Abstract
Heterochromatin is a functionally important chromosomal component, especially at centromeres. In fission yeast, conserved heterochromatin-specific modifications of the histone H3 tail, involving deacetylation of Lys 9 and Lys 14 and subsequent methylation of Lys 9, promote the recruitment of a heterochromatin protein, Swi6, a homolog of the Drosophila heterochromatin protein 1. However, the primary determinants of the positioning of heterochromatin are still unclear. The fission yeast proteins Abp1, Cbh1, and Cbh2 are homologs of the human protein CENP-B that bind to centromeric α-satellite DNA and associate with centromeric heterochromatin. We show that the CENP-B homologs are functionally redundant at centromeres, and that Abp1 binds specifically to centromeric heterochromatin. In the absence of Abp1 or Cbh1, the centromeric association of Swi6 is diminished, resulting in a decrease in silencing of the region. CENP-B-homolog double disruptants show a synergistic reduction of Swi6 at centromeric heterochromatin, indicating that the three proteins are functionally redundant in the recruitment of Swi6. Furthermore, using chromatin immunoprecipitation assays, we show that disruption of CENP-B homologs causes a decrease in heterochromatin-specific modifications of histone H3. These results indicate that the CENP-B homologs act as site-specific nucleation factors for the formation of centromeric heterochromatin by heterochromatin-specific modifications of histone tails.
Keywords: CENP-B, heterochromatin, centromere, histone modification, fission yeast
In eukaryotic cells, large blocks of heterochromatin are observed at centromeres and telomeres and function as important elements in the maintenance of chromosomal integrity. Heterochromatin is thought to be a condensed and inactive chromatin structure, as suggested by the tightly repressed transcription of euchromatic genes artificially inserted into these regions. In addition, heterochromatin occasionally spreads into adjacent regions and represses transcription from nearby genes. Once established, the repressed state is inherited stably through mitosis and sometimes even through meiosis. This aspect of heterochromatin is termed position effect variegation and is thought to represent a basis for the epigenetic regulation of gene expression (Wakimoto 1998). In heterochromatin of higher eukaryotes, a set of proteins, including chromodomain proteins such as HP1, appear to assemble on nucleosomes to form an inaccessible chromatin structure (Eissenberg and Elgin 2000; Moazed 2001). Recent studies have revealed the importance of modifications at the N-terminal ends of histones in the dynamic organization of chromatin, including the acetylation and methylation of lysine and arginine residues (Jenuwein and Allis 2001). In mammalian cells, hyperacetylation and hypoacetylation of histones H3 and H4 correlate with active and inactive chromatin states, respectively (Turner and O'Neill 1995). In particular, acetylation of the histone H3 lysine 9 residue (H3-K9) and of the lysine 14 residue (H3-K14), and methylation of H3-K4 correlate with active chromatin, whereas methylation of H3-K9 correlates with inactive chromatin or heterochromatin (Litt et al. 2001; Noma et al. 2001). The histone methyltransferase (HMTase) responsible for the H3-K9 methylation is the SUV39H1 protein, a mammalian homolog of the Drosophila Su(var)3-9 protein (Rea et al. 2000). The Su(var)3-9 gene was initially identified as a suppressor of position effect variegation (Tschiersch et al. 1994). A striking feature of methylated H3-K9 is its ability to recruit HP1 through the specific binding of the HP1 chromodomain to the histone H3 tail (Bannister et al. 2001; Lachner et al. 2001).
The heterochromatin proteins and features of their assembly, including histone tail modification, are well conserved from fission yeast to humans. Fission yeast has a HP1 homolog, Swi6, and epigenetic silencing is observed on Swi6-based heterochromatin (Ekwall et al. 1995; Nakayama et al. 2000; Partridge et al. 2000). Recently, a stepwise model for heterochromatin assembly in fission yeast, in which histone deacetylases (HDACs) and HMTases act cooperatively, has been proposed (Nakayama et al. 2001). HDACs, including Clr3 and probably Clr6 and/or Hda1, deacetylate H3-K9 and H3-K14 before the methylation of H3-K9 by the Clr4/Rik1 complex, a HMTase. Clr4 is a fission yeast homolog of SUV39H1. Swi6 then binds through its chromodomain to histone H3 methylated at Lys 9, which in turn results in the self-propagation of Swi6 to form heterochromatin.
How does heterochromatin form at specific regions such as centromeres and telomeres? In budding yeast, a role for sequence-specific DNA-binding proteins as the determinants for the positioning of heterochromatin has been well documented (Loo and Rine 1995; Lustig 1998). However, the heterochromatin of budding yeast consists of Sir proteins, which are not structurally related to chromodomain proteins like HP1/Swi6. Generally, heterochromatin is formed on repetitive sequences in higher eukaryotes (Henikoff et al. 2000). In fission yeast, there are specific regions at the mating-type locus that are important for gene silencing (Grewal 2000). These observations suggest that specific sequences and/or DNA structural motifs promote the assembly of HP1/Swi6-based heterochromatin through the binding of cis-acting DNA-binding proteins to specific sites. However, such DNA-binding proteins have not yet been identified.
Centromeres are essential for accurate chromosome segregation. The kinetochore forms on the centromere and mediates its attachment to the mitotic spindle during M phase. In higher eukaryotes, centromeres encompass long regions, are rich in repetitive sequences and transposons, and show heterochromatic characteristics. For example, human centromeres are composed of long stretches (100 kb–1 Mb) of highly repeated sequence called α-satellite (Waye and Willard 1997). The fission yeast centromeres resemble higher eukaryotic centromeres, in that they consist of large inverted repeats that contain a central core (cnt/cc) of unique sequence surrounded by inner repeats (imr/B) and outer repeats (otr/K + L; Clarke et al. 1986; Nakaseko et al. 1986, 1987; Fishel et al. 1988; Takahashi et al. 1992). The outer repeat sequences are common to all three centromeres. Histones in fission yeast centromeres are hypoacetylated, and marker genes inserted within centromeres are silenced (Allshire et al. 1994; Ekwall et al. 1997). Swi6 and Chp1, another member of the family of chromodomain proteins, are confined to the flanking outer repeats (Partridge et al. 2000). Therefore, the outer repeat regions constitute centromeric heterochromatin. On the other hand, Cnp1, a homolog of CENP-A, which is a conserved centromere-specific histone H3 variant, and Mis6, a loading factor for Cnp1 (Takahashi et al. 2000), localize to the inner repeats and central core, suggesting that the inner domain is the site of kinetochore assembly (Partridge et al. 2000). Centromeric heterochromatin is important for centromere function, because many mutations that alter silencing at the centromeric heterochromatin region impair centromere function (Allshire et al. 1995; Ekwall et al. 1995, 1999).
The human centromere protein, CENP-B, is a DNA-binding protein that specifically binds to a sequence termed the CENP-B box in α-satellite DNA (Masumoto et al. 1989; Muro et al. 1992; Yoda et al. 1992). In the human kinetochore, CENP-B localizes to the inner centromere, where α-satellite DNA and centromeric heterochromatin are localized (Cooke et al. 1990). CENP-B is thought to form a higher-order chromatin structure required for kinetochore formation by virtue of its ability to dimerize (Masumoto et al. 1989; Muro et al. 1992; Yoda et al. 1992). Supporting this idea, α-satellite DNA containing the CENP-B box can promote formation of artificial minichromosomes with higher efficiency than α-satellite sequences lacking the CENP-B box (Harrington et al. 1997; Ikeno et al. 1998).
