Abstract
The physiological role of Escherichia coli Spot 42 RNA has remained obscure, even though the 109-nucleotide RNA was discovered almost three decades ago. Structural features of Spot 42 RNA and previous work suggested to us that the RNA might be a regulator of discoordinate gene expression of the galactose operon, a control that is only understood at the phenomenological level. The effects of controlled expression of Spot 42 RNA or deleting the gene (spf) encoding the RNA supported this hypothesis. Down-regulation of galK expression, the third gene in the gal operon, was only observed in the presence of Spot 42 RNA and required growth conditions that caused derepression of the spf gene. Subsequent biochemical studies showed that Spot 42 RNA specifically bound at the galK Shine-Dalgarno region of the galETKM mRNA, thereby blocking ribosome binding. We conclude that Spot 42 RNA is an antisense RNA that acts to differentially regulate genes that are expressed from the same transcription unit. Our results reveal an interesting mechanism by which the expression of a promoter distal gene in an operon can be modulated and underline the importance of antisense control in bacterial gene regulation.
Keywords: Antisense RNA, galactose operon, riboregulation, small RNAs, Spot 42 RNA, translational regulation
Over the past two decades it has become clear that cells contain a variety of small untranslated RNA molecules (sRNAs). These RNAs play important or essential roles and regulate diverse cellular functions such as RNA processing and splicing, RNA editing and modification, protein stability and secretion, mRNA stability, and translation. Posttranscriptional control by sRNAs has been documented in bacteria and in a variety of eukaryotic cells including nematodes, plants, and mammals (Jorgensen et al. 1998; Panning and Jaenisch 1998; Eddy 1999; Wassarman et al. 1999; Adoutte 2000). The majority of these riboregulators act as antisense RNAs that pair to complementary regions on target RNAs. Antisense control was first discovered in plasmids, bacteriophages, and transposable elements. In all these cases, the antisense and target RNAs are transcribed from opposite DNA strands of the same region (i.e., cis-encoded; Wagner and Simons 1994; Zeiler and Simon 1996). In a growing number of cases, however, the antisense and target RNAs are expressed from unlinked genes, and complementarity is incomplete and relatively modest. Such trans-encoded antisense RNAs can carry separate determinants for binding/regulation, function by more than one mechanism, and affect more than one target gene (for review, see Altuvia and Wagner 2000).
Until recently only a dozen sRNAs in addition to tRNAs and 5S RNA were known to be encoded by the E. coli genome (for review, see Wassarman et al. 1999; Urbanowski et al. 2000; Majdalani et al. 2001). However, novel approaches including genomic mining have increased the number to more than 40 (Argaman et al. 2001; Rivas et al. 2001; Wassarman et al. 2001). Thus, sRNAs are much more widespread than previously imagined. At present three of the sRNAs are known to be antisense regulators (i.e., OxyS, DsrA, and MicF) that act at the level of translation initiation by base-pairing to the 5′ end of target mRNAs. They either repress translation by interfering with ribosome binding or activate translation by interfering with an inhibitory intramolecularly base-paired structure of the target RNA (Mizuno et al. 1984; Altuvia et al. 1998; Lease et al. 1998; Majdalani et al. 1998; Delihas and Forst 2001). Also, unique functions have been discovered for most of the other well-known sRNAs (Romeo 1998; Wassarman and Storz 2000; for review, see Wassarman et al. 1999); however, the role of the Spot 42 RNA has not yet been established.
Spot 42 RNA was discovered nearly three decades ago by two-dimensional (2D) gel electrophoresis as the most apparent RNA of E. coli when cells were labeled briefly with [32P]phosphate (Ikemura and Dahlberg 1973; Sahagan and Dahlberg 1979). The RNA is transcribed from the spf (spot forty-two) gene, whose expression is negatively regulated by the cAMP–CRP complex (Joyce and Grindley 1982; Rice and Dahlberg 1982; Polayes et al. 1988b). The gene includes a ρ-independent terminator site, and the mature RNA is unmodified and identical to the primary transcript. Spot 42 RNA has a half-life of 12 to 13 min at 37°C and is present in ∼200 copies per cell when cells are growing in media supplemented with glucose (Sahagan and Dahlberg 1979). This number is reduced three- to fivefold when cells are grown on nonglucose carbon sources or when cAMP is added to a glucose-grown culture (Polayes et al. 1988b). The spf gene is dispensable, and no major defects are observed in Spot 42 RNA null mutants (Hatfull and Joyce 1986; Polayes et al. 1988a). A 10-fold overproduction of Spot 42 RNA resulted in pronounced growth defects under a variety of conditions (e.g., small colony size and slow growth, inability to grow on succinate as carbon source and to respond normally to nutritional up-shift; Rice and Dahlberg 1982). However, in spite of this knowledge, the function of Spot 42 RNA has remained elusive.
Here we show that the spf gene is absolutely required for discoordinate expression of the gal operon cistrons. Moreover, in vitro studies show that Spot 42 RNA specifically binds at the galK Shine-Dalgarno region of the gal mRNA and that the RNA interferes with the formation of galK-30S initiation complexes. The data are discussed in terms of a model wherein Spot 42 RNA acts as an antisense RNA to differentially regulate the gal genes by competing with 30S ribosomes for binding to the galK translational initiation region.
Results
What is the function of Spot 42 RNA?
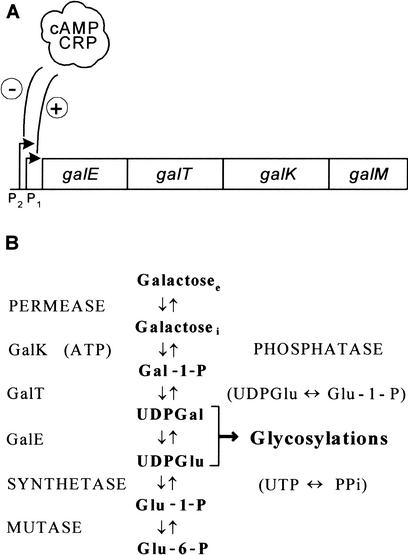
The E. coli cAMP receptor protein (CRP) is a global transcriptional regulator that can serve as an activator, repressor, coactivator, and corepressor (Kallipolitis et al. 1997; Busby and Ebright 1999). Moreover, work on the galactose operon has revealed that the cAMP–CRP complex, directly or indirectly, influences synthesis of the products of this operon differentially (Ullmann et al. 1979; Joseph et al. 1981; Adhya 1987). The E. coli galETKM operon is unusual in two respects. First, it possesses two overlapping promoters (Fig. 1). The cAMP–CRP complex stimulates transcription from the downstream promoter P1 and inhibits transcription from the upstream promoter P2. Second, although the genes are translated from a polycistronic mRNA, the relative synthesis of the enzymes UDP-galactose-4-epimerase (GalE), galactose-1-phosphate uridyl transferase (GalT), and galactokinase (GalK) has been found to vary under different metabolic conditions, a phenomenon that has been termed discoordinate expression. When cAMP levels are reduced by mutations (cya−) or catabolite repression (glucose as carbon source), the levels of GalK and GalT decline (the GalK level, in particular), but the GalE level remains high, resulting in an approximately fourfold increase in the GalE-to-GalK ratio. We note, however, that there are minor disagreements as to how cAMP levels or carbon source affect galE expression (Ullmann et al. 1979; Joseph et al. 1981; Adhya 1987). The discoordinate expression is more severe in the absence of a functional cAMP–CRP complex and can be suppressed in glucose-grown cells or in cya− strains by the addition of cAMP to the growth medium. It has been proposed that transcription termination at intercistronic regions, preferential degradation of the promoter distal portion of the mRNA, or different translational efficiencies of the two gal transcripts could account for the regulation (Ullmann et al. 1979; Joseph et al. 1981; Queen and Rosenberg 1981; Adhya 1987).
