Abstract
A nonsense mutation in the fibrillin-1 (FBN1) gene of a Marfan syndrome (MFS) patient induces in-frame exon skipping of FBN1 exon 51. We present evidence, based on both in vivo and in vitro experiments, that the skipping of this exon is due to the disruption of an SC35-dependent splicing enhancer within exon 51. In addition, this nonsense mutation induces nonsense-mediated decay (NMD), which degrades the normally spliced mRNA in the patient's cells. In contrast to NMD, skipping of FBN1 exon 51 does not require translation.
Keywords: FBN1, exon skipping, Marfan syndrome, SR proteins, NMD, ESE
Mutations inducing genetic disease affect various aspects of mRNA biogenesis, including splicing, export, translational control, and stability (Cooper and Mattox 1997; Valentine 1998; Wilkinson and Shyu 2001). It is estimated that up to 30% of the mutations causing human diseases or cancer generate in-frame nonsense codons (Dietz 1997; Valentine 1998). Proteins generated by these mutated genes are defective and can be damaging to the organism. However, mRNAs containing premature termination codons (PTCs) rarely produce truncated proteins because they are degraded by a pathway called nonsense-mediated mRNA decay (NMD) (Maquat 1995, 2002; Hentze and Kulozik 1999; Maquat and Carmichael 2001; Wilusz et al. 2001). NMD is generally viewed as a cytoplasmic event, but this view is complicated by the fact that several PTCs cause exon skipping during pre-mRNA splicing in the nucleus (Dietz 1997; Valentine 1998). This phenomenon has been named nonsense-mediated alternative splicing (NAS) (Hentze and Kulozik 1999). The mechanism underlying NAS is largely unknown and its relationship to NMD is unclear, but it suggests the presence of a nuclear reading frame scanning mechanism, which operates prior to the splicing event (Hentze and Kulozik 1999; Mendell and Dietz 2001).
Exonic splicing enhancer (ESE) sequences are present in constitutively and alternatively spliced exons, are distinct from the splice site sequences, and are required for efficient splicing of certain exons (Watakabe et al. 1993; Cooper and Mattox 1997). Missense and silent mutations disrupting these sequences can cause exon skipping (Cooper and Mattox 1997; Dietz 1997; Valentine 1998). Several ESEs have been found to interact with members of the serine-arginine rich (SR) protein family (Lavigueur et al. 1993; Ramchatesingh et al. 1995; Schaal and Maniatis 1999; Liu et al. 2000). Members of the SR protein family are well studied protein factors required for general and regulated pre-mRNA splicing (Valcarcel and Green 1996; Graveley 2000). Nine authentic human SR proteins have been identified to date. In contrast to the apparently interchangeable properties of different SR proteins in constitutive splicing, distinct effects of these proteins in alternative and enhancer-dependent splicing have been reported (Graveley 2000).
Marfan syndrome (MFS) is an inherited systemic disorder of the connective tissue, caused by mutations in the fibrillin-1 (FBN1) gene. MFS shows autosomal dominant transmission and an estimated incidence of 1 in 5000 live births. Fibrillin-1 is a 350-kD calcium binding glycoprotein, which is the major component of the 10–12-nm microfibrils of the extracellular matrix. Classic MFS manifestations primarily involve the cardiovascular, ocular, and skeletal systems (Pyeritz 2000). Analysis of the FBN1 gene in an MFS patient identified an allele with a T → G transversion at nucleotide +26 of exon 51 (Dietz et al. 1993). This mutation creates an amber (TAG) nonsense mutation, substituting a termination codon (X) for a tyrosine (Y) at codon 2113 (Y2113X), and induces skipping of exon 51 in the MFS patient (Dietz et al. 1993). Skipping of the 66-nt exon results in the in-frame removal of 22 residues and the disruption of an epidermal growth factor-like Ca2+ binding domain (Dietz et al. 1992, 1993). This leads to the production of a mutated fibrillin-1 protein that appears to disrupt the correct assembly of the 10–12-nm microfibrils (Liu et al. 1996).
