The upper tier, or apical region, gives rise to the shoot meristem and most of the flanking cotyledon primordia. The lower tier, or central region, contributes to the cotyledon primordia but mainly produces the hypocotyl, the embryonic root, and the upper tier of root meristem stem cells. The basal daughter cell of the zygote produces a file of 7–9 initially extraembryonic cells of which the uppermost cell adjacent to the proembryo adopts an embryonic fate. This so-called hypophysis generates the basal region of the embryo that gives rise to the quiescent center and the lower tier of stem cells of the root meristem. Thus, the root meristem is of mixed clonal origin, with the hypophysis initiating root meristem formation during early embryogenesis (Jürgens 2001).
Mutations in two genes, MONOPTEROS (MP) and BODENLOS (BDL), affect the formation of the hypophysis in a seemingly noncell autonomous manner. Both bdl and mp mutations initially affect the orientation of the division plane of the apical daughter of the zygote, resulting in double-octant proembryos (Berleth and Jürgens 1993; Hamann et al. 1999; see Fig. 3K). Subsequently, the presumptive hypophysis fails to undergo the asymmetric division that gives the precursors of the quiescent center and the lower-tier stem cells of the root meristem. The same mutant phenotype was observed in bdl mp double mutant embryos, suggesting that the two proteins act in the same developmental pathway (Hamann et al. 1999). The MP gene encodes ARF5, a transcription factor of the ARF (auxin response factor) family that activates auxin-responsive target genes (Hardtke and Berleth 1998; Ulmasov et al. 1999a). bdl mutant seedlings are insensitive to the auxin analog 2,4-D, implicating BDL in auxin response (Hamann et al. 1999).
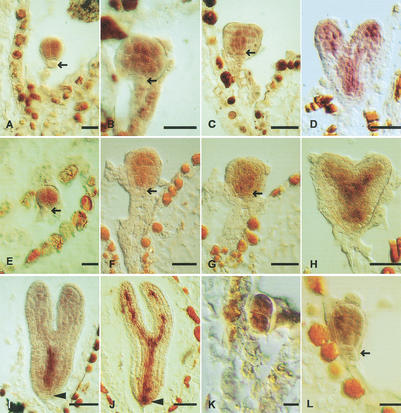
Figure 3.
mRNA in situ expression analysis of BDL and MP in developing embryos. (A–D,I) BDL, (E–H,J–L) MP mRNA accumulation in wild-type (E–J) and bdl mutant (K, L) embryos. Developmental stages (Jürgens and Mayer 1994): (A,E) octant; (B,F,L) early-globular; (C,G) globular with lens-shaped cell; (D,H) heart; (I,J) torpedo; (K) four-cell. Arrows mark the hypophysis in (A,B,E,F,L) and the lens-shaped cell in (C,G), arrowheads mark the central root cap in (I,J). Bar, 20 μm.
The Arabidopsis genome encodes 23 ARF transcription factors, of which three have been identified by mutant phenotypes: ETT/ARF3 in flower development, MP/ARF5 in embryogenesis, and NPH4/ARF7 in hypocotyl phototropism (Sessions et al. 1997; Hardtke and Berleth 1998; Harper et al. 2000; Guilfoyle and Hagen 2001). In each case, the mutant phenotypes result from loss-of-function mutations. ARF transcription factors interact with IAA (indole-3-acetic acid) proteins (Kim et al. 1997; Ulmasov et al. 1999b; Ouellet et al. 2001; for review, see Guilfoyle and Hagen 2001; Reed 2001; Rogg and Bartel 2001). The Arabidopsis genome encodes ∼28 IAA proteins (Reed 2001; Rogg and Bartel 2001). Most of the IAA proteins have recently been shown to repress auxin-responsive gene activation in transient expression assays (Tiwari et al. 2001). Mutations have been identified in eight IAA genes, all of which affect exclusively postembryonic development (Rouse et al. 1998; Tian and Reed 1999; Nagpal et al. 2000; Rogg et al. 2001; Fukaki et al. 2002; for review, see Reed 2001; Rogg and Bartel 2001). In each case, the mutant phenotype results from a single amino acid exchange in the conserved domain II that is involved in protein degradation via the ubiquitin-proteasome pathway (Worley et al. 2000; Gray et al. 2001; Oellet et al. 2001; Ramos et al. 2001; Tiwari et al. 2001; Zenser et al. 2001). These observations have led to the idea that IAA proteins form complexes with ARF transcription factors, from which the latter are released in response to auxin. It was hypothesized that specific pairs of ARF/IAA complexes may mediate specific developmental responses to auxin; however, no such pair has been identified to date.
