Abstract
Gene silencing by double-stranded RNA is a widespread phenomenon called RNAi, involving homology-dependent degradation of mRNAs. Here we show that RNAi is established in the Drosophila female germ line. mRNA transcripts are translationally quiescent at the arrested oocyte stage and are insensitive to RNAi. Upon oocyte maturation, transcripts that are translated become sensitive to degradation while untranslated transcripts remain resistant. Mutations in aubergine and spindleE, members of the PIWI/PAZ and DE-H helicase gene families, respectively, block RNAi activation during egg maturation and perturb translation control during oogenesis, supporting a connection between gene silencing and translation in the oocyte.
Keywords: RNAi, oogenesis, Drosophila, aubergine, spindle-E
Gene expression is specifically silenced by introducing double-stranded RNA (dsRNA) into an organism. First discovered in the nematode Caenorhabditis elegans (Fire et al. 1998), RNA interference (RNAi) also works effectively in insects (Kennerdell and Carthew 1998), and mammals (Elbashir et al. 2001a). Introduction of dsRNA causes rapid and prolonged degradation of mRNAs synthesized from genes with sequence identity to the dsRNA. RNAi is related to other posttranscriptional gene silencing (PTGS) phenomena that occur in response to invasion of repetitive genes into a variety of eukaryotic organisms (Carthew 2001). Based on several lines of evidence, it has been proposed that PTGS functions as a host mechanism of defense against infection and spread of foreign genomes such as transposons and viruses.
A predominant theme in PTGS by several species is the necessity for small interfering RNA products (siRNAs) 21–25 nucleotides (nt) in length to carry out silencing (Hamilton and Baulcombe 1999; Hammond et al. 2000; Zamore et al. 2000; Elbashir et al. 2001b). In Drosophila, dsRNA is processed into siRNAs in a reaction requiring an RNase III enzyme called Dicer (Bernstein et al. 2001). A Dicer-like gene, dcr-1, is also active in RNAi-mediated gene silencing by C. elegans (Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001). Subsequently, siRNAs act as adaptor molecules, which facilitate interaction of a ribonuclease complex (RISC) with complementary mRNA transcripts and their cleavage (Hammond et al. 2000; Elbashir et al. 2001b).
In addition to Dicer and siRNAs, other factors have been identified as important components of PTGS. Specific members of a protein family are necessary for PTGS or RNAi (Tabara et al. 1999; Fagard et al. 2000; Hammond et al. 2001). These proteins contain a C-terminal PIWI domain and a central PAZ domain whose biochemical activities are unknown. Although most of these proteins were identified by mutations in their corresponding genes, biochemical purification of RISC from Drosophila somatic cells identified a PIWI-PAZ protein, Ago2, as a component of the RISC complex (Hammond et al. 2001). Thus, members of the PIWI-PAZ family play a direct role in the RNAi pathway.
Several lines of evidence indicate that factors involved in RNAi are also involved in control of development. Two 22-nt endogenous RNAs in C. elegans encoded by the lin-4 and let-7 genes regulate the timing of developmental stages in the nematode life cycle, and these activities have been linked to their inhibiting expression of genes such as lin-14 and lin-41 (Olsen and Ambros 1999; Reinhart et al. 2000). Dicer is required for processing of the let-7 and lin-4 small RNAs, and dcr-1 mutants are defective in developmental timing and oogenesis (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001). It appears that let-7 and lin-4 encode two of possibly hundreds of micro RNAs (miRNAs) that are endogenous to many organisms, including nematodes, flies, and humans (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). The function of these miRNAs is not clear.
Here we show that RNAi is established in the Drosophila female germ line. mRNA transcripts are translationally quiescent at the arrested oocyte stage and are insensitive to RNAi. Upon oocyte maturation, transcripts that become activated for translation also become sensitive to degradation while untranslated transcripts remain resistant. Mutant oocytes that are defective for translation control during oogenesis fail to become RNAi-competent. These mutants correspond to the aubergine (aub) and spindle-E (spn-E) genes that encode a PIWI/PAZ domain protein and an RNA helicase, respectively. Together, our data suggest that regulation of RNAi and translational control during oogenesis may be mediated in part by similar mechanisms.