Figure 1.
The E. coli galactose operon. (A) Schematic diagram of the E. coli galactose operon. Relative positions of the four genes are shown. The genes encode: galE, epimerase (GalE); galT, transferase (GalT); galK, kinase (GalK); and galM, mutarotase. The two promoters (P1 and P2) are modulated negatively by Gal repressor, and the cAMP–CRP complex abolishes transcription from P2 and is required for P1 transcription. (B) The pathway for galactose metabolism. Note that the GalE, GalT, and GalK enzymes are part of an amphibolic pathway that produces substrates for biosynthetic glycosylations (UDP-glucose and UDP-galactose). Moreover, the pathway generates galactose from UDP-galactose when cells are growing in other carbon sources. Subscripts: (e) extracellular; (i) intracellular.
Several observations suggested to us that Spot 42 RNA is involved in mediating discoordinate expression of the gal operon. First, BLAST searches revealed that Spot 42 RNA has significant complementarity to the region surrounding the translation initiation region of galK. Second, Spot 42 RNA was predicted to form three stem–loop structures in which the first hairpin contains a seven-nucleotide single-stranded loop presented by a loosely paired top stem of interrupted helicity. Such features are often observed in antisense RNA regulatory units (Wagner and Brantl 1998). Third, the opposing regulation of spf and galK by the cAMP–CRP complex was striking.
Effects of Spot 42 RNA on plasmid-encoded GalE, GalT, and GalK proteins
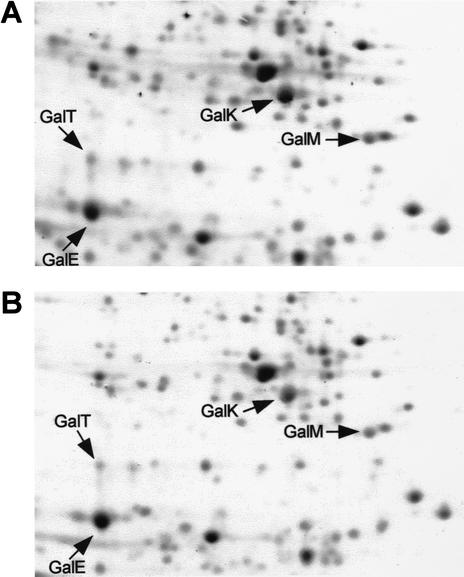
To assess the role of Spot 42 RNA on gal operon regulation, we first investigated the effect of controlled Spot 42 RNA expression on the GalE, GalT, GalK, and GalM protein synthesis pattern by 2D gel electrophoresis. To this end we constructed a low-copy-number plasmid (pSpf33) that expressed spf under the control of the inducible araBAD promoter. Moreover, we constructed a Δspf strain (SØ928Δspf) carrying the entire gal operon on plasmid pBR322 (pGal4). This strain was transformed with pSpf33 and as a control with pBAD33. Uninduced and induced cultures of the transformed strains were pulse-labeled with [35S]methionine, and their proteins were analyzed by standard 2D gel electrophoresis and autoradiography (Fig. 2). Induction of spf expression had little effect on the net synthesis rate of the GalE, T, and M polypeptides (∼10% increase of GalE, ∼10% decrease of GalT, and ∼15% decrease of GalM synthesis), whereas a significant reduction of the net rate of synthesis (∼twofold) was observed for GalK (Fig. 2, cf. panel A, uninduced culture, with panel B, induced culture; see legend to Fig. 2). Addition of arabinose to the control strain (SØ928Δspf/pGal4, pBAD33) had no effect on the net synthesis rates of the four Gal polypeptides (data not shown). These results showed that expression of spf specifically affected GalK synthesis, supporting the idea that Spot 42 RNA might be responsible for discoordinate expression of the gal operon.
Figure 2.
Effects of controlled spf expression on the Gal enzyme pattern. The figure shows the relevant part of two-dimensional gels of SØ928 Δspf cells harboring pGal4 (a derivative of pBR322 carrying the galETKM genes) and pSpf33 (a derivative of pBAD33 carrying the spf gene under the control of the inducible araBAD promoter). Uninduced and induced cultures (induced with 1 mM arabinose for 20 min) were labeled with [35S]methionine for 1 min and concentrated. Following this, their proteins were analyzed by two-dimensional gel electrophoresis and autoradiography. (A) Uninduced cells. (B) Induced cells. Arrows mark the positions of the four Gal enzymes. After quantification of the Gal protein spots from uninduced cells (A) and induced cells (B), the following ratios were obtained: GalEB/GalEA = 1.09, GalTB/GalTA = 0.92, GalKB/GalKA = 0.47, and GalMB/GalMA = 0.83 (all normalized to standard spots that were not affected by Spot 42 RNA).
Effect of deleting the spf gene on gal operon expression
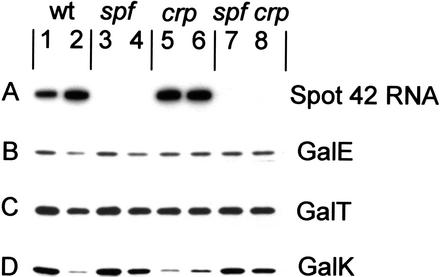
We next examined the consequences of deleting the spf gene. For these studies, we constructed isogenic spf+ and Δspf strains in gal constitutive (i.e., galR−) crp+ and Δcrp backgrounds. The various strains were grown in minimal medium with glucose or glycerol as carbon and energy source to an OD600 of 0.4. Following this, Spot 42 RNA levels and GalE, GalT, and GalK levels were determined by Northern blot and Western blot analysis, respectively. The results of the experiments are presented in Figure 3. We emphasize the following observations. Expression of galE and galT was not significantly altered by deleting spf (Fig. 3, cf. lanes 1–2 with lanes 3–4). In accordance with previous work, the expression of spf was enhanced (Sahagan and Dahlberg 1979; Polayes et al. 1988b) and the expression of galK was reduced in cells lacking the cAMP–CRP complex (Fig. 3, lanes 5,6) or deficient in cAMP and CRP (Fig. 3, lane 2, glucose grown; Ullmann et al. 1979; Joseph et al. 1981; Queen and Rosenberg 1981; Adhya 1987). Strikingly, this down-regulation of galK was eliminated in the Δspf strains (Fig. 3, cf. lane 2 with lane 4, and lanes 5–6 with lanes 7–8). For the spf mutant, expression of the three gal genes was slightly higher in glycerol-grown cells (Fig. 3, lane 3) than in glucose-grown cells (Fig. 3, lane 4), and a similar expression was observed for galE and galT in the wild- type strain (Fig. 3, cf. lanes 1–2 with lanes 3–4). Accordingly, previous work has shown that the addition of cAMP to a Δcya, galOc81 mutant stimulated galE expression 40%–70% (Joseph et al. 1981), and that galT expression in gal constitutive strains (galR− or galOc81) was 40%–70% higher in glycerol-grown cells than in glucose-grown cells (Adhya and Echols 1966). On this basis we conclude that cAMP–CRP also stimulates galE expression. Furthermore, the levels of the three Gal enzymes increased coordinately in the spf mutant when glycerol was used as carbon source (Fig. 3, lanes 3,4; see legend), indicating that transcription initiation is more efficient from the cAMP–CRP-dependent P1 promoter than from the cAMP–CRP-repressed P2 promoter (Fig. 1). Finally, we note that a similar effect of Spot 42 RNA and carbon source on gal operon expression was observed with galR+ strains (data not shown). Taken together our results corroborate the hypothesis that discoordinate expression of the gal operon is dependent on the presence of Spot 42 RNA. Moreover, the data suggest that the cAMP effect on galET expression is transcriptional.
