Abstract
Prior work has indicated that D-type cyclin–cdk4 complexes, which are only active in proliferating cells, can suppress the skeletal muscle differentiation program in proliferating myoblasts. In this study, we show that cyclin D–cdk activity can block the activity of the MEF2 family of transcriptional regulators, which are crucial regulators of skeletal muscle gene expression. We have found that cyclin D–cdk activity blocks the association of MEF2C with the coactivator protein GRIP-1 and thereby inhibits the activity of MEF2. During skeletal muscle differentiation, GRIP-1 is localized to punctate nuclear structures and can apparently tether MEF2 to such structures. Cotransfection of GRIP-1 can both potentiate the transcriptional activity of a Gal4–MEF2C construct and induce MEF2C localization to punctate nuclear structures. Consistent with the absence of punctate nuclear GRIP-1 in proliferating myoblasts, we have found that ectopic cyclin D–cdk4 expression disrupts the localization of both GRIP-1 and MEF2C to these punctate subnuclear structures. Our findings indicate that cyclin D–cdk4 activity represses skeletal muscle differentiation in proliferating cells by blocking the association of MEF2 with the coactivator GRIP-1 and concomitantly disrupts the association of these factors with punctate nuclear subdomains within the cell.
Keywords: Differentiation, myogenesis, cyclin D, MEF2, GRIP-1
Mammalian muscle differentiation involves a carefully ordered pattern of gene expression that is coordinated with terminal cell cycle exit. Two families of transcription factors control the differentiation of skeletal muscle cells: the myogenic bHLH family (including MyoD, Myf-5, myogenin, and MRF-4) and the MEF2 family. The myogenic bHLH transcription factors are expressed solely in skeletal muscle and heterodimerize with ubiquitously expressed bHLH proteins, termed E proteins, to form a transcriptional complex capable of activating muscle-specific genes containing a specific DNA sequence (CANNTG) termed the E box (Blackwell and Weintraub 1990; Lassar et al. 1991). The myogenic bHLH transcription factors are able, when overexpressed, to induce the transdifferentiation of fibroblasts into myocytes (Davis et al. 1987; Weintraub 1993). Induction of myogenesis by the myogenic bHLH proteins induces the synthesis and functional activation of MEF2C (Cserjesi and Olson 1991; Lassar et al. 1991; Wang et al. 2001). The MEF2 family of transcription factors bind to A/T-rich DNA sequences present in the regulatory regions of many muscle-specific genes (Naya and Olson 1999). Although MyoD, Myf-5, and MEF2D are expressed in proliferating myoblasts (Tapscott et al. 1988; Montarras et al. 1991; Breitbart et al. 1993), these proteins fail to engage the differentiation program unless the cells are exposed to low mitogens and withdraw from the cell cycle. Thus, mitogen signaling regulates the activity of these transcription factors and coordinates skeletal muscle differentiation with cell cycle withdrawal.
Several hypotheses have been proposed to explain how mitogenic signals block skeletal muscle differentiation. These include PKC-mediated phosphorylation of myogenic bHLH proteins (Li et al. 1992), induction of dominant-negative HLH proteins that block E protein heterodimerization (Benezra et al. 1990), MEK1-mediated inactivation of the transcriptional activation domain of the myogenic bHLH proteins (Perry et al. 2001), and cyclin D–cdk4-mediated inhibition of the differentiation program (Rao et al. 1994; Rao and Kohtz 1995; Skapek et al. 1995, 1996). In the case of cyclin D–cdk4, it has been suggested that this kinase, which is active specifically in proliferating cells, can associate with the C-terminal region of MyoD and block its interaction with DNA (J.M. Zhang et al. 1999). However, it seems likely that cyclin D–cdk4 regulates muscle differentiation via other pathways, as ectopic expression of cyclin D–cdk4 can also block the activity of myogenin (Skapek et al. 1996), which fails to directly interact with the cyclin D–cdk4 complex (Wei and Paterson 2001). Another potential target for cyclin D–cdk4 is the tumor suppressor protein pRB, whose function is required to promote the latter stages of the skeletal muscle differentiation program (Novitch et al. 1996). However, because expression of a form of pRB which lacked all cdk phosphorylation sites was unable to rescue cyclin D–cdk4-mediated inhibition of muscle differentiation, we have postulated that the cyclin D–cdk4 complex must block muscle differentiation by targeting other proteins in addition to pRB (Skapek et al. 1996).
In this work we explore the effects of cyclin D–cdk on MEF2 function. A number of recent studies have elegantly shown that MEF2 function is modulated by a network of transcriptional coactivators and repressors. The Histone Acetyl-Transferases (HATs) p300 and GRIP-1 have been found to associate with MEF2 (Sartorelli et al. 1997; Chen et al. 2000) and are thought to activate gene expression by inducing histone acetylation (Goodman and Smolik 2000; Leo and Chen 2000). GRIP-1/TIF2/SRC-2 is a member of the p160-Steroid Receptor Coactivator (SRC) family, which also includes SRC-1 and TRAM1/RAC3/ACTR/pCIP/AIB-1/SRC-3 (Leo and Chen 2000). These coactivators bind to transcription factors such as nuclear hormone receptors, to other coactivators such as p300/CBP, and to the basal transcriptional machinery such as TBP and TFIIB (Leo and Chen 2000). GRIP-1 binds directly to both myogenin and MEF2C, via their bHLH and MADS/MEF2 domains, respectively (Chen et al. 2000). Repression of endogenous GRIP-1 expression by antisense RNA has been reported to block differentiation in myogenic cell lines (Chen et al. 2000).
MEF2 function is repressed by interaction with class II Histone De-Acetylases (HDACs; Miska et al. 1999; Sparrow et al. 1999; Lemercier et al. 2000; Lu et al. 2000a,b; Dressel et al. 2001), which are thought to block transcription by deacetylating histones and thereby inducing a repressive chromatin conformation (Goodman and Smolik 2000; Marmorstein and Roth 2001; McKinsey et al. 2001a). Class II HDACs (which include HDAC4, HDAC5, and HDAC7) contain a large N-terminal domain, absent from class I HDACs (i.e., HDAC1, HDAC2, HDAC3, and HDAC8), which promotes interaction with the MADS/MEF2 domain of MEF2 factors (McKinsey et al. 2001a). Class II HDACs inhibit MEF2 transcriptional activity but do not impair the ability of these transcription factors to either bind DNA or dimerize (Lu et al. 2000a,b). Ectopic expression of either HDAC4 or HDAC5 inhibits C2C12 myoblast differentiation and MyoD-induced transdifferentiation of fibroblasts into muscle (Lu et al. 2000a). This inhibition is a result of MEF2 inactivation, as MyoD-dependent transcription is not affected significantly by ectopic expression of class II HDACs (Lu et al. 2000a). Recent findings from the Olson lab have shown that during the process of skeletal myogenesis, the Calcium/Calmodulin-dependent protein Kinase (CaMK) directly binds to and phosphorylates class II HDACs, inducing their accumulation in the cytoplasm (McKinsey et al. 2001b). Phosphorylation of conserved serines near the nuclear localization signal of HDAC4, HDAC5, and HDAC7 promote the binding of these HDACs to 14-3-3 proteins, resulting in the sequestration of these HDACs in the cytoplasm and consequent derepression of MEF2 activity (McKinsey et al. 2001b).
In this work we show that cyclin D–cdk4 activity can block MEF2 function. Cyclin D–cdk4 activity fails to alter the cellular localization of class II HDACs, suggesting that cyclin D–cdk4 blocks MEF2 function independently of modulating the interaction of MEF2 with class II HDACs. Consistent with this idea, we have found that the interaction of MEF2C with the SRC family member GRIP-1 is blocked by ectopic cyclin D–cdk4 expression. GRIP-1 is localized to punctate nuclear structures in differentiated skeletal muscle cells (Chen et al. 2001) and is absent from these structures in proliferating myoblasts. We have found that cotransfected GRIP-1 drives accumulation of MEF2C into such punctate nuclear structures and that cyclin D–cdk4 activity inhibits the translocation of both MEF2C and GRIP-1 into these nuclear structures. Our results suggest that cyclin D–cdk4 activity in proliferating myoblasts acts to repress the skeletal muscle differentiation program by blocking the interaction of MEF2 family members with the SRC family coactivator GRIP-1 and consequently inhibits the localization of both MEF2 and GRIP-1 to punctate nuclear structures.