Results and Discussion
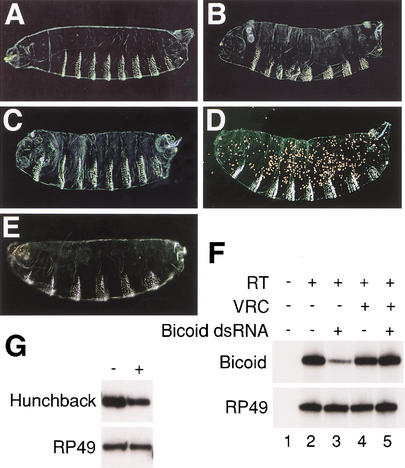
To analyze the effects of dsRNA on mRNA stability in Drosophila oocytes, we used dsRNAs corresponding to the maternally expressed genes bicoid and hunchback. We chose these genes because their mRNAs are synthesized, processed, and localized to the cytoplasm of oocytes during mid- to late oogenesis (Tautz 1988; St Johnston et al. 1989). To test the sensitivity of bicoid and hunchback to RNAi, we initially injected fertilized eggs with dsRNA. bicoid dsRNA reduced the expression of Bicoid protein and induced a bicoid loss-of-function phenotype in which embryos have partial transformation of anterior structures to posterior identities (Fig. 1A–C). The effect was robust enough that dsRNA-coated gold particles randomly introduced into fertilized eggs by a gene gun generated mutant phenotypes (Fig. 1D). hunchback dsRNA induced phenotypes in which embryos were missing thoracic and head segments (Fig. 1E). These phenotypes resembled mutant embryos generated when maternal and zygotic hunchback gene activity is reduced. To determine if dsRNA injection caused mRNA degradation, we measured endogenous mRNA levels using a semiquantitative RT-PCR assay (Kennerdell and Carthew 2000). The level of bicoid mRNA was reduced about fourfold 40 min after injection of bicoid dsRNA (Fig. 1F). Likewise, injection of hunchback dsRNA resulted in a reduction of hunchback mRNA levels (Fig. 1G). Coinjection of a pan-specific ribonuclease inhibitor, vanadyl-ribonucleoside, with bicoid dsRNA resulted in no reduction of bicoid mRNA (Fig. 1F), indicating the effect required a ribonuclease activity.
Figure 1.
bicoid and hunchback are sensitive to RNAi. (A–E) Embryonic phenotypes with anterior to the left. (A) Cuticle phenotype of an embryo that had been injected with buffer at its anterior pole at stage 2. Its morphology resembles wild type. (B) Embryo from a bicoidE1 homozygous mother. Head and thoracic segments are missing in bicoid mutants, and the acron is partially or fully transformed into telson. (C,D) Cuticle phenotypes of embryos that were injected with bicoid dsRNA (C) or bombarded with gold particles coated with bicoid dsRNA (D). The cuticle in D is speckled in appearance owing to the embedded gold particles. The RNAi phenotypes are similar to the bicoid mutant. The penetrance of phenotype by injection is 100% (n = 42); the penetrance of phenotype by bombardment is 66% (n = 36). (E) Cuticle phenotype of embryo that was injected with hunchback dsRNA. The phenotype is similar to that of an embryo mutant for both maternal and zygotic hunchback. The penetrance of phenotype is 82% (n = 34). (F) Semiquantitative RT-PCR assay for bicoid and housekeeping gene RP49 mRNA levels. Pools of stage 2 embryos were coinjected with indicated combinations of bicoid dsRNA and vanadyl ribonucleoside complexes (VRC) in buffer, and total RNA was extracted at stage 4. RNA was reverse-transcribed (RT) as indicated, and bicoid or RP49 cDNA was amplified by PCR. The number of embryos per treatment is (lane 1) n = 13; (lane 2) n = 9; (lane 3) n = 17; (lane 4) n = 9; (lane 5) n = 15. There is a noticeable absence of bicoid protein/function but only a fourfold decrease in mRNA level. This has also been observed for other genes. One reason is that the RT-PCR assay only detects RNAi if the mRNA is cleaved within the region being amplified. Transcripts cleaved outside of the amplified region are counted as full length, even though they are null for protein/function. (G) RT-PCR for hunchback and RP49 mRNA levels from stage 4 embryos after injection with buffer (−) or hunchback dsRNA (+). The number of embryos is n = 18, n = 16, respectively.
