Abstract
In Caenorhabditis elegans, Ras/ERK and Wnt/β-catenin signaling pathways cooperate to induce P12 and vulval cell fates in a Hox-dependent manner. Here we describe eor-1 and eor-2, two new positively acting nuclear components of the Ras and Wnt pathways. eor-1 and eor-2 act downstream or in parallel to ERK and function redundantly with the Mediator complex gene sur-2 and the functionally related gene lin-25, such that removal of both eor-1/eor-2 and sur-2/lin-25 mimics the removal of a main Ras pathway component. Furthermore, the eor-1 and eor-2 mutant backgrounds reveal an essential role for the Elk1-related gene lin-1. eor-1 and eor-2 also act downstream or in parallel to pry-1 Axin and therefore act at the convergence of the Ras and Wnt pathways. eor-1 encodes the ortholog of human PLZF, a BTB/zinc-finger transcription factor that is fused to RARα in acute promyelocytic leukemia. eor-2 encodes a novel protein. EOR-1/PLZF and EOR-2 appear to function closely together and cooperate with Hox genes to promote the expression of Ras- and Wnt-responsive genes. Further studies of eor-1 and eor-2 may provide insight into the roles of PLZF in normal development and leukemogenesis.
Keywords: eor-1, eor-2, PLZF, Ras, Wnt
The Ras and Wnt signaling pathways control many aspects of animal development, and hyperactivation of one or both of these pathways is a frequent cause of human cancers. A combination of biochemical studies in vertebrate cells and genetic studies in model organisms such as Drosophila melanogaster and Caenorhabditis elegans has identified the major components of each signaling pathway, as well as several transcription factors whose activities are controlled by these pathways. Growth factors stimulate receptor tyrosine kinases (RTKs) to activate the Ras GTPase and the downstream kinases Raf, MEK, and MAP kinase/ERK, ultimately regulating the activities of transcription factors such as the Ets-domain protein Elk-1 (Campbell et al. 1998). Wnt ligands stimulate Frizzled (Fz) receptors to antagonize axin and GSK3 and stabilize β-catenin, ultimately regulating the activities of transcription factors of the TCF/LEF family (Cadigan and Nusse 1997). Both Ets-domain proteins and TCF/LEF can function as either transcriptional activators or repressors (Bienz 1998; Yordy and Muise-Helmericks 2000) and are thought to act combinatorially with other factors to generate the diverse patterns of gene expression elicited by Ras or Wnt signaling in different tissues (Simon 2000). Here we provide evidence that C. elegans eor-1 and eor-2 positively regulate transcriptional responses downstream of both the Ras and Wnt pathways.
In C. elegans, a Ras pathway involving lin-3 EGF, let-23 RTK, let-60 Ras, lin-45 Raf, mek-2 MEK, and mpk-1 ERK is required for multiple developmental processes (Sternberg and Han 1998), three of which are relevant to our studies. First, Ras signaling is required for excretory system development and hence larval viability. Ras pathway loss-of-function mutants arrest as rod-like larvae with massive fluid accumulation in the pseudocoelom. One cause of rod-like lethality is failure to specify the excretory duct cell fate (Yochem et al. 1997). Second, Ras signaling controls the choice between the P11 and P12 ectodermal blast cell fates (Jiang and Sternberg 1998). In wild-type animals, P11 and P12 divide in different patterns to generate morphologically distinguishable hypodermal descendants, P11.p and P12.pa. In Ras pathway loss-of-function mutants, P12 sometimes adopts the fate of P11 (2 P11.p phenotype), whereas in Ras pathway gain-of-function mutants, P11 sometimes adopts the fate of P12 (0 P11.p phenotype). Finally, Ras signaling controls the choice between vulval and nonvulval cell fates. In wild-type animals there are six initially equipotent Vulval Precursor Cells (VPCs), only three of which (P5.p, P6.p, and P7.p) are induced to adopt vulval fates. In Ras pathway loss-of-function mutants, less than three VPCs adopt vulval fates (Vulvaless or Vul phenotype), whereas in Ras pathway gain-of-function mutants more than three VPCs adopt vulval fates (Multivulva or Muv phenotype). Most known components of the Ras pathway have been identified based on their roles in vulval development (Sternberg and Han 1998).
In addition to core components of the Ras pathway, screens for Muv and Vul mutants have identified several transcriptional regulators that affect vulval fate choice. Negatively acting gene products (defined by Muv mutants) include LIN-1 (an Elk1-related Ets-domain transcription factor; Beitel et al. 1995) and LIN-31 (an HNF3/forkhead-like transcription factor; Miller et al. 1993), as well as LIN-35 Rb and other components of the Synthetic Multivulva (SynMuv) pathways (Ferguson and Horvitz 1989; Lu and Horvitz 1998). Both LIN-1 and LIN-31 are substrates of murine ERK, and phosphorylation is thought to inactivate LIN-1 and convert LIN-31 into a positively acting transcription factor (Jacobs et al. 1998; Tan et al. 1998). Other positively acting gene products (defined by Vul mutants) include SUR-2 (a conserved component of the Mediator coactivator complex; Singh and Han 1995; Boyer et al. 1999) and LIN-25 (a novel protein thought to function closely with SUR-2; Tuck and Greenwald 1995; Nilsson et al. 1998). However, LIN-31, SUR-2, and LIN-25 are not absolutely required for all vulval cell fates, and play no role or only minor roles in other Ras-mediated processes such as excretory system development and P12 fate specification. Thus, additional genes that promote Ras/ERK-dependent transcription remain to be identified.
A Wnt pathway involving pry-1 Axin and bar-1 β-catenin cooperates with the Ras pathway to specify both vulval and P12 cell fates (Eisenmann et al. 1998; Eisenmann and Kim 2000; Gleason et al. 2002; Korswagen et al. 2002). The mechanisms through which the Ras and Wnt pathways cooperate are not known. However, both the Ras and Wnt pathways up-regulate expression of the Hox genes lin-39 and egl-5 and require lin-39 and egl-5 to specify vulval and P12 fates, respectively (Clandinin et al. 1997; Eisenmann et al. 1998; Jiang and Sternberg 1998; Maloof and Kenyon 1998). Therefore, Ras-regulated and Wnt-regulated transcription factors may converge on Hox gene promoters, or may act in cooperation with Hox proteins to promote cell-type-appropriate gene expression.
We identified multiple loss-of-function alleles of eor-1 and eor-2 (egl-1 suppressor, Di-O uptake defective, raf enhancer) in a genetic screen for enhancers of the excretory system and egg-laying defects of hypomorphic lin-45 raf mutants (Rocheleau et al. 2002). All of the other lin-45 Raf enhancer mutations identified in our screen were in known components or regulators of the Ras pathway, which strongly supports the specificity of the screen and therefore a close involvement of eor-1 and eor-2 in Ras signaling. However, unlike previously characterized Ras pathway regulators, eor-1 and eor-2 only weakly affect vulval development but more strongly affect excretory system development and P12 fate specification. We show here that EOR-1/PLZF and EOR-2 function redundantly with LIN-25 and the SUR-2 Mediator component and positively regulate both Ras and Wnt signaling output.
