Abstract
We have examined the role of checkpoint pathways in responding to a yku70Δ defect in budding yeast. We show that CHK1, MEC1, and RAD9 checkpoint genes are required for efficient cell cycle arrest of yku70Δ mutants cultured at 37°C, whereas RAD17, RAD24, MEC3, DDC1, and DUN1 play insignificant roles. We establish that cell cycle arrest of yku70Δ mutants is associated with increasing levels of single-stranded DNA in subtelomeric Y‘ regions, and find that the mismatch repair-associated EXO1 gene is required for both ssDNA generation and cell cycle arrest of yku70Δ mutants. In contrast, MRE11 is not required for ssDNA generation. The behavior of yku70Δ exo1Δ double mutants strongly indicates that ssDNA is an important component of the arrest signal in yku70Δ mutants and demonstrates a link between damaged telomeres and mismatch repair-associated exonucleases. This link is confirmed by our demonstration that EXO1 also plays a role in ssDNA generation in cdc13-1 mutants. We have also found that the MAD2 but not the BUB2 spindle checkpoint gene is required for efficient arrest of yku70Δ mutants. Therefore, subsets of both DNA-damage and spindle checkpoint pathways cooperate to regulate cell division of yku70Δ mutants.
Keywords: Checkpoint, telomere, KU, EXO1, CDC13, ssDNA
The telomere is a DNA-protein complex at the end of eukaryotic chromosomes. If telomeric DNA, which has many properties of a double strand break (DSB), was perceived as a DSB by DNA repair machinery and underwent recombination, then harmful telomere fusions and dicentric chromosomes would be generated. Similarly, if telomeric DNA were perceived as a DSB by DNA damage checkpoint machinery, it would be harmful, because in budding yeast a single unrepaired DSB elsewhere in the genome can inhibit cell cycle progression for many generation times (Sandell and Zakian 1993). Therefore, it is essential for chromosome stability and cell cycle progression that telomeres hide the DSB-like structures that they contain. An important function of some of the large number of telomere binding proteins, such as Cdc13p (essential in budding yeast) and Ku70/Ku80 heterodimer (conserved from yeast to mammalian cells), is to hide telomeric DNA from repair and checkpoint pathways.
Budding yeast mutants defective in telomere binding proteins are useful tools to address the mechanisms by which checkpoint pathways recognize damaged DNA, because in these cells telomeres become potent activators of DNA damage checkpoint pathways in a conditional manner. For example, at 23°C, a permissive temperature for cdc13-1 mutants, telomeres are not recognized as damaged DNA, but at 36°C temperatures, they are potent activators of cell cycle arrest (Weinert and Hartwell 1993). Cell cycle arrest of cdc13-1 mutants is associated with accumulation of single-stranded DNA (ssDNA) at telomeres (Garvik et al. 1995). Furthermore, not only do checkpoint pathways recognize cdc13-1-induced damage, but they also affect the rate at which ssDNA arises (Lydall and Weinert 1995).
The Ku heterodimer is an evolutionarily conserved protein complex involved in the nonhomologous end-joining (NHEJ) pathway of DNA repair (Smith and Jackson 1999). Interestingly and paradoxically, the Ku heterodimer is important for telomere stability. For example, there is evidence that the Ku heterodimer protects mammalian chromosomes from telomere fusions (Bailey et al. 1999; Hsu et al. 2000; Samper et al. 2000; d’Adda di Fagagna et al. 2001). In one study, 62% of ku80−/− mouse embryonal fibroblasts contained telomere fusions, a level 30 times higher than that seen in Ku-proficient fibroblasts (Hsu et al. 2000).
The budding yeast homolog of the Ku heterodimer is the Yku70p/Yku80p heterodimer. Mutants with deletions of YKU70 or YKU80 contain short telomeres (Boulton and Jackson 1996, 1998; Porter et al. 1996), have single-stranded DNA (ssDNA) in their repetitive telomeric TG sequences (Gravel et al. 1998; Polotnianka et al. 1998), display decreased telomeric silencing (Boulton and Jackson 1998; Mishra and Shore 1999; Pryde and Louis 1999), and altered telomere localization (Laroche et al. 1998). Furthermore, there is evidence that the KU heterodimer is able to bind to the telomerase RNA directly (Peterson et al. 2001) and is localized at telomeres (Martin et al. 1999).
While both yku70Δ and yku80Δ mutants are viable at permissive temperatures such as 30°C, they are unable to form colonies at 37°C (Feldmann and Winnacker 1993; Barnes and Rio 1997). This temperature-sensitive phenotype appears to be due specifically to a telomere defect, rather than a more generalized DNA repair defect, because the temperature-sensitive phenotype can be partially suppressed by overexpression of telomerase subunits (Nugent et al. 1998; Teo and Jackson 2001; Lewis et al. 2002) or rarely (1 × 10−7/cell) by amplification of subtelomeric repeats (Fellerhoff et al. 2000). By combining the yku70Δ mutation with checkpoint mutations and culturing the cells at high temperatures, we have been able to examine the role of different checkpoint genes in responding to (sub) telomeric defects in yku70Δ mutants.
Checkpoint pathways consist of proteins that interact with damaged DNA and signal transduction cascades that inhibit cell division (Lowndes and Murguia 2000; Caspari and Carr 2002). Here we show that some, but not all DNA damage checkpoint genes contribute to the inhibition of cell division of yku70Δ mutants. Interestingly, a subset of spindle checkpoint pathways also contributes to arrest. Furthermore, there is a correlation between cell cycle arrest and the accumulation of ssDNA in subtelomeric sequences in yku70Δ mutants. Finally, we show that the mismatch repair-associated exonuclease EXO1 is essential for ssDNA generation in yku70Δ mutants, while MRE11 is not, and that EXO1 is also required to generate ssDNA in cdc13-1 mutants.
Results
CHK1, MEC1, and RAD9 are required for a yku70Δ-induced checkpoint
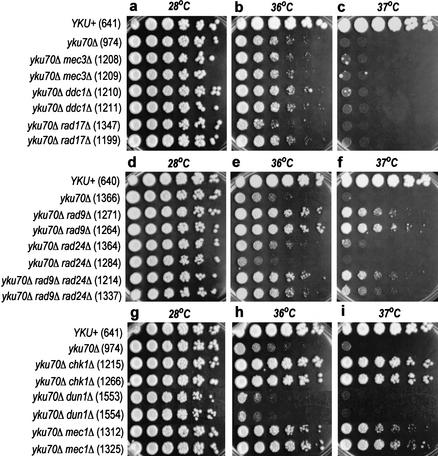
To determine how checkpoint pathways interact with yku70Δ-induced damage in budding yeast, we generated a panel of double and triple mutants. Weinert and Hartwell showed previously that checkpoint mutations allow cdc13-1 mutant strains, defective in a telomere binding protein, to form colonies at higher temperatures than checkpoint-proficient cdc13-1 strains (Weinert and Hartwell 1993; Weinert et al. 1994). This is presumably because loss of checkpoint control allows cells with nonlethal levels of DNA damage to divide and form colonies. Figure 1 shows the growth of serial dilutions of yku70Δ and checkpoint mutant cells at 28°C, 36°C, and 37°C. At 28°C, a permissive temperature for yku70Δ mutants, all strains grew at similar rates and formed similarly sized colonies. At the restrictive temperatures of 36°C and 37°C, different strains formed colonies with different efficiencies. A chk1Δ mutation had the most profound effect and significantly increased the ability of yku70Δ mutants to form colonies at both 36°C and 37°C (Fig. 1g–i). rad9Δ and mec1Δ also increased yku70Δ colony size, but the colonies were smaller than the yku70Δ chk1Δ colonies (Fig. 1d–i). In contrast, mec3Δ, ddc1Δ, rad17Δ, rad24Δ, and dun1Δ mutations had minor effects on the growth of yku70Δ mutants (Fig. 1a–i). The growth of rad9Δ rad24Δ yku70Δ triple mutants at 36°C and 37°C was most similar to that of yku70Δ rad9Δ mutants, indicating that the strong growth phenotype was epistatic (Fig. 1d–f). These experiments suggested that a CHK1, MEC1, and RAD9-dependent, but DDC1, MEC3, RAD17, RAD24, and DUN1-independent mechanism is responsible for the poor growth of yku70Δ mutants at 36°C and 37°C.
Figure 1.