In an FBN1 chimeric minigene, two different PTC mutations (TAG and TAA) at codon 2113 induce exon skipping, whereas a silent mutation (TAC) does not (Dietz and Kendzior 1994). Furthermore, a frameshift mutation in the upstream exon, which places the nonsense mutation out of frame, significantly reduces exon skipping, suggesting that the alternative splicing is frame-dependent. Thus, a nuclear scanning mechanism that recognizes the reading frame of the pre-mRNA and triggers exon skipping was proposed (Dietz and Kendzior 1994). However, an alternative mechanism was suggested by the finding that a silent mutation located at +41 in exon 51 also induces exon skipping in another MFS patient (Liu et al. 1997).
In the present study, we found that FBN1 transcripts carrying the nonsense mutation Y2113X were subject to NMD in primary fibroblasts derived from the MFS patient. In contrast to NMD, NAS (FBN1 exon 51 skipping) was not affected by inhibitors of translation, We also observed that PTCs inserted at different positions in exon 51 did not induce exon skipping in a chimeric minigene. Finally, we demonstrated that the central portion of FBN1 exon 51 functions as an SC35-dependent ESE when inserted into a heterologous splicing reporter.
Results and Discussion
Inhibition of protein synthesis and NMD has no effect on exon skipping
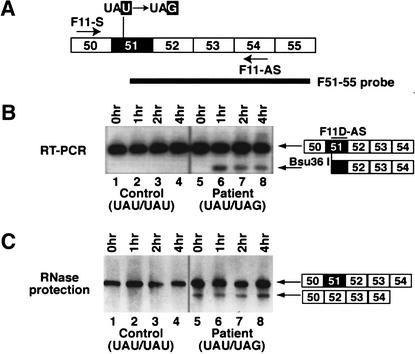
First, we wanted to determine whether NMD was induced in FBN1 transcripts, which harbored the Y2113X nonsense mutation that induced exon skipping (Dietz et al. 1993). Primary fibroblasts from this MFS patient, carrying the PTC in one FBN1 allele, and from a control individual were exposed to anisomycin. This protein synthesis inhibitor has been shown to inhibit NMD (Carter et al. 1995). Both patient and control fibroblasts were exposed to 100 μg/mL anisomycin for 0 (control), 1, 2, and 4 h. This level of anisomycin was shown to inhibit incorporation of [35S]methionine into total cellular protein by 93% after 1 h and 97% after 4 h (data not shown). RT–PCR was performed on total cellular RNA, utilizing primers complementary to FBN1 exons 50 and 54 (Fig. 1A). The amplified DNA was then digested with Bsu36 I, which specifically cleaves the mutant DNA at a new site generated by the nonsense mutation. The samples were Southern blotted and probed with an oligonucleotide complementary to a portion of exon 51 (Fig. 1B). After 1 h of anisomycin treatment, the Bsu36 I restriction product was detected in samples from the patient's cells (Fig. 1B, lanes 6–8). This product represents normally spliced mRNA transcribed from the mutant FBN1 allele harboring the PTC and includes exon 51. This RNA species was not detected in the absence of anisomycin treatment (Fig. 1B, lane 5) or in the control cells (Fig. 1B, lanes 1–4). This result indicates that normally spliced FBN1 mRNAs carrying the PTC are subject to NMD.
Figure 1.