Results
eor-1 and eor-2 function together and act downstream or in parallel to mpk-1 ERK and the SynMuv genes to positively regulate Ras signaling
eor-1 and eor-2 single mutants have several weakly penetrant defects that resemble those of Ras pathway mutants, including rod-like larval lethality, an egg-laying defective phenotype, and a 2 P11.p phenotype, but have normal vulval development (Table 1; Rocheleau et al. 2002). The lethal and 2 P11.p defects are maternally rescued in eor-1 or eor-2 homozygotes segregating from heterozygous mothers (data not shown). The rod-like lethal and 2 P11.p defects are strongly enhanced by other mutations that reduce Ras pathway activity (Table 1). For example, eor-1 and eor-2 show synthetic interactions with sur-8, a gene that promotes signaling between let-60 Ras and lin-45 Raf (Sieburth et al. 1998), and show even stronger synthetic interactions with a gain-of-function (gf) allele of lin-1 Ets (Jacobs et al. 1998). eor-1 and eor-2 also partially suppress the Muv phenotype caused by hyperactive let-60 Ras or mpk-1 ERK, and more strongly suppress the 0 P11.p defect caused by hyperactive mpk-1 ERK or the SynMuv mutation lin-15 (Table 2). These data substantiate that eor-1 and eor-2 positively regulate Ras signaling in multiple tissues. These data also suggest that eor-1 and eor-2 act at a step downstream or in parallel to mpk-1 and the SynMuv genes.
Table 1.
eor-1 and eor-2 mutations enhance defects associated with reduced Ras, Wnt, and Hox activities
| Genotypea
|
Rod-like lethal (%)
|
(n)
|
Vul (%)
|
Avg. no. VPCs induced
|
(n)
|
2 P11.p (%)
|
(n)
|
|---|---|---|---|---|---|---|---|
| + | 0 | 0 | 3.0 | 0 | |||
| eor-1b | 7 | (109) | 0 | 3.0 | (46) | 21 | (52) |
| eor-2b | 7 | (257) | 0 | 3.0 | (30) | 13 | (24) |
| eor-1; eor-2 | 8 | (162) | 0 | 3.0 | (20) | 25 | (51) |
| Ras pathway | |||||||
| sur-8(RNAi) | 0 | (309) | 0 | 3.0 | (74) | 0 | (74) |
| sur-8 | 0 | (221) | 0 | 3.0 | (25) | 0 | (25) |
| eor-1 sur-8(RNAi) | 35** | (243) | 0 | 3.0 | (49) | 52** | (52) |
| sur-8; eor-2 | 38** | (72) | 14* | 2.94 | (49) | 63** | (46) |
| lin-1(gf) | 0 | (227) | 3 | 2.97 | (33) | 0 | (33) |
| lin-1(gf)eor-1 | 66** | (154) | 9 | 2.94 | (23) | 68** | (22) |
| lin-1(gf); eor-2 | 68** | (84) | 6 | 2.97 | (33) | 73** | (33) |
| sur-2 | 1 | (78) | 100 | 0.78 | (28) | 0 | (27) |
| sur-2; eor-1 | 100 | (many) | nd | nd | nd | nd | nd |
| sur-2; eor-2 | 100 | (many) | nd | nd | nd | nd | nd |
| lin-25c | 15 | (114) | 100e | 0.84 | (22) | 0 | (28) |
| eor-1; lin-25c | 100 | (many) | 100e | 0.24 | (22) | 15g* | (26) |
| Wnt pathway | |||||||
| bar-1 | nd | nd | 74 | 2.0 | (65) | 99 | (68) |
| eor-1; bar-1 | nd | nd | 90* | 1.5 | (68) | 96 | (54) |
| bar-1 eor-2 | nd | nd | 94* | 1.0 | (47) | 99 | (79) |
| Hox genes | |||||||
| lin-39 | nd | nd | 56f | 2.58 | (38) | 0 | (21) |
| lin-39; eor-1 | nd | nd | 100f** | 1.17 | (32) | 32 | (56) |
| lin-39; eor-2 | nd | nd | 100** | 1.23 | (15) | 24 | (50) |
| egl-5/+d | nd | nd | nd | nd | nd | 0 | (50) |
| +; eor-1d | nd | nd | nd | nd | nd | 24 | (51) |
| egl-5/+; eor-1d | nd | nd | nd | nd | nd | 72** | (64) |
n, Number of animals scored. nd, Not determined. For bar-1 and lin-39 strains, Vul (%) includes animals that had Pn.p fusion defects as well as induction defects.
Alleles used were eor-1(cs28), eor-2(cs30), sur-8(ku167), lin-1(n2515gf), sur-2(ku9), lin-25(e1446), bar-1(ga80), lin-39(n709), and egl-5(n486). Since linkage made it difficult to construct an eor-1 sur-8 double mutant, RNAi was used to reduce sur-8 function in the eor-1 background.
Lethality was scored in animals segregating from homozygous mothers, whereas vulval and P11.p phenotypes were scored in animals segregating from +/DnT1; lin-25/DnT1 or eor-1/DnT1; lin-25/DnT1 heterozygous mothers.
Animals were heterozygous for the balancer hT2[qIs48], which allowed egl-5 heterozygotes to be recognized as GFP(+) segregants.
14% of lin-25 and 59% of eor-1; lin-25 animals completely lacked vulval cells (P = 0.004, Fisher's Exact Test).
In lin-39 single mutants, P6.p always adopted a normal vulval fate (n = 20). In contrast, in lin-39; eor-1 double mutants, P6.p did not divide, and we presume adopted a fused fate 4/14 times, adopted a nonvulval (3°) fate 3/14 times, and adopted a hybrid fate 6/14 times.
eor-1 homozygotes segregating from heterozygous mothers have 0% 2 P11.p (n = 96).
P < 0.01, Fisher's Exact Test, compared with single mutant controls.
0.01 < P < 0.05, Fisher's Exact Test, compared with single mutant controls.
Table 2.
Epistatic relationships between eor-1 and eor-2 and Ras, SynMuv, and Wnt pathway genes
| Genotypea
|
Rod-like lethal (%)
|
(n)
|
Muv (%)
|
Vul (%)
|
Avg. no. VPCs induced
|
(n)
|
0 P11.p (%)
|
2 P11.p (%)
|
(n)
|
|---|---|---|---|---|---|---|---|---|---|
| Ras pathway | |||||||||
| let-60(gf) | 1 | (146) | 75 | 0 | 3.81 | (20) | 0 | 0 | (40) |
| eor-1 let-60(gf) | 7 | (106) | 7** | 0 | 3.06 | (28) | 0 | 10 | (41) |
| let-60(gf); eor-2 | 4 | (141) | 16** | 0 | 3.09 | (43) | 0 | 6 | (53) |
| [hs-mpk-1(+); D-mek(gf)]b | nd | nd | 53 | 0 | 3.69 | (36) | 26 | 0 | (74) |
| eor-1; [hs-mpk-1(+); D-mek(gf)]b | nd | nd | 11** | 0 | 3.13 | (36) | 2** | 3 | (61) |
| [hs-mpk-1(+); D-mek(gf)]; eor-2b | nd | nd | 19** | 0 | 3.15 | (43) | 0** | 12 | (78) |
| lin-31 | 0 | (326) | 95e | 5e | 4.45 | (19)f | 0 | 0 | (30) |
| lin-31; eor-1 | 18 | (155) | 100e | 12e | 4.79 | (17)f | 0 | 7 | (28) |
| lin-31; eor-2 | 10 | (367) | 100e | 6e | 4.82 | (17)f | 0 | 22 | (23) |
| lin-1(el275)c | 0 | (183) | 100 | 0 | 4.1 | (20) | 0 | 0 | (23) |
| lin-1(el275) eor-1c | 100 | (many) | 56e** | 19e | 3.26 | (27) | 0 | 0 | (38) |
| lin-1(el275); eor-2c | 100 | (many) | 67* | 8 | 3.71 | (12) | 0 | 9 | (22) |
| lin-1(n304)c | 3 | (101) | 100 | 0 | 5.16 | (21) | 0 | 0 | (25) |
| lin-1(n304) eor-1c | 100 | (many) | 100e | 14e | 4.38 | (21) | 0 | 0 | (41) |
| lin-1(n304); eor-2c | 100 | (many) | nd | nd | nd | nd | nd | nd | nd |
| SynMuv pathway | |||||||||
| lin-15(n765) | nd | nd | 95 | 0 | 4.86 | (21) | 20 | 0 | (25) |
| eor-1; lin-15(n765) | nd | nd | 90 | 0 | 4.73 | (20) | 0* | 15 | (34) |
| eor-2 lin-15(n765) | nd | nd | 74 | 0 | 3.98 | (23) | 0** | 19 | (37) |
| lin-15(n309) | nd | nd | 100 | 0 | 5.82 | (21) | 33 | 0 | (30) |
| eor-1; lin-15(n309) | nd | nd | 100 | 0 | 5.82 | (26) | 5* | 21 | (19) |
| Wnt pathway | |||||||||
| pry-1d | nd | nd | 27 | 0 | 3.23 | (30) | 96 | 0 | (45) |
| pry-1; eor-1d | nd | nd | 0** | 0 | 3.0 | (29) | 6** | 0 | (33) |
| pry-1; eor-2d | nd | nd | 4* | 0 | 3.02 | (27) | 7** | 0 | (28) |
| pry-1; lin-25d | nd | nd | 45 | 23 | 3.32 | (22) | 95 | 0 | (22) |
n, Number of animals scored. nd, Not determined.