RAD9, MEC1, and CHK1 inhibit growth of yku70Δ strains at high temperatures. Small aliquots of fivefold dilution series of several yeast strains were transferred to plates and incubated at the temperatures indicated for 2 d before being photographed. The relevant genotypes of the strains are indicated on the left, and the strain numbers shown in parentheses. The mec1Δ strains also carried an sml1Δ mutation. To ensure reproducibility we routinely examined two independent strains with the same genotypes. All strains grouped together were grown on the same plates, except for the yku70Δ dun1Δ strains, which were grown on different plates. The photographs of the yku70Δ dun1Δ strains were superimposed in g,h,i.
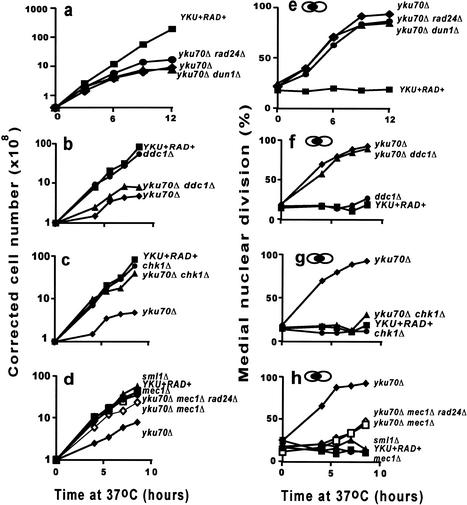
To determine whether checkpoint pathways are activated in yku70Δ mutants, we examined the growth and cell cycle distribution of yku70Δ and checkpoint mutants in liquid cultures (Fig. 2). In four separate experiments, the growth of yku70Δ mutants at 37°C was much slower than YKU70+ cells, such that by 8.5 h their cell number had increased about 4–8-fold, instead of 80–100-fold as observed in the YKU70+ cells (Fig. 2a–d). In addition, in three of four experiments, the growth of yku70Δ cells began to plateau after about 6 h in liquid culture (Fig. 2a–d; data not shown). The poor growth of yku70Δ mutants correlates with an increasing fraction of cells at the medial nuclear division stage of the cell cycle (Hartwell 1974), increasing from ∼20% at the beginning of the experiments, to over 85% during 8.5 h culture at 37°C (Fig. 2e–h). This accumulation of cells at medial nuclear division suggests they are accumulating before the metaphase/anaphase transition, and is consistent with an earlier study which showed that the large budded cells that accumulated in yku70Δ (hdf1Δ) mutant cultures at 37°C contained short mitotic spindles and a nucleus at the neck between the mother and daughter cells (Barnes and Rio 1997). The slow kinetics of arrest of yku70Δ mutants is in contrast to the behavior of cdc13-1 mutants, because 94% of cdc13-1 mutants arrest in the first cell cycle (within 2 h) at restrictive temperature (Weinert and Hartwell 1993).
Figure 2.
RAD9, MEC1, and CHK1 cause G2/M arrest of yku70Δ strains at 37°C. A series of yeast strains dividing exponentially at 23°C were placed at 37°C and their growth and cell cycle distribution were monitored. At indicated time points, cells density was determined by hemocytometer (a–d) and cell cycle distribution was determined by staining with DAPI (e–h). The yeast stains used were YKU+ RAD+: DLY640 in a,d,e,h, and DLY641 in b,c,e,f; yku70Δ: DLY1412 in a,e, and DLY1366 in b,c,d,f,g,h; yku70Δ rad24Δ: DLY 1505; yku70Δ dun1Δ: DLY 1553; ddc1Δ: DLY883; yku70Δ ddc1Δ: DLY1220; chk1Δ: DLY1095; yku70Δ chk1Δ: DLY1215; sml1Δ: DLY1248; mec1Δ: DLY1249; yku70Δ mec1Δ rad24Δ: DLY1327; and yku70Δ mec1Δ: DLY1325.
At 37°C, the growth and cell cycle distribution of yku70Δ chk1Δ, yku70Δ rad9Δ, and yku70Δ mec1Δ mutants was most like YKU70+ RAD+ strains because they grew exponentially and did not accumulate at medial nuclear division (Fig. 2c,d,g,h; data not shown). Therefore, it appears that the poor growth of yku70Δ mutants at 37°C and the accumulation in medial nuclear division is due to a CHK1, MEC1, and RAD9-dependent checkpoint pathway. In contrast, the growth and cell cycle distributions of yku70Δ ddc1Δ, yku70Δ rad24, and yku70Δ dun1Δ mutants were most similar to yku70Δ strains, suggesting that yku70Δ-induced checkpoint pathways are intact in rad24Δ, ddc1Δ, and dun1Δ mutants (Fig. 2a,b,e,f). Both yku70Δ mec3Δ and yku70Δ rad17Δ mutants behaved similarly to yku70Δ rad24Δ mutants (data not shown). We found that rad53Δ sml1Δ single mutants, as well as yku70Δ rad53Δ sml1Δ triple mutants grew poorly in liquid culture at 37°C, which made it difficult to determine the role of RAD53 in cell cycle arrest (data not shown). This may be because RAD53 has an essential function at 37°C that is unrelated to checkpoint control (Gardner et al. 1999; Sanchez et al. 1999).
We noted that, despite their initial checkpoint defective phenotype, yku70Δ mec1Δ and yku70Δ chk1Δ mutants begin to slow cell division and start to accumulate at medial nuclear division, after long periods (8 h) at 37°C (Fig. 2 c,d,g,h). This suggests that another checkpoint pathway, independent of CHK1 and MEC1, can arrest cell division of yku70Δ mutants after long periods at 37°C. This arrest is due to the activation of spindle checkpoint pathways (see Fig. 4, below). Both yku70Δ rad24Δ rad9Δ and yku70Δ rad24Δ mec1Δ triple mutants showed the exponential growth phenotype of yku70Δ rad9Δ and yku70Δ mec1Δ mutants respectively, indicating that the exponential growth phenotype is epistatic to the poor growth phenotype (Fig. 2d,h; data not shown). In summary, these liquid culture experiments suggest that a checkpoint pathway that arrests yku70Δ mutants at medial nuclear division at 37°C is dependent on CHK1, RAD9, and MEC1, but independent of RAD17, RAD24, MEC3, DDC1, and DUN1.
Figure 4.
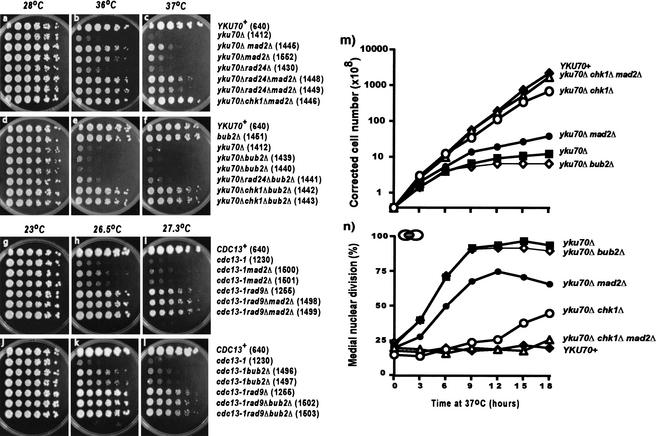
MAD2 inhibits the growth of yku70Δ mutants, while BUB2 inhibits growth of cdc13-1 mutants. (a–l) Small aliquots of fivefold dilution series of several yeast strains were transferred to plates and incubated for 2 d (a–f) or 3 d (g–l) before being photographed. The relevant genotypes of the strains are indicated on the left and the strain numbers shown in parentheses. (m,n) A series of yeast strains dividing exponentially at 23°C was placed at 37°C and their growth and cell cycle distribution were monitored. At indicated time points, cell density was determined by hemocytometer and cell cycle distribution was determined by staining with DAPI. The yeast stains used were YKU+ RAD+: DLY640; yku70Δ chk1Δ mad2Δ: DLY1446; yku70Δ chk1Δ: DLY1215; yku70Δ mad2Δ: DLY1445; yku70Δ: DLY1412; and yku70Δ bub2Δ: DLY1440.