The fission yeast proteins Abp1, Cbh1, and Cbh2 share significant homology (∼25% identity and 45% similarity) with human CENP-B (Murakami et al. 1996; Lee et al. 1997; Irelan et al. 2001). Abp1 and Cbh1 were originally isolated as autonomous replicating sequence (ars)-binding proteins and were found to preferentially bind AT-rich sequences with relatively low sequence specificity (Murakami et al. 1996; Lee et al. 1997). As fission yeast centromeres are highly AT-rich and rich in ars elements (Takahashi et al. 1992), CENP-B homologs are potentially able to bind to the centromere region. Indeed, Abp1 and Cbh1 were shown to bind to multiple distinct sites in the centromere region in vitro (Halverson et al. 1997; Lee et al. 1997). In addition, the three proteins are redundant with respect to centromere function (Baum and Clarke 2000; Irelan et al. 2001). However, the precise roles of these proteins at centromeres are unclear.
In this paper, we confirm that the three CENP-B homologs have redundant functions at the centromere. We show that the CENP-B homologs recruit Swi6 to centromeric heterochromatin through the deacetylation of H3-K9 and H3-K14 and/or methylation of H3-K9 in centromeric heterochromatin. This is the first indication that sequence-specific DNA-binding proteins can nucleate HP1/Swi6-based heterochromatin at a specific locus.
Results
Phenotypes of mutants with single and double disruptions of CENP-B homologs
To analyze the function of the CENP-B homologs in fission yeast, we constructed strains that have single or double disruption of these genes and examined their growth and morphology. These features are essentially identical to those reported for the singly disrupted strains (Halverson et al. 1997; Baum and Clarke 2000; Irelan et al. 2001). Among these strains, only Δabp1 cells show growth defects and irregularities in shape, especially at low temperatures. In contrast, deletion of either cbh1 or cbh2 does not affect the growth rate or cell morphology. Among the doubly disrupted strains, Δcbh1 Δcbh2 cells are similar to wild-type cells, as reported (Irelan et al. 2001). Δabp1 Δcbh2 cells and Δabp1 cells have the same phenotype. Irelan et al. (2001), however, reported that their Δabp1 Δcbh2 strain had a markedly slower growth rate than either single-deletion strain. The reason for this discrepancy is not clear. The Δabp1 Δcbh1 cells show severe growth defects, indicating that Abp1 and Cbh1 are functionally redundant, as reported (Baum and Clarke 2000).
Abp1 and Cbh1 are involved in centromeric function
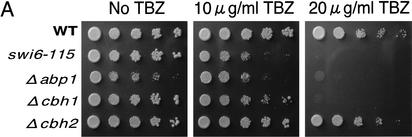
As CENP-B is implicated in centromeric function in human and mouse cells, we asked whether fission yeast CENP-B homologs have similar functions. We examined the sensitivity of single disruptants to the microtubule-destabilizing drug thiabendazol (TBZ; Fig. 1A), because mutations of genes involved in centromere function, such as the swi6-115 mutation, frequently confer hypersensitivity to microtubule-destabilizing drugs (Fig. 1A; Allshire et al. 1995; Ekwall et al. 1995). Among the three single disruptants, Δabp1 and Δcbh1 cells are hypersensitive to TBZ. Δabp1 mutants are more similar to swi6-115 cells, in that they are more sensitive to TBZ than are Δcbh1 cells (Fig. 1A). We also observed an increase in the number of Δabp1 and Δcbh1 cells that undergo abnormal nuclear division, including lagging chromosomes in the presence of TBZ (data not shown).
Figure 1.
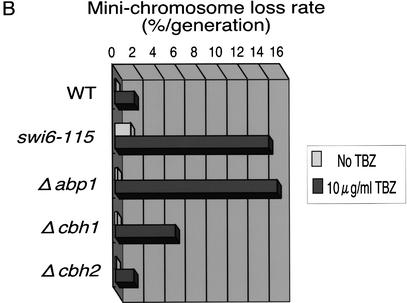
Disruption of CENP-B homologs impairs accurate chromosome segregation. (A) Exponentially growing cells of the indicated genotype were adjusted to 1.0 × 107 cells/mL and then serially diluted (1:5). Equal numbers of cells were spotted on YES plates containing the indicated concentrations of the spindle assembly inhibitor TBZ and incubated at 30°C for 4 d. The spot of highest density contained 1 × 104 cells. (B) Minichromosome loss rates were measured using the cen3-derived minichromosome ch16m23(ade6-m216)-ura4-Tel. Cells were grown for several generations in the absence or presence (10 μg/mL) of TBZ. The rate of minichromosome loss was measured as described in Materials and Methods.
Next, we examined the stability of minichromosomes in cells with a disruption of only one CENP-B homolog, because minichromosome stability reflects centromere function more clearly. We used the minichromosome ch16m23 (ade6-m216)-ura4-Tel, which is derived from the linear minichromosome ch16 and contains centromere sequences of Chromosome III (Niwa et al. 1986; Nimmo et al. 1994). Under normal conditions, the stability of the minichromosome is not affected by any single disruption of CENP-B homologs. In contrast, the swi6-115 mutation causes approximately a fivefold increase in the rate of minichromosome loss (Fig. 1B). Because Δabp1 and Δcbh1 cells are hypersensitive to TBZ, we measured minichromosome stability at a lower concentration of TBZ (10 μg/mL). This low level of TBZ decreases minichromosome stability ∼10-fold in wild-type and Δcbh2 cells. However, the same concentration of TBZ destabilizes the minichromosome ∼60-fold and ∼25-fold in Δabp1 cells and Δcbh1 cells, respectively (Fig. 1B). We obtained essentially similar results with another linear minichromosome, ch10, which is also derived from Chromosome III (data not shown).
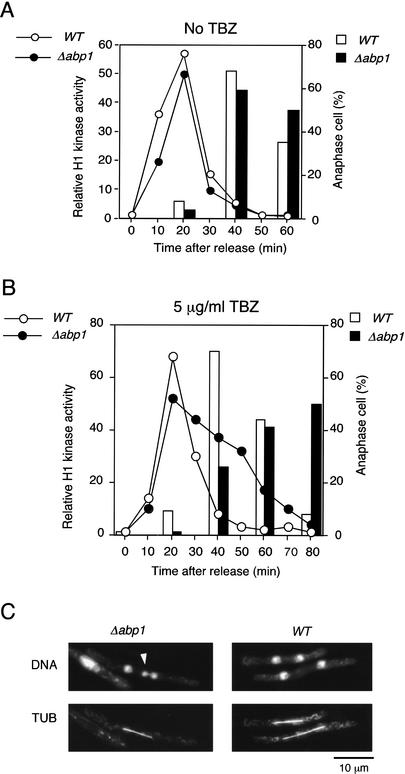
To analyze chromosome segregation in detail, we measured the progression of M phase in Δabp1 cells. abp1+ or Δabp1 cells were synchronously released from a late G2 arrest imposed by the cdc25-22 temperature-sensitive mutation in the presence or absence of a low concentration (5 μg/mL) of TBZ. The progression of M phase was assessed by monitoring H1 kinase activity and the proportion of anaphase binuclear cells (Fig. 2). When cells enter prophase, H1 kinase activity increases rapidly and then decreases sharply at the metaphase–anaphase transition (Fig. 2A), thereby triggering anaphase. In the absence of TBZ, the alteration of H1 kinase activity as well as the proportion of Δabp1 cells in anaphase are almost identical to those of the wild-type cells (Fig. 2A), indicating that the progression from prophase to anaphase is unaffected by deletion of abp1. The presence of 5 μg/mL TBZ has no effect on M-phase progression of wild-type cells. Δabp1 cells show the same kinetics of increase of H1 kinase activity as wild-type cells in the presence of TBZ. However, the loss of H1 kinase activity in Δabp1 cells is much slower than in wild-type cells. Consistent with the slow inactivation of H1 kinase, Δabp1 cells enter anaphase more slowly than do wild-type cells (Fig. 2B). For synchronized Δabp1 cells in the presence of low levels of TBZ, we observed a high incidence of chromosome missegregation, predominantly lagging chromosomes and unequal segregation; <1% and 12% of wild-type and Δabp1 cells, respectively, show abnormal nuclear divisions. However, a seemingly normal mitotic spindle forms in these cells (Fig. 2C). The results indicated in Figures 1 and 2 strongly suggest that Abp1 and Cbh1 are required for proper centromere function.