Alleles used were eor-1(cs28), eor-2(cs30), let-60(n1046gf), gaIs36[hs-mpk-1(+); EF1a-D-mek(gf); unc-30(+)], lin-31(n301), and pry-1(mu38). lin-1(e1275) was linked to unc-24. Similar results were obtained when lin-1(e1275) was marked with unc-3. eor-1 was linked to unc-24 in lin-1(e1275) background. eor-2 was linked to unc-3 in lin-1(e1275) background.
gaIs36 strains were grown at 20°C and switched to 25°C (the temperature at which the gaIs36 strain is Muv) at the L4 stage because double-mutant strains were too sick to propagate at 25°C. Progeny of mothers grown at 20°C were scored at 25°C.
Lethality was scored in animals segregating from homozygous mothers, whereas vulval and P11.p phenotypes were scored in animals segregating from lin-1(e1275) unc-24/lin-1(e1275)+, lin(e1275) eor-1 unc-24/lin-1(e1275)++, lin-1(e1275); eor-2 unc-3/++, lin-1(n304)/DnT1 or lin-1(n304) eor-1/DnT1 mothers.
pry-1 strains were grown continuously at 25°C because double-mutant strains were too sick to propagate at lower temperatures.
Some animals were both Muv and Vul, and these animals were included in both the Muv and Vul columns. Animals were considered Muv and Vul if P3.p, P4.p, and/or P8.p were induced, but P5.p, P6.p, and/or P7.p were not induced.
Vulva cells in lin-31; eor-1 and lin-31; eor-2 double mutants were less uniform in size and less organized compared with lin-31 single mutants, which could explain why the double mutants are much more Egg-laying defective (data not shown).
P < 0.01, Fisher's Exact Test, compared with single mutant controls.
0.01 < P < 0.05, Fisher's Exact Test, compared with single mutant controls.
Whereas eor-1 and eor-2 show strong genetic interactions with various Ras pathway components, eor-1 and eor-2 do not show genetic interactions with each other. eor-1; eor-2 double mutants resemble either single mutant (Table 1). This finding, combined with the fact that eor-1 and eor-2 both have similar mutant phenotypes and act downstream or in parallel to mpk-1, suggests that eor-1 and eor-2 function closely together. This hypothesis is further supported by the identical genetic behavior of eor-1 and eor-2 in all subsequent genetic tests (see below).
eor-1 and eor-2 cooperate with lin-1, sur-2, and lin-25 to positively regulate Ras signaling output
To determine more precisely at what step eor-1 and eor-2 act, we tested their epistatic relationship to other genes that act downstream of mpk-1. The lin-31 forkhead gene functions specifically during vulval development and plays both positive and negative roles (Miller et al. 1993). lin-31(n301) null mutants are viable and have a mixed Vul and Muv phenotype, and neither eor-1 nor eor-2 appreciably affects this phenotype (Table 2). Therefore eor-1 and eor-2 do not appear to act downstream of lin-31. In contrast, both eor-1 and eor-2 show dramatic genetic interactions with lin-1, an Ets gene that is negatively regulated by Ras signaling in multiple tissues (Beitel et al. 1995). lin-1(e1275) hypomorphic mutants and lin-1(n304) null mutants are viable and Muv. Surprisingly, however, lin-1 eor-1 and lin-1; eor-2 double mutants show fully penetrant maternal effect lethality (Table 2). Although the specific cause of lethality is not known, the dying larvae are filled with fluid and have a distinctive rod-like appearance that closely resembles that of let-60 Ras mutants. Furthermore, when segregating from heterozygous mothers, maternally rescued lin-1 eor-1 and lin-1; eor-2 double mutants have a mixed Vul and Muv phenotype resembling that of lin-31 mutants (Table 2). These data imply that, like lin-31, lin-1 plays both positive and negative roles in Ras signaling (see Discussion). These data also suggest that eor-1 and eor-2 cooperate with lin-1 to promote Ras signaling.
eor-1 and eor-2 also show dramatic genetic interactions with sur-2 and lin-25, two genes that positively regulate Ras signaling in multiple tissues but have a primary role in vulval development (Singh and Han 1995; Tuck and Greenwald 1995; Nilsson et al. 2000). Most sur-2(ku9) null or lin-25(e1446) null single mutants are viable and partially Vul. However, double mutants with eor-1 or eor-2 show fully penetrant maternal effect lethality (Table 1). Again, dying larvae arrest with a distinctive rod-like appearance resembling that of let-60 Ras mutants. Furthermore, when segregating from heterozygous mothers, maternally rescued eor-1; lin-25 double mutants have a more expressive Vul and a more penetrant 2 P11.p phenotype than maternally rescued lin-25 or eor-1 single mutants (Table 1). These data indicate that eor-1 and eor-2 cooperate with sur-2 and lin-25 to positively regulate Ras signaling output. Because jointly removing both eor-1 (or eor-2) and lin-25 (or sur-2) causes defects similar to those caused by removing a main Ras pathway component, eor-1 and eor-2 have the genetic properties expected for additional positive transcriptional regulators downstream of mpk-1 ERK.
One known transcriptional readout of Ras signaling is the expression of the reporter gene egl-17::gfp in the vulval precursor cell P6.p and its descendants (Burdine et al. 1998). Whereas 95% (n = 41) of eor-1(cs28) mutants and 57% (n = 81) of lin-25(e1446) mutants showed normal EGL-17::GFP expression in P6.p or its daughters, only 16% (n = 80) of maternally rescued eor-1(cs28); lin-25(e1446) double mutants expressed this reporter, indicating that eor-1 and lin-25 function redundantly to promote reporter expression (p = 0.000002, Fisher's Exact Test, compared with additive effect, which would be 52%). These data further support the model that EOR-1 cooperates with known transcriptional regulators to promote Ras target gene expression.
eor-1 and eor-2 positively regulate Wnt signaling output
We tested the epistatic relationship between eor-1 and eor-2 and the Wnt pathway by analyzing double mutants with pry-1 Axin. eor-1 and eor-2 mutations suppress the pry-1 Muv and 0 P11.p phenotypes (Table 2). This genetic behavior is similar to that of bar-1 β-catenin mutations but in contrast to that of Ras pathway mutations or a lin-25 mutation, which do not suppress pry-1 defects (Table 2; Gleason et al. 2002). These data indicate that eor-1 and eor-2 not only promote Ras signaling output, but also promote Wnt signaling output.