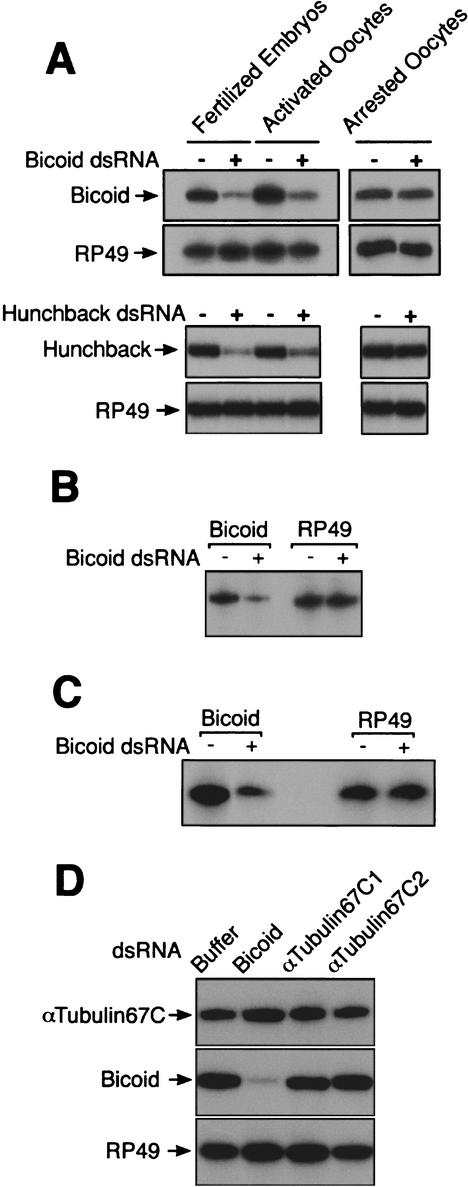
We determined if and when transcripts become sensitive to dsRNAs during oogenesis. We injected dsRNA into staged oocytes and examined their consequent levels of bicoid and hunchback mRNAs. Although we were technically unable to successfully microinject oocytes earlier than stage 14 (data not shown), we were able to examine stage 14 oocytes for RNAi activity (Fig. 2A). Levels of bicoid and hunchback mRNAs were unchanged in stage 14 oocytes after injection of dsRNA, indicating that oocytes at this stage are unable to carry out RNAi.
Figure 2.
Germ-line RNAi is established during oocyte maturation. Levels of bicoid, hunchback, αTubulin67C, and RP49 mRNA as measured by RT-PCR. (A) Fully grown oocytes arrested at stage 14, activated mature oocytes from virgin females, and fertilized eggs were mock-injected (−) or injected with dsRNA (+). Total RNA was extracted 50–80 min after injection and subjected to RT-PCR. Top panels show Bicoid and RP49 levels after injection with bicoid dsRNA. Lower panels show Hunchback and RP49 levels after injection with hunchback dsRNA. The number of eggs per treatment ranged between 8 and 18. (B) Immature oocytes arrested at stage 14 were activated in vitro for 90 min and were either mock-injected (−) or injected with bicoid dsRNA (+). Levels of bicoid and RP49 mRNAs were measured 60 min after injection. The number of oocytes per treatment ranged between 8 and 9. (C) Immature oocytes arrested at stage 14 were activated in vitro in the presence of 100 μg/mL cycloheximide and were either buffer-injected (−) or injected with bicoid dsRNA (+). Levels of bicoid and RP49 mRNAs were measured 90 min after injection. The number of oocytes per treatment ranged between 11 and 13. (D) Activated mature oocytes were injected with buffer, bicoid dsRNA, αTubulin67C1 dsRNA, or αTubulin67C2 dsRNA. Levels of RP49, bicoid, and αTubulin67C mRNAs were measured 90 min after injection. The number of treated oocytes per sample ranged between 18 and 22.