Figure 2.
Δabp1 cells show a delay in the metaphase–anaphase transition in the presence of a low concentration of TBZ. Exponentially growing abp1+ cdc25-22 and Δabp1 cdc25-22 cells were shifted to 36°C for 4 h and 4.5 h, respectively, to induce arrest in late G2. The cells were released from arrest by transfer to 25°C in the absence (A) or presence (B) of 5 μg/mL TBZ. Mitotic progression was monitored by measuring H1 kinase activity and calculating the frequency of anaphase cells as described previously (Moreno et al. 1989); these results are plotted as a line graph and a bar graph, respectively. (C) Micrographs were taken at 60 min after release from arrest in the presence of 5 μg/mL TBZ. Chromatin DNA was stained by DAPI (DNA), and the spindle was stained by an anti-tubulin antibody (TUB).
Abp1 and Cbh1 are required for gene silencing at centromeric heterochromatin
Previous studies have shown that transcription of genes inserted into the inner or the outer domain of fission yeast centromeres is silenced and mutations affecting gene silencing in either domain also disturb centromeric functions (Allshire et al. 1995; Ekwall et al. 1999; Partridge et al. 2000). Therefore, we examined whether disruption of the CENP-B homologs might influence centromeric gene silencing.
We assayed silencing of the ura4 gene inserted at three different loci in the centromere of Chromosome I: the outer repeat (otr::ura4), the innermost repeat (imr::ura4), and the center region (cnt::ura4) (Fig. 3). At the innermost repeat, the ura4 gene was inserted just outside of the tRNA gene cluster that demarcates the border between the inner and the outer domains (Partridge et al. 2000). We monitored the expression of the inserted ura4 gene by plating serial dilutions of cells from each strain on nonselective (N/S), selective (−URA), or counterselective (FOA; 5-fluoro-orotic acid) plates. (FOA is a toxigenic substrate for the Ura4 protein.) When the inserted ura4 gene is fully expressed, the cells can grow on −URA plates but not on FOA plates. In wild-type cells, the ura4 gene is strongly repressed at the otr::ura4 insertion site, but only moderately repressed at the cnt::ura4 and imr::ura4 insertion sites (Fig. 3).
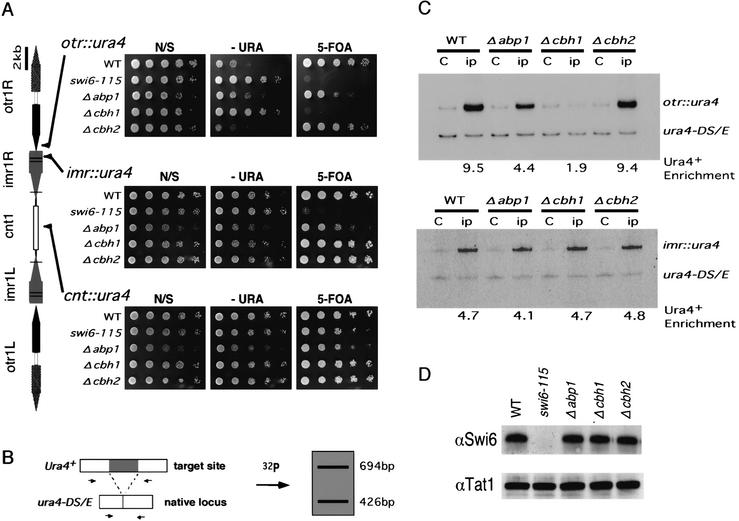
Figure 3.
Centromeric silencing at the outer repeat region is specifically derepressed in Δabp1 and Δcbh1 cells; silencing correlates with Swi6 dissociation. (A, left panel) A schematic representation of the cen1 and the ura4 insertion sites within cen1. Horizontal lines represent the positions of tRNA genes. Each strain was grown to 1.0 × 107 cells/mL in YES. Serial dilutions (1:5) of the indicated cultures were spotted onto nonselective (N/S), selective (−URA), or counterselective (A, right panel) plates and incubated at 30°C for 4 d. The highest-density spots contained 1 × 104 cells. (B) The competitive PCR strategy used for quantitating the amount of the centromeric inserted ura4+ versus the endogenous ura4 (ura4-DS/E) locus is shown. (C) Quantitative estimation of Swi6 levels at centromeric sites with ura4 inserts in strains of the indicated genotype. DNA isolated from crude extracts (C), or anti-Swi6 immunoprecipitated chromatin (ip) was used as a template for PCR. PCR products of 426 bp and 694 bp from the ura4-DS/E locus and the centromeric ura4+ insert, respectively, were resolved on polyacrylamide gel and then quantified using a PhosphorImager. The relative enrichment of centromeric ura4+ sequence is shown below each lane. (D) Crude extracts of the indicated strains were separated on SDS–polyacrylamide gels and subjected to Western blot analysis with anti-Swi6 antibodies. The anti-tubulin antibody, which recognizes β-tubulin, was used as a loading control.
The swi6-115 mutation causes a marked decrease of silencing at the otr::ura4 and imr::ura4 insertion sites, but the mutation does not affect silencing at the cnt::ura4 insertion site (Fig. 3A). These results are consistent with previous reports that the Swi6 protein is present mainly at the outer domain and that it is required for the silencing of genes inserted in this domain (Partridge et al. 2000).
Deletion of either abp1 or cbh1 causes a decrease in silencing at the otr::ura4 insertion site, as shown by the ability of these strains to form colonies on −URA plates (Fig. 3A). Deletion of cbh1 causes a more severe effect than does deletion of abp1, because Δabp1 but not Δcbh1 cells can grow on FOA. Both disruptions, however, have little effect on ura4 expression at the imr::ura4 and cnt::ura4 insertion sites. Therefore, Abp1 and Cbh1 are required for efficient gene silencing in centromeric heterochromatin. The deletion of cbh2 does not affect silencing at any of the sites tested, probably because the other two homologs can compensate for any defect of cbh2 (see below).