eor-1 and eor-2 differ from bar-1 and other Wnt pathway genes in several important respects. First, unlike bar-1, eor-1 and eor-2 mutations do not suppress all aspects of the pry-1 phenotype such as frequent herniation and general sickness (Table 2). Second, unlike eor-1 and eor-2, bar-1 mutations do not cause rod-like larval lethality either alone or in lin-45, lin-1, or lin-25 mutant backgrounds (Eisenmann et al. 1998; S. Tuck, pers. comm.; R.M. Howard and M.V. Sundaram, unpubl.). Finally, eor-1 and eor-2 mutations slightly enhance the bar-1 Vul phenotype (Table 1). These results support the argument that eor-1 and eor-2 do not act solely to regulate or respond to bar-1. Instead, we favor a model in which eor-1 and eor-2 positively regulate downstream responses to both Ras and Wnt signaling.
eor-1 encodes a BTB/zinc-finger protein similar to PLZF
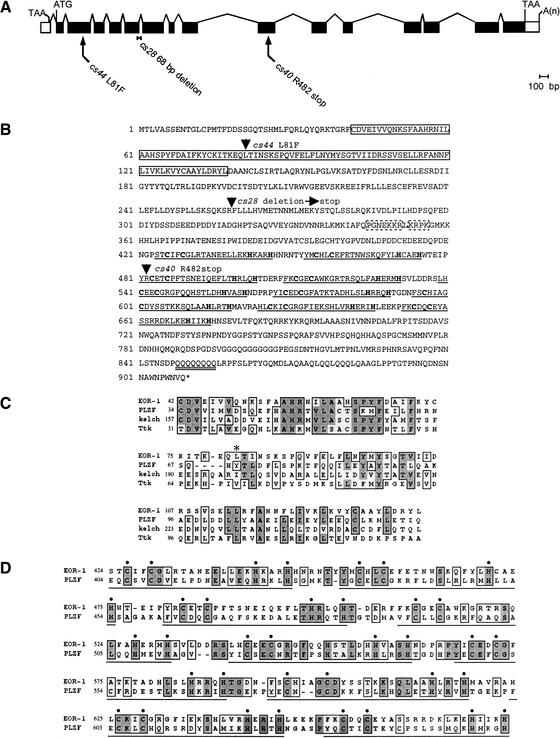
We positionally cloned eor-1 by transformation rescue (Materials and Methods) and found that it corresponds to the predicted gene R11E3.6. Analysis of eor-1 cDNAs (Fig. 1A) indicates that eor-1 encodes a 909-amino-acid protein with an N-terminal BTB domain (or POZ domain), followed by nine closely spaced C2H2 zinc fingers and a polyglutamine stretch (Fig. 1B–D). We identified the lesions in all three of our mutant eor-1 alleles. eor-1(cs28) contains a 68-bp deletion that leads to a frameshift, and is predicted to encode a truncated protein lacking all nine zinc fingers (Materials and Methods). eor-1(cs40) contains a nonsense mutation between the second and third zinc fingers. eor-1(cs44) contains a missense mutation that affects a conserved residue in the BTB domain (Fig. 1B,C). All three alleles behave similarly and appear to severely reduce or eliminate eor-1 function (Rocheleau et al. 2002). Consistent with eor-1(cs28) representing the loss-of-function phenotype, RNA-mediated interference of eor-1 caused few defects in a wild-type background but caused significant rod-like lethality in the lin-1(e1275) background (Materials and Methods).
Figure 1.
eor-1 encodes a PLZF-related BTB/zinc-finger protein. (A) Gene structure of eor-1 based on analysis of cDNA clone yk87e7. Coding sequences are shaded black. (B) Amino acid sequence of EOR-1. Arrowheads indicate the sites of the cs28, cs40, and cs44 lesions. The BTB domain is boxed with a solid line. The nine C2H2 zinc-finger domains are underlined, and the cysteines and histidines are in bold. The polyglutamine stretch is underlined twice. Potential nuclear localization sequences are boxed with dashed lines. (C) EOR-1 BTB domain alignment. Conserved regions are boxed. Shaded areas are identical residues. An asterisk indicates a conserved leucine changed to a phenylalanine in eor-1(cs44). The EOR-1 BTB domain is 31% identical and 45%–55% similar to the BTB domains of mammalian PLZF, Drosophila kelch, and Drosophila Tramtrack (Ttk). (D) Alignment of zinc-finger domain regions of EOR-1 and PLZF. EOR-1 and PLZF are 30% identical and 45% similar over this region. The nine C2H2 zinc-finger domains are underlined, and the cysteines and histidines are indicated with dots.
The molecular identity of EOR-1 suggests a role in transcriptional regulation. BTB domains mediate homodimerization as well as heterotypic interactions with other proteins such as the corepressors N-CoR and SMRT (Collins et al. 2001). Most C2H2 zinc fingers bind DNA (Nelson 1995), and the BTB/zinc-finger family of proteins includes many known transcription factors, including both transcriptional activators and repressors. Polyglutamine stretches are also common features of transcriptional regulators (Gerber et al. 1994). Because EOR-1 facilitates Ras/ERK signaling, it could be a transcriptional activator of genes stimulated by ERK or a transcriptional repressor of genes inhibited by ERK.
EOR-1 was previously identified as the C. elegans ortholog of mammalian promyelocytic leukemia zinc-finger protein (PLZF), a transcription factor that is fused to RARα in retinoic acid-resistant forms of acute promyelocytic leukemia in humans (Lin et al. 1999; Zhang et al. 1999). Like EOR-1, PLZF also contains an N-terminal BTB domain (Fig. 1C) and nine similarly spaced C2H2 zinc fingers (Fig. 1D). Although the normal role of PLZF is poorly understood, EOR-1 and PLZF have several intriguing functional similarities (see Discussion).
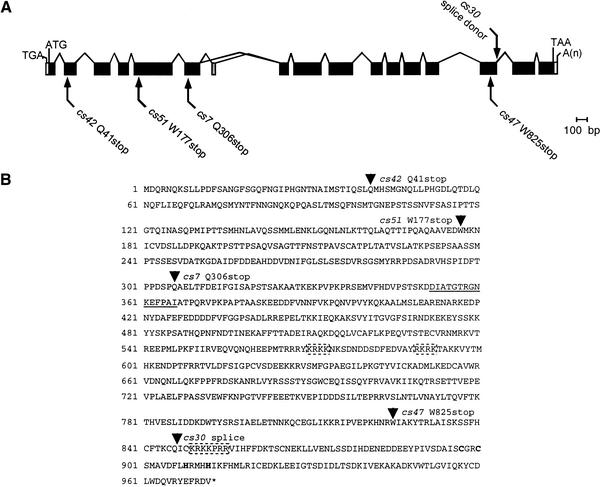
eor-2 encodes a novel protein
We also positionally cloned eor-2 by transformation rescue (Materials and Methods) and found that it corresponds to the predicted gene C44H4.7. Analysis of eor-2 cDNAs (Fig. 2A) indicates that eor-2 encodes a novel protein with several potential nuclear localization signals (Fig. 2B). Alternative splicing of eor-2 leads to the generation of two highly related isoforms (A and B) that differ only in that EOR-2A contains 15 amino acids that are not present in EOR-2B (Fig. 2A,B). EOR-2 does not contain any recognizable motifs (as assessed by SMART) and is not highly related to any other proteins in the databases. However, the C-terminal region does have moderate similarity to an uncharacterized Drosophila gene product (Materials and Methods). It is possible that analogous proteins exist in other organisms but share too limited an amino acid identity to be detected by standard BLAST searches.
Figure 2.
eor-2 encodes a novel protein. (A) Gene structure of eor-2 based on analysis of cDNA clone yk257c2 (eor-2a) and RT-PCR products (eor-2b). Coding sequences common to eor-2a and eor-2b are shaded black. Coding sequences unique to eor-2a are shaded gray. Numbering is according to EOR-2A. (B) Amino acid sequence of EOR-2. Arrowheads indicate the site of the splice junction affected by cs30 and the sites of cs7, cs42, cs47, and cs51 lesions. The portion of EOR-2 unique to the EOR-2A isoform is underlined. Cysteines and histidines that comprise a C2H2-like sequence (which does not conform to zinc-finger spacing requirements) are in bold. Potential nuclear localization sequences are boxed with dashed lines. Numbering is according to EOR-2A.