Results
D-type cyclins require cdk activity to repress MEF2-dependent transcription
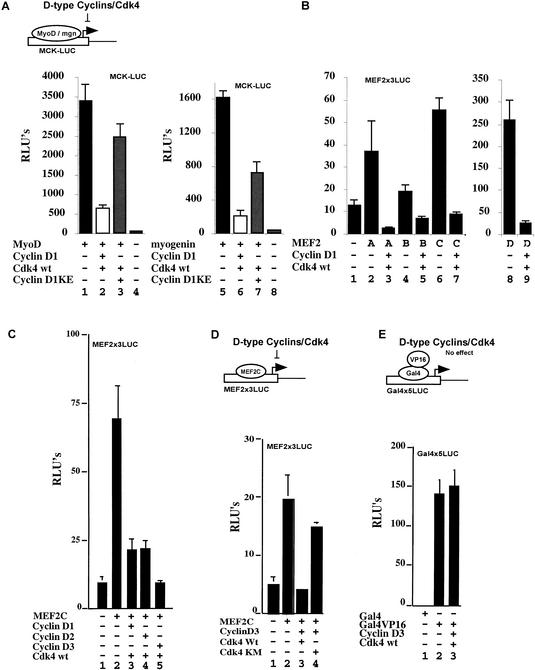
Cotransfection of cyclin D1 and cdk4 can block the ability of either MyoD or myogenin to activate the expression of muscle reporter constructs such as muscle creatine kinase-CAT (Fig. 1A, cf. lanes 1 and 2, and lanes 5 and 6; Rao et al. 1994; Skapek et al. 1995, 1996; J.M. Zhang et al. 1999). Maximal repression of either MyoD or myogenin activity by cotransfected cyclin D1–cdk4 requires the kinase activity of this complex as cotransfection of cdk4 together with cyclin D1-KE, a mutant form of cyclin D1 that fails to activate the cdk4 kinase (Hinds et al. 1994), restored significant expression of the MCK reporter construct (Fig. 1A, cf. lanes 2 and 3, and lanes 6 and 7). Because Paterson and colleagues have shown that the cyclin D1–cdk4 complex can bind directly to MyoD but not myogenin (J.M. Zhang et al. 1999), we speculated that there must be another cyclin D–cdk4 target other than MyoD that is required for the muscle differentiation program. One common factor that acts downstream and is presumably required for both MyoD and myogenin-mediated myogenic differentiation is the transcription factor MEF2. To evaluate whether D-type cyclins could affect the activity of MEF2 family members, we first monitored the effects of cyclin D1–cdk4 on the ability of exogenous MEF2A, MEF2B, MEF2C, or MEF2D to activate expression of a luciferase reporter driven by reiterated MEF2 binding sites (MEF2x3LUC). Although the various MEF2 family members activated the expression of MEF2x3LUC to varying levels in 10T1/2 cells (Fig. 1B, lanes 2,4,6,8), in all cases cotransfection with cyclin D1–cdk4 markedly blocked this activation (Fig. 1B, lanes 3,5,7,9). Apparently all D-type cyclins can block MEF2 activity as cotransfection of cyclin D1, D2, or D3 together with cdk4 blocked the ability of exogenous MEF2C to activate expression of the MEF2x3LUC reporter in 10T1/2 cells (Fig. 1C, cf. lane 2 with lanes 3,4,5). Again, cdk4 kinase activity is necessary for this inhibition as cotransfection of cyclin D3 together with a catalytically inactive mutant form of cdk4 (cdk4KM; van den Heuvel and Harlow 1993) failed to repress the activity of MEF2C (Fig. 1D, cf. lanes 3 and 4). We believe that ectopic expression of cyclin D–cdk4 was specifically repressing MEF2 activity, as opposed to repressing all transcriptional activation, because this cyclin–cdk combination failed to repress the transcriptional activity of a Gal4–VP16 fusion construct (Fig. 1E, cf. lanes 2 and 3).
Figure 1.
Cyclin D–cdk4 complexes can repress MEF2 transcription in a kinase-dependent manner. (A) Luciferase activity was monitored from whole-cell extracts of 10T1/2 fibroblasts cotransfected with a muscle creatine kinase (MCK)–luciferase reporter plus either empty expression vehicles (lanes 4,8), plasmids encoding MyoD (lanes 1–3), or myogenin (lanes 5–7) either in the absence (lanes 1,5) or presence (lanes 2,3,6,7) of cdk4 and either cyclin D1-WT (lanes 2,6) or the cyclin D1-KE mutant, which fails to activate cdk4 (lanes 3,7). (B) Luciferase activity was monitored from whole-cell extracts of 10T1/2 fibroblasts cotransfected with a MEF2x3-luciferase reporter plus either empty expression vehicles (lane 1) or plasmids encoding MEF2A (lanes 2,3), MEF2B (lanes 4,5), MEF2C (lanes 6,7), or MEF2D (lanes 8,9) either in the absence (lanes 2,4,6,8) or the presence (lane 3,5,7,9) of cyclin D1 and cdk4. (C) 10T1/2 cells were cotransfected with a MEF2x3 luciferase reporter construct plus either empty expression vehicles (lane 1) or plasmids encoding MEF2C (lanes 2–5), either in the absence (lane 2) or the presence of cdk4 (lanes 3–5) and cyclin D1 (lane 3), cyclin D2 (lane 4), or cyclin D3 (lane 5). (D) 10T1/2 cells were cotransfected with a MEF2x3 luciferase reporter construct plus either empty expression vehicles (lane 1) or plasmids encoding MEF2C (lanes 2–4), either in the absence (lane 2) or presence of cdk4-WT and cyclin D3 (lane 3) or a kinase dead mutant of cdk4 (cdk4-KM) and cyclin D3 (lane 4). (E) 10T1/2 cells were cotransfected with a Gal4x5 reporter construct plus plasmids encoding the Gal4 DNA-binding domain (lane 1), Gal4VP16 (lanes 2,3), either in the absence (lane 2) or in the presence of cdk4-WT and cyclin D3 (lane 3). The (−) sign indicates that an empty expression vehicle has been added instead of the corresponding expression plasmid.
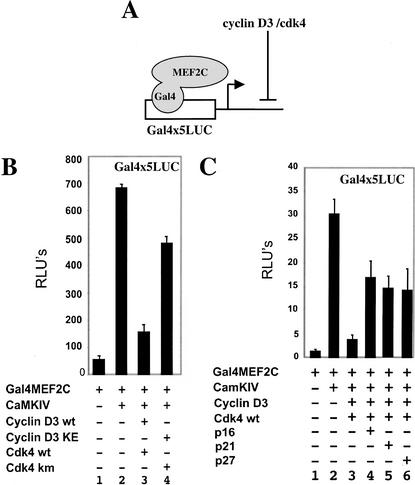
Coexpression of both cyclin D3 and cdk4 can block the ability of a constitutively active form of CaMKIV to augment MEF2C-driven transcription
The activity of MEF2C can be boosted significantly by cotransfection with an activated form of CaMK (McKinsey et al. 2000a), which induces the retention of class II HDACs in the cytoplasm (McKinsey et al. 2000b, 2001b; Kao et al. 2001). To evaluate if cyclin D–cdk activity would counter the effects of CaMKIV on MEF2 function, we assayed the effects of this kinase on the ability of a Gal4–MEF2C fusion protein (Gal4MEF2C) to activate a Gal4x5–luciferase reporter (diagramed in Fig. 2A). Although activated CaMKIV boosts the activity of Gal4MEF2C (Fig. 2B, cf. lanes 1 and 2), cotransfection of cyclin D3–cdk4 significantly dampens this effect (Fig. 2B, lane 3). Cdk4 kinase activity is required to maximally repress the effects of CaMKIV on Gal4MEF2C, because cotransfection of cyclin D3KE, which fails to activate cdk4 function, together with a catalytically inactive mutant of cdk4 (cdk4KM; van den Heuvel and Harlow 1993) only partially repress the activity of Gal4MEF2C (Fig. 2B, cf. lanes 3 and 4). Consistent with the notion that cdk4 catalytic activity is necessary to completely block MEF2 function, cotransfection of the cdk inhibitors p16, p21, or p27 partially reversed the inhibitory affects of cyclin D3–cdk4 on Gal4MEF2 activity (Fig. 2C, lanes 3–6).