Figure 3.
Effects of Spot 42 RNA on the cellular levels of the Gal enzymes. (A) Northern blot analysis of Spot 42 RNA levels and (B–D) immunoblots of GalE, GalT, and GalK levels in isogenic gal constitutive spf+ and Δspf strains. The RNA and the proteins from equal amounts of cells growing to early-exponential phase were electrophoretically separated on a urea–8% polyacrylamide gel and on an SDS–10% polyacrylamide gel, respectively, and probed as described in Materials and Methods. (Lanes 1,3,5,7) Glycerol-grown cells; (lanes 2,4,6,8) glucose-grown cells. In all experiments casamino acids (0.05%) were included in the growth medium. After quantification of the Gal proteins from glycerol- and glucose-grown ΔgalR and ΔgalR, Δspf cells, the following ratios were obtained for GalE, GalT, and GalK, respectively: ΔgalR strain (lane 1/lane 2) 1.5, 1.4, and 4.9; ΔgalR, Δspf strain (lane 3/lane 4) 1.4, 1.4, and 1.4. The results are the average of three independent experiments.
The secondary structure of Spot 42 RNA
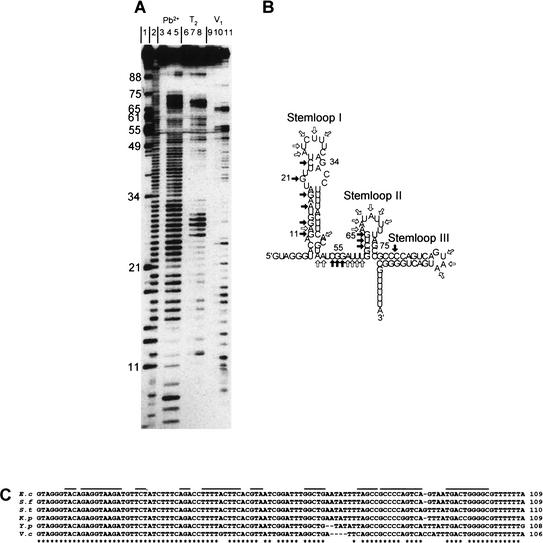
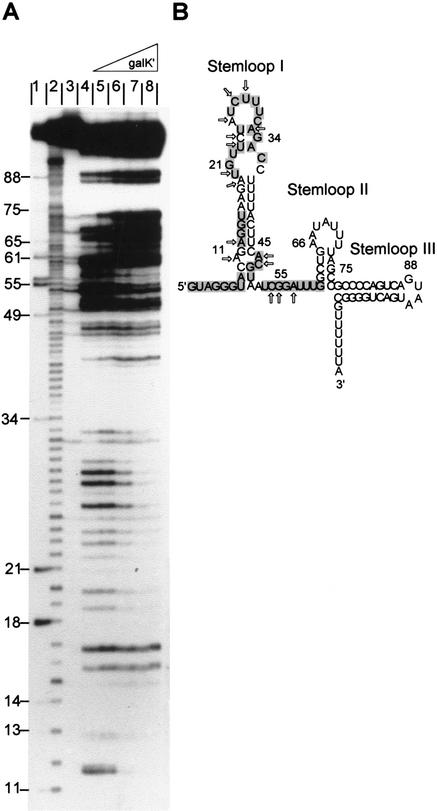
To learn more about the mechanism of Spot 42 RNA action, we carried out structural studies. The secondary structure of the RNA, predicted by the folding program Mfold, contains three stem–loop structures (Fig. 4B). To evaluate this structure, the conformation of Spot 42 RNA in solution was deduced by structural probing using RNase T2, RNase V1 and lead (II) acetate, and 5′-end-labeled Spot 42 RNA. The results are presented in Figure 4A, and the structural information derived from these analyses is summarized in Figure 4B. Partial cleavage with the single-strand-specific endonuclease RNase T2 confirmed the presence of three loop regions and an unstructured segment between the first and second hairpins (Fig. 4A, lanes 7,8). Strikingly, the predicted double-stranded region of hairpin I was also accessible to cleavage by RNase T2 (e.g., positions 12 and 16–17), indicating that nucleotides in this region of Spot 42 RNA are only loosely paired. Probing with lead (II) acetate, which preferentially cleaves in single-stranded regions, supported this interpretation (Fig. 4A, lanes 4,5).
Figure 4.
Secondary structure of Spot 42 RNA. (A) Nuclease and lead (II) acetate probing of 5′-end-labeled Spot 42 RNA. (Lane 1) RNase T1 cleavage of Spot 42 RNA under denaturing condition (G-specific cleavage); (lane 2) alkaline hydrolysis ladder; (lanes 3,6,9) control (untreated Spot 42 RNA); (lanes 4,5), partial cleavage by lead (II) acetate; (lanes 7,8) partial cleavage by RNase T2; (lanes 10,11) partial cleavage by RNase V1. (B) Structure predictions of Spot 42 RNA based on thermodynamic criteria and summary of the structural probing. Single-stranded-specific and double-stranded-specific nuclease susceptibility are indicated by open and filled arrows, respectively. (C) Phylogenetic structure analysis of Spot 42 RNA. The DNA sequences of Spot 42 RNA from E. coli, Shigella flexneri, Salmonella typhimurium, Klebsiella pneumoniae, Yersinia pestis and Vibrio cholerae were aligned. A star indicates identity. The inverted repeats predicted to fold into stem structures are overlined. Sequences for Shigella, Salmonella, Klebsiella, and Yersinia were obtained from a BLAST search of the E. coli sequence with the unfinished microbial genome database at the Institute for Genomic Research Web site at http://www.tigr.org. Shigella and Yersinia sequencing data were from the University of Wisconsin–Madison Genome Project. The Washington University Consortium sequencing project obtained the data for the Salmonella and Klebsiella sequences.