The amount of Swi6 bound to the otr::ura4 insertion site is decreased in Δabp1 and Δcbh1 cells
The extent of silencing in heterochromatin correlates well with the amount of Swi6 protein bound to an indicator gene in fission yeast (Nakayama et al. 2000). Therefore, we examined the amount of Swi6 bound to the ura4 gene at the otr::ura4 and the imr::ura4 insertion sites using the chromatin immunoprecipitation (ChIP) assay. In this assay, the cross-linked Swi6–DNA complex is immunoprecipitated with affinity-purified anti-Swi6 antibodies, and the amount of precipitated DNA is analyzed by a competitive PCR method (Fig. 3B; Materials and Methods). This method takes advantage of the fact that the strains used for silencing analysis also contain the ura4 minigene (ura4DS/E) with a 268-bp deletion at the endogenous ura4 locus, in addition to the inserted ura4 gene (Fig. 3B; Ekwall et al. 1997; Nakayama et al. 2000). The primer pairs used produce 426- and 694-bp-long DNA fragments by amplification of the euchromatic ura4DS/E gene and the inserted ura4 gene, respectively. By measuring the levels of the two products, the relative enrichment of the inserted ura4 gene in the immunoprecipitated fractions can be calculated (Materials and Methods). The calculated relative enrichment presumably represents the relative amount of Swi6 bound to the inserted ura4 gene.
In wild-type cells, a 9.5-fold enrichment of Swi6 was observed at the otr::ura4 insertion site, as reported (Nakayama et al. 2000). Disruption of abp1 or cbh1 decreases the extent of Swi6 binding: a 4.4-fold and 1.9-fold enrichment were observed in Δabp1 cells and Δcbh1 cells, respectively. No change in the extent of Swi6 binding was observed in Δcbh2 cells (Fig. 3C, upper panel). In contrast, no deletions of CENP-B homologs affect the enrichment of Swi6 at the imr::ura4 insertion site (Fig. 3C, lower panel) as well as at the cnt::ura4 insertion site (data not shown). In each strain, the level of the Swi6 enrichment correlates well with the extent of silencing at each site (Fig. 3A,C).
A decrease in the level of the Swi6 protein could explain the observed decrease of Swi6 at the otr::ura4 insertion site in Δabp1 or Δcbh1 cells. This, however, was not the case, because the overall amount of cellular Swi6 is the same in both wild-type cells and in mutants with disruptions of CENP-B homologs (Fig. 3D). Therefore, we conclude that Abp1 and Cbh1 are required for the efficient recruitment of Swi6 to the outer repeat region.
Abp1 binds to the centromeric heterochromatin region
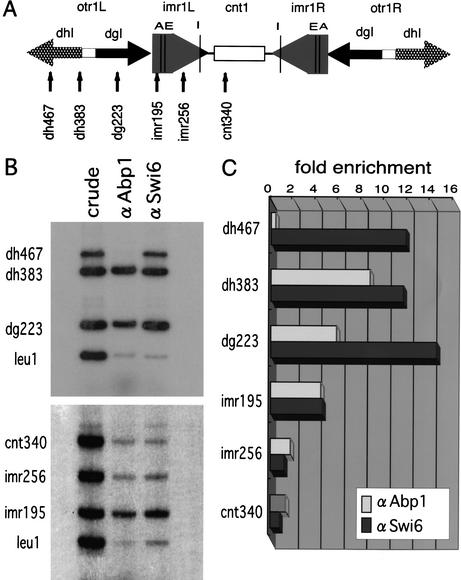
The results described above indicate that Abp1 and Cbh1 function in centromeric heterochromatin but not at the center region. Using the ChIP assay, Irelan et al. (2001) reported that Abp1 binds to both the outer and center domains as well as to single-copy euchromatic gene sequences such as leu1. However, they expressed a C-terminally GST-tagged version of Abp1 from a heterologous promoter on a multicopy plasmid and immunoprecipitated the tagged protein with an anti-GST antibody. Thus, the function and the level of expression of the tagged Abp1 might be different from those of endogenous Abp1, and this difference might affect the chromosomal distribution of Abp1. Indeed, overexpression of Abp1 impairs cell growth as well as chromosomal segregation in a DNA-binding domain-dependent manner (K. Tanaka and Y. Murakami, unpubl.), suggesting that overexpressed Abp1 binds to sites to which it normally does not bind at lower concentration. For this reason, we examined the localization of native Abp1 in the centromeric region by ChIP assay with an affinity-purified polyclonal antibody raised against Abp1. As a control, we performed ChIP assays with an anti-Swi6 antibody. For PCR, we used the primer sets shown in Figure 4A to amplify centromeric DNA from the precipitated DNA. In addition, we used a primer set to amplify the leu1 locus as a euchromatic control. Because multiple copies of the dg and dh sequences, two copies of the imrI sequence, and a single copy of the cnt1 sequence are present in these cells, we used conditions in which each primer set produced approximately the same amount of PCR products with DNA prepared from whole-cell extracts (Fig. 4B). We then calculated the enrichment in DNA derived from each locus relative to the amount of DNA derived from the leu1 locus in samples of immunoprecipitated DNA (Fig. 4C). The extent of enrichment presumably represents the relative amount of each protein.
Figure 4.
Abp1 associates with the centromeric heterochromatin region in vivo. (A) Schematic representation of cen1 and the corresponding positions recognized by PCR primer sets. Vertical lines indicate tRNA gene (I,E,A). Because of sequence diversity within the otr sequences, including segmental insertions or substitutions, the otr primer sets (dg223, dh383, dh467) were chosen to produce only a single band as the major PCR product. (B) The primer sets described in A were used for ChIP analysis to quantify the extent of localization of Abp1 and Swi6 to cen1, imr1, and otr sequences. DNA isolated from crude extracts (crude), anti-Abp1 immunoprecipitated chromatin (αAbp1), or anti-Swi6 immunoprecipitated chromatin (αSwi6) was used as the PCR template for each measurement. The PCR products were separated on polyacrylamide gels. The results of two sets of multiplex PCR reactions using the primer sets corresponding to otr and imr1 + cnt1 are indicated. (C) The intensity of the bands was quantified using a PhosphorImager and the relative enrichment of each centromeric band versus a euchromatic control leu1 band in the precipitates was plotted.
Swi6 strongly associates with sequences in the outer repeat regions (the dg223, dh383, and dh467 sites) and moderately binds to the imr195 site located just outside the tRNA cluster. Low levels of binding were observed at the inner repeat sites (the imr256 site) and at the central core site (the cnt340 site; Fig. 4B,C), as reported previously (Partridge et al. 2000). Under the same conditions, Abp1 shows the same pattern of distribution as Swi6, except at the dh467 site, where Swi6 but not Abp1 strongly associates (Fig. 4B,C). From these data, we conclude that native Abp1 primarily localizes to the centromeric outer domain, which is comprised of centromeric heterochromatin, but not at the center region nor at leu1.
CENP-B homologs are functionally redundant for the assembly of Swi6 on centromeric heterochromatin
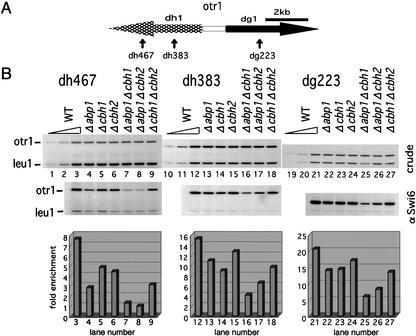
From the results described above, we assumed that Abp1 and Cbh1 bind to the centromeric heterochromatin region and affect the localization of the heterochromatin protein Swi6. In Figure 3C, we monitored the spreading of Swi6 from neighboring heterochromatin onto the inserted ura4 gene. We tested for a direct association of Swi6 with outer repeat sequences in the CENP-B-homolog single and double disruptants using the ChIP assay (Fig. 5). At the dg223 and dh383 sites, disruption of abp1 or cbh1 results in a 30%–40% decrease in Swi6 binding, whereas disruption of cbh2 results in a decrease of ∼15%. Doubly disrupted strains show a more severe reduction of Swi6 binding than do single disruptants. In Δabp1 Δcbh1 cells, the association of Swi6 is most strongly decreased, ∼70%, whereas in Δabp1 Δcbh2 cells and in Δcbh1 Δcbh2 cells the level of binding is decreased ∼60% and ∼40%, respectively. At the dh467 site, the disruption of CENP-B homologs causes a more pronounced decrease in Swi6 binding. The association of Swi6 with the dh467 site is decreased in Δabp1 cells, although Abp1 is not detected at this site by the ChIP assay (Fig. 4). We speculate that Abp1 may bind at some distance from the dh467 site and contribute to the localization of Swi6 at this site through the ability of Swi6 to propagate itself (Partridge et al. 2000). Moreover, among the CENP-B homologs, Abp1 appears to make the greatest contribution to promoting the binding of Swi6 to centromeric heterochromatin, because the doubly disrupted Δcbh1 Δcbh2 cells, which contain only Abp1, show the highest enrichment of Swi6 at all sites examined (Fig. 5, lanes 7–9, 16–18, and 25–27).