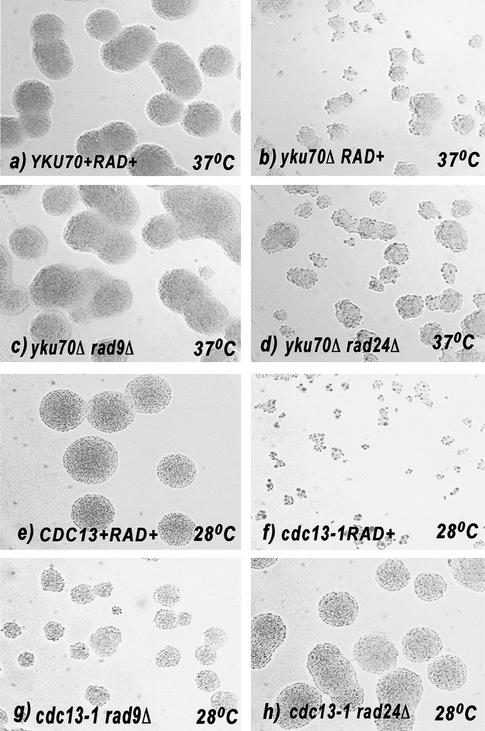
Microcolony assays were used to confirm that yku70Δ mutants are able to divide several times before ceasing growth at 37°C, and that rad9Δ and rad24Δ mutations had different effects on cell division. MATa cells were first arrested in G1 using the mating pheromone alpha factor, the pheromone was removed, and single cells incubated on plates for 20 h at 37°C. After 20 h at 37°C, YKU70+ RAD+ and yku70Δ rad9Δ cells had formed much larger and more uniformly sized colonies (>1000 cells) than yku70Δ and yku70Δ rad24Δ cells (2–200 cells) (Fig. 3a–d). This shows that deletion of RAD9, but not RAD24, allows yku70Δmutants to grow over 20 h, at 37°C, at rates indistinguishable from the rate of YKU70+ cells. The behavior of yku70Δ checkpointΔ double mutants cultured at 37°C is in contrast to the behavior of cdc13-1 checkpointΔ double mutants cultured at a similar temperature, as cdc13-1 rad9Δ mutants formed smaller microcolonies at 36°C compared to the cdc13-1 rad24Δ mutants (Lydall and Weinert 1997).
Figure 3.
RAD9 and RAD24 have different effects on the growth of yku70Δ and cdc13-1 colonies. Single G1-arrested cells were spread on YEPD (ade) plates and photographed after 20-h growth. The yeast strains were (a) DLY640, (b) DLY1412, (c) DLY1271, (d) DLY1364, (e) DLY640, (f) DLY1230, (g) DLY1255, and (h) DLY1257.
A model that might explain the different roles of RAD9 and RAD24 in responding to yku70Δ and cdc13-1-induced damage is that RAD9 is required for the primary checkpoint response in yku70Δ and cdc13-1 mutants, and that RAD24 is required for arrest only when damage becomes more extensive. If true, this could explain why cdc13-1 mutants cultured at 36°C, 10°C higher than their maximum permissive temperature, depend on both RAD9 and RAD24 for cell cycle arrest (Lydall and Weinert 1995), whereas yku70Δ mutants, cultured at 37°C, 2°C higher than their maximum permissive temperature, depend on RAD9, but not on RAD24. Wild-type yeast strains do not form colonies above 38°C, and so it was not possible to test whether arrest of yku70Δ mutants at higher temperatures depends on RAD24 as well as RAD9. However, it was possible to test whether arrest of cdc13-1 mutants at marginally permissive temperatures depended more on RAD9 than RAD24. We used the microcolony assay to test whether RAD9 was required for the primary checkpoint pathways in cdc13-1strains cultured at the moderately restrictive temperature of 28°C. At this temperature, cdc13-1 cells formed colonies in the range of 2 to 20 cells, compared with 2–6 cells at 36°C (Lydall and Weinert 1997); cdc13-1 rad9Δ cells formed medium-sized colonies (20–200 cells), whereas cdc13-1 rad24Δ and cdc13-1 cells formed large-sized colonies (1000–3000 cells) (Fig. 3e–h). Therefore, at both moderately (28°C) and strongly (36°C) restrictive temperatures, cdc13-1 rad9Δ mutants form smaller colonies than cdc13-1 rad24Δ mutants. In contrast, yku70 rad9Δ mutants form larger colonies than yku70Δ rad24Δ mutants at 37°C. Therefore, we conclude that RAD9- and RAD24-dependent checkpoint pathways play different roles in responding to yku70Δ or cdc13-1-induced DNA damage.
MAD2 contributes to the arrest of yku70Δ mutants and BUB2 to the arrest of cdc13-1 mutants
Despite their initial checkpoint-defective phenotype, yku70Δ mec1Δ and yku70Δ chk1Δ mutants began to slow cell division, and started to accumulate at medial nuclear division, after long periods (8 h) at 37°C (Fig. 2g,h). The MAD2-dependent spindle checkpoint arrests cells at a stage of cell division similar to the RAD9-dependent DNA damage checkpoint, just prior to the metaphase/anaphase transition, when the APC (anaphase promoting complex) is activated. The MAD2-dependent checkpoint inhibits APC activation by inhibiting Cdc20p, an essential factor for APC activation (Hwang et al. 1998). To determine whether spindle checkpoint pathways might be responsible for the residual cell cycle arrest observed in yku70Δ chk1Δ mutants, we examined the effect of mad2Δ and bub2Δ mutations on the growth of yku70Δ strains at 37°C (MAD2 and BUB2 belong to different arms of the spindle checkpoint pathways (Gardner and Burke 2000). Whereas a mad2Δ mutation increased the growth of yku70Δ mutants (Fig. 4a–c), a bub2Δ mutation had no effect on the growth of yku70Δ mutants (Fig. 4d–f). Interestingly, simultaneous disruption of both RAD24 and MAD2 increased the growth of yku70Δ mutants more than either single mutation (Fig. 4b,c), suggesting that perhaps RAD24 plays a small role in the arrest of yku70Δ mutants, a role that can be unmasked by deletion of MAD2. A yku70Δ chk1Δ mad2Δ triple mutant grew nearly as well as YKU70+ cells (Fig. 4c).
To determine whether the MAD2 spindle checkpoint also contributes to inhibiting the growth of cdc13-1 mutants, we combined mad2Δ and bub2Δ mutations with cdc13-1. Curiously, and once again, cdc13-1 and yku70Δ mutants showed different, almost opposite interactions with checkpoint pathways. A bub2Δ deletion had a moderate effect on the growth of cdc13-1 mutants (Fig. 4j–l), whereas a mad2Δ deletion had less effect (Fig. 4g–l). The effect of the bub2Δ mutation was not as strong as a rad9Δ DNA damage checkpoint mutation, and the rad9Δ bub2Δ double mutant behaved like the single rad9Δ mutant (Fig. 4j–l). Similarly, the cdc13-1 rad9Δ mad2Δ triple mutants formed colonies with efficiency similar to that of the cdc13-1 rad9Δ double mutants.
To confirm that MAD2 was responsible partially for arrest of yku70Δ mutants and to produce evidence that it contributed to the residual arrest observed in yku70Δ chk1Δ mutants at 37°C, we performed liquid culture experiments. Figure 4m shows that the decrease in growth observed in yku70Δ chk1Δ mutants after several hours at 37°C (vs. YKU70+ cells) could be overcome by a mad2Δ deletion, because yku70Δ chk1Δ mad2Δ triple mutants grew almost as well as the YKU70+ strain. Figure 4n shows that the increase of yku70Δ chk1Δ muants at medial nuclear division, during 9–18 h of incubaion at 37°C, did not occur in the yku70Δ chk1Δ mad2Δ triple mutants. The growth of yku70Δ bub2Δ mutants is most similar to yku70Δ cells, indicating that BUB2 does not play a role in the arrest of yku70Δ mutants at 37°C. Consistent with the hypothesis that yku70Δ damage induces a MAD2-dependent arrest, yku70Δ mad2Δ mutants reached an approximately fivefold higher cell density than yku70Δ cells over an 18-h period (Fig. 4m). In addition, a maximum of 75% of yku70Δ mad2Δ mutants arrested at medial nuclear division, whereas about 95% of yku70Δ cells arrested (Fig. 4n).