FBN1 mRNAs carrying the Y2113X nonsense mutation are subject to NMD. (A) Schematic of the FBN1 exon 51 region. The Y2113X nonsense mutation is indicated, the primers used in the RT–PCR assay are indicated by arrows, and the probe utilized in the RNase protection assay is indicated by the heavy black line. (B) RT–PCR analysis of FBN1 transcripts in the presence of anisomycin, an inhibitor of protein synthesis. Fibroblasts from an MFS patient, harboring the mutant allele causing exon skipping (lanes 5–8), and from a healthy individual as control (lanes 1–4) were exposed to 100 μg/mL anisomycin for the indicated times (0 h, untreated). RT–PCR amplification was carried out with primers F11S and F11-AS. The amplified DNA was cleaved with Bsu36 I, which cleaves at the site of the nonsense mutation. The DNA products were separated by electrophoresis, Southern blotted, and hybridized with the oligonucleotide probe F11D-AS. (C) RNase protection analysis shows that exon skipping does not require protein synthesis. RNA isolated from patient and control fibroblasts after exposure to anisomycin was subjected to RNase protection analysis with the F51–55 probe shown in A.
Next we asked whether inhibition of protein synthesis, and thus NMD, would alter exon 51 skipping in the patient's cells. To distinguish between normally spliced and exon 51-skipped FBN1 mRNAs, an RNase protection assay was performed using a riboprobe spanning exons 51 through 55 (Fig. 1A). The amount of exon skipping remained constant over the time course of anisomycin exposure (Fig. 1C, lanes 5–8). No aberrant splicing was detected in normal fibroblasts assayed as a control (Fig. 1C, lanes 1–4). These results indicate that inhibition of protein synthesis had no effect on exon skipping. In contrast, PTC-containing mRNAs are protected from NMD by anisomycin treatment (Carter et al. 1995). Thus, NMD and exon skipping, although induced by the same PTC, appear to be activated by two independent mechanisms.
A nuclear scanning mechanism that recognizes the reading frame of pre-mRNAs and triggers exon skipping was previously proposed to explain the aberrant splicing of the FBN1 Y2113X transcript (Dietz and Kendzior 1994). A frameshift mutation in the upstream exon, which placed the nonsense mutation out of frame, significantly reduced exon skipping, suggesting that the alternative splicing mechanism was frame-dependent (Dietz and Kendzior 1994). However, we have shown here that NMD is actively degrading the RNAs carrying the PTC mutation in the patient's cells (Fig. 1B). Thus, the frameshift mutation, which reestablished the exon 51 open reading frame, would be expected to eliminate NMD of the mutant RNAs. As a consequence, exon 51-included RNAs would be increased, and the relative amount of skipped mRNA reduced, after amplification by RT–PCR as was observed by Dietz and Kendzior (1994).
Exon skipping is dependent on the position of the PTC and on other exonic sequences
Since exon skipping still occurs when translation is inhibited (Fig. 1C) and thus seems to be independent of NMD, we next asked whether it was dependent on the position of the PTC within exon 51. If a nuclear scanning mechanism were causing exon skipping, the position of the PTC within the exon would not be expected to affect the splicing pattern. To test this, we utilized the chimeric minigene represented in Figure 2A. The parental construct, pFBN-WT, contains the wild-type FBN1 exon 51, flanked by the native introns and portions of the upstream and downstream exons, inserted in-frame into an ornithine δ-aminotransferase (OAT) cDNA expression construct driven by a cytomegalovirus (CMV) promoter (Dietz and Kendzior 1994). Skipping of exon 51 had been shown previously to occur in human fibroblasts transiently transfected with the FBN1-OAT minigene, when nucleotide +26 in exon 51 was mutated to either a G or an A, creating the PTC UAG or UAA, respectively (Dietz and Kendzior 1994).
Figure 2.
Mutations causing exon skipping in vivo. (A) Schematic of the OAT-FBN chimeric minigene used in this study. Mutations introduced in exon 51 are indicated below the sequence. (*) The site of the PTC at +26, which caused exon skipping in an MFS patient. (B) RT–PCR analysis of transcripts of the OAT-FBN minigenes, transiently transfected into human fibroblasts. Exon 51 skipping and inclusion was assayed with the wild-type construct (lane 2) and with constructs carrying PTCs at different positions: codon 2113 (lane 3), upstream (US) (lane 5), and downstream (DS) (lane 6). RNAs with missense mutations in a polypurine element (PPE) were assayed in lane 4. In lane 1, cells were mock transfected as a control.