Figure 2.
Cyclin D–cdk4 coexpression can repress CaMKIV-mediated augmentation of MEF2-driven transcription. (A) Schematic indicating that cyclin D3–cdk4 blocks Gal4MEF2C transcriptional activity. (B) Luciferase activity was monitored from 10T1/2 cells cotransfected with the Gal4x5 luciferase reporter construct plus plasmids encoding Gal4MEF2C (lanes 1–4), either in the absence (lane 1), or in the presence of activated CaMKIV (lanes 2–4), plus either cyclin D3 and cdk4 (lane 3) or cyclin D3KE and cdk4KM (lane 4). (C) 10T1/2 cells were cotransfected with the Gal4x5 luciferase reporter plus plasmids encoding Gal4MEF2C (lanes 1–6), either in the absence (lane 1) or in the presence of activated CaMKIV (lanes 2–6), plus cyclin D3 and cdk4 (lanes 3–6) and p16 (lane 4), p21 (lane 5), p27 (lane 5). The (−) sign indicates that an empty expression vehicle has been added instead of the corresponding expression plasmid.
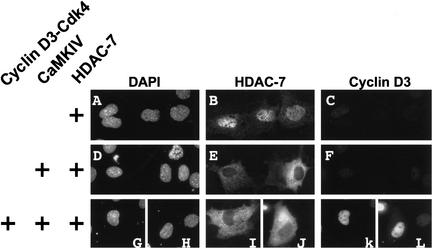
Cyclin D3–cdk4 does not affect the ability of CaMKIV to promote the nuclear-to-cytoplasmic shuttling of class II HDAC molecules
Class II HDAC molecules are actively shuttled from the nucleus to the cytoplasm in the presence of activated CaMKI or CaMKIV (McKinsey et al. 2000b). Given that coexpression of cyclin D3 and Cdk4 can inhibit the ability of CaMKIV to potentiate Gal4MEF2C-driven transcription (Fig. 2), we were interested in determining whether active cyclin D3–cdk4 complexes could inhibit the ability of CaMKIV to shuttle class II HDACs from the nucleus into the cytoplasm. 10T1/2 cells were initially transfected with HDAC7 alone. Consistent with prior work of others (Dressel et al. 2001; Kao et al. 2001), HDAC7 was predominantly detected in the nucleus (Fig. 3B). As expected, when HDAC7 was coexpressed with an activated form of CaMKIV, HDAC7 was relocalized to the cytoplasm in 100% of the cells that expressed both proteins (Fig. 3E). When cotransfected with exogenous cyclin D3–cdk4, CaMKIV still directed the retention of HDAC7 in the cytoplasm (Fig. 3I,J). We similarly found that activated CaMK-induced cytoplasmic retention of HDAC4 was also not altered by ectopic expression of D-type cyclins together with cdk4 (data not shown). These findings suggest that cyclinD–cdk4 complexes repress MEF2-dependent transcription by a mechanism that does not affect the nuclear export of class II HDACs by CaMK signaling.
Figure 3.
Cyclin D3–Cdk4 does not inhibit the ability of CaMKIV to exclude class II HDAC molecules from the nucleus. 10T1/2 fibroblasts were transfected with plasmids encoding HDAC7-HA alone (A–C), or in combination with either activated CaMKIV (D–F) or activated CaMKIV plus cyclin D3 and cdk4 (G–L). Then, 48 h after transfection, the cells were fixed and stained for HDAC7-HA (B,E,I,J) and cyclin D3 (C,F,K,L). Nuclei were visualized by staining with DAPI (A,D,G,H).
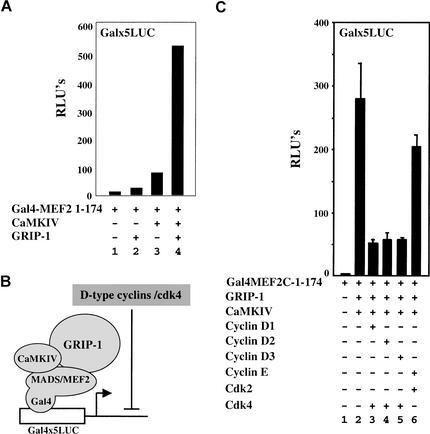
CaMKIV and GRIP-1 synergistically activate MEF2-dependent transcription by enhancing the association of GRIP-1 with MEF2
Because GRIP-1, a member of the SRC family of coactivators, has been shown to bind to MEF2 and thereby potentiate its transcriptional activity (Chen et al. 2000), we wondered if CaMKIV and GRIP-1 would function synergistically to activate MEF2-dependent transcription. Indeed, we found that the transcriptional activity of a fragment of MEF2C containing the MADS/MEF2 domain fused to the DNA-binding domain of GAL4 (Gal4–MEF2C-(1–174); Molkentin et al. 1995) was enhanced only slightly by cotransfection with either GRIP-1 or activated CaMKIV (Fig. 4A, lanes 1–3). In contrast, cotransfection of Gal4–MEF2C-(1–174) with both GRIP-1 and activated CaMKIV led to robust transcriptional activity of this fusion protein (Fig. 4A, lane 4). Whereas activated CaMKIV could induce the activity of both Gal4–MEF2C-(1–174) or Gal4–MEF2C-full length, synergistic activation by cotransfected GRIP-1 and CaMKIV was most evident with Gal4–MEF2C-(1–174) (data not shown).
Figure 4.
Cyclin D–cdk4 blocks synergistic activation of MEF2-driven transcription by CaMKIV and GRIP-1. (A) Luciferase activity was monitored from whole-cell extracts of 10T1/2 fibroblasts transfected with the Gal4x5 luciferase reporter construct and plasmids encoding Gal4MEF2C-1–174 (lanes 1–4), either alone (lane 1) or in the presence of GRIP-1 (lanes 2,4) and activated CaMKIV (lanes 3,4). (B) Model of the synergistic activation of Gal4MEF2C-1–174 transcriptional activity by CaMKIV and GRIP-1, and its inhibition by cyclin D–cdk4. (C) 10T1/2 fibroblasts were transfected with the Gal4x5 luciferase reporter construct plus plasmids encoding Gal4MEF2C-1–174 (lanes 1–6), either alone (lane 1) or in the presence of GRIP-1 and CaMKIV (lanes 2–6), plus cyclin D1 (lane 3), cyclin D2 (lane 4), cyclin D3 (lane 5), cyclin E (lane 6), cdk4 (lanes 3–5), and cdk2 (lane 6).
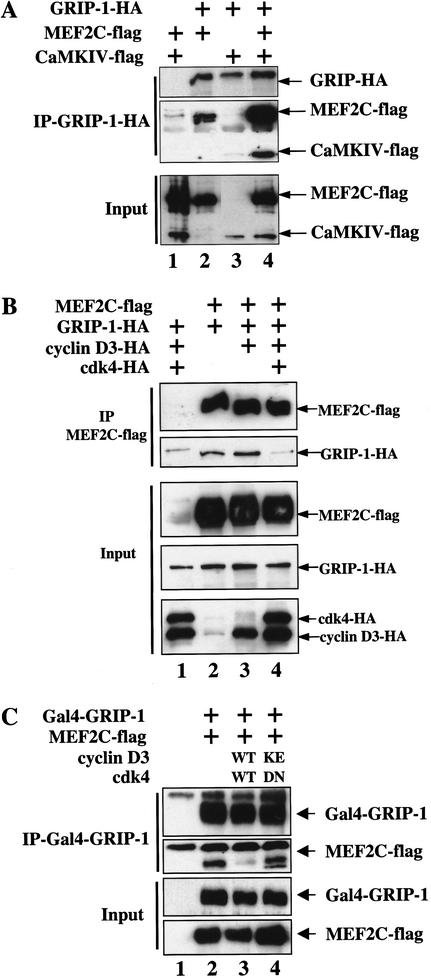
To evaluate whether CaMKIV could augment the association of MEF2C with GRIP-1, we examined the interaction of these proteins by coimmunoprecipitation in transfected COS cells (Fig. 5A). COS cells were cotransfected with flag-tagged MEF2C (Chen et al. 2000), flag-tagged activated CaMKIV (McKinsey et al. 2000b), and HA-tagged GRIP-1 (Ma et al. 1999). GRIP-1–HA and associated proteins were coimmunoprecipitated with an anti-HA monoclonal antibody (NM11). When GRIP-1–HA and MEF2C-Flag were coexpressed in COS cells, MEF2C-Flag was readily detected in the GRIP-1–HA immunoprecipitation (Fig. 5A, lane 2). When these same proteins were cotransfected in the presence of an activated form of CaMKIV, the apparent association between MEF2C and GRIP-1 was augmented significantly as increased levels of MEF2C-Flag were detected in the GRIP-1–HA immunoprecipitation (Fig. 5A, lane 4). Interestingly, CaMKIV-Flag was also present in the GRIP-1–HA IP, when cotransfected in the presence (Fig. 5A, lane 4) but not in the absence (Fig. 5A, lane 3) of MEF2C-Flag. Importantly, when MEF2C-Flag and CaMKIV-Flag were cotransfected in the absence of GRIP-1–HA, they each failed to be immunoprecipitated by the anti-HA monoclonal antibody (Fig. 5A, lane 1). These findings suggest that activated CaMKIV can induce MEF2 transcriptional activity by augmenting the association of GRIP-1 with MEF2. In addition, CaMKIV may facilitate the association of GRIP-1 with MEF2 by both directing the cytoplasmic localization of class II HDACs as well as by stabilizing a ternary complex between MEF2, GRIP-1, and CaMKIV (schematically shown in Fig. 4B).