Here, we show that BDL encodes IAA12, which is thus the first IAA protein involved in embryogenesis. Whereas the failure of mp mutant embryos to establish the hypophysis is caused by loss-of-function mutations of the ARF5 transcription factor, the similar bdl mutant phenotype is due to a gain-of-function mutation in the conserved degradation domain. In addition, BDL and MP proteins interact in yeast, and both genes are expressed in the proembryo cells but not the presumptive hypophysis. Our results suggest that the hypophysis is established in response to an auxin-dependent signal from the adjacent proembryo.
Results and Discussion
The bodenlos phenotype is due to a gain-of-function mutation in the auxin response regulator IAA12
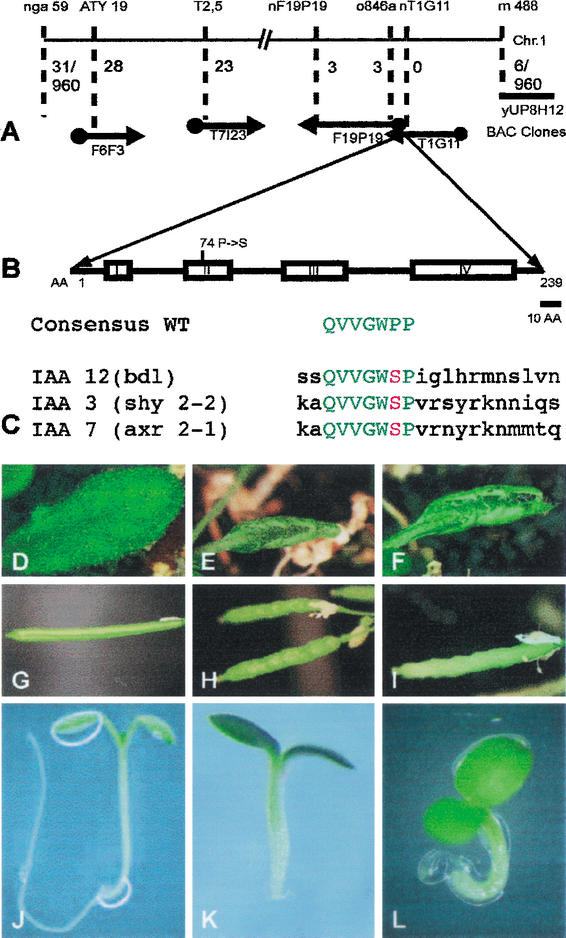
The BDL gene was mapped to a genomic interval of ∼200 kb that includes the IAA12 gene involved in auxin response (Fig. 1A). The IAA12 gene was amplified by PCR from both the parental line, Landsberg erecta, and the bdl mutant line. Sequence analysis revealed a single G to A transition that results in a predicted amino acid exchange of Pro 74 to Ser in the highly conserved domain II (Fig. 1B). Identical mutations have been identified in dominant mutant alleles of IAA3 (shy2-2; Tian and Reed 1999) and IAA7 (axr2-1; Nagpal et al. 2000) (Fig. 1C). Comparable mutations affecting the same or the adjacent proline residue are known for IAA14 (slr-1; Fukaki et al. 2002), IAA17 (axr3-1; Rouse et al. 1998), IAA19 (msg2-1; Nagpal et al. 2000) and IAA28 (iar2-1; Rogg et al. 2001). Recent analyses of half-life times for axr3 mutant protein in vitro and in vivo indicate that these amino acid replacements render the mutant proteins several-fold more stable than the wild-type protein (Worley et al. 2000; Ouellet et al. 2001; Ramos et al. 2001; Zenser et al. 2001). Furthermore, the IAA7 protein from the axr2-1 mutant line, in contrast to the wild-type protein, does not interact with the SCFTIR1 complex involved in IAA protein degradation (Gray et al. 2001).