We identified the lesions in all five of our mutant eor-2 alleles. eor-2(cs30) affects a splice donor site, whereas the other four eor-2 alleles, cs7, cs42, cs47, and cs51, are nonsense mutations (Fig. 2B). The earliest of these mutations, eor-2(cs42), is predicted to encode a truncated protein of only 40 amino acids. All five alleles behave similarly and appear to severely reduce or eliminate eor-2 function (Rocheleau et al. 2002).
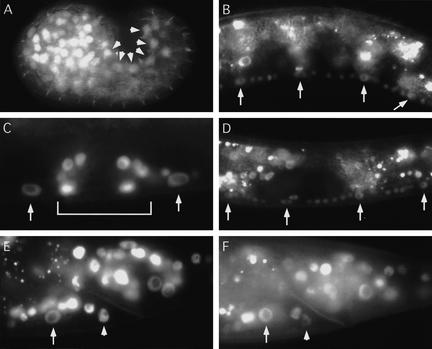
EOR-1::GFP and EOR-2::GFP are widely expressed and nuclear-localized
To analyze the expression patterns of EOR-1 and EOR-2, we generated functional GFP reporters capable of rescuing eor-1 or eor-2 mutant defects (Materials and Methods). EOR-1::GFP and EOR-2::GFP are expressed in most cells throughout development and are both nuclear-localized, consistent with the postulated role of EOR-1 and EOR-2 in transcriptional regulation. Of particular relevance, the reporters are expressed in the precursors to the VPCs and P11 and P12 (Fig. 3A), in VPCs that adopt vulval and nonvulval fates and their descendants (Fig. 3B–D), and in both P11.p and P12.pa (Fig. 3E,F). Thus, EOR-1::GFP and EOR-2::GFP are expressed in VPCs and in P12 at the time of cell fate determination. However, EOR-1::GFP and EOR-2::GFP expression and localization do not appear to change in response to Ras or Wnt signaling.
Figure 3.
(A–C,E) EOR-1::GFP and (D,F) EOR-2::GFP localize to the nuclei of many cell types. Anterior is left. (A) P1/2, P3/4, P5/6, P7/8, P9/10, and P12 (arrowheads) during embryogenesis before P cell fate specification. P cells are the precursors to the VPCs, P11.p and P12.pa. ajm-1::GFP (Mohler et al. 1998) was used to aid in P-cell identification. The right side of the embryo is shown. P cells on the left side also express EOR-1::GFP (data not shown). (B) P5.p–P8.p (arrows) at late L2 stage during inductive signaling. (C) Nonvulval cells P4.pp and P8.pa (arrows) and vulval cells (in brackets) at L4 stage. (D) P5.p–P8.p (arrows) at early L2 stage before inductive signaling. (E,F) P11.p (arrow) and P12.pa (arrowhead) at L4 stage.
eor-1 does not act simply to up-regulate Hox gene expression
The Ras, Wnt, and SynMuv pathways regulate vulval and P12 cell fates at least in part by regulating expression of the Sex combs reduced-like Hox protein LIN-39 in VPCs and the Abdominal-B-like Hox protein EGL-5 in P12 (Eisenmann et al. 1998; Jiang and Sternberg 1998; Maloof and Kenyon 1998; Chen and Han 2001; Gleason et al. 2002). Like Hox genes, eor-1 and eor-2 appear to act downstream of the Ras, Wnt, and SynMuv pathways at the level of transcriptional regulation. Furthermore, eor-1 and eor-2 show strong genetic interactions with lin-39 and egl-5 (Table 1). Therefore, we wanted to test if eor-1 mutant defects could be explained by a reduction in Hox gene expression.
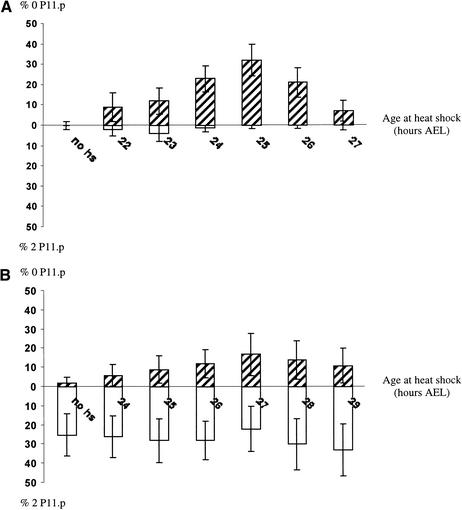
Because eor-1 most strongly affects the P11/P12 cell fate decision, we examined the relationship between eor-1 and egl-5 in those cells. We first tested whether an eor-1 mutation would reduce expression of an egl-5::lacz reporter gene but did not see an obvious effect (data not shown). We next tested whether we could rescue the eor-1 2 P11.p defect by heat-shock-promoter (hs)-driven expression of egl-5. The hs-egl-5 transgene syEx178 (Jiang and Sternberg 1998) causes a moderate 0 P11.p phenotype under our heat-shock conditions (Fig. 4A). This transgene did not rescue the 2 P11.p defect of eor-1 mutants, and eor-1 only slightly suppressed the 0 P11.p defect of hs-egl-5 (Fig. 4B). This additive phenotype of eor-1; hs-egl-5 animals indicates that eor-1 cooperates with egl-5 to promote the P12 fate and does not simply up-regulate egl-5 expression.
Figure 4.
egl-5 overexpression does not rescue eor-1 mutant 2 P11.p phenotype. (A) syEx178[hsEGL-5 + dpy-20(+)]; dpy-20. (B) eor-1(cs28) dpy-20; syEx178[hsEGL-5 + dpy-20(+)]. Larvae were heat-shocked for 1 h in a 33°C waterbath at the indicated hours after egg-lay (AEL) (X-axis). Open bars indicate percentage 2 P11.p. Hatched bars indicate percentage 0 P11.p. eor-1; syEx178 animals develop slightly slower than syEx178 animals. Therefore, the percentage 0 P11.p peaks at 27 h AEL for syEx178; eor-1 versus 25 h AEL for syEx178. Error bars are 95% confidence limits based on the limiting normal distribution.
Discussion
We have shown that eor-1 and eor-2 act downstream or in parallel to both the Ras and Wnt pathways to promote P12 and vulval cell fates. eor-1 and eor-2 appear to function closely together because they have identical mutant phenotypes and expression patterns, show identical genetic interactions with other signaling pathway components, yet do not interact genetically with each other. eor-2 encodes a novel nuclear protein. eor-1 encodes a BTB/zinc-finger protein orthologous to PLZF, a transcription factor that is altered in retinoic acid-resistant forms of promyelocytic leukemia in humans. Our data suggest that EOR-1 and EOR-2 are transcriptional regulators that cooperate with Mediator components, LIN-1 Ets, and Hox proteins to promote the expression of Ras- and Wnt-responsive genes.
eor-1 and eor-2 cooperate with sur-2 and lin-25 to positively regulate Ras signaling output
The sur-2 Mediator component and lin-25 function together as positive regulators of Ras/ERK signaling output in C. elegans (Singh and Han 1995; Tuck and Greenwald 1995; Nilsson et al. 1998, 2000). However, because sur-2 and lin-25 are not required for the full extent or spectrum of Ras/ERK-mediated processes, and because the Mediator complex itself does not bind DNA, other positively acting transcriptional regulators remain to be identified. eor-1 and eor-2 have the genetic properties expected for such factors. First, eor-1 and eor-2 mutations cause several weakly penetrant ras-like defects and enhance defects caused by other mutations that reduce Ras signaling. Second, eor-1 and eor-2 mutations suppress defects caused by activated mpk-1 ERK. Third, double mutants between eor-1/eor-2 and sur-2/lin-25 show fully penetrant synthetic rod-like lethality, and removing both eor-1 and lin-25 zygotic contributions closely phenocopies the Vul defects caused by loss of a core Ras pathway component. Finally, eor-1 encodes a nuclear-localized BTB/zinc-finger protein and hence is likely to function directly as a transcriptional regulator. Therefore, we propose that eor-1 and eor-2 cooperate with sur-2 and lin-25 and function as global regulators of Ras/ERK-dependent gene transcription (Fig. 5A). Although both sets of genes facilitate multiple Ras-dependent processes, sur-2 and lin-25 are primarily involved in vulval fate specification, whereas eor-1 and eor-2 are primarily involved in excretory system development and P12 fate specification. It will be interesting to determine why different tissues have different requirements for sur-2/lin-25 versus eor-1/eor-2.