We have shown that arrest of yku70Δ mutants at 37°C is due to CHK1- and MAD2-dependent pathways; Figure 4n allows us to estimate their respective contributions. A CHK1-dependent pathway is responsible for 75% of arrest (determined from the percentage of yku70Δ mad2Δ mutants arrested at the time point of maximum arrest, 12 h), whereas a MAD2-dependent pathway is responsible for 20% of the arrest (determined from the percentage of yku70Δ mutants arrested minus the percentage of yku70Δ mad2Δ cells arrested at 12 h). In the case of yku70Δ chk1Δ cells, the percentage of cells arrested by MAD2 only noticeably increased at later time points (over 15 h), consistent with the idea that at early times arrested cells were diluted by the large mass of dividing cells. Thus, the effects of the CHK1-dependent DNA damage and MAD2-dependent spindle checkpoint pathways are additive and together they contribute to all (95%) of the arrest observed in yku70Δ mutants.
yku70Δ mutants accumulate ssDNA in subtelomeric Y‘ sequences at 37°C
There is much evidence that single-stranded DNA is an important stimulus for DNA damage checkpoint pathways. For example, cdc13-1 mutants accumulate ssDNA up to 20kb from their telomeres, when cultured at restrictive temperatures (Garvik et al. 1995). If ssDNA is an important component of the signal that activates checkpoint pathways in yku70Δ mutants at 37°C, then increased levels should be observed at restrictive temperatures. It was known that yku70Δ and yku80Δ mutants contain more ssDNA in their repetitive TG telomeric sequences than do YKU+ cells (Gravel et al. 1998; Polotnianka et al. 1998; Teo and Jackson 2001). However, yku80Δ mutants appear to contain as much single-stranded TG DNA at their permissive temperatures of 23°C and 30°C as at their restrictive temperature of 37°C (Gravel et al. 1998; Teo and Jackson 2001), suggesting that ssDNA at telomeric sequences is not necessarily an important stimulus for cell cycle arrest (Teo and Jackson 2001). We reasoned that the ssDNA in yku70Δ mutants might extend beyond the telomeres, as it does in cdc13-1 mutants, and that there may be a better correlation between the appearance of ssDNA in subtelomeric repeats and cell cycle arrest.
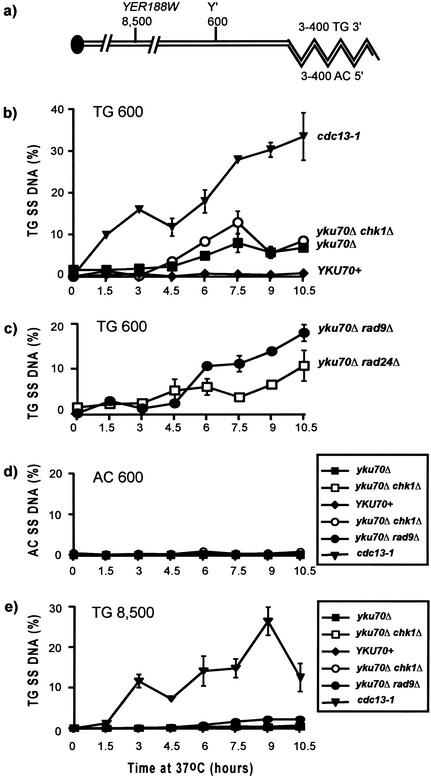
Quantitative amplification of ssDNA (QAOS) (Booth et al. 2001) was used to examine the appearance of ssDNA in telomere proximal sequences of yku70Δ mutants at 37°C (Fig. 5). This quantitative PCR-based method can be used to measure ssDNA levels in the range 0.2% to 100% at single-copy loci in the genome. We found that yku70Δ mutants cultured at the restrictive temperature of 37°C generated increasing amounts of ssDNA at subtelomeric loci. We measured ssDNA at a locus situated 600 bp from the telomeric end of the Y‘ subtelomeric repeat. In telomeres that contain Y‘ repeats, this locus is about 900–1000 bp from the very end of the chromosome (Fig. 5a). At these positions, the amount of ssDNA in yku70Δ mutants increased from 1.6% at the beginning of the experiment to values between 5% and 8%, after 6–10 h of incubation at 37°C (Fig. 5b). The increase in ssDNA is less rapid and less extensive than that observed in cdc13-1 mutants during the same experiment, but the amount of ssDNA is significantly higher than that observed in control YKU70+ cells (Fig. 5b). No ssDNA was detected on the AC strand in any strains (Fig. 5d). Therefore, the ssDNA observed in yku70Δ mutants is on the TG (3′) strand of (sub) telomeres and is presumably caused by loss of the AC (5′) strand, as in cdc13-1 mutants. The increase in the levels of ssDNA in Y‘ sequences with time at 37°C is consistent with the hypothesis that the subtelomeric ssDNA in yku70Δ mutants contributes to activation of the RAD9, CHK1, and MEC1-dependent checkpoint pathway. There is a clear correlation between the accumulation of ssDNA at the Y‘ 600 locus, after a 6-h incubation at 37°C (Fig. 5b) and the accumulation of cells at medial nuclear division in checkpoint-proficient yku70Δ cells (Figs. 2 and 4n).
Figure 5.
yku70Δ mutants accumulate ssDNA in subtelomeric sequences. A series of yeast strains was cultured at 37°C, and the amount of ssDNA at their telomeres was measured by quantitative amplification of ssDNA (QAOS). The yeast stains used were cdc13-1: DLY1230; yku70Δ chk1Δ: DLY1215; yku70Δ: DLY1412; YKU70+: DLY640; and yku70Δ rad9Δ: DLY1271. The results shown for yku70Δ rad24Δ are the average amount of ssDNA observed in two independent strains DLY1364 and DLY1430. The error bars indicate the standard error of the mean derived from three independent measurements of the amount of ssDNA in a sample, except for the yku70Δ rad24Δ, where they indicate the difference in the amount of ssDNA in the two strains. (a) A schematic model of the telomere of chromosome V in budding yeast. (b,c) Detection of ssDNA on the TG strand 600 bases from the telomeric end of the Y‘ sequence. (d) Detection of ssDNA on the AC strand, 600 bases from the telomeric end of the Y‘ sequence. (e) Detection of ssDNA on the TG strand at YER188W, 8500 bases from the right end of chromosome V.
Despite the fact that yku70Δ rad9Δ and yku70Δ chk1Δ mutants grow at 37°C, whereas yku70Δ and yku70Δ rad24Δ did not, all of these mutants accumulated comparable amounts of ssDNA, arguing that their different growth phenotypes are not due to differences in the amount of DNA damage among these strains. Interestingly, the strains that did not grow at 37°C, the yku70Δ and yku70Δ rad24Δ strains, had marginally less ssDNA than the strains that grew (yku70Δ rad9Δ and yku70Δ chk1Δ; Fig. 5b,c). This observation, together with the cell cycle distribution results (Fig. 2) argues that checkpoint genes differ in their ability to induce arrest, rather than in their ability to affect the production of ssDNA in Y‘ sequences of yku70Δ mutants.
The effect of checkpoint genes on the production of ssDNA in cdc13-1 mutants is easily detected at a locus 12 kbp from the telomere, cdc13-1 rad9Δ mutants generate ssDNA more rapidly than cdc13-1 cells, which in turn generate ssDNA more rapidly than cdc13-1 rad24Δ cells (Lydall and Weinert 1995). To investigate whether yku70Δ mutants generate ssDNA beyond the Y‘ sequences, we examined ssDNA production at the YER188W locus, 8,500 bp from the telomere. This is the first unique gene close to the right telomere of chromosome V. Figure 5e shows that yku70Δ mutants generate considerably less ssDNA at this locus compared to cdc13-1 mutants, and compared to the amount of ssDNA they generate at their Y‘ sequences. Interestingly, yku70Δ rad9Δ mutants generated some ssDNA at YER188W, which suggests that RAD9 may inhibit ssDNA production in yku70Δ mutants, as it does in cdc13-1 mutants (Lydall and Weinert 1995).
In summary, we find that yku70Δ mutants contain significantly more subtelomeric ssDNA at restrictive temperatures than at permissive temperatures. This suggests that the subtelomeric ssDNA is an important stimulus for activation of checkpoint control pathways in yku70Δ mutants.
EXO1 is required for ssDNA generation and arrest of yku70Δ mutants
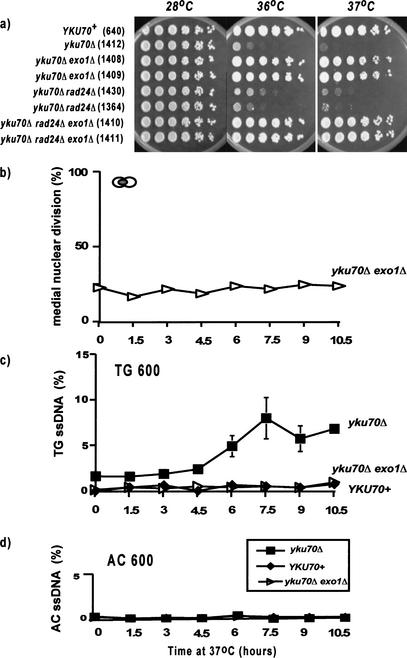
If ssDNA contributes to the signal that arrests cell division of yku70Δ mutants, then mutations that reduce the amount of ssDNA should alleviate arrest. To test this hypothesis, we examined the effect of an exo1Δ mutation on arrest of yku70Δ mutants. EXO1 encodes an exonuclease that is normally recruited to DNA by the mismatch repair machinery (Tishkoff et al. 1997) and is involved in the resection of meiotic DSBs (Tsubouchi and Ogawa 2000), but does not appear to affect telomere length (Tsubouchi and Ogawa 2000). The resection of meiotic DSBs by 5′ to 3′ exonucleases, to generate 3′ ssDNA tails, is in many ways similar to the processes that occur at damaged telomeres, which also produce 3′ ssDNA tails. Figure 6a shows that an exo1Δ mutation strongly increases the ability of yku70Δ mutants to form colonies at 36°C and 37°C. The effect is as strong as that seen with a chk1Δ mutation (cf. Figs. 6 and 1). To determine whether the strong growth is due to alleviation of checkpoint control, we examined the cell cycle distribution of yku70Δ exo1Δ double mutants in liquid cultures and found that yku70Δ exo1Δ mutants do not accumulate in medial nuclear division (Fig. 6b).