Oocytes of most animals arrest at species-specific stages of meiosis while differentiation of the oocytes occurs. Drosophila oocytes arrest transiently in prophase I while the oocytes are loaded with RNAs and proteins (van Eeden and St Johnston 1999). Some of these molecules are differentially localized within the oocyte, imparting positional information to be used for embryonic axis formation. When Drosophila oocytes reach stage 14, they undergo meiotic arrest once more, this time at metaphase I. These arrested oocytes remain translationally quiescent in the ovary, potentially for weeks (Mahowald et al. 1983). Arrest is relieved as in most animal eggs by the process of maturation or activation that precedes fertilization. In the case of Drosophila, it appears that ovulation triggers activation of the oocyte to resume meiosis (Heifetz et al. 2001). When oocytes are activated, meiosis is completed and translation of maternal RNAs is dramatically elevated (Mahowald et al. 1983; Driever and Nusslein-Volhard 1988; Page and Orr-Weaver 1997). Shortly thereafter, the oocyte is fertilized as it passes into the uterus.
RNAi-like effects were not detected in arrested stage 14 oocytes injected with dsRNA. Was this a general feature of the female germ line? To explore this issue, we injected dsRNA into mature activated oocytes. Injection of dsRNA caused reduction in bicoid and hunchback mRNA levels comparable to those seen in embryos (Fig. 2A). To confirm that mRNA sensitivity to dsRNA was strictly coincident with oocyte maturation, we isolated arrested stage 14 oocytes from dissected ovaries and activated the oocytes in vitro. This maturation procedure reactivates meiosis, mRNA translation, and vitelline membrane cross-linking (Page and Orr-Weaver 1997). After maturation, oocytes were injected with bicoid dsRNA and assayed for bicoid mRNA levels. These oocytes showed a decrease in bicoid mRNA (Fig. 2B). Thus, immature Drosophila oocytes that are coordinately blocked for meiosis and translation are resistant to RNAi, and the block to these processes can be released by maturation or activation of oocytes.
There are several possible ways in which RNAi could be blocked in arrested oocytes. One possibility is that an essential component of the RNAi machinery might be missing at this stage. Oocyte maturation would then involve synthesis of the component. To address if synthesis of a missing component is responsible, we activated oocytes in the presence of the protein synthesis inhibitor cycloheximide. Arrested stage 14 oocytes were preincubated with cycloheximide and then activated in vitro in the presence of cycloheximide. This treatment inhibits >95% of the protein synthesis that occurs during maturation (Page and Orr-Weaver 1997). These oocytes were injected with bicoid dsRNA and, strikingly, they showed a decrease in bicoid mRNA levels that was comparable to that of normal mature oocytes (Fig. 2C). RNAi was established during oocyte maturation even when protein synthesis was blocked. Thus, RNAi establishment during oocyte maturation does not likely occur by synthesis of an essential protein component of the RNAi machinery.