Figure 1.
Molecular identification of the BDL gene. (A) Molecular mapping. Molecular markers and BAC clones are shown at the top and bottom, respectively. Numbers indicate recombinants between markers and the BDL gene on BAC clone T1G11 (960 gametes tested). (B) BDL protein domain structure showing the four conserved domains of IAA proteins with the Pro 74 → Ser amino acid exchange in bdl mutant protein. (C) Homologous amino acid exchanges in the conserved domain II of IAA12 (bdl), IAA3 (shy2-2) and IAA7 (axr2-1) mutant proteins. The consensus sequence of wild-type is shown for comparison. (D–L) Phenotypic analysis of transgenic progeny from wild-type plants transformed with the bdl mutant genomic fragment. (D,G,J) Wild-type (BDL/BDL) control; (E,H,K) bdl heterozygous (E,H) and homozygous (K) mutant controls; (F,I,L) transgenic BDL/BDL with one (F,I) or two copies (L) of the bdl transgene. (D–F) Leaves, (G–I) siliques, (J–L) seedlings.
The bdl mutant has a semidominant phenotype during postembryonic development but a recessive embryonic phenotype (Hamann et al. 1999). To analyze the effect of the bdl mutant allele, we performed reciprocal plant transformation experiments, using homologous BDL 4-kb genomic fragments from wild-type and bdl mutant lines. The BDL wild-type transgene did not rescue the bdl mutant phenotypes (Table 1). In contrast, nine wild-type plants independently transformed with the bdl mutant transgene gave transgenic progeny that displayed the bdl/BDL heterozygous plant phenotypes (Fig. 1D–I). Three of these transgenic plants were analyzed further. Upon selfing, they produced about 22% bdl mutant seedlings, a proportion comparable to that of selfed bdl/BDL heterozygous plants (Fig. 1J–L; Table 1). Reciprocal crosses between bdl/BDL heterozygous plants and wild-type plants carrying the bdl mutant transgene also resulted in a similar proportion of bdl mutant seedling progeny (Table 1). These results suggest that the copy number of the bdl mutant allele is critical, both in the absence and in the presence of wild-type alleles.
Table 1.
Analysis of progeny from transgenic plants bearing bdl alleles
| Parental genotypesa
|
Seedling progeny phenotypesb
|
||||
|---|---|---|---|---|---|
| No. analyzed
|
bdl (R)c
|
bdl (s)d
|
WT (R)c
|
WT (s)d
|
|
| bdl/BDL; T(BDL)/− (selfing) | 630 | 18% | 5% | 57% | 20% |
| BDL/BDL; T(bdl)/− (selfing) | 3,672 | 22% | 0 | 53% | 25% |
| bdl/BDL;−/− × BDL/BDL; T(bdl)− (reciprocal crosses) | 295 | 29% | 0 | 26% | 45% |
| bdl/BDLe (selfing) | 1,203 | 0 | 18% | 0 | 82% |
Endogenous alleles at BDL locus and transgenic alleles (T): (bdl) mutant; (BDL) wild type.
See Figure 1. Phenotypes: (bdl) mutant; (WT) wild type.
(R) Resistant to Basta due to transgene.
(s) Sensitive to Basta due to lack of transgene.
Data from Hamann et al. (1999).