Figure 5.
Kinase-dependent inhibition of GRIP-1–MEF2C association by cyclin D3–cdk4 in vivo. (A) Activated CaMKIV augments the association between MEF2 and GRIP-1. COS cells were cotransfected with plasmids encoding MEF2C–Flag (lanes 1,2,4), GRIP-1–HA (lanes 2–4), and CaMKIV-Flag (lanes 1,3,4). Cells were lysed, and anti-GRIP-1–HA immunoprecipitations were performed. Western blot analysis for the indicated proteins is shown for either the input cell lysate or for material immunoprecipitated by the anti-HA antibody. (B) Cyclin D3–cdk4 disrupts the interaction between MEF2 and GRIP-1. COS cells were transfected with plasmids encoding MEF2C–Flag (lanes 2–4), GRIP-1–HA (lanes 1–4), cyclin D3 (lanes 1,3,4), and cdk4 (lanes 1,4). Cells were lysed and anti-MEF2C–Flag immunoprecipitations were performed. Western blot analysis for the indicated proteins is shown for either the input cell lysate or for material immunoprecipitated by the anti-Flag antibody. (C) Cyclin D3–cdk4 catalytic activity is necessary to disrupt the interaction between MEF2 and GRIP-1. COS cells were transfected with plasmids encoding MEF2C–Flag and Gal4GRIP-1 (lanes 2–4), cyclin D3WT and cdk4WT (lane 3), or cyclin D3KE and cdk4DN (lane 4). Cells were lysed and anti-Gal4GRIP-1 immunoprecipitations were performed. Western blot analysis for the indicated proteins is shown for either the input cell lysate or for material immunoprecipitated by the anti-Gal4 antibody.
Cyclin D–cdk4 inhibits GRIP-1-mediated coactivation of MEF2C transcription by blocking the association of GRIP-1 and MEF2C
Because cyclin D–cdk4 blocks MEF2 function independently of altering CaMKIV-induced cytoplasmic sequestration of class II HDACs (Fig. 3), we wondered if cyclin D–cdk4 disrupts the ability of GRIP-1 to potentiate the activity of MEF2. 10T1/2 cells were cotransfected with the Gal4x5 luciferase reporter plus Gal4–MEF2C-(1–174), activated CaMKIV and GRIP-1 in either the absence or presence of the various D-type cyclins or cyclin E together with their cognate cdk (i.e., cdk4 or cdk2, respectively; schematically outlined in Fig. 4B). Whereas cotransfection of each of the D-type cyclins and cdk4 significantly reduced the expression of the Gal4 reporter by Gal4–MEF2C-(1–174) (Fig. 4C, lanes 2–5), cotransfection with cyclin E/cdk2 only slightly dampened expression of the reporter construct (Fig. 4C, lane 6). Thus D-type cyclin/cdk4 complexes specifically disrupt the synergistic activation of Gal4–MEF2C-(1–174) function by activated CaMKIV and the coactivator GRIP-1.
To evaluate if cyclin D–cdk activity disrupts the association of MEF2C with GRIP1, we cotransfected COS cells with MEF2C-Flag and GRIP-1–HA in either the absence or presence of cyclin D3 or cyclin D3–cdk4 and monitored the association of MEF2C with GRIP-1 by coimmunoprecipitation. Immunoprecipitation of MEF2C-Flag with an anti-Flag monoclonal antibody led to coimmunoprecipitation of cotransfected GRIP-1–HA (Fig. 5B, lane 2). Although cotransfection of cyclin D3, in the absence of cdk4, failed to affect the amount of GRIP-1 that was coimmunoprecipitated with MEF2C-Flag (Fig. 5B, lane 3), the addition of cyclin D3 together with cdk4 led to a significant decrease in coimmunoprecipitated GRIP-1 (Fig. 5B, lane 4). Importantly, the total amounts of either MEF2C-Flag or GRIP-1–HA expressed in cells were not significantly affected by the presence of cyclin D3–cdk4. Because cotransfection of the catalytically inactive cdk4DN together with cyclin D3KE failed to block Gal4–MEF2C function (Fig. 2B, lane 4), we investigated if cyclin D3KE and cdk4DN would affect the binding of GRIP-1 with MEF2C, like their wild-type counterparts. Cotransfection of a Gal4–GRIP-1 fusion construct and MEF2C-Flag into COS cells led to a detectable Gal4-GRIP-1–MEF2C-Flag complex as detected by the presence of MEF2C in an anti-Gal4–GRIP-1 immunoprecipitation (Fig. 5C, lane 2). As described above, the GRIP-1–MEF2C complex was disrupted by cotransfection of cyclin D3WT and cdk4WT, as evidenced by the loss of MEF2C-Flag from the Gal4–GRIP-1 immunoprecipitation (Fig. 5C, lane 3). In contrast, cotransfection of cdk4DN and cyclin D3KE failed to block the association of Gal4–GRIP-1 with MEF2C-Flag (Fig. 5C, lane 4). Thus, these findings indicate that an active cyclin D–cdk4 complex can block the association of the coactivator GRIP-1 with MEF2C and may thereby repress the transcriptional activity of the latter.
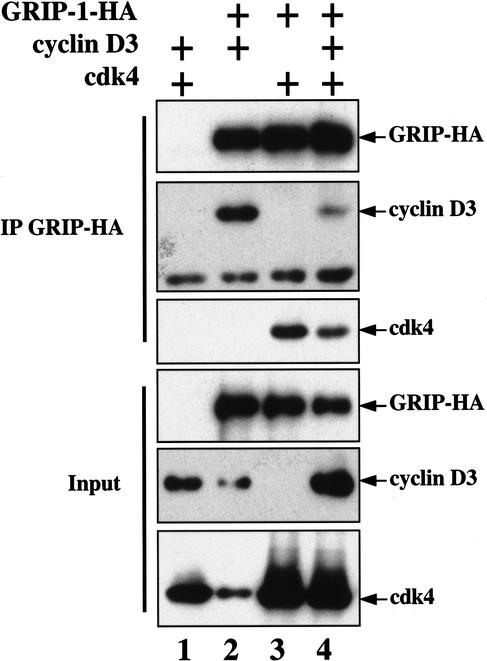
GRIP-1 interacts with both cyclin D3 and cdk4
We wondered if cyclin D–cdk4 activity inhibits the association of GRIP-1 with MEF2 by either directly binding to or phosphorylating either of these proteins. Indeed, other SRC family members (i.e., SRC1 and SRC3) have been found to specifically associate with cyclin D1 (Neuman et al. 1997; Zwijsen et al. 1998). Interaction of GRIP-1–HA with exogenous cyclin D3 and/or cdk4 was assayed by coimmunoprecipitation of GRIP-1–HA-associated proteins in extracts made from cotransfected COS cells. We found that when cotransfected with GRIP-1–HA, both cyclin D3 and cdk4 could be coimmunoprecipitated with GRIP-1–HA (Fig. 6, lanes 2–4). Because kinase activity is necessary for cyclin D–cdk4 to efficiently block MEF2 function (Fig. 1B), we speculate that cyclin D–cdk4 may block the association of GRIP-1 with MEF2 by directly binding to GRIP-1 and phosphorylating sites within either this protein or MEF2.