Figure 5.
The three CENP-B homologs, Abp1, Cbh1, and Cbh2, are functionally redundant for the recruitment of Swi6 to the outer repeat region. (A) Schematic representation of otr1 and the positions of the PCR primer set used in B are indicated. (B) The ChIP assays were performed using strains of the indicated genotype and the indicated primer set to monitor the association of Swi6 with otr. The PCR products amplified from DNA isolated from crude extracts (crude) or anti-Swi6 immunoprecipitated chromatin (αSwi6) were separated on polyacrylamide gels (upper panel). DNA isolated from crude extracts of wild-type cells was serially diluted (1:3) to monitor each band for amplification in a linear range (lanes 1–3, 10–12, 19–21). The relative enrichment of immunoprecipitated chromatin was determined as in Fig. 4B and plotted (bottom panel).
These results indicate that the three CENP-B homologs are functionally redundant for the localization of Swi6 to centromeric heterochromatin. Among the three proteins, Abp1 and Cbh1 likely play a major role in promoting the association of Swi6 with centromeric heterochromatin.
CENP-B homologs are required for the efficient heterochromatin-specific modifications of histone H3 in centromeric heterochromatin
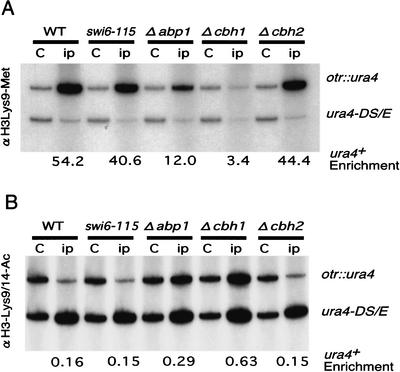
In fission yeast, deacetylation of H3-K9 and H3-K14 by HDACs including Clr3 appears to precede the methylation of H3-K9 by the HMTase Clr4 (Nakayama et al. 2001). This acetylation/methylation transition of the histone H3 tail is required for the localization of Swi6 to heterochromatin (Nakayama et al. 2001). To test if CENP-B homologs are involved in the deacetylation and methylation steps, we measured the modification status of histone H3 at the otr::ura4 insertion site by the ChIP assay (Fig. 6). We used H3-K9 methyl-specific and H3-K9/14 diacetyl-specific antibodies for this assay.
Figure 6.
Abp1 and Cbh1 are required for the heterochromatin-specific modification of the histone H3 tail. (A) Quantitative estimation of the levels of methylation of H3-K9 at the otr::ura4 insertion site in strains of the indicated genotype. DNA isolated from crude extracts (C), or chromatin immunoprecipitated with an anti-histone H3-K9 methyl-specific antibody (ip) was used as template. PCR products of 426 bp and 694 bp, amplified from the ura4-DS/E and centromeric inserted ura4+ loci, respectively, were resolved on polyacrylamide gels and quantified with a PhosphorImager. The relative enrichment of centromeric ura4 sequence is shown below each lane. (B) Quantitative estimation of the level of acetylation of H3-K9/14 at the otr::ura4 insertion site in strains of the indicated genotype. The same ChIP assays as described in A were carried out except that an anti-histone H3-K9/14 diacetyl-specific antibody was used to precipitate chromatin.
H3-K9 residues are highly methylated (increased ∼50-fold) at the otr::ura4 insertion site in wild-type cells, as compared with H3-K9 residues at the euchromatic ura4DS/E locus. The swi6-115 mutation, which severely reduces the level of Swi6 protein (Fig. 3D), has little effect on methylation (Fig. 6, lanes 1–4), confirming the previous notion that Swi6 acts downstream of H3-K9 methylation (Nakayama et al. 2001). Disruption of abp1 or cbh1 but not cbh2 significantly reduces H3-K9 methylation (Fig. 6A). This reduction is more pronounced in Δcbh1 cells than in Δabp1 cells: a 12-fold enrichment was observed in Δabp1 cells, whereas only a threefold enrichment was observed in Δcbh1 cells. This level of methylation correlates well with the reduction of bound Swi6 in these strains (cf. Figs. 3C and 6A).
The anti-H3-K9/14 diacetyl-specific antibody precipitates only a small amount of otr::ura4 DNA, showing that Lys 9/14 residues are acetylated at low levels at this site in wild-type cells (Fig. 6B). Consistent with the assumption that histone tail modifications precede Swi6 association, the swi6-115 mutation does not affect H3-K9/14 deacetylation. In contrast, increases of about two- and fourfold in the extent of H3-K9/14 diacetylation were observed in Δabp1 and Δcbh1 cells, respectively, whereas no increase was detected in Δcbh2 cells. These results show that Abp1 and Cbh1 are required to maintain the deacetylated state of H3-K9 and H3-K14 in centromeric heterochromatin, which, in turn, allows an increase in the extent of H3-K9 methylation and subsequent association of histone H3 with Swi6.
Discussion
The CENP-B homologs bind to centromeric heterochromatin to recruit Swi6
Using the ChIP assay with a specific antibody, we showed that one of the CENP-B homologs, Abp1, binds to the region of centromeric heterochromatin but not to the inner domain, which coincides with the centromeric distribution of Swi6. This pattern is consistent with a role for Abp1 as a Swi6 nucleation factor. A strain that expresses a GFP-tagged Cbh1 as the only form of cellular Cbh1 was used to show by ChIP assay that Cbh1 also binds to the outer repeat of the centromere (Baum and Clarke 2000). However, Cbh1 binds as well to noncentromeric regions, and its association with the inner domain has not been examined (Baum and Clarke 2000). Cbh2 was shown to associate with the inner core region. In this case, however, GFP-tagged Cbh2 was expressed from a multicopy plasmid (Irelan et al. 2001). A more detailed analysis will be required to confirm the chromosomal distribution of native Cbh1 and Cbh2. We speculate that Cbh1 and Cbh2 also localize to the centromeric heterochromatin region, because our results indicate that all three CENP-B homologs contribute to the association of Swi6 with centromeric heterochromatin, as shown in Figure 5. Interestingly, each single disruption has a different effect on the localization of Swi6 to different sites (Figs. 3C,5). This difference may reflect the difference in binding specificity for each protein. Indeed, Abp1 and Cbh1 show different binding affinities in vitro (Halverson et al. 1997; Lee et al. 1997). Therefore, the localization of Swi6 to a particular site may be, at least in part, determined by the amount of CENP-B homologs bound at or near that site.