We have confirmed and extended these results, using RT–PCR to assay the splicing products generated from transiently transfected human fibroblasts. As reported previously, no exon skipping was detected with the wild-type pFBN WT construct (Fig. 2B, lane 2), and a skipped band was generated with a PTC generated by a U → A mutation at nucleotide +26 of exon 51 (the position of the patient's Y2113X mutation) (Fig. 2B, lane 3). We have extended these results to analyze the effects of PTCs at other positions in exon 51. We generated an in-frame PTC four codons upstream (US) of codon 2113 by a C → U mutation at +12. No exon skipping was detected with this construct, called pFBN US-PTC (Fig. 2B, lane 5). Normal splicing was also observed from a pre-mRNA with a C → G mutation at nucleotide 58, generating a PTC 11 codons downstream (DS) of codon 2113 in pFBN DS-PTC (Fig. 2B, lane 6).
These results indicate that exon skipping is dependent on the position of the PTC within the exon; this is not consistent with a general nuclear PTC scanning mechanism. This raises the alternative possibility that the PTC at +26 is disrupting an ESE, which is not affected by the PTCs at the other positions tested. If so, it seems likely that silent or missense mutations near +26 would also disrupt the ESE and cause exon skipping. To test this, we generated a missense mutation in the pFBN PPE construct by substituting pyrimidines for purines at positions +30, +31, and +33 (AGUG → UCUC) in a polypurine-rich element (PPE). Remarkably, RT–PCR analysis showed that exon 51 was completely skipped in the mRNAs transcribed from the pFBN PPE construct (Fig. 2B, lane 4).
These data strongly suggest that disruption of an ESE sequence, rather than a frame-dependent nuclear scanning mechanism, activates exon skipping in the mutant FBN1 gene. Further evidence supporting the presence of a functional ESE in this exon comes from the report of a silent point mutation (C → U) at nucleotide +41, which induces exon 51 skipping in another MFS patient (Liu et al. 1997). We observed that a missense mutation in the central portion of exon 51 in the pFBN PPE construct (Fig. 2, lane 4) caused more exon skipping than the nonsense mutation in pFBN PTC (Fig. 2, lane 3). This can be explained if an ESE sequence is present in the central portion of exon 51, and the 3-nt substitution in pFBN PPE disrupts this sequence more drastically than the single point mutation in pFBN PTC. Similarly, the absence of detectable exon skipping with a T → C missense mutation at +26 (Dietz and Kendzior 1994) may be due to the fact that a wild-type pyrimidine is maintained at this position, together with the failure of this mutation to induce NMD.
The Y2113X nonsense mutation disrupts an SC35-dependent ESE
Next we sought to determine whether sequences distinct from the splice sites in FBN1 exon 51 are acting as an SR-dependent splicing enhancer. To this end, we inserted a 46-nt sequence derived from the central portion of exon 51 (nucleotides 13–58) into the downstream exon of an enhancer-dependent splicing reporter substrate derived from the D. melanogaster dsx gene (Fig. 3A). The intron in the dsx system is efficiently spliced only when an SR-dependent splicing enhancer is present in the downstream exon (Tian and Maniatis 1992, 1994; Graveley et al. 1998). Splicing of the parental dsx substrate (dsx-ΔE) is extremely weak (Fig. 3, lane 1) because of a nonconsensus 3′ splice site. Insertion of an SR-dependent splicing enhancer activates splicing because SR proteins bound to the ESE can stabilize the interaction of splicing factors with the weak splice site (Tian and Maniatis 1994; Graveley et al. 1998). As a control, a strong SR-dependent splicing enhancer derived from the avian sarcoma-leukosis virus (ASLV) was inserted into the downstream exon of the dsx substrate, generating dsx-ASLV which was spliced efficiently (Fig. 3B, lane 2).