Figure 6.
GRIP-1 interacts with both cyclin D3 and cdk4 in vivo. COS cells were transfected with plasmids encoding GRIP-1–HA (lanes 2–4), cyclin D3 (lanes 1,2,4), and cdk4 (lanes 1,3,4). Cells were lysed, and anti-GRIP-1–HA immunoprecipitations were performed. Western blot analysis for the indicated proteins is shown for either the input cell lysate or for material immunoprecipitated by the anti-HA antibody.
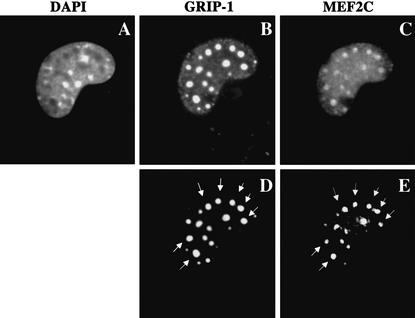
GRIP-1 and MEF2C colocalize to identical subnuclear domains
Members of the SRC family have been shown previously to migrate into punctate subnuclear structures either upon association with ligand-bound nuclear hormone receptors (Stenoien et al. 2000a,b) or during the process of skeletal muscle differentiation (Chen et al. 2000). Because GRIP-1 and MEF2 apparently associate within the cell, we wondered if both these proteins would be colocalized to the same subnuclear structures. 10T1/2 cells were cotransfected with GRIP-1–HA plus MEF2C-Flag, and their cellular localization was assayed by immunocytochemistry. In ∼30% of transfected cells, both GRIP-1 and MEF2C were found to be colocalized to identical subnuclear domains (Fig. 7, cf. D and E). In the remaining 70% of transfected cells, these proteins were present in a diffuse pattern throughout the nuclei.
Figure 7.
GRIP-1 and MEF2C colocalize to identical punctate subnuclear domains. 10T1/2 fibroblasts were transfected with plasmids encoding GRIP-1–HA and MEF2C–Flag. Then, 48 h after transfection, the cells were fixed and stained for GRIP-1–HA (B,D) and MEF2C–Flag (C,E). Nuclei were visualized by staining with DAPI (A). D and E are highly contrasted image acquisitions of B and C. The arrows indicate punctate subnuclear domains where GRIP-1 and MEF2C colocalize.
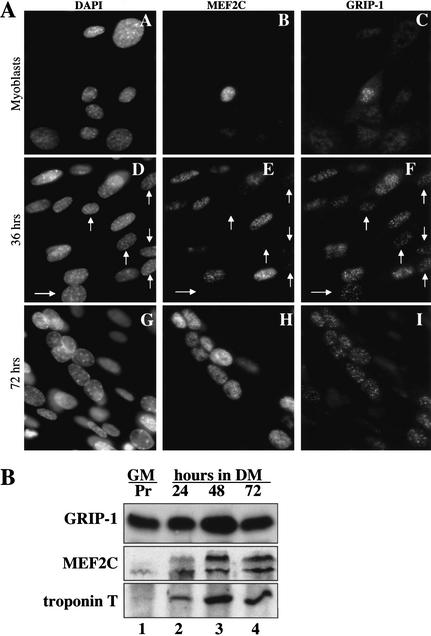
GRIP-1 localization to punctate nuclear structures proceeds that of MEF2C during skeletal muscle differentiation
Because GRIP-1 and MEF2C colocalized to identical subnuclear structures in transfected 10T1/2 cells (Fig. 7), we wanted to evaluate if these proteins colocalized to such structures during skeletal muscle differentiation. Immunofluorescence was used to analyze the localization of endogenous MEF2C and GRIP-1 in both proliferating C2C12 myoblasts and in these cells after exposure to differentiation conditions (i.e., low mitogens) for either 36 or 72 h. Although both MEF2C and GRIP-1 were undetectable in most proliferating myoblasts, an occasional cell expressed relatively low levels of each protein (Fig. 8A, panels A–C). After incubation in low mitogen medium for 48 h, both MEF2C and GRIP-1 levels were observed to rise by Western analysis (Fig. 8B, lanes 1–4), and by 36 h in differentiation conditions, each accumulated in punctate nuclear structures (Fig. 8A, panels D–F). Interestingly, GRIP-1 was observed in punctate nuclear structures in cells containing only trace levels of MEF2C (arrows in Fig. 8A, panels D–F). In contrast, MEF2C association with these structures only occurred in cells showing readily detectable GRIP-1. After 72 h in differentiation conditions, while GRIP-1 remained localized exclusively to the punctate nuclear structures (Fig. 8A, panel I), MEF2C was localized diffusely throughout the nuclei as well as in the punctate structures (Fig. 8A, panel H). Although we failed to detect GRIP-1 in punctate nuclear structures in the majority of proliferating myoblasts by immunofluorescence, the protein is present in these cells as it could be readily detected by Western analysis in both proliferating myoblasts and in differentiated myotubes (Fig. 8B, lanes 1–4). These findings suggest that mitogen signaling in proliferating myoblasts somehow excludes GRIP-1 association with the punctate nuclear structures. After 36 h in differentiation medium, GRIP-1 association with the punctate nuclear structures proceeds that of MEF2C. After 72 h in differentiation medium, MEF2C expression continues to rise to levels that may saturate MEF2C binding sites in the punctate nuclear structures; consequently, MEF2C is present in both the punctate nuclear structures as well as being distributed diffusely throughout the nucleus.
Figure 8.
Endogenous GRIP-1 and MEF2C colocalize to punctate nuclear structures during C2C12 differentiation. (A) C2C12 cells were fixed during proliferation (panels A–C) or after 36 h (panels D–F) or 72 h (panels G–I) of incubation in differentiation medium. The cells were then stained for MEF2C (panels B,E,H) and GRIP-1 (panels C,F,I). Nuclei were visualized by staining with DAPI (panels A,D,G). The arrows indicate C2C12 cells where GRIP-1 is present in punctate nuclear structures, and MEF2C is almost undetectable in these structures, suggesting that GRIP-1 accumulates in punctate nuclear structures prior to MEF2C during myogenic differentiation. (B) GRIP-1 is present in both C2C12 myoblasts and differentiated myotubes. C2C12 cells were harvested and lysed for immunoblotting analysis, during proliferation (Pr) in Growth Medium (GM; lane 1), or after 24, 48, or 72 h of incubation in Differentiation Medium (DM; lanes 2,3,4, respectively). Immunoblotting analysis was performed for GRIP-1, MEF2C, and Troponin T expression.
GRIP-1 cotransfection is necessary to localize MEF2 to the punctate subnuclear domains
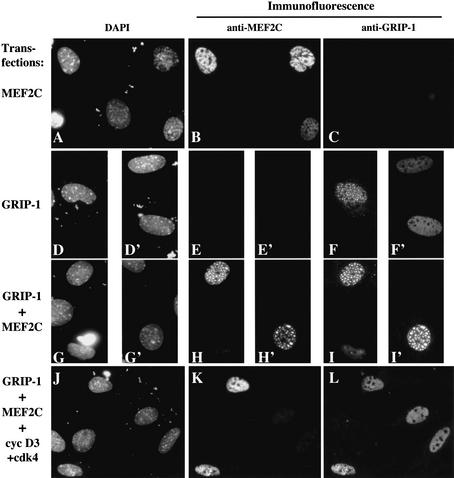
Although we have found that both GRIP-1 and MEF2C can be sequestered into nuclear subdomains, it is unclear which component of this complex is driving this cellular localization. In the instance of steroid hormone receptors, it is the ligand-bound receptor that translocates into these nuclear subdomains and brings the SRC family member (i.e., GRIP-1) along for the ride (Schaufele et al. 2000; Stenoien et al. 2000a,b). Thus, we wondered whether translocation of either GRIP-1 or MEF2C into such nuclear subdomains relied upon the presence of the other protein. To evaluate this, we transfected 10T1/2 cells with either MEF2C–Flag alone, GRIP-1–HA alone, or the combination of MEF2C–Flag plus GRIP-1–HA, and analyzed the cellular localization of transfected MEF2C and GRIP-1 by immunofluorescence. When transfected individually, MEF2C–Flag was present in a diffuse pattern throughout the nuclei (Fig. 9B), but GRIP-1–HA was localized to punctate nuclear subdomains in ∼30% of transfected cells (Fig. 9F,F‘). In contrast, when cotransfected together, MEF2C was now colocalized with GRIP-1 in the punctate nuclear structures (Fig. 9H,H‘,I,I‘). Importantly, MEF2C was only located in punctate nuclear structures in that 30% of transfected cells that contained GRIP-1 in these same nuclear subdomains. These results suggest that trafficking of MEF2C into the punctate nuclear structures requires interaction of this transcription factor with GRIP-1. Consistent with our findings that MEF2C translocates to punctate nuclear subdomains in a GRIP-1-dependent manner, we have found that during the process of skeletal muscle differentiation, GRIP-1 is first observed to localize to the punctate nuclear structures prior to the appearance of MEF2C in these subnuclear domains (see arrows in Fig. 8A, panels D–F).