BODENLOS/IAA12 interacts with MONOPTEROS/ARF5, and the two genes are coexpressed in early embryogenesis
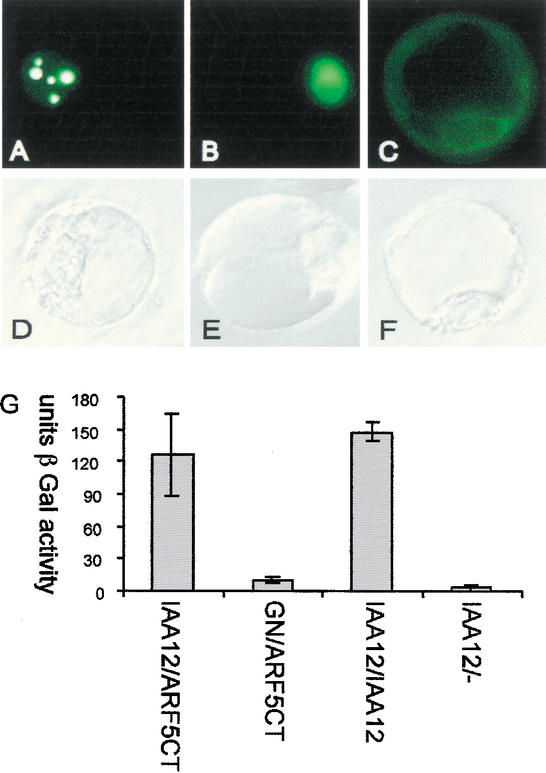
To determine its subcellular localization, we transiently expressed BDL protein fused to green fluorescent protein (GFP) in parsley protoplasts (Kircher et al. 1999). The GFP-BDL protein strongly accumulated in the nucleus in a compartmentalized manner (Fig. 2A–F), whereas BDL proteins truncated at amino acids 66 or 72 were equally partitioned between nucleus and cytoplasm (data not shown). Domains III and IV of IAA proteins, which are also present at the C terminus of ARFs, mediate homomeric and heteromeric interactions with other IAAs and ARFs in the yeast two-hybrid system (Kim et al. 1997; Ulmasov et al. 1999b). In this assay, BDL interacted both with itself and with the C terminus of MP/ARF5 (Fig. 2G). Thus, BDL can interact directly with MP.
Figure 2.
Nuclear localization and interactions of BDL protein. (A–F) Transient expression of 35S∷GFP-BDL in parsley protoplasts. (A–C) GFP fluorescence, (D–F) corresponding bright-field illumination images of (A,D) GFP-BDL, (B,E) GFP with nuclear localization signal, and (C,F) GFP with nuclear export signal. Note strong accumulation of BDL-GFP in patches within the nucleus. (G) Yeast two-hybrid interaction assays. The strength of interaction is shown in activity units of the β-galactosidase reporter (β-Gal). BDL (IAA12) interacts with itself and with the C terminus of MP (ARF5) comprising domains III and IV (ARF5Ct). Negative controls: BDL tested against the empty prey vector (IAA12/−), and GNOM tested against the C terminus of MP (GN/ARF5Ct).
A prerequisite for in vivo interaction to occur is coexpression of the interacting partners. We compared the spatial and temporal accumulation patterns of BDL and MP mRNAs during embryogenesis (Fig. 3). At the octant stage, BDL and MP mRNAs accumulated in the proembryo cells derived from the apical daughter cell of the zygote (Fig. 3A,E). Subsequently, both mRNA species were gradually confined to subepidermal cells, eventually being restricted to the vascular precursor cells (Fig. 3B–D,F–H). At the basal end of early embryos, the hypophysis adjacent to the proembryo did not express either mRNA species (Fig. 3A,B,E,F, arrows). After the asymmetric division of the hypophysis, both BDL and MP mRNAs were observed in its smaller lens-shaped daughter cell that gives rise to the quiescent center of the root meristem (Fig. 3C,G, arrows). Thus, there was a coordinated basal shift in the expression domains of both genes at the globular stage. Late in embryogenesis, MP but not BDL was expressed in the central root cap, which was the only conspicuous difference between the two gene expression patterns (Fig. 3I,J, arrowheads). Since the bdl stabilizing mutation mimics the mp early-embryo phenotype, we examined the possibility that bdl might interfere with MP transcription. However, MP was expressed in the proembryo cells of bdl mutant early embryos (Fig. 3K,L), indicating that the bdl mutation does not abolish expression of the MP gene.