Figure 3.
In vitro splicing of dsx-FBN substrates. (A) Schematic representation of dsx-FBN splicing substrates. Dsx exonic sequences (dark boxes), ASLV and exon 6D sequences (light boxes) are shown. PTC PPE and US-PTC mutations indicated here correspond to the mutations in Figure 2A. (B) In vitro splicing reactions. The indicated radiolabeled pre-mRNAs were incubated in in vitro splicing reactions containing HeLa cell nuclear extract. RNA precursors and spliced products are indicated by the schematics at the left of the autoradiogram.
Insertion of the wild-type central portion of FBN1 exon 51 into the dsx-FBN WT substrate increased splicing efficiency to a level similar to that obtained with the ASLV enhancer (Fig. 3, lane 3). Furthermore, insertion of the FBN1 sequence bearing a PTC at codon 2113, to generate dsx-FBN PTC, resulted in greatly decreased splicing enhancement (Fig. 3B, lane 4). The PPE missense mutation (Fig. 2A) completely disrupted the ESE function (Fig. 3B, lane 5). Finally, a point mutation generating a PTC 14 nt upstream of the Y2113X nonsense mutation did not affect the ESE activity (Fig. 3B, lane 6). This same upstream PTC did not promote exon skipping in vivo (Fig. 2B, lane 5).
We conclude that the central portion of FBN1 exon 51 acts as a strong ESE in the dsx enhancer reporter system. A point mutation at +26, which induced exon skipping in vivo in the OAT-FBN1 minigene (Fig. 2B, lane 3), decreased the splicing efficiency of the dsx-FBN PTC splicing substrate as well (Fig. 3B, lane 4). Furthermore, the ESE function of the exon 51 sequence was completely inactivated by the PPE mutation, which resulted in only exon skipping of mRNAs from the OAT-FBN1 minigene (cf. Fig. 2, lane 4 with Fig. 3, lane 5). Interestingly, the PTC mutation at +26 had a greater effect on splicing efficiency in vitro than in vivo (cf. Fig. 2, lanes 2 and 3 to Fig. 3, lanes 3 and 4). This may be due to additional enhancer sequences in the extended FBN1 fragment present in the OAT-FBN construct (Fig. 2A), which are not present in the shorter FBN1 sequence inserted into the dsx construct.
Krainer and colleagues predicted that the Y2113X mutation, inducing FBN1 exon 51 skipping, down-regulates binding of SRp40 and SRp55 (Liu et al. 2001). To determine whether these or other individual SR proteins promote splicing of the dsx-FBN1 transcripts, we complemented splicing-deficient HeLa S100 cytoplasmic extracts with single SR proteins. SR proteins are absent from the S100 extracts, and the addition of SR proteins can complement this deficiency to allow splicing to occur (Krainer et al. 1991). Total and individual SR proteins were purified from HeLa cells and calf thymus (Zahler 1999). The 35-kD SR protein fraction purified from calf thymus contains >95% SC35, with only a trace of ASF/SF2 detectable (data not shown). When incubated in HeLa S100 splicing-deficient extract, splicing of β-globin pre-mRNA was activated equivalently by each of the SR proteins tested in this assay (Zahler et al. 1993). As a first step, we normalized the amount of the different SR proteins added to the HeLa S100 splicing-deficient extract, so that equal amounts of spliced human β-globin RNA were generated with each SR protein added (Fig. 4, lanes 3–7). The splicing efficiencies obtained were comparable to that seen in HeLa nuclear extract (Fig. 4, lane 1).
Figure 4.
In vitro splicing of the dsx-FBN WT substrate in HeLa S100 extract complemented with SR proteins. The dsxFBN WT radiolabeled pre-mRNA was incubated in in vitro splicing reactions containing HeLa S100 extract (lanes 2–7 and 9–14) or HeLa nuclear extracts (lanes 1,8). Roughly 200 ng of each SR protein, purified from calf thymus, or 500 ng of a total SR protein preparation from HeLa cells was added to the splicing mixture as indicated.