We further characterized the secondary structure of Spot 42 RNA by treatment with RNase V1, which can cleave double-stranded RNA of 4–6 bp and may also cleave stacked nucleotides in single-stranded regions (Fig. 4A, lanes 10,11). The cleavage pattern suggested the existence of several regions of double-stranded nature within Spot 42 RNA (stems I and II). The nucleotides (52A, 55G, 56G, and 57A) that are accessible to RNase V1, Pb2+, and RNase T2 cleavage are likely to be stacked. Taken together, the probing data are consistent with the conformation predicted for Spot 42 RNA. Furthermore, the data suggest a dynamic or loosely paired character of hairpin I.
Support for the predicted structure was also obtained by comparative studies. Spot 42 RNA is highly conserved in Klebsiella, Shigella, Salmonella, Yersinia, and Vibrio species (Fig. 4C). Sequence variations mostly occur in regions predicted to be single-stranded (i.e., loops II and III and the linker region between hairpins I and II) or, when in regions predicted to be double-stranded, are compensated by alternative pairing or by second-site mutations that restore base-pairing. Hence, the phylogenetic studies support the structural analyses.
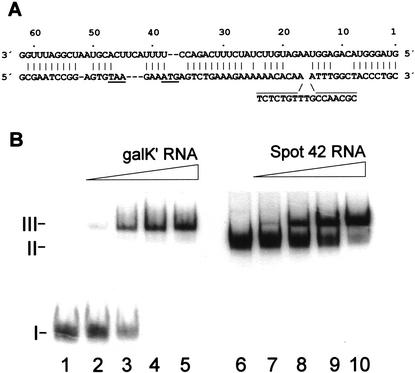
Spot 42 RNA binds to the translational initiation region of galK
Spot 42 RNA has significant sequence complementarity to the gal mRNA in the region surrounding the translational initiation region of galK (Fig. 5A), and, strikingly, the galK complementary regions reside within exposed regions of Spot 42 RNA (Fig. 6B). To test whether Spot 42 RNA might bind the complementary regions, we performed binding experiments with 5′-end-labeled Spot 42 RNA and an unlabeled galK‘ RNA substrate that carried the galK Shine–Dalgarno sequence in the central portion of the molecule. Samples containing a fixed amount of Spot 42 RNA, a 500-fold excess of tRNA, and increasing amounts of galK‘ RNA were incubated on ice; formation of complexes was assayed in gel-retardation assays. The results, presented in Figure 5B, show that Spot 42 RNA and the galK‘ RNA formed a single retarded complex. A similar complex was observed by adding unlabeled Spot 42 RNA to incubations containing a fixed amount of 5′-end-labeled galK‘ RNA. Addition of excess unlabeled galT‘ RNA substrate, which carried the distal part of galE and the proximal part of galT (i.e., the galT translational initiation region), did not affect formation of the retarded complex (data not shown). We infer that Spot 42 RNA specifically binds the galK‘ RNA, supporting the idea that regulation of galK expression occurs via RNA–RNA interaction.
Figure 5.
Spot 42 RNA–galK mRNA interaction. (A) The complementarity between Spot 42 RNA (top sequence) and the galK translation initiation region (bottom sequence). The stop codon of galT and the start codon of galK are underlined. (B) Gel mobility shift assays of Spot 42 RNA binding to galK‘ RNA. (Lanes 1–5) 5′-End-labeled transcript of Spot 42 RNA (0.05 pmole) and 500-fold molar excess tRNA were incubated with increasing amounts of unlabeled galK‘ RNA (0, 1, 5, 10, and 20 pmole) to allow complex formation and then resolved on a native polyacrylamide gel. (Lanes 6–10) 5′-End-labeled galK‘ RNA (0.05 pmole) and 500-fold molar excess of tRNA were incubated with increasing amounts of unlabeled Spot 42 RNA (0, 1, 5, 10, and 20 pmole) to allow complex formation and then resolved on a native polyacrylamide gel.
Figure 6.
Nuclease probing of Spot 42 RNA–galK‘ RNA complexes. (A) In vitro synthesized 5′-end-labeled Spot 42 RNA (0.05 pmole) was mixed with increasing concentrations of unlabeled galK‘ mRNA (1, 5, 10, and 20 pmole), and the incubations were treated with RNase T2. (Lane 1) RNase T1 cleavage of Spot 42 RNA under denaturing condition (G-specific cleavage); (lane 2) alkaline hydrolysis ladder; (lane 3) control (untreated Spot 42 RNA); (lanes 4–8) RNase T2 footprinting reactions in absence (lane 4) and in presence (lanes 5–8) of galK‘ RNA. (B) Summary of changes in the cleavage pattern induced by galK‘ RNA. The galK complementary regions are shown on a gray background. Arrows indicate reduced cleavage.
We used structural probing for further characterization of galK–Spot 42 RNA interaction. Here, 5′-end-labeled Spot 42 RNA was mixed with increasing concentrations of unlabeled galK‘ RNA, and the incubations were treated with RNase T2. The structural probing is presented in Figure 6 and shows that additions of galK‘ RNA reduced or abolished cleavage by RNase T2 in the 5′ tail, loop I, and at positions 47–48 and 53–58. Thus, three short antisense sequences exposed in single-stranded regions of Spot 42 RNA appear to interact with separate target sites located at or near the translational initiation region of galK (Fig. 5A).
Spot 42 RNA prevents ribosome binding
To test whether Spot 42 RNA binding to galK mRNA might interfere with ribosome binding, we used toeprinting assays (Hartz et al. 1989). Briefly, in the presence of initiator tRNA (fMet-tRNA), the ribosomal 30S subunits bind to the Shine–Dalgarno sequence of mRNAs and block for reverse transcription primed downstream. The inhibition of primer extension generates a stop signal at the 3′ end of the bound 30S subunit (usually 15–17 nt downstream of the start codon). The strength of this stop signal provides a measure for the formation of the ternary complexes.
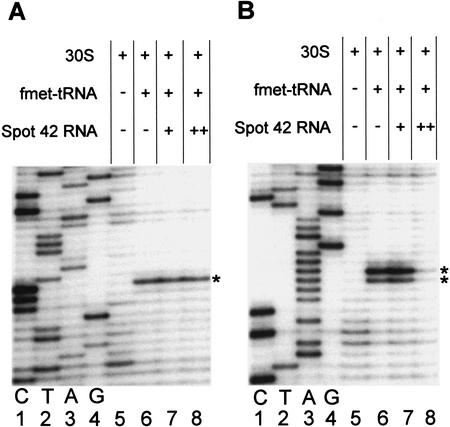
For the toeprint analysis we used the galT‘ and galK‘ RNA substrates. The probes were annealed with a 5′-end-labeled primer complementary to a region downstream of the translation start site and incubated with 30S subunits in the presence or absence of fMet-tRNA. As shown in Figure 7A, the addition of Spot 42 RNA at different molar ratios to galT‘ RNA before the addition of 30S subunits and fMet-tRNA did not interfere with ternary complex formation. In contrast, when the experiments were performed with the galK‘ RNA (Fig. 7B), the presence of Spot 42 RNA resulted in a diminished toeprint signal, and an almost complete inhibition of ternary complex formation was observed at a molar excess of 5:1 of Spot 42 RNA to galK‘ RNA (Fig. 7B, lane 8). A diminished toeprint signal, however, was not detected if 30S subunits and fMet-tRNA were added prior to Spot 42 RNA to incubations containing galK‘ RNA (data not shown). These assays show that Spot 42 RNA binding interferes directly with the formation of galK–30S initiation complexes.