The stage 14 oocyte is coordinately blocked in both translation and RNAi. The two processes are released near simultaneously from this block, suggesting perhaps that a shared mechanism links their regulation. To test this possibility, we examined the effectiveness of dsRNA against a transcript that is present but not translated after oocyte maturation. The αTubulin67C gene encodes one of three α-tubulin proteins synthesized during oogenesis and embryogenesis (Kalfayan and Wensink 1982). Transcript accumulates and is actively translated in early immature oocytes (Matthews et al. 1989). However, after oocyte maturation, no translation of αTubulin67C mRNA occurs, even though transcripts at this stage are associated with ribosomes and are competent to drive translation in vitro (Kalfayan and Wensink 1982; Matthews et al. 1989). The stable pool of αTubulin67C mRNA is comparable to levels of bicoid and hunchback mRNA in mature oocytes (Fig. 2D). When two nonoverlapping dsRNAs against αTubulin67C transcript were independently injected into mature activated oocytes, no destruction of mRNA was detected (Fig. 2D). This suggests that the ability of dsRNAs to destroy transcripts during oogenesis is coupled to the translation activity of the transcript. Successful translation of transcripts is possibly necessary to link a transcript to dsRNA-triggered degradation.
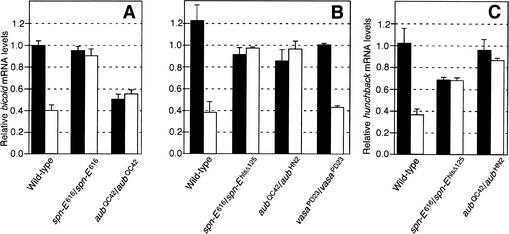
Several Drosophila genes have been identified that affect translation of maternal mRNAs during oogenesis (van Eeden and St Johnston 1999). One of these genes, aubergine (aub), encodes a protein with a PIWI and PAZ domain (Harris and Macdonald 2001). To determine whether Aub has any role for RNAi in oocytes, we examined the effect of aub mutations on RNAi activity. We injected bicoid and hunchback dsRNAs into aub mutant oocytes that were activated in vitro (Fig. 3A). Degradation of bicoid and hunchback mRNAs was not observed in aub mutants, indicating that Aub is necessary for germ-line RNAi. Two independent aub alleles in heteroallelic combination produced the same result, indicating that the effect was not due to the influence of linked modifiers (Fig. 3B,C).
Figure 3.
Impaired RNAi in aub and spn-E mutant oocytes. bicoid mRNA levels were measured by RT-PCR and were normalized to corresponding RP49 mRNA levels for each sample. (A) bicoid dsRNA was injected into oocytes after in vitro activation (open bars). Control oocytes were mock-injected (filled bars). RNA was extracted 100 min after injection. The strains used were wild-type, spn-E616/spn-E616, and aubQC42/aubQC42. The error bars show standard error values. (B) bicoid dsRNA was injected into mature activated oocytes that had been collected from virgin females (open bars). Control oocytes were mock-injected. RNA was extracted 60 min after injection. The strains were wild-type, spn-E616/spn-Ehls-Δ125, aubQC42/aubHN2, and vasaPD23/vasaPD23. The error bars show standard error values. (C) hunchback dsRNA was injected into mature activated oocytes that had been collected from virgin females (open bars). Control oocytes were mock-injected. RNA was extracted 80 min after injection. The strains were wild-type, spn-E616/spn-Ehls-Δ125, and aubQC42/aubHN2. The error bars show standard error values.
The aub gene is a member of a family of genes implicated in RNAi and PTGS. Indeed, aub has been implicated in PTGS regulation of the Stellate repeats and Su(Ste) genes on X and Y chromosomes (Aravin et al. 2001). Another member of the family, piwi, has been implicated in PTGS within somatic cells (Pal-Bhadra et al. 2002). A third family member, Ago2, is a subunit of the mRNA-cleaving complex that mediates RNAi in Drosophila embryonic cells (Hammond et al. 2001). Thus, several members of this gene family in Drosophila have been implicated in RNAi and PTGS at various steps.
It was of interest to determine if other translational regulatory genes are involved in RNAi. To test this possibility, we examined two genes that possibly act through interactions with RNA. vasa and spindle-E (spn-E) encode DexH-box RNA helicases (Lasko and Ashburner 1988; Gillespie and Berg 1995). When activated spn-E mutant oocytes were injected with bicoid or hunchback dsRNAs, no reduction in cognate mRNA levels occurred (Fig. 3A–C). In contrast, activated vasa mutant oocytes injected with bicoid dsRNA were found to show transcript degradation comparable to wild type (Fig. 3B). We conclude that activation of RNAi in oocytes is dependent on the activity of Spn-E but not Vasa.