Next, we tested these normalized amounts of SR proteins for activation of splicing of the dsx-FBN WT splicing reporter substrates. Only the total SR protein preparation and purified SC35 were able to efficiently complement the HeLa S100 extract and to activate splicing of the dsx-FBN WT substrate (Fig. 4, lanes 10,11). SRp70, SRp55, and SRp40 were unable to stimulate splicing of the dsx-FBN WT substrate (Fig. 4, lanes 12–14) even though they complemented the S100 extract to activate splicing of the β-globin substrate with the same efficiency as SC35 (Fig. 4, lanes 3–7). These results indicate that the central portion of FBN1 exon 51 contains a strong SC35-dependent splicing enhancer that can be disrupted by a nonsense mutation at +26 or by missense mutations downstream.
The phenomenon of exon skipping induced by nonsense mutations has been associated with a number of diseases (Valentine 1998). Although hypothesized, a nuclear scanning mechanism able to discriminate pre-mRNAs harboring a PTC and induce exon skipping has never been characterized (Hentze and Kulozik 1999; Mendell and Dietz 2001). Furthermore, a general mechanism for NAS seems unlikely because most PTCs do not induce alternative splicing (Fig. 2B, lanes 5 and 6; Willing et al. 1996). A simple explanation for NAS is the chance disruption of an ESE by the same mutation that generated the PTC (Cooper and Mattox 1997; Shiga et al. 1997; Liu et al. 2001). Consistent with this, it has been shown that exon skipping associated with a nonsense mutation in the BRCA1 gene is due to the disruption of an ESE, rather than to the interruption of the reading frame (Liu et al. 2001). Furthermore, utilizing scoring matrices for SR protein binding sequences, it was shown that in more than one-half of the genes undergoing NAS, nonsense mutations also disrupt SR protein binding consensus sequences (Liu et al. 2001). In contrast to the prediction that the mutation at +26, inducing FBN1 exon 51 skipping, down-regulates binding of SRp40 and SRp55 (Liu et al. 2001), we have experimentally shown that exon 51 has an ESE, which is SC35-dependent. Indeed, only SC35 activated splicing of the dsx-FBN WT substrate in an S100 cytosolic extract. SRp40 and SRp55 do not appear to interact functionally with the ESE because they did not activate splicing of the substrate even in amounts fourfold higher than SC35 (data not shown). The discrepancy between the predicted and the functional results can be explained because single SR protein binding sequences have similar characteristics, and several SR protein binding sites are predicted to overlap in the same region of the transcript. Indeed, the silent point mutation at +41 in exon 51, which also induces exon skipping in an MFS patient (Liu et al. 1997), was predicted to disrupt an SC35 consensus sequence (Liu et al. 2001). It is conceivable that only one of the predicted binding sequences, possibly the one with the strongest affinity, is functionally recognized. Furthermore, since scoring matrices do not take into account the secondary structure of the RNA, it is possible that protein binding sites become unavailable when the RNA is folded.
Nonsense mutations typically behave like loss-of-function alleles. However, in the case of the FBN1 Y2113X mutation, a PTC confers a dominant negative phenotype when the NMD system is bypassed by exon skipping. However, PTCs at other positions in FBN1 exon 51 do not induce NAS. The data presented here argue against the role of a nuclear scanning mechanism regulating NAS in the FBN1 gene. On the contrary, it appears that the nonsense mutation Y2113X in an MFS patient plays two roles. It disrupts an SR-dependent splicing enhancer, which leads to exon skipping, and induces NMD, which degrades the PTC-containing mRNA containing exon 51.