Figure 7.
Toeprinting analysis of 30S ribosomal subunit binding to galT‘ and galK‘ RNA. (A) Ternary complex formation on galT‘ RNA. The toeprint signal is at position +15 relative to A of the start codon (marked by a star). (B) Ternary complex formation on galK‘ RNA. The toeprint signal is at positions +16 and +17 relative to A of the start codon (marked by stars). In all reactions the concentration of mRNA and 30S subunits was 0.04 μM. fMet-tRNA was in molar excess over 30S subunits. When present, Spot 42 RNA was added prior to the addition of 30S subunits and fMet-tRNA. Spot 42 RNA was added at the molar ratios to mRNA, 1:2 (lane 7) and 5:1 (lane 8). (Lanes 1–4) The DNA sequence reactions (G A T C) were carried out with the same end-labeled oligonucleotide used in the toeprinting assays.
Discussion
In this work we have provided insight into two longstanding questions concerning a physiological function of Spot 42 RNA and the regulation of discoordinate gene expression of the E. coli galactose operon. Our results establish that Spot 42 RNA is a riboregulator that acts to regulate the gal operon genes differentially under certain physiological conditions. Our results further show that Spot 42 RNA inhibits expression of galK, the third gene in the operon, by an antisense mechanism. Three short antisense sequences exposed in single-stranded regions of Spot 42 RNA appear to interact with three separate target sites located at or near the translational initiation region. Finally, toeprinting experiments with 30S ribosomal subunits revealed that galK–Spot 42 RNA interactions interfere with the formation of translational initiation complexes. Taken together, our results suggest that Spot 42 RNA inhibits galK translation by sterically hindering ribosome binding. As Spot 42 RNA regulation only interfered weakly with the expression of the two proximal genes and the distal gene of the gal operon (Figs. 2, 3), it seems reasonable to suppose that the stability of the gal message is not grossly altered by Spot 42 RNA interaction. This view is further supported by gene array analysis of the expression of the individual genes of the gal operon in spf+ and Δspf cells (T. Møller, unpubl.). Therefore, we favor the idea that Spot 42 RNA acts primarily at the level of translation initiation.
Previously, the stimulatory effect of cAMP on the GalT levels (∼1.5) and on the GalK levels (∼fourfold) led to the idea that cAMP–CRP acts, directly or indirectly, at the transcriptional level by modulating polarity (e.g., by interfering with the transcription terminator ρ; Ullmann et al. 1979). However, the hypothesis was based on the assumptions that the gal operon carried a single cAMP–CRP-independent promoter and that expression of the promoter proximal gene (galE) was unaffected by carbon source and/or cAMP. Moreover, the authors did not take into account the fact that galE expression was stimulated (40%–70%) by cAMP in a Δcya, galOc81 strain (Joseph et al. 1981). Also, when the amount of gal mRNA corresponding to promoter proximal and distal genes was measured in a Δcya strain, grown in the absence or in presence of cAMP, it was found that cAMP could only partly restore coordinate expression (Guidi-Rontani et al. 1984). Therefore, it was concluded that a translational control was probably involved in the discoordinate regulation of the gal operon (Guidi-Rontani et al. 1984). The present results on gal operon expression, consistent with data for gal operator constitutive mutants (Adhya and Echols 1966; Joseph et al. 1981), suggest that the cAMP effect on galE and galT expression is transcriptional. Thus, we showed that the expression of galE and galT in a ΔgalR strain and the expression of galE, galT, and gal K in a ΔgalR Δspf strain was affected to a very similar extent by carbon source (i.e., 40%–50% higher in glycerol-grown cells than in glucose-grown cells; see Fig. 3), indicating that transcription initiation is more efficient from the cAMP–CRP-dependent P1 promoter than from the cAMP–CRP-repressed P2 promoter (Fig. 1).
A second interesting aspect of Spot 42 RNA regulation concerns ongoing translation. Because sequences encoding the end of the galT gene overlap with sequences involved in ribosome binding and initiation of galK translation as well as with sequences involved in Spot 42 RNA binding (Fig. 5), a terminating ribosome would be expected to interfere with Spot 42 RNA binding. We speculate that the proposed interaction between Spot 42 RNA and separated target regions could be crucial for an efficient control. Thus, the three binding regions of Spot 42 RNA are all exposed, and each of these may possibly facilitate bimolecular RNA–RNA interaction. Moreover, the separated target regions within the coding region of galK may allow formation of initial duplex formation in the presence of a terminating ribosome. Such binding features could lead to an increase in the local concentration of Spot 42 RNA, and facilitate rapid formation of repression complexes.
Finally, we note that recent work has shown that Spot 42 RNA regulation of gal operon expression relies on the global posttranscriptional regulator Hfq, which shares functional and structural features with the eukaryotic Sm and Sm-like proteins (Møller et al. 2002; Zhang et al. 2002). In hfq− strains, the half-life of Spot 42 RNA is strongly reduced and Hfq binds specifically to the RNA. Moreover, in vitro Hfq strongly cooperates in complex formation between Spot 42 RNA and galK mRNA (Møller et al. 2002). Because Hfq also increases OxyS RNA interaction with target mRNAs (Zhang et al. 2002), is required for riboregulation of rpoS by DsrA and RprA RNA (Majdalani et al. 2001; Sledjeski et al. 2001), and binds to several other sRNAs (Wassarman et al. 2001), it has been proposed that Hfq acts as a general cofactor for antisense RNAs that rely on short stretches of base pairing (Wassarman et al. 2001; Møller et al. 2002; Zhang et al. 2002).