Promoter sequences of BODENLOS/IAA12 and SHY2/IAA3 confer different expression patterns
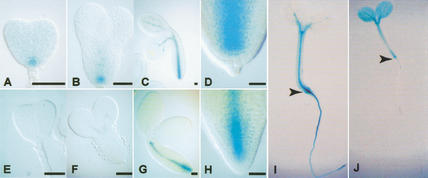
The bdl mutation causes an embryonic phenotype, whereas the identical mutation in the IAA3 gene only leads to a postembryonic defect that was opposite to its loss-of-function phenotype (Tian and Reed 1999). This raised the possibility that transcriptional regulation of IAA genes may limit their in vivo interactions with ARF proteins. We therefore compared the expression patterns of BDL and IAA3 promoter sequences that were transcriptionally fused to the β-glucuronidase (GUS) reporter (Reintanz et al. 2001). In contrast to BDL∷GUS (Fig. 4A–D), IAA3∷GUS expression was not detectable before the mid-torpedo stage of embryogenesis (Fig. 4E–H). Different expression patterns were also observed in seedlings (Fig. 4, cf. I and J; see also Tian et al. 2002 for IAA3∷GUS expression). Thus, transcriptional regulation can account for the phenotypic difference between identical mutations in two different IAA genes by limiting the choices of potential interaction between IAA and ARF proteins.
Figure 4.
IAA gene promoter-GUS reporter expression in embryos and seedlings. (A–D,I) pBDL∷GUS, (E–H,J) pIAA3∷GUS. Developmental stages (Jürgens and Mayer 1994): (A,E) triangular; (B,F) early-torpedo; (C,G) bent-cotyledon. (D,H) Root end of bent-cotyledon embryos shown at higher magnification. (I,J) Four-day-old seedlings; arrowheads mark the boundary between hypocotyl and root. Bar for A–H, 20 μm.
A model for BDL/MP action in root meristem initiation
Our results suggest that BDL and MP are interacting IAA and ARF proteins that are required for root meristem initiation in early embryogenesis. They act genetically in the same pathway, can interact physically, and are expressed in the same cells. Most IAA proteins, including IAA12, were shown to repress auxin response in the carrot protoplast transient expression assay (Tiwari et al. 2001). Furthermore, mutations in domain II that stabilize IAA proteins resulted in the inhibition of auxin-responsive reporter gene expression (Worley et al. 2000; Ouellet et al. 2001; Ramos et al. 2001; for review, see Rogg and Bartel 2001). Conversely, ARF5 encoded by the MP gene acts as an activator of auxin-responsive genes in carrot protoplasts (Hardtke and Berleth 1998; Ulmasov et al. 1999a), suggesting that loss-of-function mutations in the MP gene eliminate the capacity of ARF5 to activate auxin-responsive genes (Hardtke and Berleth 1998). Thus, failure to initiate the root meristem in early embryogenesis results from either of two genetic alterations: a stabilizing mutation of the auxin-response inhibitor BDL, or loss of the auxin-response activator MP. These observations suggest a simple model for their developmental action in early embryogenesis. BDL and MP may form a complex that prevents MP from activating target genes. BDL protein would be degraded in response to auxin, thus releasing MP.
Both BDL and MP are expressed in the proembryo cells but not in the adjacent hypophysis which normally initiates root meristem formation. This observation suggests that an auxin-dependent signal needs to be relayed from the proembryo to the adjacent extraembryonic cell, which in response switches to the hypophyseal cell fate (Hamann et al. 1999). It remains to be determined whether this apparent noncell autonomous effect of MP involves activation of auxin-response target genes that would then mediate signaling to the adjacent cell. Alternatively, MP protein may be transported into the presumptive hypophysis, which would be analogous to the movement of the SHORT ROOT transcription factor from the provascular cells to the adjacent ground tissue layer (Nakajima et al. 2001).