We note that the distribution of Abp1, as determined by immunofluorescence labeling using an anti-Abp1 antibody, overlaps with the pattern of DAPI staining (K. Tanaka and Y. Murakami, unpubl.). Thus, Abp1 may not be limited to regions of heterochromatin. Consistent with this, we have detected Abp1 binding to ars3002 in a euchromatic region that was originally used to detect Abp1 binding in vitro (Murakami et al. 1996). Interestingly, we also found a weak but significant association of Swi6 with ars3002 and observed that this association is diminished in Δabp1 cells (H. Nakagawa, unpubl.). Therefore, Abp1 may bind some euchromatic AT-rich regions and recruit Swi6 to these sites. The bound Swi6 might be involved in repressing the expression of nearby genes, similar to what has been shown for human HP1, which was found recently to be involved in the silencing of euchromatin genes (Nielsen et al. 2001).
The CENP-B homologs recruit Swi6 to the centromeric heterochromatin region through the histone tail modification
We have shown that Abp1 and Cbh1 are required for heterochromatin-specific modifications of the N-terminal tail of histone H3, which include deacetylation of the H3-K9 and H3-K14 residues and methylation of the H3-K9 residue. In the stepwise assembly model, deacetylation is a prerequisite for methylation (Nakayama et al. 2001). The CENP-B homologs may thereby promote efficient deacetylation of H3-K9/14, which then results in H3-K9 methylation and subsequent assembly of Swi6. Alternatively, the CENP-B homologs might promote H3-K9 methylation rather than H3-K9/14 deacetylation. A decrease in methylation of the H3-K9 residue caused by disruption of the CENP-B homologs would promote H3-K9/14 acetylation as well as cause a reduction in the level of bound Swi6. It is also possible that the CENP-B homologs promote both deacetylation and methylation. In any event, these proteins determine the positioning of centromeric heterochromatin by promoting specific histone H3 modifications.
In fission yeast, HDACs, including Clr6, Hda1, and Clr3, and HMTases, including Clr4, appear to be responsible for the heterochromatin-specific modifications of histone H3 (Nakayama et al. 2001). Therefore, the CENP-B homologs somehow assist these HDACs and/or HMTases to function in heterochromatic regions. How might they do so? One possibility is that the CENP-B homologs recruit HDACs and/or HMTases to their binding sites. Some DNA-binding transcription factors have been shown to recruit HDACs to their binding sites, where they act as corepressors. The mammalian retinoblastoma protein (Rb) acts as a corepressor for some transcription factors, including E2F, at least partly through the recruitment of HDAC activities (Brehm and Kouzarides 1999). Recently, Rb was shown to attract the SUV39H1–HP1 complex to the cyclin E promoter, which could methylate H3-K9 and thereby provide an HP1 binding site (Nielsen et al. 2001). Similarly, the CENP-B homologs themselves, or protein(s) interacting with them, may recruit HDACs and/or HMTases to provide an association site for Swi6. An attractive speculation is that the CENP-B homologs recruit a protein complex(es) containing Swi6 and histone-modifying enzymes, because HP1 has been shown to form a complex with SUV39 (Aagaard et al. 1999; Nielsen et al. 2001).
Another possible mechanism is that CENP-B homologs create a chromatin environment that facilitates HDAC/HMTase activity. Human CENP-B is thought to organize higher-order chromatin structures in α-satellite DNA through dimer formation and DNA-bending activity (Muro et al. 1992; Yoda et al. 1992, 1998; Tanaka et al. 2001). In addition, analysis of CENP-B-containing chromatin assembled on α-satellite DNA in vitro shows that CENP-B might contribute to the formation of the regular nucleosome array that is a characteristic of heterochromatin (Yoda et al. 1998). Because fission yeast CENP-B homologs form homodimers (Murakami et al. 1996; Lee et al. 1997; Irelan et al. 2001), they might be able to form higher-order chromatin structures like CENP-B. Such structures and/or nucleosome arrangements promoted by CENP-B homologs may enhance the heterochromatin-specific modification of the histone H3 tail by HMTase and/or HDACs. In this regard, it is noteworthy that human pericentromeric heterochromatin consists of a higher-order structure involving a distinct pattern of histone modification (Maison et al. 2002).
We should note that there are other mechanisms that we cannot rule out at this stage, including the possibility that the CENP-B homologs somehow affect HMTase/HDAC activities or protein levels. Further experiments will clarify this matter.
The role of CENP-B homologs and CENP-B in centromere function
The importance of centromeric heterochromatin for centromere function has been well documented. The removal of flanking heterochromatin from centromeres reduces chromosomal stability, and a region of centromeric heterochromatin has been identified as an enhancer of centromeric function (Matsumoto et al. 1990; Hahnenberger et al. 1991; Takahashi et al. 1992; Baum et al. 1994). Mutations that disturb silencing at centromeric heterochromatin, such as swi6, rik1, clr3, clr4, and clr6, impair the faithful segregation of chromosomes (Allshire et al. 1995; Ekwall et al. 1999). Recent studies indicate that heterochromatin is a prerequisite for the establishment of centromeric sister-chromatid cohesion, which is important for proper centromere function (Bernard et al. 2001; Nonaka et al. 2002). There is some correlation between the amount of Swi6 in centromeric heterochromatin and the extent of centromere dysfunction, as observed in single CENP-B homolog disruptants (Figs. 1, 5). In addition, we found that the swi6-115 strain shows a higher rate of minichromosome loss than does any single CENP-B homolog disruptant (Fig. 1). This could be explained by the fact that single disruptants retain Swi6 in centromeric heterochromatin (Fig. 5), whereas the swi6-115 mutant has no detectable Swi6 (Fig. 3D). Altogether, we propose that the CENP-B homologs contribute to centromere function through their influence on the formation of centromeric heterochromatin.
Based on the results presented here, we assume that one of the roles of CENP-B is to promote centromeric heterochromatin formation in human cells, like its fission yeast counterparts. Considering that more than 95% of CENP-B proteins localize to centromeric heterochromatin (Cooke et al. 1990) and that α-satellite DNA containing the CENP-B box efficiently forms artificial chromosomes (Harrington et al. 1997; Ikeno et al. 1998), we speculate that CENP-B plays an active and specialized role in such centromeric functions as the recruitment of kinetochore proteins, in addition to general heterochromatin assembly. Indeed, in α-satellite DNA, half of the total CENP-B takes part in forming the chromatin complexes containing the kinetochore proteins CENP-A and CENP-C (Ando et al. 2002).
Some evidence, such as the apparently normal phenotype of cenp-b null mice, argues against a role for CENP-B in centromere function in human and mouse cells (Hudson et al. 1998; Kapoor et al. 1998; Perez-Castro et al. 1998). How can this controversy be resolved? The existence of the three fission yeast CENP-B homologs with redundant functions suggests that there may be other functionally redundant proteins in human and mouse cells. Another explanation, which is not mutually exclusive with the first one, is that there is an epigenetic maintenance mechanism for centromeres (Karpen and Allshire 1997). Therefore, in human and mouse cells, CENP-B may strongly contribute to the formation of an active centromere, but once established, centromere function is maintained in the absence of CENP-B by this epigenetic mechanism.
In this paper, we have shown that the CENP-B homologs in fission yeast may be missing links that connect specific DNA sequences and heterochromatin. Further analysis of these proteins will shed light on the still obscure mechanism of heterochromatin formation in fission yeast, as well as in higher eukaryotes.