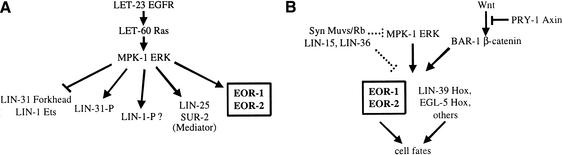
Figure 5.
Model for EOR-1 and EOR-2 function. (A) EOR-1 and EOR-2 act together and function downstream of MPK-1 ERK and in parallel to SUR-2, LIN-25, and LIN-1 to positively regulate Ras/ERK-dependent gene transcription. The positively acting form of LIN-1 may be phosphorylated (P), by analogy to findings with other Ets-domain proteins and with LIN-31 (see text). (B) EOR-1 and EOR-2 function with Hox proteins at a convergence point of the Ras, Wnt, and SynMuv/Rb pathways. EOR-1 and EOR-2 could function as cofactors to facilitate Hox-mediated transcription, or could specify cell fates independently of Hox genes. Dotted bars indicate the two alternative hypotheses for where the SynMuvs function.
lin-1 Ets both positively and negatively regulates Ras signaling output
The Elk-1 related gene lin-1 mainly functions as a negative regulator of Ras signaling output (Beitel et al. 1995), but our studies of eor-1 and eor-2 have provided the first evidence that lin-1 may also function as a positive regulator. We found that lin-1 loss-of-function mutations cause dramatic rod-like lethal and partial Vul defects in eor-1 or eor-2 mutant backgrounds, suggesting that lin-1 mutations partially reduce Ras signaling output. Notably, lin-1 mutations do not cause similar synthetic defects in sur-2 or lin-25 mutant backgrounds (Singh and Han 1995; Tuck and Greenwald 1995), whereas lin-1, sur-2, and lin-25 all interact similarly with eor-1 and eor-2. We therefore propose that, in its capacity as a positive regulator, lin-1 functions together with sur-2 and lin-25 but in parallel to eor-1 and eor-2 (Fig. 5A). Such a model is consistent with the recent report that mammalian Sur-2 specifically links Elk-1 to the Mediator complex to allow transcriptional activation (Stevens et al. 2002).
Ets-domain proteins can function as either transcriptional activators or repressors, and these activities are regulated by ERK phosphorylation. For example, in Drosophila, the transcriptional activator Pointed is stimulated by ERK phosphorylation (O'Neill et al. 1994), but the transcriptional repressor Yan/Aop is down-regulated by ERK phosphorylation (Rebay and Rubin 1995). In mammalian cells, the Elk-1-related protein Net is converted from a transcriptional repressor to a transcriptional activator by ERK phosphorylation (Maira et al. 1996). We speculate that LIN-1 could also be converted from a transcriptional repressor to a transcriptional activator by ERK phosphorylation (Fig. 5A). Such a model could explain why eor-1 and eor-2 interact similarly with the lin-1(n304) null mutation and with the lin-1(n2515gf) mutation that interferes with ERK phosphorylation (cf. Tables 1 and 2; Jacobs et al. 1998).
Relationship of eor-1 and eor-2 to the SynMuv/Rb pathway
The SynMuv Class A and Class B genes define two redundant pathways or complexes that inhibit both vulval and P12 fates (Ferguson and Horvitz 1989). SynMuv Class B genes include lin-35 Rb, lin-53 RbAp48, and hda-1 histone deacetylase, and therefore are thought to maintain an inactive chromatin structure and repress transcription (Lu and Horvitz 1998). Because SynMuv mutant defects are suppressed by loss-of-function mutations in the Ras pathway, SynMuv genes formally act upstream of let-23 RTK. However, an alternative interpretation of the epistasis is that SynMuv genes antagonize basal levels of Ras/ERK-dependent gene transcription (Lu and Horvitz 1998). In either case, the fact that eor-1 and eor-2 mutations suppress some defects of lin-15 SynMuv mutants is consistent with the model that EOR-1 and EOR-2 act downstream or in parallel to the SynMuv/Rb pathway to promote Ras/ERK-dependent gene transcription (Fig. 5B).
Interestingly, EOR-1 was previously isolated in a yeast two-hybrid screen with the SynMuv B pathway component LIN-36 (Thomas and Horvitz 1999; Walhout et al. 2000), and we were able to confirm this interaction using full-length proteins (data not shown). Although the significance of this interaction is presently unknown, it suggests that SynMuv gene products could antagonize Ras signaling in part by binding to and inhibiting the activity of positively acting transcriptional regulators such as EOR-1. Alternatively, the LIN-36–EOR-1 interaction could be involved in the recently reported positive role of some SynMuv B genes (Chen and Han 2001).
eor-1 and eor-2 function with Hox genes at a convergence point of the Ras and Wnt pathways
The Ras pathway cooperates with a Wnt pathway to promote vulval and P12 fates (Jiang and Sternberg 1998; Eisenmann and Kim 2000; Gleason et al. 2002). Because we have shown that eor-1 and eor-2 facilitate signaling through both the Ras and Wnt pathways, eor-1 and eor-2 appear to function at a convergence point of these pathways (Fig. 5B). Other genes previously placed at this convergence point are the Hox genes lin-39 and egl-5. Our epistasis experiments do not support models in which eor-1 and eor-2 are required primarily for Hox gene expression, and instead suggest that eor-1 and eor-2 cooperate with Hox genes to control the expression of genes required for vulval and P12 fates (Fig. 5B).
EOR-1 is a BTB/zinc-finger protein related to mammalian PLZF
EOR-1 is a member of the BTB-domain/zinc-finger family of proteins, several of which have been shown to regulate Ras/ERK signaling. Drosophila Tramtrack is a transcriptional repressor that functions antagonistically to Ras/ERK and is down-regulated in response to Ras/ERK signaling (S. Li et al. 1997; Tang et al. 1997). Mammalian BCL6 is also a transcriptional repressor but cooperates with Ras/ERK to promote cyclin D expression and oncogenic transformation (Shvarts et al. 2002). Drosophila Trithorax-like/GAGA is a transcriptional activator that facilitates Ras/ERK signaling (Maixner et al. 1998), possibly by counteracting chromatin repression and/or up-regulating Hox gene expression (Wilkins and Lis 1997). Although EOR-1 does not appear to be orthologous to any of the above proteins, our findings do suggest that BTB/zinc-finger proteins may be common regulators of Ras/ERK-dependent gene transcription in many organisms.
EOR-1 is the C. elegans ortholog of mammalian PLZF (Zhang et al. 1999). Reciprocal translocations between human PLZF and the retinoic acid receptor (RARα) are found in retinoic acid-resistant forms of acute promyelocytic leukemia. Retinoic acid resistance correlates with the ability of PLZF to bind corepressor complex components and thereby recruit histone deacetylases to RARα target promoters, preventing myeloid gene transcription (Lin et al. 1999). Transfection studies have suggested that intact PLZF functions as a transcriptional repressor (J.Y. Li et al. 1997); however, the normal targets of PLZF are not known. Interestingly, although PLZF has not been previously implicated as a target of Ras or Wnt signaling, it does influence developmental patterning and Hox gene expression in the mouse (Barna et al. 2000). Given that, like PLZF, EOR-1 interacts with a possible histone deacetylase complex component, LIN-36 (Walhout et al. 2000), and regulates Hox-dependent patterning (this work), these proteins could function in a related manner. Further studies of EOR-1 and EOR-2 may provide insight into the function and regulation of PLZF in both normal development and leukemogenesis.