Figure 6.
EXO1 is required for arrest and ssDNA generation in yku70Δ mutants. (a) Small aliquots of fivefold dilution series of several yeast strains were transferred to plates and incubated at 28°C, 36°C, or 37°C for 2 d before being photographed. The relevant genotypes of the strains are indicated on the left, and the strain numbers shown in parentheses. (b) A yku70Δ exo1Δ (DLY1408) yeast strain dividing exponentially at 20°C was placed at 37°C, and its cell cycle distribution was monitored by staining with DAPI (see Figs. 2 and 4n for the behavior of control strains). (c) Detection of ssDNA on the TG strand 600 bases from the telomeric end of the Y‘ sequence. The yeast strains were yku70Δ: DLY1412; yku70Δ exo1Δ: DLY1408; and YKU70+: DLY640. (d) Detection of ssDNA on the AC strand 600 bases from the telomeric end of the Y‘ sequence. The yeast strains are as in c.
To determine whether EXO1 was required for the production of ssDNA in yku70Δ mutants, we used QAOS to measure ssDNA production in Y‘ sequences during growth at 37°C (Fig. 6c). It is clear that yku70Δ exo1Δ mutants contain extremely low levels of ssDNA at subtelomeric sequences at both 20°C and 37°C. The levels are indistinguishable from the levels in YKU70+ cells. No ssDNA was detected on the AC strand at telomeres (Fig. 6d). Thus, it appears that EXO1 plays an important role in the accumulation of ssDNA in yku70Δ mutants and that in the absence of this EXO1-dependent ssDNA, yku70Δ mutants do not arrest cell division at 37°C.
EXO1 contributes to ssDNA production in cdc13-1 mutants
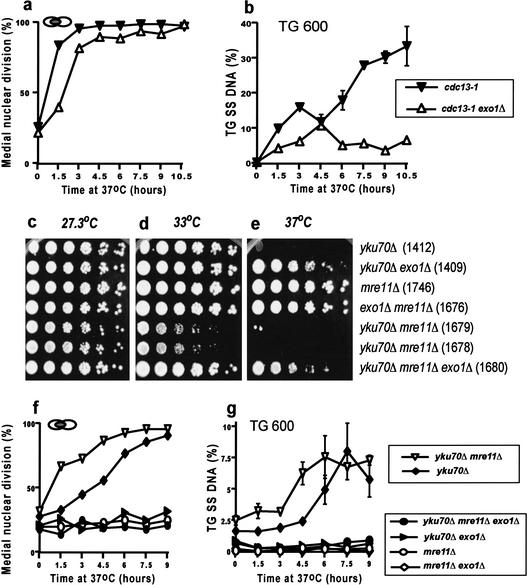
Since EXO1 is required to generate ssDNA at the telomeres of yku70Δ mutants, we asked whether EXO1 is also required to generate ssDNA and induce cell cycle arrest in cdc13-1 mutants. Figure 7a shows that cdc13-1 exo1Δ double mutants arrest at medial nuclear division, as do cdc13-1 mutants, when cultured at 37°C, but with slower kinetics. Therefore, EXO1 contributes to, but is not completely required for, the arrest of cdc13-1 mutants grown at 37°C. When we examined the effect of EXO1 on the appearance of ssDNA at the Y‘ 600 locus in cdc13-1 mutants, we found that ssDNA did appear in cdc13-1 exo1Δ mutants, reaching a level of about 6% after 1.5 h at 37°C and largely staying at this level for the rest of the experiment. This level of ssDNA was considerably less than the 30% level of ssDNA observed in cdc13-1 EXO1 strains (Fig. 7b). We conclude that EXO1 contributes to ssDNA generation in cdc13-1 mutants, but that another exonuclease (ExoX) must also contribute to the production of ssDNA in cdc13-1 mutants.
Figure 7.
EXO1 contributes to cell cycle arrest and ssDNA production in cdc13-1 and yku70Δ mre11Δ mutants. (a) Yeast strains 1230 (cdc13-1) and 1296 (cdc13-1 exo1Δ) dividing exponentially at 20°C were placed at 37°C, and the fraction of cells in medial nuclear division was monitored by staining with DAPI. (b) Detection of ssDNA on the TG strand 600 bases from the telomeric end of the Y‘ sequence. The yeast strains were as in a. (c,d,e) Small aliquots of fivefold dilution series of several yeast strains were transferred to plates and incubated for 2 d before being photographed. The relevant genotypes of the strains are indicated on the right, and the strain numbers are shown in parentheses. (f) Yeast strains dividing exponentially at 20°C were placed at 37°C, and their cell cycle distribution was monitored. The strains were yku70Δ: DLY1412; yku70Δ mre11Δ: DLY1679; yku70Δ mre11Δ exo1Δ: DLY1680; yku70Δ exo1Δ: DLY1408; Δ mre11Δ: DLY1746; and mre11Δ exo1Δ: DLY1676. (g) Detection of ssDNA on the TG strand 600 bases from the telomeric end of the Y‘ sequence. The yeast strains were as in f.
MRE11 protects telomeres in yku70Δ mutants
EXO1 functions redundantly with MRE11 to process DSBs to create 3′ ssDNA tails (Tsubouchi and Ogawa 2000) and in other aspects of DNA damage metabolism (Moreau et al. 2001; Lewis et al. 2002). Therefore, it was possible that MRE11 also played a role in generating ssDNA in yku70Δ mutants. It was shown previously that yku80Δ mre11Δ double mutants display a synthetic poor growth phenotype (Nugent et al. 1998), which is opposite to the phenotype observed in yku70Δ exo1Δ strains (Fig. 6a), suggesting that MRE11 does not have EXO1-type properties when combined with a yku70Δ defect. To test this directly, we combined yku70Δ, exo1Δ, and mre11Δ mutations and examined their effects on growth, cell cycle arrest, and ssDNA production. Figure 7c–e shows that yku70Δ mre11Δ double mutants are more temperature-sensitive than yku70Δ mutants. Interestingly, this temperature-sensitive growth phenotype is dependent on EXO1 (Fig. 7e). Liquid culture experiments demonstrated that yku70Δ mre11Δ double mutants arrested at medial nuclear division more rapidly than yku70Δ single mutants, arguing that MRE11 functions to maintain telomere structure in yku70Δ mutants, rather than to degrade telomere structure, as EXO1 does. The mre11Δ yku70Δ exo1Δ triple mutant did not arrest cell division at 37°C over a 9-h time course, suggesting that EXO1-dependent ssDNA is required for the cell cycle arrest of yku70Δ mre11Δ mutants cultured at 37°C.
Accumulation of telomeric ssDNA in yku70Δ mre11Δ mutants provides an explanation for their rapid arrest at medial nuclear division at 37°C. Even at 20°C (at the beginning of the experiment), yku70Δ mre11Δ double mutants contain more ssDNA than yku70Δ mutants in their Y‘ sequences (Fig. 7g). Furthermore, the amount of ssDNA increases more rapidly in yku70Δ mre11Δ double mutants than in yku70Δ mutants, which is consistent with the more rapid arrest observed in these strains. Finally, all of the ssDNA in yku70Δ mre11Δ double mutants appears to be dependent on EXO1.