Materials and methods
Schizosaccharomyces pombe media, genetic procedures, and cytological procedures
Media and genetic procedures were essentially as described (Moreno et al. 1991). Tubulin staining was performed as described (Hagan and Hyams 1998). Comparative plating and serial dilution experiments for measuring silencing at centromeric heterochromatin were performed as described previously (Allshire et al. 1994).
S. pombe strains
All strains used in this study are listed in Table 1. To generate the disruptions of abp1, cbh1, and cbh2, homologous recombination was used to replace these genes with the Saccharomyces cerevisiae LEU2 gene or the S. pombe ura4 gene. Other strains were obtained by genetic cross.
Table 1.
S. pombe strains used in this study
| Strains
|
Genotype
|
|---|---|
| TP5A | h-, ura4-D18,leu1-32,ade6-m210 |
| TP10 | h+, ura4-D18,leu1-32,ade6-m216,his2 |
| HN35 | h−, ura4-D18,leu1-32,ade6-m216, Δadpl::LEU2 |
| JL112 | h−, ura4-D18,leu1-32,ade6-m216, Δcbhl::LEU2 |
| HN3 | h−, ura4-D18,leu1-32,ade6-m216, Δcbh2::LEU2 |
| HN20 | hA, ura4-DS/E,leu1-32,ade6-m210,swi6-115 |
| HN5 | h−, ura4-D18,leu1-32, Δadpl::Ura4, Δcbh2::LEU2 |
| HN6 | h−, ura4-D18,leu1-32,ade6-m210, Δcbh1::hisG-Ura4-hisG, Δcbh2::LEU2 |
| HN12 | h−, ura4-D18,leu1-32,ade6-m210, Δadpl::Ura4, Δcbh1::LEU2 |
| YM501 | h−, cdc25-22,leu1-32 |
| YM502 | h−, cdc25-22,Δadp1::ura4,leu1-32 |
| FY336 | h−, ura4-DS/E,leu1-32,ade6-m210,cnt1/TM(NcoI)-Ura4 |
| FY1158 | hA, ura4-DS/E,leu1-32,ade6-m210,cnt1/TM(NcoI)-Ura4,swi6-115 |
| HN30 | h−, ura4-DS/E,leu1-32,ade6-m210,cnt1/TM(NcoI)-Ura4, Δadp1::LEU2 |
| JL227 | h−, ura4-DS/E,leu1-32,ade6-m210,cnt1/TM(NcoI)-Ura4, Δadp1::LEU2 |
| HN21 | h−, ura4-DS/E,leu1-32,ade6-m210,cnt1/TM(NcoI)-Ura4, Δcbh2::LEU2 |
| FY498 | h+, ura4-DS/E,leu1-32,ade6-m210,imr1R(NcoI)::Ura4 |
| FY699 | hA, ura4-DS/E,leu1-32,ade6-m210,imr1R(NcoI)::Ura4, swi6-115 |
| JL225 | h−, ura4-DS/E,leu1-32,ade6-m210,imr1R(NcoI)::Ura4, Δadp1::LEU2 |
| JL222 | h−, ura4-DS/E,leu1-32,ade6-m210,imr1R(NcoI)::Ura4, Δcbh1::LEU2 |
| HN15 | h−, ura4-DS/E,leu1-32,ade6-m210,imr1R(NcoI)::Ura4, Δcbh2::LEU2 |
| FY648 | h+, ura4-DS/E,leu1-32,ade6-m210,otr1R(SphI)::Ura4 |
| FY711 | hA, ura4-DS/E,leu1-32,ade6-m210,otr1R(SphI)::Ura4,swi6-115 |
| JL223 | h−, ura4-DS/E,leu1-32,ade6-m210,otr1R(SphI)::Ura4, Δadp1::LEU2 |
| JL220 | h−, ura4-DS/E,leu1-32,ade6-m210,otr1R(SphI)::Ura4, Δcbh1::LEU2 |
| HN25 | h−, ura4-DS/E,leu1-32,ade6-m210,otr1R(SphI)::Ura4, Δcbh2::LEU2 |
| FY520 | h+, ura4-DS/E,leu1-32,ade6-m210/ch16m23(ade6-m216)-ura-Tel |
| FY611 | hA, ura4-DS/E,leu1-32,ade6-m210,swi6-115/ch16m23(ade6-m216)-ura4-Tel |
| JL212 | h−, ura4-DS/E,leu1-32,ade6-m210, Δadp1::LEU2/ch16m23(ade6-m216)-ura4-Tel |
| JL209 | h−, ura4-DS/E,leu1-32,ade6-m210, Δcbh1::LEU2/ch16m23(ade6-m216)-ura4-Tel |
| HN16 | h−, ura4-DS/E,leu1-32,ade6-m210, Δcbh2::LEU2/ch16m23(ade6-m216)-ura-Tel |
Antibodies
Anti-Abp1 antisera were prepared from rabbits by injecting N-terminal histidine-tagged Abp1 protein (amino acids 30– 466). Crude rabbit antisera were affinity-purified against His-tagged Abp1 immobilized on Ni-NTA agarose (Amersham). In the Western blotting analysis, this affinity-purified anti-Abp1 antibody detects a 60-kD protein in crude extracts from wild-type cells, but does not detect any protein in crude extracts from Δabp1 cells, indicating that the antibody is specific for Abp1. Preparation of affinity-purified anti-Swi6 polyclonal antibody has been described previously (Nakayama et al. 2000). Anti-dimethyl-histone H3-K9 antibody and anti-acetyl-histone H3-K9/14 antibodies were purchased from UpState Biotechnology. The TAT1 antibody (Woods et al. 1989) was used for staining of tubulin.
Minichromosome stability assay
Strains carrying the minichromosome ch16m23(ade6-m216)-ura4-tel were cultured for several generations in complete medium in the presence or absence of 10 μg/mL TBZ. Aliquots were taken from the cultures before and after the cultivation, appropriately diluted, and then plated onto YE plates and incubated at 30°C for several days to allow colonies to form. The fully red adee− phenotype was used as an indication of minichromosome loss. Minichromosome loss rates were calculated using the following formula: loss rate = 1 − (F/I)1/N, where F is the final percentage of cells carrying the minichromosome, I is the initial percentage of cells carrying the minichromosome, and N is the number of generations between I and F (Kipling and Kearsey 1990).
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed as described (Ekwall and Partridge 1999) with some modifications. Cells were grown at 30°C in YES to 5 × 106 cells/mL and then shifted to 18°C for 2 h. Cells were fixed for 30 min in 3% formaldehyde, and soluble chromatin was immunoprecipitated with affinity-purified anti-Abp1 antibodies (this study) or affinity-purified anti-Swi6 antibodies (Nakayama et al. 2000). For precipitation with anti-dimetyl-histone H3-K9 antibodies and anti-acetyl-histone H3-K9/14 antibodies, growing cells were fixed with 1% formaldehyde at 30°C for 5 min. Recovered DNA was assayed by PCR in the presence of [α-32P]dCTP and separated on 5% native polyacrylamide gels, which were then dried. Bands were quantified using a PhosphorImager (Fuji BAS2000, Fuji Medical System). The primer sets used were: for competitive PCR of Ura4+/ura4DS/E, 5′-GAGGGGATGAAAAATC CCAT-3′, 5′-TTCGACAACAGGATTACGACC-3′; for multiplex PCR, leu1–158bp, 5′-CCTCCTAGGAGCATTCAAC-3′, 5′-CGGTGTGGTAGGAAAATCAG-3′; for dg-223bp, 5′-TGG TAATACGTACTAGCTCTCG-3′, 5′-AACTAATTCATGGTG ATTGATG-3′; for dh-383bp, 5′-TGCTGTCATACTACACT GCA-3′, 5′-TTCTGAATAATTGGGATCGC-3′; for dh-467bp, 5′-GTCCTACTCCTACACCTACT-3′, 5′-AAACGAGTCGAG ATGAGAGG-3′; for imr-195bp, 5′-GTATCGCAACGATTGA ACTG-3′, 5′-CTCCATAACCAACACATGCTC-3′; for imr-256bp, 5′-GATACTATGACATATTCTCA-3′, 5′-AGCATTGC CAGCTCATATTA-3′; for cnt-340bp, 5′-AGTTAAGCGGTAT TATCACG-3′, 5′-GAATTGACATATACTCTGTC-3′. Using the signal from the euchromatic ura4DS/E locus or from leu1–158bp as the euchromatic control, the enrichment of immunoprecipitated DNA relative to control DNA was calculated and normalized to ratios obtained for crude DNA. Note that in the absence of the antibody in the immunoprecipitation reaction, no detectable PCR products were detected.