Materials and methods
Plasmids
The parent construct pFBN WT (originally pFib WT) (Dietz and Kendzior 1994) was modified by site-directed mutagenesis to obtain the constructs pFBN PTC (A), pFBN PPE, pFBN US-PTC, and pFBN DS-PTC. The central region (nucleotides 13–58) of FBN1 exon 51 was inserted into the BstBI site of the dsx-ΔE plasmid (Tian and Maniatis 1994), using the following pairs of kinased complementary DNA oligonucleotides: Plasmids dsx-FBN WT: 51.1 cgaaagatatgtccttatggaagtgggatcatcgtgggacctga tgatttt, 51.2 cgaaaatcatcaggtcccacgatgatcccacttccataaggacatatcttt; dsx-FBN PTC: 51.3 cgaaagatatgtccttagggaagtgggatcatcgtgggacctgatgatttt, 51.4 cgaaaatcatcaggtcccacgatgatcccacttccctaaggacatatcttt; dsx-FBN PPE: 51.5 cgaaagatatgtccttatggatctcggatcatcgtgggacctgatgatttt, 51.6 cgaaaatcatcag gtcccacgatgatccgagatccataaggacatatcttt; dsx-FBN US-PTC: 51.7 cgaatag atatgtccttatggaagtgggatcatcgtgggacctgatgattt, 51.8 cgaaatcatcaggtcccac gatgatcccacttccataaggacatatctatt.
Transient transfection assay
Human fibroblasts at 80% confluence were transfected with 10 μg of expression vector using DOTAP (Boehringer Mannheim). Total RNA was extracted and reverse transcribed using random hexamers (Pharmacia) and oligo dT (Life Tech). The resulting cDNA was PCR-amplified with primers F11C-S and OATBE9-AS (Dietz and Kendzior 1994) for 20 cycles; 0.5 μL of the primary reaction served as template for a second amplification of 20 cycles using nested primers F11EA-S and OATE9-AS (Dietz and Kendzior 1994).
Inhibition of protein synthesis
Human primary fibroblasts from a patient with MFS described previously (Dietz et al. 1992) and from a control individual were grown to 80% confluence. Then, 100 μg/mL anisomycin (Sigma) was added to the media and total RNA was extracted after 0, 1, 2, and 4 h incubations. RNA was reverse transcribed, and the resulting cDNA was amplified with primers F11s and F11A-S for 30 cycles. The PCR product was cut with Bsu36 I. Southern blotting using labeled F11D-AS primer as probe was performed as described (Dietz and Kendzior 1994). Total cellular RNA was also subjected to RNase protection assay. Transcription of antisense RNA probe and RNase digestion were performed as described (Barker and Beemon 1991). The FBN1 probe was a PCR-amplified fragment of cDNA spanning exons 51–55 inserted into pCR II (Invitrogen). Quantitation was carried out on an InstantImager (Packard).
In vitro pre-mRNA splicing assays and preparation of SR proteins
Capped, 32P-labeled run-off transcripts were synthesized by in vitro transcription using T7 RNA polymerase. HeLa cell and S100 extracts were prepared as described (Mayeda and Krainer 1999). Splicing reactions were performed in a total volume of 25 μL, containing 15 μL of HeLa cell nuclear extract or S100 as described (Mayeda and Krainer 1999). The reaction mixtures were incubated at 30°C for 2 h. RNAs recovered from the splicing reaction mixtures were separated on an 8 M Urea–6% polyacrylamide gel and visualized with an InstantImager (Packard). SR proteins were prepared from HeLa cells and calf thymus as described (Zahler 1999) and added to the splicing reactions as indicated.
Acknowledgments
We thank H.C. Dietz for the OAT-FBN1 minigene and the MFS patient fibroblasts; T. Maniatis and B.R. Graveley for the dsx in vitro splicing constructs; and A.M. Zahler for the purified SR proteins. We also thank Raymond Fernalld for technical assistance. This work was supported by NIH grant CA48746 to K.L.B. R.J.K. was supported by NIH predoctoral training grant 52T32G07231.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL maxcaputi@earthlink.net; FAX (410) 516-7292.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.997502.