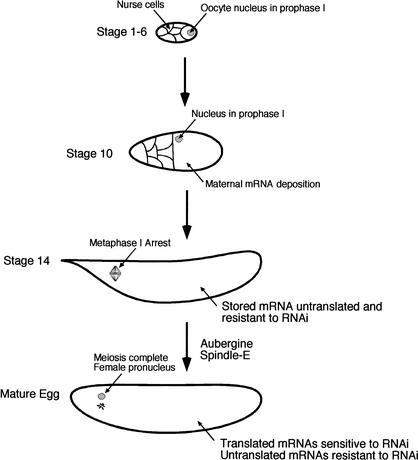
Arrested Drosophila oocytes are unable to generate RNAi silencing of endogenous maternal mRNAs, but selectively establish this capability upon egg maturation (Fig. 4). How is RNAi activated by egg maturation? We would argue that it is linked in some way to translation of maternal mRNAs, which is also specifically activated by egg maturation. We have shown that establishment of RNAi is probably not caused by translation of a missing RNAi component. Rather, the complete RNAi apparatus may be present and poised for action but is unable to target homologous substrate mRNAs until egg maturation. Translational masking of mRNAs, a mechanism that operates on maternal Drosophila gene expression, may conceivably be one way in which mRNA is blocked from RNAi attack. Alternatively, targeting of mRNA might require transcripts be assembled onto active polysomes. This may be the case, because siRNA-containing RISC complexes physically fractionate with polysomes (Hammond et al. 2001), and siRNAs associate with polysomes in Trypanosoma brucei (E. Ullu, pers. comm.). There is no evidence to indicate that dsRNA-targeting requires ribosome translocation on transcripts, because we find that cycloheximide inhibition of ribosome translocation does not block RNAi activity in activated mature oocytes.
Figure 4.
Model of oocyte development and onset of RNAi competence. Early in oogenesis, the oocyte chromosomes condense into a compact karyosome while arrested at prophase I. Later, regulated mRNA deposition and translation set up axial polarity within the oocyte while it is still arrested at prophase I. Aub and Spn-E, among other genes, coordinate these processes. By stage 14, the oocyte nucleus arrests in metaphase I, mRNA translation is blocked, and the oocyte is not competent to degrade mRNA by RNAi. This quiescent stage is maintained until ovulation signals the oocyte to undergo maturation. Meiosis is rapidly completed, forming a female pronucleus and three other coalesced meiotic producers (rosette). Select oocyte mRNAs such as bicoid and hunchback begin to be translated, and these transcripts become competent to be degraded by an RNAi mechanism. Transcripts that are not translated such as αTubulin67C are still not sensitive to degradation. Aub and Spn-E are required for the activation of the RNAi mechanism, possibly by activating translation.
Coupling RNAi to translated mRNA might facilitate base-pairing interactions between siRNAs and an unfolded mRNA target, or it might simply be a means to mark RNAs to be scanned for destruction. The key evidence suggesting that transcript translation is linked to transcript degradation by RNAi comes from experiments in which we tested dsRNA against the αTubulin67C message. We find that dsRNA is ineffective against the untranslated αTubulin67C transcript in mature activated oocytes, which are nevertheless competent to carry out RNAi against translated bicoid and hunchback transcripts. Thus, there is a correlation between the ability of a transcript to be translated and its ability to be destroyed by dsRNA.