Physiological significance of Spot 42 RNA regulation
Despite the great understanding of D-galactose metabolism in E. coli, the physiological significance of the uncoordinated expression of the gal operon genes is still unclear. It is intriguing, however, that the GalE, T, and K enzymes form part of an amphibolic pathway that produces substrates for biosynthesis of lipopolysaccharides (i.e., UDP-glucose and UDP-galactose; see Fig. 1; for review, see Adhya 1987). When the cell derives its energy from galactose, all three enzymes are required. The net result is the conversion of galactose to the glycolytic intermediate glucose-1-phosphate. When the cell derives its energy from other carbon sources, a relatively high basal level of epimerase (GalE) is required to produce UDP-galactose for biosynthetic glycosylations. Under such metabolic conditions, the amphibolic pathway, together with a galactose-1-phosphatase activity, generates galactose from UDP-galactose (i.e., GalT is also required). However, the galactose generated intracellularly does not accumulate to a level that is high enough to induce the gal operon because it is either excreted or recycled. Excretion takes place in the presence of glucose, the preferred carbon source, whereas the intracellular galactose is recycled when the cell uses less favorable carbon sources. In accordance with this metabolism, discoordinate expression of the gal operon is operative when there is no need for GalK (glucose-grown cells), and we suggest that Spot 42 RNA repression of galK expression may help facilitate excretion of galactose. It is also possible that Spot 42 RNA regulation may be important for optimal utilization of carbon sources. These explanations assign a physiological significance to Spot 42 RNA regulation and reveal a form of cellular regulation that may play a decisive role in the fine-tuning of gene expression. We expect that additional operons required for appropriate carbon-source utilization are regulated in a similar fashion and note that one likely candidate is the sucABCD operon encoding subunits for two enzymes of the tricarboxylicacid cycle—ketoglutaratedehydrogenase (sucAB) and succinylcoenzymeA synthetase (sucCD). Thus, Spot 42 RNA has extended complementarity to sequences near the start of sucC, and one of the reported phenotypes associated with overproducing the sRNA is the inability to grow on succinate as energy and carbon source.
Materials and methods
Bacterial strains
All strains used were E. coli K-12 derivatives: SØ928 (Δdeo, Δlac; Valentin-Hansen et al. 1978); SØ2928 (Δdeo, Δlac, Δcrp-96zdh-732:Tn10; Søgaard-Andersen et al. 1991); MW130 (ΔgalR::CmR; Weickert and Adhya 1993); KM22 [Δ(recC, ptr, recD)::Plac-bet, exo, kan] (Murphy 1998), and MC1000Δssr [F−, araD139, Δ(argF-lac)U169, Δssr::apha]. The ΔgalR derivatives of SØ928 and SØ2928 were constructed by P1-transduction from MW130. The Δspf4 derivatives of SØ928ΔgalR and SØ2928ΔgalR strains were constructed by P1-transduction from KM22Δspf4. This strain was constructed by transforming strain KM22 with a SmaI-linearized pSpfΔ4 plasmid as described in Murphy (1998). PCR and Northern blot analysis verified gene replacement and the absence of Spot 42 RNA.
Plasmid construction
To construct pSpfΔ4, the DNA regions flanking the spf gene were amplified from SØ928 chromosomal DNA using primers ospfdel1 (5′-CCCGGATCCGCCGGGTGGCGCGCCGTC-3′) plus ospfdel2 (5′-GCCGGAATCCCGGACTCGAGTTGTTCA GTTCAGAAAGCAC-3′); and Ospfdel3 (5′-GCCGCTCGAGG GTACCTCACGAGCTCTAGCCGCCCCAGTCAGTAATG-3′) plus ospfdel4 (5′-GCCGGAATTCGTTAGCGTCCTCGCTCT ATGG-3′), respectively. The upstream DNA fragment was digested with EcoRI and BamHI and cloned into EcoRI/BamHI-restricted pUC19, generating pUspf. Subsequently, pUDspf was constructed by replacing the XhoI–EcoRI segment of pUspf with the downstream DNA fragment (digested with XhoI and EcoRI). The aphA gene was amplified from chromosomal DNA of strain MC1000Δssr using ossr1 (5′-GCCGGAGCTCCAGTTGGTC TTCATTGCCGCATTG-3′) and ossr2 (5′-CGGCGAGCTCAC GGGTACCGGGATTTACGATGGCAGGGCAGC-3′) as primers. The aphA fragment was digested with KpnI and cloned into the unique KpnI site of pUDspf, generating pSpfΔ1 and pSpfΔ4 (with aphA in the opposite or the same orientation as the original spf gene, respectively). Concerning construction of spf knockout strains, we did not observe any importance of the orientation of the kanamycin cassette.
To create pSpf33, two PCR products were generated using (1) pBAD33 as template and ospfara (5′-CATCTTACCTCTGTAC CCTACATGGAGAAACAGTAGAGAGTTG-3′) and o33mluI (5′-CAAAAACGCGTAACAAAAGTG-3′) as primers; and (2) SØ928 chromosomal DNA as template and oaraspf (5′-CAAC TCTCTACTGTTTCTCCATGTAGGGTACAGAGGTAAGA TG-3′) and ospfpstI (5′-CGCCTGCAGCGTTGGCCTGATA AACATAGCGC-3′) as primers. A third round of amplification with these two products as template and o33mluI and ospfpstI as primers generated a DNA fragment carrying the spf gene fused to the arabinose promotor. Finally, pSpf33 was constructed by replacing the MluI–PstI fragment of pBAD33 with the restricted araP–spf fragment.
Plasmid pSpf3 was constructed by replacing the EcoRI–BamHI fragment of pUC18 with a PCR-generated fragment, prepared using oT7spf (5′-CCGGATCCTGTAATACGACTCAC TATAGTAGGGTACAGAGGTAAG-3′) and o3spf (5′-CCGAA TTCGGATGTTCGCCCAATAAAAAACGC-3′) as primers and SØ928 chromosomal DNA as template, followed by digestion with EcoRI and BamHI.
Plasmid pFps was constructed by replacing the EcoRI–BamHI fragment of pGem4 (Promega) with a PCR-generated fragment, prepared using ospf1 (5′-GTTAGAATTCGCCTGAACATGCC 3′) and ospf2 (5′-CCGGATCCGTTGGCCTGATAAACATA GCGC-3′) as primers and SØ928 chromosomal DNA as template, followed by digestion with EcoRI and BamHI.
Plasmid pGal, containing galETK cloned between the EcoRI and PvuII sites of pBR322, was constructed as described (Busby and Dreyfus 1983). Plasmid pGal4 encoding the entire gal operon was generated by replacing the SapI–SapI fragment of pGal with a PCR-generated fragment, prepared using ogalM1 (5′-CGTCGCGCTGATCCCGGAAGAGCTGG-3′) and ogalM2 (5′-GCGCGCAGCGGAAGAGCAAAAAGCCCTCATGACGA GGGCGT-3′) as primers and SØ928 chromosomal DNA as template, followed by digestion with SapI.
Plasmids pH8-E, pH8-T, and pH8-K, which encode N-terminally 8xhistidine-tagged GalE, GalT, and GalK proteins from the IPTG-inducible phage T7-promoter derivative PA1/O4/O3, were constructed by replacing the EcoRI–BamHI fragment of pUHE24-2 (obtained from Hermann Bujard, University of Heidelberg, Germany) with PCR-generated fragments using pGal as template. The primers used were galE, oh8E (5′-CGC CCGAATTCATAAGGAGTTTTATAAATGCATCACCATCA CCATCACCATCACAGAGTTCTGGTTACCGGTGGTAGC3′) and oE (5′-GCCCCGGATCCGGCGATGTGGATGATCA ACGGG-3′); galT, oh8T (5′-CGCCCGAATTCATAAGAGT TTTATAAATGCATCACCATCACCATCACCATCACACGC AATTTAATCCCGTTGATCATC-3′) and oT (5′-GCCCCG GATCCCGCGGCCAGGCGCCTGAATGGTG-3′); galK, oh8K (5′-CGCCCGAATTCATAAGGAGTTTTATAAATGCATCACC ATCACCATCACCATCACAGTCTGAAAGAAAAAACACAA TCTC-3′) and oK (5′-GCCCCGGATCCCAGCGACAGCTTG CTGTACGGC-3′).