Figure 9.
GRIP-1-induced recruitment of MEF2C into punctate subnuclear domains is inhibited by cyclin D3–cdk4. 10T1/2 fibroblasts were transfected with plasmids encoding MEF2C (A–C, G–L), GRIP-1 (D–L), and cyclin D3–cdk4 (J–L). Then, 48 h after transfection, the cells were fixed and stained for MEF2C–Flag (B,E,E‘,H,H‘,K) and GRIP-1 (C,F,F‘,I,I‘,L). Nuclei were visualized by staining with DAPI (A,D,D‘,G,G‘,J).
D-type cyclin/cdk4 overexpression affects the subnuclear distribution of GRIP-1
Because SRC family members translocate into punctate subnuclear domains when bound to ligand-bound steroid hormone receptors (Stenoien et al. 2000a,b), we wondered if colocalization of MEF2C and GRIP-1 into these same structures might correlate with the transcriptional activity of MEF2. To begin to address this issue, we monitored whether the subnuclear localization of GRIP-1 would be affected by cotransfection with cyclin D3–cdk4. Indeed, we found that although GRIP-1 accumulated in subnuclear structures of 25%–36% of cells transfected with GRIP-1 alone, this protein was present in such subnuclear domains in only 2%–5% of cells cotransfected with cyclin D3/cdk4 (See Table 1). We noted that the few cells that expressed GRIP-1 in subnuclear structures following cyclin D3–cdk4 cotransfection failed to express ectopic cyclin D3, suggesting that cyclin D3–cdk4 drives complete exclusion of GRIP-1 from such subnuclear structures. Cotransfection of MEF2C with GRIP-1 did not significantly alter the percentage of cells that showed punctate subnuclear staining of GRIP-1 (31%–41%; Table 1). However, cotransfection of cyclin D3–cdk4 plus MEF2C and GRIP-1 reduced the subnuclear localization of both MEF2C and GRIP-1 to 5%–8% of transfected cells (Fig. 9J–L; Table 1). These findings indicate that exogenous GRIP-1 is localized to punctate subnuclear structures in the cell and that this localization is disrupted by cyclin D3–cdk4 overexpression. Consistent with our findings that cdk catalytic activity is necessary to disrupt MEF2C function, we observed that cotransfection of cyclin D3 with cdk4DN failed to disrupt the subnuclear localization of GRIP-1 (data not shown). Thus, the presence of GRIP-1 in punctate subnuclear structures correlates with the functional activity of MEF2C within the cell.
Table 1.
Cyclin D3–cdk4 blocks the localization of GRIP-1 to punctate nuclear subdomains
| Plasmid transfected
|
No. of cells counted
|
C3H10T1/2 cells showing a GRIP-1-HA nuclear staining
|
|
|---|---|---|---|
| Punctate nuclear
|
Diffuse nuclear
|
||
| GRIP-1-HA + vehicles | Exp 1 = 200 | 35% | 65% |
| Exp 2 = 200 | 25% | 75% | |
| Exp 3 = 100 | 36% | 64% | |
| GRIP-1-HA Cyclin D3, Cdk4 + vehicles | Exp 1 = 200 | 2% | 98% |
| Exp 2 = 200 | 4% | 96% | |
| Exp 3 = 100 | 5% | 95% | |
| GRIP-1-HA MEF2C + vehicles | Exp 1 = NDa | — | — |
| Exp 2 = 200 | 31% | 69% | |
| Exp 3 = 100 | 41% | 59% | |
| GRIP-1-HA Cyclin D3, Cdk4 MEF2C | Exp 1 = NDa | — | — |
| Exp 2 = 200 | 5% | 95% | |
| Exp 3 = 100 | 8% | 92% | |
10T1/2 cells were cotransfected with the indicated plasmids and subsequently fixed and immunostained for the localization of transfected GRIP-1–HA. Cells showing either punctate or diffuse nuclear staining of GRIP-1–HA were tailed in each experiment.
Discussion
Cyclin D–cdk4 activity inhibits the association of MEF2 with GRIP-1 and disrupts the localization of these proteins to punctate nuclear structures
We have found that cyclin D–cdk4 activity can block MEF2C activity without altering CaMK-induced cytoplasmic retention of class II HDACs. These findings suggest that cyclin D–cdk4 blocks MEF2C function independently of modulating the interaction of MEF2C with class II HDACs. Consistent with this idea, we have found that the interaction of MEF2C with the SRC family member GRIP-1 is inhibited by ectopic cyclin D–cdk4 expression. Because the MADS/MEF2 domain, which is conserved among all MEF2 family members (Black and Olson 1998), is able to support interaction with GRIP-1 (Chen et al. 2001), it seems likely that cyclin D–cdk4 activity can block the interaction of all MEF2 family members with this coactivator. As cyclin D1/cdk4 activity is limited to proliferating myoblasts (Wang and Walsh 1996), which are known to contain MEF2D (Breitbart et al. 1993), it seems likely that the transcriptional activity of this isoform of MEF2 is restrained in proliferating myoblasts by cyclin D1–cdk4 activity. Furthermore, because MyoD activates the expression of other MEF2 family members, including MEF2C (Wang et al. 2001) during the differentiation process, the presence of active cyclin D1–cdk4 in proliferating myoblasts would serve as a failsafe mechanism to ensure that such isoforms of MEF2 would not be functional if they were promiscuously expressed in proliferating cells.
GRIP-1 nuclear localization is modulated during the skeletal muscle differentiation process. Although the protein is present in both proliferating myoblasts and differentiated myotubes, we could detect this protein in punctate nuclear structures only in the differentiated myotubes. Muscat and colleagues have found that GRIP-1 can also be detected in punctate subnuclear structures in proliferating myoblasts (Chen et al. 2001), but both this prior work and our own findings suggest that the localization of GRIP-1 to these subnuclear structures is more prominent in differentiated myotubes (Chen et al. 2001; this work). During skeletal muscle differentiation, GRIP-1 can be detected in the punctate nuclear structures prior to MEF2C, suggesting that GRIP-1 may somehow initiate the association of MEF2 with these structures. Indeed, we have found that GRIP-1 is capable of tethering MEF2C to these punctate nuclear structures and that cyclin D–cdk4 activity inhibits the accumulation of both MEF2C and GRIP-1 into this nuclear subdomain. Our results suggest that cyclin D1–cdk4 activity in proliferating myoblasts acts to repress the muscle differentiation program by blocking both the interaction of MEF2 family members with the coactivator GRIP-1 and localization of both these proteins to the punctate nuclear subdomains. As cyclin D–cdk4 complexes can bind to SRC family members in vivo (Neuman et al. 1997; Zwijsen et al. 1998; this work), it seems plausible that phosphorylation of GRIP-1 by cyclin D–cdk4 may induce a conformation of GRIP-1 that is unable to associate with both MEF2 and the punctate nuclear structures.