Acknowledgments
We especially thank Y. Nakaseko and M. Yanagida for helping us technically at the initial stage of this research and providing strains. We are grateful to H. Masumoto for critical reading of the manuscript. This research was sponsored by Grant-in-Aid for Scientific Research on Priority Areas (C) from the Ministry of Education, Cultures, Sports, Science, and Technology to Y.M.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yota@virus.kyoto-u.ac.jp; FAX 81-75-751-3975.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.997702.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersonrger A, Singh PB, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes & Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. CENP-A, -B, and -C chromatin complex that contains the I-type α-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Baum M, Clarke L. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol. 2000;20:2852–2864. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Ngan VK, Clarke L. The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol Biol Cell. 1994;5:747–761. doi: 10.1091/mbc.5.7.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Brehm A, Kouzarides T. Retinoblastoma protein meets chromatin. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- Clarke L, Amstutz H, Fishel B, Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Bernat RL, Earnshaw WC. CENP-B: A major human centromere protein located beneath the kinetochore. J Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Elgin SC. The HP1 protein family: Getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Partridge JF. Fission yeast chromosome analysis: Fluorescence in-situ hybridization (FISH) and chromatin immunoprecipitation (CHIP). Oxford, UK: Oxford University Press; 1999. [Google Scholar]
- Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire RC. The chromodomain protein Swi6: A key component at fission yeast centromeres. Science. 1995;269:1429–1431. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Cranston G, Allshire RC. Fission yeast mutants that alleviate transcriptional silencing in centromeric flanking repeats and disrupt chromosome segregation. Genetics. 1999;153:1153–1169. doi: 10.1093/genetics/153.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel B, Amstutz H, Baum M, Carbon J, Clarke L. Structural organization and functional analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol. 1988;8:754–763. doi: 10.1128/mcb.8.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI. Transcriptional silencing in fission yeast. J Cell Physiol. 2000;184:311–318. doi: 10.1002/1097-4652(200009)184:3<311::AID-JCP4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hahnenberger KM, Carbon J, Clarke L. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol Cell Biol. 1991;11:2206–2215. doi: 10.1128/mcb.11.4.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halverson D, Baum M, Stryker J, Carbon J, Clarke L. A centromere DNA-binding protein from fission yeast affects chromosome segregation and has homology to human CENP-B. J Cell Biol. 1997;136:487–500. doi: 10.1083/jcb.136.3.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 1977;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Heterochromatin function in complex genomes. Biochim Biophys Acta. 2000;1470:1–8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- Hudson DF, Fowler KJ, Earle E, Saffery R, Kalitsis P, Trowell H, Hill J, Wreford NG, de Kretser DM, Cancilla MR, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill N, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- Irelan JT, Gutkin GI, Clarke L. Functional redundancies, distinct localizations and interactions among three fission yeast homologs of centromere protein-B. Genetics. 2001;157:1191–1203. doi: 10.1093/genetics/157.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Montes de Oca Luna R, Liu G, Lozano G, Cummings C, Mancini M, Ouspenski I, Brinkley BR, May GS. The cenpB gene is not essential in mice. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kipling D, Kearsey SE. Reversion of autonomously replicating sequence mutations in Saccharomyces cerevisiae: Creation of a eucaryotic replication origin within procaryotic vector DNA. Mol Cell Biol. 1990;10:265–272. doi: 10.1128/mcb.10.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lee JK, Huberman JA, Hurwitz J. Purification and characterization of a CENP-B homologue protein that binds to the centromeric K-type repeat DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci. 1997;94:8427–8432. doi: 10.1073/pnas.94.16.8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Loo, S. and Rine, J. 1995. Silencing and heritable domains of gene expression. Annu. Rev. Cell Dev. Biol. 11519–11548. [DOI] [PubMed]
- Lustig AJ. Mechanism of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/s0959-437x(98)80146-9. [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Murakami S, Niwa O, Yanagida M. Construction and characterization of centric circular and acentric linear chromosomes in fission yeast. Curr Genet. 1990;18:323–330. [Google Scholar]
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001;8:489–498. doi: 10.1016/s1097-2765(01)00340-9. [DOI] [PubMed] [Google Scholar]
- Moreno S, Hayles J, Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989;58:361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Huberman JA, Hurwitz J. Identification, purification, and molecular cloning of autonomously replicating sequence-binding protein 1 from fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci. 1996;93:502–507. doi: 10.1073/pnas.93.1.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T. Centromere protein B assembles human centromeric α-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahasi S, Niwa O, Yanagida M. Chromosome walking shows a highly repetitive sequence present in all the centromere regions of Schizosaccharomyces pombe chromosomes. EMBO J. 1986;5:1011–1021. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Kinoshita N, Ynagida M. A novel sequence common to the centromere regions of Schizosaccharomyces pombe. Nucleic Acids Res. 1987;15:4705–4715. doi: 10.1093/nar/15.12.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–317. doi: 10.1016/s0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nimmo ER, Cranston G, Allshire RC. Telomere-associated chromosome breakage in fission yeast results in variegated expression of adjacent genes. EMBO J. 1994;13:3801–3811. doi: 10.1002/j.1460-2075.1994.tb06691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Yanagida M. Construction of a minichromosome by deletion and its mitotic and meiotic behaviour in fission yeast. Mol Gen Genet. 1986;203:397–405. [Google Scholar]
- Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes & Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- Perez-Castro AV, Shamanski FL, Meneses JJ, Lovato TL, Vogel KG, Moyzis RK, Pedersen R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev Biol. 1998;201:135–143. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun Z-W, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nureki O, Kurumizaka H, Fukai S, Kawaguchi S, Ikuta M, Iwahara J, Okazaki T, Yokoyama S. Crystal structure of the CENP-B protein–DNA complex: The DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, O'Neill LP. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Nucleotide sequence heterogeneity of α satellite repetitive DNA: A survey of alphoid sequences from different chromosomes. Nucleic Acids Res. 1997;157:7549–7569. doi: 10.1093/nar/15.18.7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T. A human centromere protein, CENP-B, has a DNA binding domain containing four potential α helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol. 1992;119:1413–1427. doi: 10.1083/jcb.119.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K, Ando S, Okuda A, Kikuchi A, Okazaki T. In vitro assembly of the CENP-B/α-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells. 1998;3:533–548. doi: 10.1046/j.1365-2443.1998.00210.x. [DOI] [PubMed] [Google Scholar]