Aub and Spn-E are required for RNAi in Drosophila oocytes (Fig. 4). They also regulate several features of germ-cell development, including translation of certain maternal mRNAs. Germ-line and stem-cell functions have been reported for orthologs of piwi and ago1 in a variety of species (Bohmert et al. 1998; Cox et al. 1998; Kataoka et al. 2001). The ego-1 gene of C. elegans is required for both germ-line development and germ-line PTGS (Smardon et al. 2000). Finally, mutations in dcr-1, the C. elegans gene encoding Dicer, disrupt oogenesis in an unspecified manner (Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001). The developmental defects associated with mutations in these genes, including aub and spn-E, might reflect a loss of gene silencing important for oocyte development. Thus, Aub and Spn-E might play a specific role in gene silencing mechanisms, including RNAi, that nevertheless have a widespread impact on many features of development. Alternatively, Aub and Spn-E could be required for RNAi because they activate translation of germ-line transcripts including those for bicoid and hunchback. Although there is no evidence for translational control of bicoid mRNA in aub mutants (Wilson et al. 1996), these mutants may perturb steps in the translation of transcripts that are essential for triggering RNAi. Future experiments should define the specific roles for Aub and Spn-E in dsRNA-mediated destruction and its relationship to translation control.
Materials and methods
Strains
The following strains have been described: bicoidE1, aubQC42, aubHN2, spn-E616, spn-Ehls-Δ125, and vasaPD23 (Lasko and Ashburner 1988; Gillespie and Berg 1995; Harris and Macdonald 2001). Homozygous or trans-heterozygous mutant females were used to generate mutant eggs.
Synthesis and introduction of dsRNA into Drosophila
bicoid RNA from positions 706 to 1405 (GenBank accession no. X14459), hunchback RNA from positions 9176 to 9920 (GenBank accession no. U17742), αTubulin67C1 RNA from positions 995 to 1491 (GenBank accession no. M14646), and αTubulin67C2 RNA from positions 1530 to 2045 (GenBank accession no. M14646) were transcribed in vitro on both strands simultaneously using PCR fragments as templates (Kennerdell and Carthew 1998). The complementary RNA products annealed during the transcription reactions to form dsRNAs, as assayed by agarose gel electrophoresis. The transcribed regions did not include sequences coding for conserved protein domains. Microinjection of dsRNA into fertilized eggs at embryonic stage 2 was as described (Kennerdell and Carthew 1998). Injection of 3 × 10−3 to 3 × 10−2 fmole of dsRNA generated loss-of-function phenotypes at a uniform frequency, indicating that even with 3 × 10−3 fmole, the dsRNA was saturating the RNAi machinery. Injection of 3 × 10−2 fmole was routinely performed. The site of bicoid dsRNA injection was the anterior pole, where bicoid mRNA is localized. Buffer-only injection into the anterior pole had no effect on generating bicoid-like phenotypes. hunchback and αTubulin67C dsRNAs were injected midventrally. To deliver dsRNA into embryos by particle bombardment, a Helios Gene Gun System (Bio-Rad) was used. Preparation of dsRNA-coated gold particles was as described in the manufacturer's instructions for DNA. RNA dissolved in TE buffer was precipitated in the presence of 1-μm diameter gold particles and 20 mM spermidine by the addition of CaCl2 to a final concentration of 400 mM. The ratio of dsRNA to gold was 2 μg RNA per milligram of gold particles. After dehydration and coating with 0.05 mg/mL PVP in ethanol, 0.5 mg of gold was loaded into each cartridge. Embryos collected 0–10 min AEL were dechorionated, adhered to tape-glued coverslips, and mildly desiccated 5 min in open air before they were shot with a single cartridge from the gun positioned 2.5 cm above the coverslip at a helium pressure of 300–450 psi. Treated embryos were immediately covered with halocarbon oil. Approximately two-thirds of embryos that were shot with only gold survived to hatching.