Discussion
In this study, we examined the interactions of checkpoint pathways with the damaged telomeres that are present in yku70Δ mutant cells. We found that yku70Δ mutants, like cdc13-1 mutants cultured at 37°C, contain increased levels of ssDNA in subtelomeric sequences. However, the amount of ssDNA observed in yku70Δ mutants is considerably less than in cdc13-1 mutants. We demonstrated that EXO1 but not MRE11 is required for the production of this ssDNA and for cell cycle arrest. The correlation between the amount of ssDNA and cell cycle arrest in yku70Δ mutant cells is in many ways analogous to the situation observed with DSBs, when strains with more ssDNA arrest cell division for longer (Lee et al. 1998).
Interestingly, the damage induced in yku70Δ mutants activates a RAD9, CHK1, and MEC1-dependent checkpoint pathway, but is independent of RAD17, RAD24, MEC3, DDC1, and DUN1, whereas arrest of cdc13-1 mutants is dependent on all eight genes. yku70Δ-induced damage is the first type of DNA damage demonstrated to have these properties. A complementary pathway appears to exist in meiosis, because prophase arrest of dmc1Δ mutants, which cannot complete meiotic recombination, depends on RAD24 but is independent of RAD9 and CHK1 (Bishop et al. 1992; Lydall et al. 1996; Roeder and Bailis 2000).
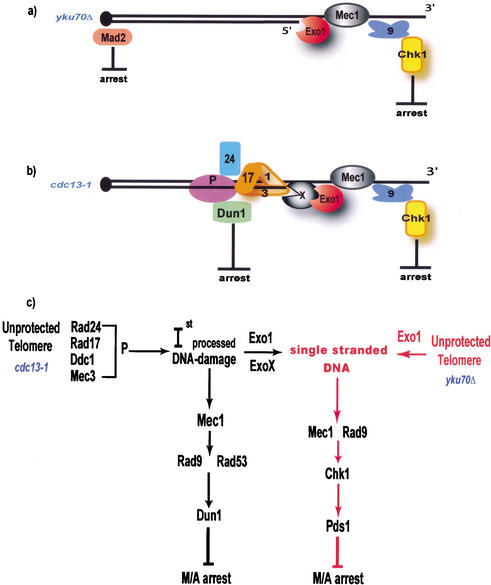
We propose a model to explain the functions of checkpoint proteins in responding to (sub) telomere defects in cdc13-1 and yku70Δ mutants (Fig. 8). According to this model, in cdc13-1 mutants, unprotected telomeres are perceived as DSBs with a short 3′ overhang. The Rad24p/Rfc2-5p clamp loader complex (Green et al. 2000) recognizes this structure and loads the Ddc1p, Mec3p, Rad17p sliding clamp (Venclovas and Thelen 2000). The sliding clamp tethers an unknown protein, “P” (e.g., a helicase), which processes the telomeric termini to generate the “1st processed DNA damage”. The “1st processed DNA damage” is the substrate that activates a Mec1p/Rad53p/Dun1p-dependent checkpoint pathway. Rad9p might also participate in Rad53p activation (Gilbert et al. 2001). Rad53p and Dun1p are known to be responsible for 50% of the arrest observed in cdc13-1 mutants (Gardner et al. 1999). We suggest that protein “P” is then replaced by Exo1p and by another 5′ to 3′ exonuclease ExoXp, which have affinity for the “1st processed DNA damage.” The ssDNA generated by Exo1p and ExoXp activates a Rad9p/Mec1p/Chk1p- and Pds1p-dependent pathway. Chk1p/Pds1p are required for 50% of the arrest observed in cdc13-1 mutants. Thus, together Rad53p/Dun1p and Chk1p/Pds1p pathways are responsible for 100% arrest of cdc13-1 cells (Gardner et al. 1999; Sanchez et al. 1999). In yku70Δ mutants, unprotected telomeres are also perceived as DSBs with a short 3′ overhang. But in cells lacking Yku70p, Exo1p can be recruited independently of Rad24p and the Rad17p, Mec3p, Ddc1p sliding clamp. Exo1p generates ssDNA that activates the Rad9p/ Mec1p/Chk1p- and Pds1p-dependent checkpoint pathway. In yku70Δ mutants, this checkpoint pathway is responsible for the arrest of 75% of cells, and together with a MAD2-dependent pathway, results in arrest of 95% of cells (see Fig. 4n).
Figure 8.
A model for the roles of checkpoint proteins in responding to cdc13-1 and yku70Δ-induced DNA damage. (a) A representation of the proteins that are responsible for the arrest of yku70Δ mutants. (b) A representation of proteins that are responsible for the arrest of cdc13-1 mutants. (c) A schematic model of the DNA damage checkpoint pathways responsible for metaphase/anaphase arrest of cdc13-1 and yku70Δ mutants.
Rad24p, the sliding clamp, and protein P might play a minor role in responding to yku70Δ-induced DNA damage, but the “1st processed DNA damage” does not form to a sufficient extent to activate Rad53p and Dun1p. Teo and Jackson (2001) showed that Rad53p kinase is activated at 37°C in yku80Δ mutants, but at very low levels. Pellicioli et al. (1999) showed that cells released from hydroxyurea arrest contain residual Rad53 kinase activity, even when the cell cycle has restarted. Therefore, a threshold of Rad53p kinase activity may be necessary to cause cell cycle arrest, and this threshold may not be reached in yku70Δ mutants.
This model raises many questions, most importantly: Why do DDC1, MEC3, RAD17, RAD24, and DUN1 play insignificant roles in the arrest of yku70Δ mutants? We suggest the following, not mutually exclusive, hypotheses: (1) Rad24p cannot bind to telomeres in yku70Δ mutants; (2) The “1st processed DNA damage” is not generated in yku70Δ mutants; (3) “ExoXp” is not recruited to telomeres in yku70Δ mutants; and (4) Exo1p competes with “ExoXp” for substrates and is preferentially recruited to telomeres in yku70Δ mutants.
Our experiments reveal for the first time the role of the spindle checkpoint in arresting yku70Δ mutants at 37°C. MAD2 contributes to the arrest of yku70Δ mutants, whereas BUB2 has no significant effect. Other studies have also shown interactions between mitotic spindle checkpoint pathways and cells that contain DNA damage. For example, all of the thymic lymphomas that developed in Brca2 knockout mice, defective in a protein thought to be involved in DNA repair (Venkitaraman 2002), had generated mutations in spindle checkpoint pathways Bub1 and Mad3 (Lee et al. 1999). Drosophila double parked mutants, defective in a homolog of CDT1, a gene whose product is required for DNA replication in fission yeast and Xenopus, depend on both DNA damage and spindle checkpoint genes to block cell division (Garner et al. 2001). bub2 mutations, but not mad2 mutations, allow cdc13-1 mutants to rebud and reduplicate their DNA, without completing anaphase (Wang et al. 2000). Finally, recent experiments demonstrated that Rad53p is required to modify Bfa1p, the partner of Bub2p in response to cdc13-1-induced DNA damage (Hu et al. 2001).
In our present experiments, the fraction or cells arrested by the MAD2-spindle checkpoint pathway was about fourfold lower than that arrested by the DNA damage checkpoint (Fig. 4n). In a culture of yku70Δ chk1Δ cells, missing DNA damage checkpoint control, the fraction of cells arrested at the spindle checkpoint increased to about 20% after 18 h. This shows that the spindle checkpoint can substitute for the DNA damage checkpoint and stop cells with damaged telomeres from dividing. In a culture of yku70Δ mad2Δ cells, missing a spindle checkpoint pathway, the fraction of cells arrested at the DNA damage checkpoint reached a maximum of 75% (after 12 h; Fig. 4n). Why did the remaining 25% of cells fail to arrest? Presumably these cells were not arrested by the DNA damage checkpoint because they did not contain DNA damage. Instead, they appear to have generated another defect that triggers arrest by the MAD2-dependent spindle checkpoint pathway. It is interesting that during the time course of our experiment the fraction of dividing cells did not significantly increase, suggesting that the cells that are not arrested at medial nuclear division carry lesions (perhaps chromosome losses?) that limit cell division. Consistent with this interpretation, only 18% of yku70Δ mad2Δ cells were able to form colonies after 18 h at 37°C (data not shown).
Why should cells with damaged telomeres activate spindle checkpoint pathways? One explanation is that cells with damaged telomeres generate telomere fusions and dicentric chromosomes at high rates. Indeed, it is known that mammalian cells lacking Ku suffer from high levels of telomere fusions (Bailey et al. 1999; Hsu et al. 2000; Samper et al. 2000; d’Adda di Fagagna et al. 2001). In yeast, it has been shown that dicentric chromosomes, a product of telomere fusion, are activators of both spindle and DNA damage checkpoint pathways (Neff and Burke 1992).