In addition, work of others has established that mitogenic signals additionally block the activity of MyoD in proliferating myoblasts by both MEK-dependent (Perry et al. 2001) and cdk4-dependent (J.M. Zhang et al. 1999) pathways (summarized in Fig. 10). Together, these findings indicate that mitogenic signals use several distinct pathways to ensure that myogenic transcription factors are functionally inert in proliferating cells. Both inhibition of MEF2C function and release of MEF2C and GRIP-1 from the punctate nuclear structures by cyclin D–cdk4 require cdk catalytic activity. Although cyclin D3 is present at high levels in differentiated myotubes, because of the high level of cdk inhibitors in these cells (Halevy et al. 1995; Franklin and Xiong 1996; P. Zhang et al. 1999), cyclin D3–cdk4 complexes are catalytically inactive in differentiated myotubes (Cenciarelli et al. 1999) and, therefore, would not be expected to block MEF2 function in differentiated skeletal muscle.
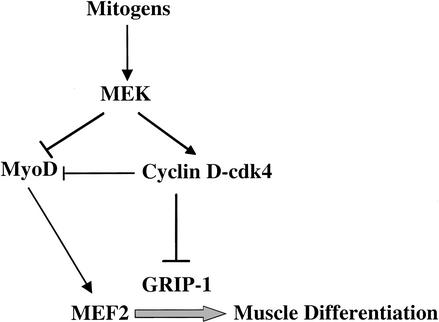
Figure 10.
Mitogens block skeletal muscle differentiation by several pathways. MyoD and MEF2C cooperate to activate skeletal-muscle-specific genes. In proliferating cells, mitogen signaling activates MEK, which, in turn, activates the expression of D-type cyclins (Ladha et al. 1998). Whereas MEK blocks the transcriptional activity of MyoD (Perry et al. 2001), D-type cyclins and cdk4 can inhibit both MyoD (J.M. Zhang et al. 1999) and GRIP-1-dependent MEF2C activities (this work).
The association of GRIP-1 and MEF2 with punctate nuclear structures may be regulated by environmental conditions
We have found that whereas ∼30% of 10T1/2 cells cotransfected with GRIP-1 and MEF2 colocalize both GRIP-1 and MEF2 to punctate nuclear structures, 100% of differentiated myotube nuclei contain GRIP-1 in these nuclear subdomains (see also Chen et al. 2001). It is unclear why only a portion of transfected 10T1/2 cells show accumulation of GRIP-1 and MEF2C in these punctate nuclear structures. However, we think it likely that physiological conditions may influence the association of GRIP-1 with the punctate nuclear structures and could potentially influence the activity of MEF2 family members. Consistent with this notion, we have found that cyclin D–cdk4 expression blocks MEF2 transcriptional activity, the association of MEF2 with the coactivator GRIP-1, and the localization of GRIP-1 and MEF2 to punctate nuclear structures. Work with nuclear steroid hormone receptors has indicated that GRIP-1 is shuttled into such nuclear subdomains upon binding to ligand-bound steroid hormone receptors, which are also shuttled into these subdomains of the nucleus when they are transcriptionally active (Schaufele et al. 2000; Stenoien et al. 2000a,b). We are currently investigating if environmental signals that promote skeletal muscle differentiation, such as IGF-1 signaling or activated p38 (Zetser et al. 1999; Wu et al. 2000), act to promote the accumulation of GRIP-1 and MEF2 into the punctate nuclear structures in differentiated myotubes.
What is the physiological significance of GRIP-1–MEF2 association with punctate nuclear structures?
We have found that whereas all detectable GRIP-1 is associated with the punctate nuclear structures in differentiated myotubes, only a portion of MEF2C is associated with such structures, the remainder being present diffusely throughout the nuclei. During the process of skeletal muscle differentiation, low levels of MEF2C initially accumulate in the punctate nuclear structures, prior to the accumulation of high levels of MEF2C in both the punctate structures and diffusely throughout the nuclei, suggesting that MEF2C-binding sites within the punctate nuclear structures may become saturated by high levels of MEF2C. As GRIP-1 is apparently necessary to tether MEF2C to these structures, it is possible that GRIP-1 or other SRC family members are the limiting MEF2C-binding component in the punctate nuclear structures. In addition, because it is known that GRIP-1 can rapidly exchange in and out of the punctate nuclear structures when bound to steroid hormone receptors (Schaufele et al. 2000), it seems possible that the GRIP-1–MEF2 complex may similarly diffuse in and out of these structures. It will be interesting to determine what domains of GRIP-1 are necessary to tether MEF2 to the punctate nuclear structures and whether the transcriptional activity of MEF2 can be uncoupled from association with these subnuclear domains.
Materials and methods
Cell culture and transient transfection
C2C12 myoblasts, C3H-10T1/2 fibroblasts, and COS7 cells were grown in Growth Medium (GM: DMEM supplemented with 10% Fetal Bovine Serum, 100 units/mL penicillin, and 0.1 mg/mL streptomycin). Differentiated C2C12 cells were obtained by substituting Growth Medium with Differentiation Medium (DM: DMEM supplemented with 2% Horse Serum, 100 units/mL penicillin, and 0.1 mg/mL streptomycin). Cells were plated and transfected in 12-well plates for promoter-luciferase assays and in 10-cm plates for immunoprecipitations and indirect immunofluorescence microscopy. The cells were transfected using the Fugene 6 (Roche) reagent, according to the manufacturer’s protocol. Each well of a 12-well plate received 0.5 μg of DNA, and each 10-cm plate received 10 μg of DNA.
Promoter assays and plasmids
The eukaryotic expression vehicles used in this study were transfected into cells as described above. Luciferase activity-based promoter assays were performed in triplicate, using the Luciferase Assay System (Promega) and the manufacturer's standard protocol. Luciferase activity was measured using a Turner Designs luminometer. The Gal4 promoter activity assays were performed with a Gal4x5LUC construct (PG5-LUC, Promega). MEF2-responsive promoter activity was monitored using MEF2x3LUC. pRC-CMV-Cyclin E, D1, D2, and D3 wild-type and mutant constructs were obtained from Phil Hinds with the exception of the pRC-CMV-Cyclin D3-K112E, which was engineered in this laboratory. pCMV-neo-bam-cdk4 wild-type and dead kinase mutant (cdk4-KM) and pCMV-neo-BAM-cdk2 were obtained from Ed Harlow and Sander Van den Heuvel. pCMX-MEF2C-Flag was obtained from Dennis Dowhan-he and George Muscat. PSG5-GRIP-1-HA was obtained from Michael Stallcup. The pGal4-MEF2C construct was obtained from Jiahuai Han (Han et al. 1997). pCS2-MEF2A, pcDNAI-MEF2B, pcDNAI-MEF2D, pGal4-MEF2C-1–174, pCMX-HDAC7, and an activated form of CaMKIV (pSG5-CaMK-Flag containing a codon stop in place of gln318) were obtained from Eric Olson. pRC/CMV-p16 is described in Skapek et al. (1995), and pCMV-p21cip1 was obtained from Wade Harper and Steve Elledge (Harper et al. 1993). pCS2-mouse-MyoD and pCS2-mouse-myogenin were obtained from M. Horwitz.
Indirect immunofluorescence microscopy
The cells were grown in 10-cm dishes, as described above, fixed in PBS-4% paraformaldehyde for 10 min at room temperature (extracted with methanol for 10 min if indicated), and subsequently incubated in PBS containing 4% Bovine Serum Albumin (BSA) for another 10 min. Primary antibodies were incubated in PBS-4% BSA at 37°C for 1 h at a 1/300 dilution. Cyclin D3 was detected using the C-16 rabbit polyclonal antibody (Santa Cruz Biotechnology). Flag- and HA-tagged proteins were detected using the M2 anti-Flag (Sigma) and anti-HA (BabCO) rabbit polyclonal antibody (HA11), respectively. Endogenous MEF2C and GRIP-1 were detected using a rabbit anti-MEF2C (Cell Signaling Technology) and a monoclonal anti-TIF2 (Transduction Laboratories), respectively. Secondary antibodies were incubated in PBS-4% BSA at 37°C for 30 min at a 1/200 dilution. We used combinations of anti-rabbit or anti-mouse goat IgGs conjugated to Alexia Fluor-488 (Molecular Probes) and anti-mouse CY3 or anti-rat CY3 IgGs (Jackson Immunochemicals). DNA was visualized by DAPI staining (Sigma).