Growth media
Strains were grown at 37°C in AB minimal medium (Clark and Maaløe 1967) containing 1 μg/mL thiamine, 0.2% carbon source (glucose or glycerol), and 0.05% casamino acids. For the growth media used for [35S]methionine labeling, casamino acids were replaced by a 1/20 volume defined amino acid mixture (concentrations: L-alanine, 1.75 g/L; L-arginine, 1.75 g/L; L-histidine, 0.78 g/L; L-lysine, 1.5 g/L; L-proline, 1.15 g/L; L-threonine, 1.20 g/L; glycine, 1.50 g/L; L-asparagine, 1.33 g/L; L-glutamine, 2.18 g/L; L-isoleucine, 1.30 g/L; L-phenylalanine, 0.60 g/L; L-tyrosine, 0.90 g/L; L-valine, 1.75 g/L; L-aspartic acid, 1.33 g/L; L-glutamic acid, 2.2 g/L; L-cysteine, 0.30 g/L; L-serine, 25.0 g/L; L-tryptophan, 0.50 g/L). Ampicillin (100 μg/mL), spectinomycin (100 μg/mL), and chloramphenicol (50 μg/mL) were added where appropriate.
2D gel electrophoresis
Samples (1 mL) of exponentially grown cells were labeled with 10 μCi of carrier-free [35S]methionine for 1 min, followed by a chase with unlabeled methionine (final concentration 200 μg/mL) for 1 min. The chase was terminated by the addition of 5 μL of chloramphenicol (50 mg/mL). Cells were collected by centrifugation at 0°C, resuspended in 50 μL of loading buffer (10% sucrose, 3% SDS, 0.16 M Tris-HCl at pH 6.8, with freshly added β-mercaptoethanol), boiled for 3 min, and mixed with 25 μL of 10% NP-40. A 30-μL sample was loaded onto the first dimensional gel. Gel electrophoresis was carried out as described by O’Farrell (1975). Quantification of protein spots was done by PhosphorImager and the Imagequant software package (Molecular Dynamics).
Protein purification, antibody production, and Western blotting
The N-terminally histidine-tagged Gal enzymes were expressed from pH8-E, pH8-T, or pH8-K, respectively, in strain SØ928 harboring pMS421 (pSC101 derivative that encodes the lacIq allele; Grana et al. 1988). Exponentially grown cells were induced for 2 h by addition of IPTG to 500 μM. Protein purification was carried out using Talon Metal Affinity Resin (Clontech) according to the manufacturer's specifications (denaturing procedure with 8 M urea). Antisera were generated by immunizing rabbits with the purified proteins. For Western blot analysis, the strains were grown to an OD600 of 0.4. Equal amounts of total cellular extracts were electrophoretically separated on SDS–10% polyacrylamide gels, and blotted to an Immobilon-P membrane (Millipore). The membrane was probed with rabbit anti-GalK, anti-Gal E, or anti-GalK polyclonal antisera, and the antibody–antigene complexes were visualized with the ECL plus system (Amersham Pharmacia Biotech).
Synthesis, purification, and labeling of RNA
In vitro synthesis of RNA was carried out with T7 RNA polymerase (Promega) as described (Franch and Gerdes 1996). FokI-digested pSpf3 was used as the DNA template for synthesis of Spot 42 RNA. Two DNA templates carrying a T7 promoter were generated by PCR for synthesis of galK‘ RNA and galT‘ RNA, prepared using oT7ktp (5′-TGTAATACGACTCACTATA GGGCCTCTGCTGCGCTCCGCCACCG-3′) plus oktp (5′-GGTGCACAACTGATCACGGTTTG-3′); and oT7ttp (5′-TG TAATACGACTCACTATAGGGCGCGTCGCGAGGGCGAC CTTCCGG-3′) plus ottp (5′-GAGGAAGCAATCTGGATCGT GCGC-3′) as primers and plasmid pGal as template. The RNAs were purified and quantified as described (Thisted et al. 1994). Transcripts were radioactively labeled at the 5′ end with T4 kinase (Roche Molecular Biochemicals) and [γ-32P]ATP.
Northern blot analysis
Preparation of total RNA from E. coli and Northern blot analyses were carried out as described elsewhere (Franch et al. 1997). The 32P-labeled RNA probe used for detection of Spot 42 RNA was generated by in vitro transcription of plasmid pFps (linearized with EcoRI) with T7 RNA polymerase.
Gel mobility shift assay
The binding assays were performed as follows: 5′-end-labeled RNA (0.05 pmole of Spot 42 RNA or galK‘ RNA), 1 μg of tRNA (Boehringer Mannheim), and unlabeled RNA (1, 5, 10, and 20 pmole of Spot 42 RNA or galK‘ RNA) were mixed in binding buffer (20 mM Na-HEPES at pH 8, 100 mM KCl, 1 mM DTT, 1 mM MgCl2) and incubated on ice for 20 min. The binding reactions (10 μL) were mixed with 5 μL of loading dye (10% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol) and analyzed on a 5% nondenaturing polyacrylamide gel in TBE buffer at 200 V at 4°C for 1.5 h. Subsequently, the gel was dried and subjected to autoradiography.
Toeprinting analysis
Purification of ribosomal 30S subunits and toeprinting assays were carried out as described previously (Franch et al. 1997). The 5′-end-labeled oligonucleotides otpk (5′-GCCTGAATG GTGTGAGTGG-3′), complementary to the galK gene, and otpt (5′-GAAACCAGAATCCATTGCCC-3′), complementary to the galT gene, were used as primers for cDNA synthesis in the toeprinting reactions.
Enzymatic and chemical probing of Spot 42 RNA
RNase T2 (Sigma) and RNase V1 (Amersham Pharmacia Biotech) digestion was performed as described previously (Franch et al. 1997). 5′-End-labeled Spot 42 RNA (0.05 pmole) was incubated in RNase buffer (20 mM Tris-acetate at pH 7.8, 100 mM NH4Cl, 10 mM Mg-acetate, 200 mM K-glutamate, and 5 μg of tRNA) at 37°C. Partial digestion was carried out with 0.001 units of RNase T2 or 0.005 units of RNase V1 for 3 min and 15 min. For lead (II) acetate probing, 5′-end-labeled Spot 42 RNA (0.05 pmole) was incubated in 50 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, and 50 mM KCl, at 37°C. Samples were treated with Pb(Ac)2 (final concentration, 5 mM) for 3 min and 15 min. Alkaline hydrolysis and RNase T1 cleavage of Spot 42 RNA were carried out as described in Franch et al. (1999).