Materials and methods
Plant material and map-based cloning of the BDL gene
Plant growth conditions and the bdl mutant allele have been described (Hamann et al. 1999). From a cross between bdl/BDL (Ler) and wild type (Nd), 480 bdl mutant F2 seedlings were selected to map the gene against CAPS or SSLP markers (www.arabidopsis.org). After the interval was narrowed down to a small region at the top of chromosome I, wild-type and mutant alleles of the candidate gene were sequenced. Genomic fragments from 2.4 kb upstream of the translation start to 0.75 kb downstream of the BDL gene were amplified by PCR from wild-type and bdl mutant genomic DNA, using a Roche High Fidelity amplification kit (primer sequences, 5′-GGTTGGACTTGC TAAGCACTG-3′ and 3′-CCATGTGGTAGTGTCGAGAAGG-5′). Fragments were subcloned in pGEM (Promega), sequenced, and transferred in pBAR vector by cutting with Eco52I, blunting and ligating via the SmaI site of the vector (Völker et al. 2001). Constructs were introduced into A. tumefaciens strain GV 3101 (Clough and Bent 1998). Plants were transformed using the floral-dip method, and transgenic seedlings were selected on plates (2.1 g MS salts per L [Duchefa], 1% bactoagar, 1% sucrose) or soil, using 4 mg/L phosphinotricin (Clough and Bent 1998).
In situ hybridization
RNA in situ hybridization was performed as described, except that Western Blue (Promega) was used for staining (Mayer et al. 1998). The fragment for the BDL antisense probe was amplified by PCR, and restriction sites for HindIII/XhoI were introduced (primer sequences, 5′-TTCT CGAGGAATTTCGAGAGCCATTGAGTGG-3′ and 3′-AACAAGCTTAGTCCACTAGCTTCT GAGGTTCC-5′). The fragment was inserted in pBssk−(MBI). Linearization of the antisense probe was done with Bsp120I and transcription with T3 RNA polymerase (hydrolysis time 37 sec; Promega transcription kit). Clones for MP antisense probes were transcribed as described (Hardtke and Berleth 1998).
Nuclear localization studies
Full-length cDNA was amplified from the BDL cDNA by PCR, and BamHI/XbaI restriction sites were introduced at the ends (5′-CGG GATCCATGCGTGGTGTGTCAGAATTG-3′ and 3′-GCTCTAGACTA AACAGGGTTGTTTCTTTGT-5′). The fragment was then inserted in frame in the transfection vector RT103 and sequenced. Nuclear (NLS-GFP) and cytosolic (NES-GFP) controls were provided by K. Harter (University of Freiburg, Germany). Parsley protoplasts were isolated and transformed as described (Kircher et al. 1999).
Yeast interaction assays
The cDNA encoding the C terminus of MP (amino acids 753–902) was amplified from a flower/young silique cDNA library (primer sequences, 5′-CCGCTCGAGTTATGAAACAGAAGTCTTAAG-3′ and 3′-CCGGA ATTCGATTTTGATGATTGTAGTCTG-5′), subcloned in pGEM (Promega) and sequenced before in-frame insertion in yeast two-hybrid vectors pJG4-5 or pEG202 via EcoRI/XhoI restriction sites (Grebe et al. 2000). BDL was amplified from a full-length BDL cDNA clone by PCR (primer sequences, 5′-CGGAATTCATGCGTGGTGTGTCAGAATTG3′ and 3′-GCCTCGAGCTAAACAGGGTTGTTTCTTTGT-5′), and EcoRI/XhoI sites were introduced. The fragment was subcloned in pGEM and sequenced before in-frame insertion in pJG4-5 or pEG202 via EcoRI/XhoI sites. The GNOM (amino acids 1–246) control construct has been described (Grebe et al. 2000). For yeast transformation, cultures, and assays, standard techniques were employed as described (Ausubel et al. 2000).