Immunoprecipitation and Western blotting
Cells were collected in PBS, pelleted by gentle centrifugation at room temperature, and lysed. Typically, the pellet of one 10-cm dish was lysed in 350 μL of lysis buffer (200 mM NaCl, 50 mM Tris-HCl at pH 7.4, 2 mM EDTA, 0.5% NP-40, supplemented with anti-protease tablets; Roche) at 4°C for 15 min. After a brief sonication, insoluble materials were pelleted, and the supernatant (lysate) was collected. For each transfection, 5% of the lysate was aliquoted as an input control. Flag-tagged, Gal4-tagged, and HA-tagged proteins were immunoprecipitated using either the M2-anti-Flag (Sigma), the RK5C1-anti-Gal4 (Santa Cruz Biotechnology), or the HA11-anti-HA monoclonal antibodies (BabCO), respectively. The lysate was incubated at 4°C for 1 h on a rotating wheel, with 5 μg of the indicated antibody. Subsequently, 25 μL of A/G protein beads was added to each lysate, and the lysate was rotated at 4°C for an additional 30 min. Finally, the beads were washed three times in lysis buffer, and both immunoprecipitates and input controls were denaturated at 95°C for 2 min in Laemmli Buffer. The proteins were separated by 10% SDS-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes (Schleicher & Schuell), and analyzed by immunoblotting using the indicated antibodies at 1/500 to 1/2000 dilutions.
Acknowledgments
We thank Dennis Dowhan-he, Steve Elledge, Jiahuai Han, Ed Harlow, Wade Harper, Phil Hinds, George Muscat, Eric Olson, Michael Stallcup, and Sander Van den Heuvel for generously providing us with reagents. This work was supported by NIH grants to A.B.L. J.-B.L. was supported by a grant from the Association pour la Recherche contre le Cancer. P.J.B. was supported by a C.J. Martin Fellowship awarded by the NHMRC of Australia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL andrew_lassar@hms.harvard.edu; FAX (617) 738-0516.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.U-9988R.
References
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix–loop–helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Breitbart RE, Liang C, Smoot LB, Laheru DA, Mahdavi V, Nadal-Ginard B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- Cenciarelli C, De Santa F, Puri PL, Mattei E, Ricci L, Bucci F, Felsani A, Caruso M. Critical role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblast differentiation. Mol Cell Biol. 1999;19:5203–5217. doi: 10.1128/mcb.19.7.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes & Dev. 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Wang SC, Hosking B, Muscat GE. Subcellular localization of the steroid receptor coactivators (SRCs) and MEF2 in muscle and rhabdomyosarcoma cells. Mol Endocrinol. 2001;15:783–796. doi: 10.1210/mend.15.5.0637. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Olson EN. Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol Cell Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dressel U, Bailey PJ, Wang SC, Downes M, Evans RM, Muscat GE. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J Biol Chem. 2001;276:17007–17013. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- Franklin DS, Xiong Y. Induction of p18INK4C and its predominant association with CDK4 and CDK6 during myogenic differentiation. Mol Biol Cell. 1996;7:1587–1599. doi: 10.1091/mbc.7.10.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes & Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- Harper J, Adami G, Wei N, Keyomarsi K, Elledge S. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Verdel A, Tsai CC, Simon C, Juguilon H, Khochbin S. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- Ladha MH, Lee KY, Upton TM, Reed MF, Ewen ME. Regulation of exit from quiescence by p27 and cyclin D1-CDK4. Mol Cell Biol. 1998;18:6605–6615. doi: 10.1128/mcb.18.11.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Davis RD, Wright WE, Kadesh T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J Biol Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- Li L, Zhou J, James G, Heller-Harrison R, Czech MP, Olson EN. FGF inactivates myogenic helix–loop–helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell. 1992;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol Cell. 2000a;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci. 2000b;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Hong H, Huang SM, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R, Roth SY. Histone acetyltransferases: Function, structure, and catalysis. Curr Opin Genet Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci. 2000a;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000b;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001a;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- ————— Identification of a signal-responsive nuclear export sequence in class II histone deacetylases. Mol Cell Biol. 2001b;21:6312–6321. doi: 10.1128/MCB.21.18.6312-6321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Montarras D, Chelly J, Bober E, Arnold H, Ott MO, Gros F, Pinset C. Developmental patterns in the expression of Myf5, MyoD, myogenin, and MRF4 during myogenesis. New Biologist. 1991;3:592–600. [PubMed] [Google Scholar]
- Naya FS, Olson E. MEF2: A transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr Opin Cell Biol. 1999;11:683–688. doi: 10.1016/s0955-0674(99)00036-8. [DOI] [PubMed] [Google Scholar]
- Neuman E, Ladha MH, Lin N, Upton TM, Miller SJ, DiRenzo J, Pestell RG, Hinds PW, Dowdy SF, Brown M, et al. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Mulligan GJ, Jacks T, Lassar AB. Skeletal muscle cells lacking the retinoblastoma protein display defects in muscle gene expression and accumulate in S and G2 phases of the cell cycle. J Cell Biol. 1996;135:441–456. doi: 10.1083/jcb.135.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RL, Parker MH, Rudnicki MA. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol Cell. 2001;8:291–301. doi: 10.1016/s1097-2765(01)00302-1. [DOI] [PubMed] [Google Scholar]
- Rao SS, Kohtz DS. Positive and negative regulation of D-type cyclin expression in skeletal myoblasts by basic fibroblast growth factor and transforming growth factor β. A role for cyclin D1 in control of myoblast differentiation. J Biol Chem. 1995;270:4093–4100. doi: 10.1074/jbc.270.8.4093. [DOI] [PubMed] [Google Scholar]
- Rao SS, Chu C, Kohtz DS. Ectopic expression of cyclin D1 prevents activation of gene transcription by myogenic basic helix–loop–helix regulators. Mol Cell Biol. 1994;14:5259–5267. doi: 10.1128/mcb.14.8.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Huang J, Hamamori Y, Kedes L. Molecular mechanisms of myogenic coactivation by p300: Direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F, Chang CY, Liu W, Baxter JD, Nordeen SK, Wan Y, Day RN, McDonnell DP. Temporally distinct and ligand-specific recruitment of nuclear receptor-interacting peptides and cofactors to subnuclear domains containing the estrogen receptor. Mol Endocrinol. 2000;14:2024–2039. doi: 10.1210/mend.14.12.0572. [DOI] [PubMed] [Google Scholar]
- Skapek SX, Rhee J, Spicer DB, Lassar AB. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- Skapek SX, Rhee J, Kim PS, Novitch BG, Lassar AB. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRb hyperphosphorylation. Mol Cell Biol. 1996;16:7043–7053. doi: 10.1128/mcb.16.12.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow DB, Miska EA, Langley E, Reynaud-Deonauth S, Kotecha S, Towers N, Spohr G, Kouzarides T, Mohun TJ. MEF-2 function is modified by a novel co-repressor, MITR. EMBO J. 1999;18:5085–5098. doi: 10.1093/emboj/18.18.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA. Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol Endocrinol. 2000a;14:518–534. doi: 10.1210/mend.14.4.0436. [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Simeoni S, Sharp ZD, Mancini MA. Subnuclear dynamics and transcription factor function. J Cell Biochem Suppl. 2000b;35:99–106. doi: 10.1002/1097-4644(2000)79:35+<99::aid-jcb1132>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng P-F, Weintraub H, Lassar AB. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128:4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- Wang J, Walsh K. Inhibition of retinoblastoma protein phosphorylation by myogenesis-induced changes in the subunit composition of the cyclin-dependent kinase 4 complex. Cell Growth Diff. 1996;7:1471–1478. [PubMed] [Google Scholar]
- Wei Q, Paterson BM. Regulation of MyoD function in the dividing myoblast. FEBS Lett. 2001;490:171–178. doi: 10.1016/s0014-5793(01)02120-2. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The MyoD family and myogenesis: Redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Wu Z, Woodring PJ, Bhakta KS, Tamura K, Wen F, Feramisco JR, Karin M, Wang JY, Puri PL. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol Cell Biol. 2000;20:3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A, Gredinger E, Bengal E. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J Biol Chem. 1999;274:5193–5200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- Zhang JM, Wei Q, Zhao X, Paterson BM. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with cdk4. EMBO J. 1999;18:926–933. doi: 10.1093/emboj/18.4.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes & Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen RM, Buckle RS, Hijmans EM, Loomans CJ, Bernards R. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes & Dev. 1998;12:3488–3498. doi: 10.1101/gad.12.22.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]