Materials and methods
General methods and alleles
General methods for the handling and culturing of nematodes were as previously described (Brenner 1974). Experiments were performed at 20°C unless otherwise noted. Bristol N2 was the wild-type strain. The following genes, alleles, and balancers were used and are described in Riddle et al. (1997) unless otherwise noted: ayIs4 (Burdine et al. 1998); pry-1(mu38) (Maloof et al. 1999); sur-2(ku9), hT2[qIs48] (I; III) (Wang and Kimble 2001) I; lin-31(n301) II; egl-5(n486), lin-39(n709) III; dpy-20(e1282), eor-1(cs28), eor-1(cs40), eor-1(cs44) (Rocheleau et al. 2002); let-60(n1046gf), lin-1(e1275), lin-1(n304) (Beitel et al. 1995); lin-1(n2515gf) (Jacobs et al. 1998); sur-8(ku167) (Sieburth et al. 1998); unc-24(e138), DnT1(IV; V) (Rogalski et al. 1988) IV; gaIs36 (Lackner and Kim 1998); lin-25(e1446) V; bar-1(ga80) (Eisenmann et al. 1998); eor-2(cs7), eor-2(cs30), eor-2(cs42), eor-2(cs47), eor-2(cs51) (Rocheleau et al. 2002); lin-15(n309), lin-15(n765), unc-3(e151) X. syEx178 (Jiang and Sternberg 1998).
Phenotypic characterizations
To score lethality, hermaphrodites were allowed to lay eggs on a plate for 2–24 h. Rod-like arrested larvae were counted and removed. Less than 5% of eor-1 or eor-2 mutants became rod-like as adults; these were not included in the lethal category. To score vulval phenotypes, the numbers of vulval and nonvulval VPC descendants were counted during the L4 stage using Nomarski Differential Interference Contrast (DIC) microscopy. In calculating the average number of VPCs induced, hybrid lineages were assigned a value of 0.5. The number of P11.p-like cells was counted in L3 or L4 larvae using DIC.
EGL-17::GFP analysis
ayIs4, an integrated egl-17::gfp transgene (Burdine et al. 1998), was crossed into various backgrounds. eor-1 and lin-25 were balanced over DnT1, which causes a dominant Unc phenotype. Non-Uncs were scored at the mid-L3 stage for the presence or absence of GFP in P6.p or P6.pa and P6.pp. We found that 93% (n = 54) of the ayIs4 animals showed EGL-17::GFP expression in P6.p or its daughters.
Cloning of eor-1 and eor-2
eor-1 maps between dpy-13 and unc-5 on Chromosome IV (Rocheleau et al. 2002). Cosmid R11E3 (15 ng/μL) rescued the Unc and non-Muv phenotypes of eor-1(cs28) let-60(n1046gf) animals. An 8.7-kb XhoI–PstI fragment, containing the entire predicted R11E3.6 coding region with 1.1 kb of upstream sequence and 1.8 kb of downstream sequence (pRH3, 15 ng/μL) also rescued the mutants. A probe corresponding to yk87e7 detected a single band of ∼3.1 kb from mixed-stage RNA by Northern Blot analysis, consistent with this cDNA clone being full-length.
eor-2 maps to the left of unc-3 on the X chromosome (Rocheleau et al. 2002). Cosmid C44H4 (15 ng/μL) rescued the mating defects of eor-2(cs30) males and the Unc and non-Muv phenotypes of let-60(n1046gf); eor-2(cs30) hermaphrodites. A genomic PCR fragment amplified with oRH33 (5′-GGGATC CTAGCCATTGTTATG-3′) and oRH34 (5′-AAATGCACGGC GGAATAATGCG-3′), containing the entire predicted C44H4.7 coding region with 3.8 kb of additional upstream sequence and 2.1 kb of downstream sequence (15 ng/μL), also rescued. The sequence of eor-2 cDNA clone yk257c2 (eor-2a) differs slightly from our RT-PCR product sequence (eor-2b). A probe corresponding to yk257c2 detected a single band of ∼3.1 kb from mixed-stage RNA by Northern Blot analysis, consistent with these cDNA clones being full-length. BLASTp searches with EOR-2 identified Drosophila SD04853p as the closest match (5e-08); the similarity is limited to the C termini of both proteins. This region of similarity contains one C2H2-like sequence (Fig. 2B).
Identification of eor-1 and eor-2 mutant lesions
All coding regions and splice junctions of eor-1 and eor-2 were PCR-amplified from eor-1 and eor-2 mutant genomic DNA and sequenced. Lesions were verified by sequencing two independently derived PCR products. eor-1(cs28) contains a 68-bp deletion that removes 10 bp from the end of exon 5 and 58 bp from the intron between exons 5 and 6. We sequenced five RT-PCR product clones from eor-1(cs28) mutants and found that, in all cases, there was no splicing between exons 5 and 6. In the resulting protein, F259 is replaced with a leucine followed by a stop codon.
RNA-mediated interference (RNAi)
sur-8 and eor-1 RNAi experiments were performed essentially as described (Fire et al. 1998). For sur-8, double-stranded RNA was prepared from a full-length cDNA template. For eor-1, double-stranded RNA was prepared from a 1.8-kb PCR product derived from the cDNA clone yk151c9. Phenotypes were scored in progeny laid 12 or more hours after injection. eor-1(RNAi) caused 0% lethality (n = 363) in N2 but 24% lethality (n = 338) in lin-1(e1275).
EOR-1::GFP and EOR-2::GFP
A rescuing eor-1::gfp reporter (pRH5) was generated by cloning the GFP coding region from pPD119.45 (provided by A. Fire, Carnegie Institution, Baltimore, MD) into a unique NcoI site of pRH3, which results in the in-frame insertion of GFP(S65C) before the first zinc finger of EOR-1. A rescuing eor-2::gfp reporter (pRH29) was made by first generating a rescuing clone (pRH27), which is a BamHI–BglII fragment containing the entire eor-2 coding region with 3.8 kb of upstream sequence and 1.1 kb of downstream sequence cloned into pBSK(+). The GFP coding region from pPD102.33 (provided by A. Fire) was then cloned into a unique NruI site in pRH27, which results in the in-frame insertion of GFP(S65C) three residues before the stop codon.
Accession numbers
Accession numbers are as follows: AF519108 (EOR-1), AF19109 (EOR-2A), AF519110 (EOR-2B).
Acknowledgments
We thank D. Garbe, L. Girard, E. Wong, and M. Volk for technical assistance; I. Greenwald, S.Tuck, M. Lazar, and members of our lab for advice and critical reading of the manuscript; D. Eisenmann, M. Hengartner, M. Herman, R. Korswagen, and S. Tuck for sharing their unpublished results; S. Kim, J. Kimble, and P. Sternberg for strains; and Y. Kohara for cDNA clones. Some strains were provided by the Caenorhabditis Genetics Center. This work was supported by a Penn-Hughes award and National Institutes of Health grant 1R01GM58540-01A1 to M.V.S. R.M.H. is a predoctoral trainee supported by National Institutes of Health Training Program in Developmental Biology grant 5-T32-HD-07516.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sundaram@mail.med.upenn.edu; FAX (215) 573-9411.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.998402.