Analysis of GUS expression
Promoter GUS fusions were generated by replacing the 35S promotor of pVKH35sGUSpA with PCR-amplified 1.76 kb or 1.96 kb fragments upstream of the translational start sites of IAA3 and IAA12 genes (IAA3 5′-CGCGAGCTCTCGTTGGTTAAGTGCTAAAAAT-3′, 3′-CGC GGATCCTTCTTCAAGAATTGCAGGAGAA-5′; IAA12 5′-CGCGAG CTCTATGCCGTATAGTTCGTATCAA-3′, 3′-CGCGGATCCGTCAAT AACAAAACCCTAGACT-5′; Reintanz et al. 2001). A BamHI site was introduced at one end while the other was cleaved with either PvuII (IAA 12) or HincII (IAA3). The promoter fragment was cloned in the plant transformation vector pVKH35sGUSpA via BamHI/Ecl136I (IAA12) or BamHI/HincII (IAA3). Agrobacterium and plant transformations were performed as described above. Transgenic embryos and 4-day-old seedlings were stained with variable concentrations of potassium ferricyanide/ferrocyanide (0.5, 1, 2 mM each) for up to 20 h (Malamy and Benfey 1997). Stained embryos were cleared in a chloralhydrate:water:glycerol solution (8:2:1), stained seedlings were cleared as described (Malamy and Benfey 1997).
Microscopic analysis, image processing, and computer work
Microscopic analysis was done on a Zeiss Axiophot or a Leica confocal laser scanning microscope with TCS-NT imaging software (Leica); pictures were taken with a Nikon Coolpix 990 camera. Primer design and sequence analysis were performed using Mac-Vector (Kodak Imaging Systems) and Vector NTI (Informax). Database searches were done using the BLAST algorithm (Altschul et al. 1990). Images were processed with Adobe Photoshop and Adobe Illustrator (Adobe Systems).
Acknowledgments
We thank C. Maulbetsch for isolating BDL cDNA clones; T. Berleth and J. Friml for providing clones for in situ probes; K. Harter for making available the parsley protoplast system; and J. Friml, N. Geldner, M. Griffith, C. Schwechheimer, D. Weigel, and D. Weijers for helpful comments and critical reading of the manuscript. This work was supported by Sonderforschungsbereich 446 “Mechanismen des Zellverhaltens bei Eukaryoten.”
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gerd.juergens@zmbp.uni-tuebingen.de; FAX 49-7071-295797.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.229402.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman GJ, Smith JA, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley & Sons; 2000. [Google Scholar]
- Berleth T, Jürgens G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUU/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld J-U, Salchert K, Koncz C, Jürgens G. A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. J Plant Growth Regul. 2001;20:281–291. [Google Scholar]
- Hamann T, Mayer U, Jürgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J. 2001;20:3609–3616. doi: 10.1093/emboj/20.14.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. Arabidopsis. In: Bard JBL, editor. EMBRYOS: Colour atlas of development. London, UK: Wolfe Publishing; 1994. pp. 7–21. [Google Scholar]
- Kim J, Harter K, Theologis A. Protein–protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rugner A, Schäfer E, Harter K. Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J Cell Biol. 1999;144:201–211. doi: 10.1083/jcb.144.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. Intercellular movement of the putative transcription factor SHR in root patterning. Nature. 2001;413:307–311. doi: 10.1038/35095061. [DOI] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3. Biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13:829–842. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J W. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001;6:420–425. doi: 10.1016/s1360-1385(01)02042-8. [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalgzyk M, Sandberg G, Godde M, Uhl R, Palme K. bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogg LE, Bartel B. Auxin signaling: Derepression through regulated degradation. Dev Cell. 2001;1:595–604. doi: 10.1016/s1534-5807(01)00077-6. [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B. A gain-of-function mutation in iaa28 suppresses lateral root development. Plant Cell. 2001;13:465–480. doi: 10.1105/tpc.13.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O. Changes in auxin response from mutations in an AUX/IAA gene. Science. 1998;279:1371–1373. doi: 10.1126/science.279.5355.1371. [DOI] [PubMed] [Google Scholar]
- Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang X-J, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci. 1999a;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999b;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Völker A, Stierhof Y-D, Jürgens G. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- Worley CK, Zenser N, Ramos J, Rouse D, Leyser O, Theologis A, Callis J. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 2000;21:553–562. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci. 2001;98:11795–11800. doi: 10.1073/pnas.211312798. [DOI] [PMC free article] [PubMed] [Google Scholar]