5′ to 3′ exonucleases are thought to play a physiological role in the replication and stability of telomeres, but the nature of the exonuclease(s) responsible for telomere replication remains unclear (Wellinger et al. 1996; Diede and Gottschling 2001; Tsukamoto et al. 2001). We have shown that EXO1 but not MRE11 is required to generate ssDNA at the telomeres of yku70Δ mutants, and so it is conceivable that EXO1 also plays a role in physiological metabolism of telomeres. MRE11, in contrast to EXO1, stablizes telomeres of yku70Δ mutants. If Exo1p does play a role in the physiology of telomeres, then other exonucleases must function redundantly with Exo1p because the telomeres of exo1Δ mutants appear normal (Tsubouchi and Ogawa 2000).
Defects in mismatch repair are associated with checkpoint defects in mammalian cells (Bellacosa 2001; Yan et al. 2001) and enhanced cellular proliferation of yeast cells that lack telomerase (Rizki and Lundblad 2001). Our demonstration that the mismatch repair-associated exonuclease, Exo1p, affects the metabolism of damaged telomeres, and checkpoint responses, suggests a mechanism by which mismatch repair affects checkpoint control and tolerance of damaged telomeres.
Materials and methods
Yeast strains
All strains used in this study are isogenic and in the W303 background; in most cases we used RAD5 rather than rad5-535 strains (Fan et al. 1996), but we observed no effect of the rad5-535 mutation in any experiments. To construct strains, standard genetic procedures of transformation and tetrad analysis were followed (Adams et al. 1997). Since W303 strains contain an ade2-1 mutation YEPD (yeast extract, peptone, and dextrose), the medium was routinely supplemented with adenine at 50 mg/L. The yku70Δ deletion strains were obtained from L. Guarente (Massachusetts Institute of Technology, Cambridge, MA) and S. Jackson (University of Arizona, Tuscon, AZ). A chk1∷HIS3 deletion was created using pYS51 (Sanchez et al. 1999). The mec1Δ and sml1Δ deletion strains were obtained from M.P. Longhese (Paciotti et al. 2000). An exo1∷LEU2 disruption was constructed using pHT246 and an mre::hisg::URA3 deletion with pHT16 (Tsubouchi and Ogawa 2000). mad2Δ and bub2Δ deletion strains were obtained from L Dirick. Dun1Δ strains were obtained from T. Weinert. Other deletions have been described elsewhere (Lydall and Weinert 1997). cdc13-1int strains contain a cdc13-1 integrated allele rather than one that was introduced by backcrossing from the A364a genetic background.
We have observed that other yku70Δmre11Δ (exo1Δ) mutants enter crisis after several generations, and therefore we assume the strains analysed in Figure 7 have escaped or avoided crisis.
Serial dilution and growth on plates
Colony-purified yeast strains were inoculated into 1mL YEPD (ade), and grown overnight with aeration. In the morning, cultures were diluted 1:10, grown for about 4 h, sonicated, counted by hemocytometer, and diluted to 1.5 × 107 cells/mL. Fivefold dilution series were set up in 96-well plates, and small aliquots of the dilution series were transferred to YEPD (ade) plates using metal prongs. Plates were incubated for 2 d before being photographed.
Liquid culture, medial nuclear division, and viability assays
Single purified colonies were inoculated directly into 50 mL of YEPD (ade) and cultured overnight, with aeration, at 23°C. In the morning, cell densities were determined by hemocytometer, and cultures were diluted to 2 × 106 cells/mL (1 × 108 cells in 50 mL, or 0.4 × 108 cells in 20 mL). The cultures were placed at the restrictive temperature of 37°C, and samples were taken at the times indicated. Cultures were maintained at a concentration that allowed exponential growth, diluting when necessary with prewarmed (37°C) medium. Cell densities were determined by hemocytometer, and the corrected cell number was calculated as a product of cell density and cumulative dilution factor. To score checkpoint arrest, samples were taken at the indicated time points and fixed in 70% EtOH, then washed twice with water. To visualize the DNA, cells were resuspended in 0.2μg/mL 4‘6‘-diamidino-2 phenylindole (DAPI), sonicated, and examined by fluorescent microscopy. At least 200 cells were counted using the multicounter, and classified as described previously (Gardner et al. 1999) as: (1) unbudded, single DAPI-stained body; (2) small budded, single DAPI-stained body, the bud <50% of the diameter of the mother cell; (3) medial nuclear division, single DAPI-stained body, bud >50% diameter of mother cell; and (4) late nuclear division, two buds, and two DAPI-stained bodies, and (5) none of these types.
Microcolony assays
Colony-purified yeast strains were inoculated into 1 mL YEPD (ade), grown overnight with aeration, at the appropriate temperature (20°C for cdc13-1 strains and 23°C for yku70Δ cells), until they reached a concentration of about 8 × 106 cells/mL. Cells were arrested in G1 with alpha-factor for about 2.5 h, and arrest was monitored microscopically. Arrested cells were washed twice with YEPD (ade), sonicated briefly, and spread on plates. The plates were incubated at the indicated temperature. After an appropriate length of time the colonies were photographed.
Single-stranded DNA measurements
Single-stranded DNA was measured as described (Booth et al. 2001) except that we calculated ssDNA levels by comparison with a PDA1 “loading control.” PDA1 is 30 kbp from the telomere and does not become single-stranded in yku70Δ mutants. The PDA1 and YER188W primers are as described (Booth et al. 2001). The sequences of the primers used to detect ssDNA in the Y‘ sequence are available on request.
Table 1.
Yeast strains
| DLY
|
Genotype
|
Origin
|
|---|---|---|
| 640 | Mata ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+ psi+ ssd1-d2 RAD5 | R. Rothstein |
| 641 | MATalpha ada2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+ psi+ ssd1-d2 RAD5 | R. Rothstein |
| 883 | MATa ddc1::KanMX4 rad5-535 | M.P. Longhese |
| 974 | MATa yku70::HIS3 RAD5 rDNA::ADE2 | L. Guarente |
| 1028 | MATalpha yku70::LEU2 rad5-535 | S. Jackson |
| 1095 | MATa chk1::HIS3 RAD5 | 640 transformation |
| 1199 | MATa yku70::HIS3 rad17::LEU2 rad5-535 | 974 × 607 |
| 1208 | MATalpha yku70::LEU2 mec3::TRP1 rad5-535 | 886 × 1028 |
| 1209 | MATa yku70::LEU2 mec3::TRP1 rad5-535 | 886 × 1028 |
| 1210 | MATa ddc1::KanMX4 yku70::LEU2 rad5-535 | 886 × 1028 |
| 1211 | MAT alpha ddc1::KanMX4 yku70::LEU2 rad5-535 | 886 × 1028 |
| 1214 | MATalpha yku70::LEU2 rad9::HIS3 rad24::TRP1 rad5-535 | 1028 × 261 |
| 1215 | MATa yku70::LEU2 chk1::HIS3 RAD5 | 1028 × 1096 |
| 1230 | Mata cdc13-1int RAD5 | 1108 transformation |
| 1248 | MATa rad5-535 sml1:KANMX4 | M.P. Longhese |
| 1249 | MATa rad5-535 sml1del::KANMX4 mec1::HIS3 | M.P. Longhese |
| 1255 | MATa cdc13-1int rad9::HIS3 RAD5 | 662 × 1218 |
| 1257 | MATa cdc13-1int rad24::TRP1 RAD5 | 662 × 1218 |
| 1264 | MATalpha yku70::LEU2 rad9::HIS3 RAD5 | 1218 × 1214 |
| 1266 | MAT a yku70::LEU2 chk1::HIS3 RAD5 | 1028 × 1096 |
| 1271 | MATa yku70::LEU2 rad9::HIS3 RAD5 | 1264 × 1285 |
| 1284 | MATa rad24::TRP1 yku70::HIS3 RAD5 | 974 × 1258 |
| 1296 | MATa exo1::LEU2 cdc13-1int RAD5 | 1272 × 1230 |
| 1312 | MATalpha yku70::LEU2 mec1::HIS3 rad5-535 sml1::KANMX4 | 1028 × 1249 |
| 1325 | MATa yku70::LEU2 mec1::HIS3 rad5-535 sml1::KANMX4 | 1028 × 1249 |
| 1327 | MATa yku70::LEU2 mec1::HIS3 rad24::TRP1 sml1del::KANMX4 | 1312 × 1285 |
| 1337 | MATA yku70::LEU2 rad9::HIS3 rad24::TRP1 rad5-535 | 1028 × 261 |