Oocyte manipulations
Unfertilized oocytes that had undergone maturation (activation) were collected from virgin females, dechorionated, and injected with dsRNA in the same manner as fertilized eggs. To collect stage 14 oocytes, ovaries were dissected from adult females in Schneider's medium. Oocytes were lined up against a slab of 5% agarose in PBS on a glass slide. The Schneider's medium bathing the oocytes was removed and immediately replaced with halocarbon oil that had been saturated with water to prevent dehydration. Stage 14 oocytes were activated 20 to 90 min in vitro as described (Page and Orr-Weaver 1997). The extent of activation was assayed by treating oocytes with 50% bleach. Activated oocytes were transferred to tape-glued coverslips, desiccated, and covered with halocarbon oil. Oocytes were injected with 0.13–0.21 fmole of dsRNA, which was distributed uniformly throughout the cytoplasm. Injected oocytes were harvested after incubation at 25°C for 1-2 h.
Histology, immunohistochemistry, and RT-PCR analysis
Cuticle analysis of RNAi-treated embryos was as described (Kennerdell and Carthew 1998). To isolate total RNA for RT-PCR, eggs or embryos were washed from tape-glued coverslips with heptane and lysed in Trizol (GIBCO-BRL) at a concentration of 1 egg/embryo per 5 μL of Trizol. After RNA was dissolved in water, quantitative RT-PCR was performed as described (Kennerdell and Carthew 2000) using published primers to amplify RP49 mRNA, two 18-base primers at positions 485 and 636 (GenBank accession no. X14459) to amplify bicoid mRNA, two 18-base primers at positions 8181 and 8323 (GenBank accession no. U17742) to amplify hunchback mRNA, and 16- and 17-base primers at positions 855 and 960 (GenBank accession no. M14646) to amplify αTubulin67C mRNA. Each 10-μL reaction contained 1 egg/embryo equivalent of cDNA. RP49 reactions were cycled 15 times, Bicoid and Hunchback reactions were cycled 19 times, and αTubulin67C reactions were cycled 20 times at a 55°C annealing temperature. These cycle numbers were chosen because they were in the middle of the geometric phases of PCR for each particular RT product. PCR product levels were quantitated with a Fuji phosphoimager.
Acknowledgments
We thank C. Berg, P. MacDonald, T. Schupbach, G. Struhl, and the Bloomington Stock Center for fly strains; G. Struhl for DNA clones; and G. Struhl and H. Lipshitz for helpful advice. This research was supported by the Pew Charitable Trust while the authors were at the University of Pittsburgh.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL r-carthew@northwestern.edu; FAX (847) 467-1380.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.990802.
References
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW. Gene silencing by double-stranded RNA. Curr Opin Cell Biol. 2001;13:244–248. doi: 10.1016/s0955-0674(00)00204-0. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes & Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Heifetz Y, Yu J, Wolfner MF. Ovulation triggers activation of Drosophila oocytes. Dev Biol. 2001;234:416–424. doi: 10.1006/dbio.2001.0246. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Kalfayan L, Wensink PC. Developmental regulation of Drosophila α-tubulin genes. Cell. 1982;29:91–98. doi: 10.1016/0092-8674(82)90093-9. [DOI] [PubMed] [Google Scholar]
- Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- ————— Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Mahowald AP, Goralski TJ, Caulton JH. In vitro activation of Drosophila eggs. Dev Biol. 1983;98:437–445. doi: 10.1016/0012-1606(83)90373-1. [DOI] [PubMed] [Google Scholar]
- Matthews KA, Miller DF, Kaufman TC. Developmental distribution of RNA and protein products of the Drosophila α-tubulin gene family. Dev Biol. 1989;132:45–61. doi: 10.1016/0012-1606(89)90203-0. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Page AW, Orr-Weaver TL. Activation of the meiotic divisions in Drosophila oocytes. Dev Biol. 1997;183:195–207. doi: 10.1006/dbio.1997.8506. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ- line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- St Johnston D, Driever W, Berleth T, Richstein S, Nusslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development. 1989;107:13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332:281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- van Eeden F, St Johnston D. The polarisation of the anterior–posterior and dorsal–ventral axes during Drosophila oogenesis. Curr Opin Genet Dev. 1999;9:396–404. doi: 10.1016/S0959-437X(99)80060-4. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Connell JE, Macdonald PM. aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122:1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]