References
- Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Tuck S, Greenwald I, Horvitz HR. The Caenorhabditis elegans gene lin-1 encodes an ETS-domain protein and defines a branch in the vulval induction pathway. Genes & Dev. 1995;9:3149–3162. doi: 10.1101/gad.9.24.3149. [DOI] [PubMed] [Google Scholar]
- Bienz M. TCF: Transcriptional activator or repressor? Curr Opin Cell Biol. 1998;10:366–372. doi: 10.1016/s0955-0674(98)80013-6. [DOI] [PubMed] [Google Scholar]
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine RD, Branda CS, Stern MJ. EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development. 1998;125:1083–1093. doi: 10.1242/dev.125.6.1083. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- Chen Z, Han M. C. elegans Rb, NuRD, and Ras regulate lin-39-mediated cell fusion during vulval fate specification. Curr Biol. 2001;11:1874–1879. doi: 10.1016/s0960-9822(01)00596-6. [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Katz WS, Sternberg PW. Caenorhabditis elegans HOM-C genes regulate the response of vulval precursor cells to inductive signal. Dev Biol. 1997;182:150–161. doi: 10.1006/dbio.1996.8471. [DOI] [PubMed] [Google Scholar]
- Collins T, Stone JR, Williams AJ. All in the family: The BTB/POZ, KRAB, and SCAN domains. Mol Cell Biol. 2001;21:3609–3615. doi: 10.1128/MCB.21.11.3609-3615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Kim SK. Protruding vulva mutants identify novel loci and Wnt signaling factors that function during Caenorhabditis elegans vulva development. Genetics. 2000;156:1097–1116. doi: 10.1093/genetics/156.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. The β-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development. 1998;125:3667–3680. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–121. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- Gleason JE, Korswagen HC, Eisenmann DM. Activation of Wnt signaling bypasses the requirement for RTK/Ras signaling during C. elegans vulval induction. Genes & Dev. 2002;16:1281–1290. doi: 10.1101/gad.981602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D, Beitel GJ, Clark SG, Horvitz HR, Kornfeld K. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics. 1998;149:1809–1822. doi: 10.1093/genetics/149.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LI, Sternberg PW. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development. 1998;125:2337–2347. doi: 10.1242/dev.125.12.2337. [DOI] [PubMed] [Google Scholar]
- Korswagen HC, Coudreuse DYM, Betist MC, van de Water S, Zivkovic D, Clevers HC. The Axin-like protein PRY-1 is a negative regulator of a canonical Wnt pathway in C. elegans. Genes & Dev. 2002;16:1291–1302. doi: 10.1101/gad.981802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Kim SK. Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics. 1998;150:103–117. doi: 10.1093/genetics/150.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, English MA, Ball HJ, Yeyati PL, Waxman S, Licht JD. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- Li S, Li Y, Carthew RW, Lai ZC. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell. 1997;90:469–478. doi: 10.1016/s0092-8674(00)80507-3. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Egan DA, Evans RM. Molecular genetics of acute promyelocytic leukemia. Trends Genet. 1999;15:179–184. doi: 10.1016/s0168-9525(99)01710-2. [DOI] [PubMed] [Google Scholar]
- Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- Maira SM, Wurtz JM, Wasylyk B. Net (ERP/SAP2), one of the Ras-inducible TCFs, has a novel inhibitory domain with resemblance to the helix–loop–helix motif. EMBO J. 1996;15:5849–5865. [PMC free article] [PubMed] [Google Scholar]
- Maixner A, Hecker TY, Phan QN, Wassarman DA. A screen for mutations that prevent lethality caused by expression of activated sevenless and Ras1 in the Drosophila embryo. Dev Genet. 1998;23:347–361. doi: 10.1002/(SICI)1520-6408(1998)23:4<347::AID-DVG9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Kenyon C. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development. 1998;125:181–190. doi: 10.1242/dev.125.2.181. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Miller LM, Gallegos ME, Morisseau BA, Kim S. lin-31, a Caenorhabditis elegans HNF-3/fork head transcription factor homolog, specifies three alternative cell fates in vulval development. Genes & Dev. 1993;7:933–947. doi: 10.1101/gad.7.6.933. [DOI] [PubMed] [Google Scholar]
- Mohler WA, Simske JS, Williams-Masson EM, Hardin JD, White JG. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Nelson HC. Structure and function of DNA-binding proteins. Curr Opin Genet Dev. 1995;5:180–189. doi: 10.1016/0959-437x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Li X, Tiensuu T, Auty R, Greenwald I, Tuck S. Caenorhabditis elegans lin-25: Cellular focus, protein expression and requirement for sur-2 during induction of vulval fates. Development. 1998;125:4809–4819. doi: 10.1242/dev.125.23.4809. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Tiensuu T, Tuck S. Caenorhabditis elegans lin-25: A study of its role in multiple cell fate specification events involving Ras and the identification and characterization of evolutionarily conserved domains. Genetics. 2000;156:1083–1096. doi: 10.1093/genetics/156.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumental T, Meyer BJ, Priess JR. C. elegans II: Cold Spring Harbor Monograph Series 33. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Rocheleau CE, Howard RM, Goldman AP, Volk ML, Girard LJ, Sundaram MV. A lin-45 raf enhancer screen identifies eor-1, eor-2, and unusual alleles of Ras pathway genes in Caenorhabditis elegans. Genetics. 2002;161:121–131. doi: 10.1093/genetics/161.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Bullerjahn AM, Riddle DL. Lethal and amanitin-resistance mutations in the Caenorhabditis elegans ama-1 and ama-2 genes. Genetics. 1988;120:409–422. doi: 10.1093/genetics/120.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Brummelkamp TR, Scheeren F, Koh E, Daley GQ, Spits H, Bernards R. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes & Dev. 2002;16:681–686. doi: 10.1101/gad.929302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth DS, Sun Q, Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94:119–130. doi: 10.1016/s0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- Simon MA. Receptor tyrosine kinases: Specific outcomes from general signals. Cell. 2000;103:13–15. doi: 10.1016/s0092-8674(00)00100-8. [DOI] [PubMed] [Google Scholar]
- Singh N, Han M. sur-2, a novel gene, functions late in the let-60 ras-mediated signaling pathway during Caenorhabditis elegans vulval induction. Genes & Dev. 1995;9:2251–2265. doi: 10.1101/gad.9.18.2251. [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Han M. Genetics of RAS signaling in C. elegans. Trends Genet. 1998;14:466–472. doi: 10.1016/s0168-9525(98)01592-3. [DOI] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 Mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell. 1998;93:569–580. doi: 10.1016/s0092-8674(00)81186-1. [DOI] [PubMed] [Google Scholar]
- Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90:459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Horvitz HR. The C. elegans gene lin-36 acts cell autonomously in the lin-35 Rb pathway. Development. 1999;126:3449–3459. doi: 10.1242/dev.126.15.3449. [DOI] [PubMed] [Google Scholar]
- Tuck S, Greenwald I. lin-25, a gene required for vulval induction in Caenorhabditis elegans. Genes & Dev. 1995;9:341–357. doi: 10.1101/gad.9.3.341. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Wang S, Kimble J. The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 2001;20:1363–1372. doi: 10.1093/emboj/20.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins RC, Lis JT. Dynamics of potentiation and activation: GAGA factor and its role in heat shock gene regulation. Nucleic Acids Res. 1997;25:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Sundaram M, Han M. Ras is required for a limited number of cell fates and not for general proliferation in Caenorhabditis elegans. Mol Cell Biol. 1997;17:2716–2722. doi: 10.1128/mcb.17.5.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- Zhang T, Xiong H, Kan LX, Zhang CK, Jiao XF, Fu G, Zhang QH, Lu L, Tong JH, Gu BW, et al. Genomic sequence, structural organization, molecular evolution, and aberrant rearrangement of promyelocytic leukemia zinc finger gene. Proc Natl Acad Sci. 1999;96:11422–11427. doi: 10.1073/pnas.96.20.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]