Acknowledgments
This work was supported by grants from the Danish Natural Science Research Council (P.V-H.) and the Carlsberg Foundation (T.F.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL valentin@bmb.sdu.dk; FAX 45-6550-2467.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.231702.
References
- Adhya S. The galactose operon. In: In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella thyphimurium: Cellular and molecular biology. Washington, DC.: American Society for Microbiology; 1987. pp. 1503–1512. [Google Scholar]
- Adhya S, Echols H. Glucose effect and the galactose enzymes of Escherichia coli: Correlation between glucose inhibition of induction and inducer transport. J Bacteriol. 1966;92:601–608. doi: 10.1128/jb.92.3.601-608.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adoutte A. Evolutionary biology. Small but mighty timekeepers. Nature. 2000;408:37–38. doi: 10.1038/35040669. [DOI] [PubMed] [Google Scholar]
- Altuvia S, Wagner EG. Switching on and off with RNA. Proc Natl Acad Sci. 2000;97:9824–9826. doi: 10.1073/pnas.97.18.9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Busby S, Dreyfus M. Segment-specific mutagenesis of the regulatory region in the Escherichia coli galactose operon: Isolation of mutations reducing the initiation of transcription and translation. Gene. 1983;21:121–131. doi: 10.1016/0378-1119(83)90154-3. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Clark B, Maalø O. DNA and the division cycle in Escherichia coli. J Mol Biol. 1967;23:99–112. [Google Scholar]
- Delihas N, Forst S. micF: An antisense RNA gene involved in response of Escherichia coli to global stress factors. J Mol Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Noncoding RNA genes. Curr Opin Genet Dev. 1999;9:695–699. doi: 10.1016/s0959-437x(99)00022-2. [DOI] [PubMed] [Google Scholar]
- Franch T, Gerdes K. Programmed cell death in bacteria: Translational repression by mRNA end-pairing. Mol Microbiol. 1996;21:1049–1060. doi: 10.1046/j.1365-2958.1996.771431.x. [DOI] [PubMed] [Google Scholar]
- Franch T, Gultyaev AP, Gerdes K. Programmed cell death by hok/sok of plasmid R1: Processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J Mol Biol. 1997;273:38–51. doi: 10.1006/jmbi.1997.1294. [DOI] [PubMed] [Google Scholar]
- Franch T, Thisted T, Gerdes K. Ribonuclease III processing of coaxially stacked RNA helices. J Biol Chem. 1999;274:26572–26578. doi: 10.1074/jbc.274.37.26572. [DOI] [PubMed] [Google Scholar]
- Grana D, Gardella T, Susskind MM. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988;120:319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi-Rontani C, Danchin A, Ullmann A. Transcriptional control of polarity in Escherichia coli by cAMP. Mol Gen Genet. 1984;195:96–100. doi: 10.1007/BF00332730. [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes & Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hatfull GF, Joyce CM. Deletion of the spf (spot 42 RNA) gene of Escherichia coli. J Bacteriol. 1986;166:746–750. doi: 10.1128/jb.166.3.746-750.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T, Dahlberg JE. Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control. J Biol Chem. 1973;248:5033–5041. [PubMed] [Google Scholar]
- Jorgensen RA, Atkinson RG, Forster RL, Lucas WJ. An RNA-based information superhighway in plants. Science. 1998;279:1486–1487. doi: 10.1126/science.279.5356.1486. [DOI] [PubMed] [Google Scholar]
- Joseph E, Danchin A, Ullmann A. Regulation of galactose operon expression: Glucose effects and role of cyclic adenosine 3′,5′-monophosphate. J Bacteriol. 1981;146:149–154. doi: 10.1128/jb.146.1.149-154.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Grindley ND. Identification of two genes immediately downstream from the polA gene of Escherichia coli. J Bacteriol. 1982;152:1211–1219. doi: 10.1128/jb.152.3.1211-1219.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallipolitis BH, Nørregaard-Madsen M, Valentin-Hansen P. Protein–protein communication: Structural model of the repression complex formed by CytR and the global regulator CRP. Cell. 1997;89:1101–1109. doi: 10.1016/s0092-8674(00)80297-4. [DOI] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Regulation of RpoS by a novel small RNA: The characterization of RprA. Mol Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: Translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, Brennan RG, Valentin-Hansen P. Hfq: A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Murphy KC. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93:305–308. doi: 10.1016/s0092-8674(00)81155-1. [DOI] [PubMed] [Google Scholar]
- Polayes DA, Rice PW, Dahlberg JE. DNA polymerase I activity in Escherichia coli is influenced by spot 42 RNA. J Bacteriol. 1988a;170:2083–2088. doi: 10.1128/jb.170.5.2083-2088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP–cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol. 1988b;170:3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C, Rosenberg M. Differential translation efficiency explains discoordinate expression of the galactose operon. Cell. 1981;25:241–249. doi: 10.1016/0092-8674(81)90249-x. [DOI] [PubMed] [Google Scholar]
- Rice PW, Dahlberg JE. A gene between polA and glnA retards growth of Escherichia coli when present in multiple copies: Physiological effects of the gene for spot 42 RNA. J Bacteriol. 1982;152:1196–1210. doi: 10.1128/jb.152.3.1196-1210.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Sahagan BG, Dahlberg JE. A small, unstable RNA molecule of Escherichia coli: spot 42 RNA. I. Nucleotide sequence analysis. J Mol Biol. 1979;131:573–592. doi: 10.1016/0022-2836(79)90008-1. [DOI] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard-Andersen L, Mironov AS, Pedersen H, Sukhodelets VV, Valentin-Hansen P. Single amino acid substitutions in the cAMP receptor protein specifically abolish regulation by the CytR repressor in Escherichia coli. Proc Natl Acad Sci. 1991;88:4921–4925. doi: 10.1073/pnas.88.11.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T, Sørensen NS, Wagner EG, Gerdes K. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 1994;13:1960–1968. doi: 10.1002/j.1460-2075.1994.tb06465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann A, Joseph E, Danchin A. Cyclic AMP as a modulator of polarity in polycistronic transcriptional units. Proc Natl Acad Sci. 1979;76:3194–3197. doi: 10.1073/pnas.76.7.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Svenningsen BA, Munch-Pedersen A, Hammer-Jespersen K. Regulation of the deo operon in Escherichia coli: The double negative control of the deo operon by the cytR and deoR repressors in a DNA directed in vitro system. Mol Gen Genet. 1978;159:191–202. doi: 10.1007/BF00270893. [DOI] [PubMed] [Google Scholar]
- Wagner EG, Brantl S. Kissing and RNA stability in antisense control of plasmid replication. Trends Biochem Sci. 1998;23:451–454. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu Rev Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes & Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert MJ, Adhya S. Control of transcription of gal repressor and isorepressor genes in Escherichia coli. J Bacteriol. 1993;175:251–258. doi: 10.1128/jb.175.1.251-258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiler BN, Simon RW. Control by antisense RNA. In: In: Lin ECC, Lynch AS, editors. Regulation of gene expression in Escherichia coli. Austin, TX: R.G. Landers Company; 1996. pp. 67–83. [Google Scholar]
- Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]