References
- Barker GF, Beemon KL. Nonsense codons within the Rous sarcoma virus gag gene decrease the stability of unspliced viral RNA. Mol Cell Biol. 1991;11:2760–2768. doi: 10.1128/mcb.11.5.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Mattox W. The regulation of splice-site selection, and its role in human disease. Am J Hum Genet. 1997;61:259–266. doi: 10.1086/514856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC. Nonsense mutations and altered splice-site selection. Am J Hum Genet. 1997;60:729–730. [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, Kendzior RJ., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet. 1994;8:183–188. doi: 10.1038/ng1094-183. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Pyeritz RE, Puffenberger EG, Kendzior RJ, Jr, Corson GM, Maslen CL, Sakai LY, Francomano CA, Cutting GR. Marfan phenotype variability in a family segregating a missense mutation in the epidermal growth factor-like motif of the fibrillin gene. J Clin Invest. 1992;89:1674–1680. doi: 10.1172/JCI115766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz HC, Valle D, Francomano CA, Kendzior RJ, Jr, Pyeritz RE, Cutting GR. The skipping of constitutive exons in vivo induced by nonsense mutations. Science. 1993;259:680–683. doi: 10.1126/science.8430317. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Lavigueur A, La Branche H, Kornblihtt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001;27:55–58. doi: 10.1038/83762. [DOI] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Qian C, Comeau K, Brenn T, Furthmayr H, Francke U. Mutant fibrillin-1 monomers lacking EGF-like domains disrupt microfibril assembly and cause severe marfan syndrome. Hum Mol Genet. 1996;5:1581–1587. doi: 10.1093/hmg/5.10.1581. [DOI] [PubMed] [Google Scholar]
- Liu W, Qian C, Francke U. Silent mutation induces exon skipping of fibrillin-1 gene in Marfan syndrome. Nat Genet. 1997;16:328–329. doi: 10.1038/ng0897-328. [DOI] [PubMed] [Google Scholar]
- Maquat LE. When cells stop making sense: Effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- ————— Nonsense-mediated mRNA decay. Curr Biol. 2002;12:R196–R197. doi: 10.1016/s0960-9822(02)00747-9. [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG. Quality control of mRNA function. Cell. 2001;104:173–176. doi: 10.1016/s0092-8674(01)00202-1. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol Biol. 1999;118:309–314. doi: 10.1385/1-59259-676-2:309. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Dietz HC. When the message goes awry: Disease-producing mutations that influence mRNA content and performance. Cell. 2001;107:411–414. doi: 10.1016/s0092-8674(01)00583-9. [DOI] [PubMed] [Google Scholar]
- Pyeritz RE. The Marfan syndrome. Annu Rev Med. 2000;51:481–510. doi: 10.1146/annurev.med.51.1.481. [DOI] [PubMed] [Google Scholar]
- Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: Identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiga N, Takeshima Y, Sakamoto H, Inoue K, Yokota Y, Yokoyama M, Matsuo M. Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Invest. 1997;100:2204–2210. doi: 10.1172/JCI119757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992;256:237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- ————— A splicing enhancer exhibits both constitutive and regulated activities. Genes & Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Valentine CR. The association of nonsense codons with exon skipping. Mutat Res. 1998;411:87–117. doi: 10.1016/s1383-5742(98)00010-6. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes & Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Shyu AB. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. BioEssays. 2001;23:775–787. doi: 10.1002/bies.1113. [DOI] [PubMed] [Google Scholar]
- Willing MC, Deschenes SP, Slayton RL, Roberts EJ. Premature chain termination is a unifying mechanism for COL1A1 null alleles in osteogenesis imperfecta type I cell strains. Am J Hum Genet. 1996;59:799–809. [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Zahler AM. Purification of SR protein splicing factors. Methods Mol Biol. 1999;118:419–432. doi: 10.1385/1-59259-676-2:419. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]