| 1347 | MATalpha yku70::HIS3 rad17::LEU2 RAD5 | 1308 × 1284 |
| 1364 | MATa rad24::TRP1yku70::HIS3 RAD5 | 1308 × 1284 |
| 1366 | MATalpha yku70::HIS3 RAD5 | 1308 × 1284 |
| 1408 | MATa yku70::HIS3 exo1::LEU2 RAD5 | 1273 × 1364 |
| 1409 | MATalpha yku70::HIS3 exo1::LEU2 RAD5 | 1273 × 1364 |
| 1410 | MATalpha yku70::HIS3 exo1::LEU2 rad24::TRP1 RAD5 | 1273 × 1364 |
| 1411 | MATa yku70::HIS3 exo1::LEU2 rad24::TRP1 RAD5 | 1273 × 1364 |
| 1412 | MATa yku70::HIS3 RAD5 | 1273 × 1364 |
| 1430 | MATa yku70::HIS3 rad24::TRP1 RAD5 | 1399 × 1364 |
| 1439 | MATalpha bub2::URA3 yku70::LEU2 rad5-535 | 1429 × 1371 |
| 1440 | MATalpha bub2::URA3 yku70::LEU2 RAD5 | 1429 × 1371 |
| 1441 | MATalpha bub2::URA3 yku70::LEU2 rad24::TRP1 RAD5 | 1429 × 1371 |
| 1442 | MATa bub2::URA3 yku70::LEU2 chk1::HIS3 rad5-535 | 1429 × 1371 |
| 1443 | MATa bub2::URA3 yku70::LEU2 chk1::HIS3 rad24::TRP1 RAD5 | 1429 × 1371 |
| 1445 | MATa mad2::URA3 yku70::LEU2 rad5-535 | 1429 × 1372 |
| 1446 | MATa mad2::URA3 yku70::LEU2 chk1::HIS3 RAD5 | 1429 × 1372 |
| 1448 | MATa mad2::URA3 yku70::LEU2 rad24::TRP1 RAD5 | 1429 × 1372 |
| 1449 | MATalpha mad2::URA3 yku70::LEU2 rad24::TRP1RAD5 | 1429 × 1372 |
| 1451 | MATalpha bub2::URA3 RAD5 | 1429 × 1371 |
| 1496 | MATalpha cdc13-1int bub2::URA3 RAD5 | 1451 × 1255 |
| 1497 | MATalpha cdc13-1int bub2::URA3 RAD5 | 1451 × 1255 |
| 1498 | MATa cdc13-1int mad2::URA3 rad9::HIS3 RAD5 | 1452 × 1255 |
| 1499 | MATalpha cdc13-1int mad2::URA3 rad9::HIS3 RAD5 | 1452 × 1255 |
| 1500 | MATalpha cdc13-1int mad2::URA3 RAD5 | 1452 × 1255 |
| 1501 | MATa cdc13-1int mad2::URA3 RAD5 | 1452 × 1255 |
| 1502 | MATa cdc13-1int bub2::URA3 RAD5 rad9::HIS3 | 1451 × 1255 |
| 1503 | MATalpha cdc13-1int bub2::URA3 RAD5 rad9::HIS3 | 1451 × 1255 |
| 1505 | MATalpha yku70::LEU2 rad24::TRP1 RAD5 | 1429 × 1371 |
| 1552 | MATa mad2::URA3 yku70::LEU2 | 1400 × 1449 |
| 1553 | MATa dun1::HIS3 yku70::LEU2 | 1400 × 1449 |
| 1554 | MATalpha dun1::HIS3 yku70::LEU2 | 1400 × 1449 |
| 1676 | MATalpha mre11::hisG::URA3 exo1::LEU2 RAD5 | 1330 × 1409 |
| 1678 | MATalpha yku70::HIS3 mre11::hisG::URA3 RAD5 | 1330 × 1409 |
| 1679 | MATalpha yku70::HIS3 mre11::hisG::URA3 RAD5 | 1330 × 1409 |
| 1680 | MATa yku70::HIS3 mre11::his G::URA3 exo1::LEU2 RAD5 | 1330 × 1409 |
| 1746 | MATalpha mre11::hisG::URA3 RAD5 | 1330 × 1409 |
The strains are in the W303 background and relevant genotypes are shown. Where strains are the products of a genetic cross, the numbers of parent strains are also indicated.
Acknowledgments
We thank L. Dirick, S. Elledge, H. Ogawa, S. Jackson, L. Guarente, M.P.Longhese, V. Lundblad, S. Piatti, R. Rothstein, Y. Sanchez, L. Symington, H. Tsubouchi, and T. Weinert for plasmids, protocols, or yeast strains. We thank all members of our lab, Keith Caldecott, and Elspeth Stewart for comments on the manuscript. L.M. was partially supported by the Vorarlberger Landesregierung. D.L. is a Wellcome Senior Research Fellow in Basic Biomedical Science.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL lydall@man.ac.uk; FAX 44-0-161-275-5600.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.225102.
References
- Adams A, Gottshcling DE, Kaiser CA, Stearns T. Methods in yeast genetics. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 131–132. [Google Scholar]
- Bailey SM, Meyne J, Chen DJ, Kurimasa A, Li GC, Lehnert BE, Goodwin EH. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc Natl Acad Sci. 1999;96:14899–14904. doi: 10.1073/pnas.96.26.14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Carr AM. Checkpoints: How to flag up double-strand breaks. Curr Biol. 2002;12:R105–R107. doi: 10.1016/s0960-9822(02)00673-5. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Hande MP, Tong W, Roth D, Lansdorp PM, Wang Z, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- Fan HY, Cheng KK, Klein HL. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- Fellerhoff B, Eckardt-Schupp F, Friedl AA. Subtelomeric repeat amplification is associated with growth at elevated temperature in yku70 mutants of Saccharomyces cerevisiae. Genetics. 2000;154:1039–1051. doi: 10.1093/genetics/154.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: Two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Gardner R, Putnam CW, Weinert T. RAD53, DUN1 and PDS1 define two parallel G(2)/M checkpoint pathways in budding yeast. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M, van Kreeveld S, Su TT. mei-41 and bub1 block mitosis at two distinct steps in response to incomplete DNA replication in Drosophila embryos. Curr Biol. 2001;11:1595–1599. doi: 10.1016/s0960-9822(01)00483-3. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Gravel S, Larrivee M, Labrecque P, Wellinger RJ. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, Li GC, Campisi J, Kohwi-Shigematsu T, Chen DJ. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes & Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: A target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Lee H, Trainer AH, Friedman LS, Thistlethwaite FC, Evans MJ, Ponder BA, Venkitaraman AR. Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol Cell. 1999;4:1–10. doi: 10.1016/s1097-2765(00)80182-3. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lewis LK, Karthikeyan G, Westmoreland JW, Resnick MA. Differential suppression of DNA repair Deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase) Genetics. 2002;160:49–62. doi: 10.1093/genetics/160.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, Murguia JR. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: Implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- ————— G2/M checkpoint genes of Saccharomyces cerevisiae: Further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MW, Burke DJ. A delay in the Saccharomyces cerevisiae cell cycle that is induced by a dicentric chromosome and dependent upon mitotic checkpoints. Mol Cell Biol. 1992;12:3857–3864. doi: 10.1128/mcb.12.9.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes & Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, DiFiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet. 2001;27:64–67. doi: 10.1038/83778. [DOI] [PubMed] [Google Scholar]
- Polotnianka RM, Li J, Lustig AJ. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr Biol. 1998;8:831–834. doi: 10.1016/s0960-9822(98)70325-2. [DOI] [PubMed] [Google Scholar]
- Porter SE, Greenwell PW, Ritchie KB, Petes TD. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki A, Lundblad V. Defects in mismatch repair promote telomerase-independent proliferation. Nature. 2001;411:713–716. doi: 10.1038/35079641. [DOI] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Smith GC, Jackson SP. The DNA-dependent protein kinase. Genes & Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP. Telomerase subunit overexpression suppresses telomere-specific checkpoint activation in the yeast yku80 mutant. EMBO Rep. 2001;2:197–202. doi: 10.1093/embo-reports/kve038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff DX, Boerger AL, Bertrand P, Filosi N, Gaida GM, Kane MF, Kolodner RD. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Taggart AK, Zakian VA. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- Yan T, Schupp JE, Hwang HS, Wagner MW, Berry SE, Strickfaden S, Veigl ML, Sedwick WD, Boothman DA, Kinsella TJ. Loss of DNA mismatch repair imparts defective cdc2 signaling and G(2) arrest responses without altering survival after ionizing radiation. Cancer Res. 2001;61:8290–8297. [PubMed] [Google Scholar]