Abstract
In addition to creating the DNA double strand breaks that initiate V(D)J recombination, the RAG proteins are thought to play a critical role in the joining phase of the reaction. One such role, suggested by in vitro studies, might be to ensure the structural integrity of postcleavage complexes, but the significance of such a function in vivo is unknown. We have identified RAG1 mutants that are proficient in DNA cleavage but defective in their ability to interact with coding ends after cleavage and in the capture of target DNA for transposition. As a result, these mutants exhibit severe defects in hybrid joint formation, hairpin coding end opening, and transposition in vitro, and in V(D)J recombination in vivo. Our results suggest that the RAG proteins have an architectural function in facilitating proper and efficient V(D)J joining, and a protective function in preventing degradation of broken ends prior to joining.
Keywords: RAG1, RAG2, V(D)J recombination, transposition, DNA double strand break
The variable portion of the genes encoding immunoglobulins and T cell receptors are assembled from component V, D, and J DNA segments by a site-specific recombination reaction termed V(D)J recombination (Tonegawa 1983). V(D)J recombination is targeted to specific sites on the chromosome by recombination signal sequences (RSSs) that flank antigen receptor gene segments. The RSS consists of a conserved heptamer (consensus, 5′-CACAGTG-3′) and nonamer (consensus, 5′-ACAAAAACC-3′) separated by a spacer of either 12 or 23 bp. Efficient recombination occurs between a 12-RSS and a 23-RSS, a restriction known as the 12/23 rule (for review, see Lewis 1994).
Conceptually, V(D)J recombination can be divided into two phases, DNA cleavage and DNA joining. DNA cleavage requires two lymphocyte-specific factors, the products of the recombination activating genes, RAG1 and RAG2 (Schatz et al. 1989; Oettinger et al. 1990), which together recognize the RSSs and create double strand breaks at the RSS-coding segment junctions (for review, see Fugmann et al. 2000a). RAG-mediated DNA cleavage occurs in a synaptic complex termed the paired complex, which is constituted from two distinct RSS-RAG complexes, a 12-SC and a 23-SC (where SC stands for signal complex) (Hiom and Gellert 1998; Mundy et al. 2002). The DNA cleavage reaction involves two distinct enzymatic steps, initial nicking that creates a 3′-OH between a coding segment and its RSS, followed by hairpin formation in which the newly created 3′-OH attacks a phosphodiester bond on the opposite DNA strand. This generates a blunt, 5′ phosphorylated signal end containing all of the RSS elements, and a covalently sealed hairpin coding end (McBlane et al. 1995).
The second phase of V(D)J recombination, in which broken DNA fragments are processed and joined, is less well characterized. Signal ends are typically joined precisely to form a signal joint, whereas joining of the coding ends requires the hairpin structure to be opened and typically involves nucleotide addition and deletion before formation of the coding joint. The factors involved in these processes include ubiquitously expressed proteins involved in the repair of DNA double strand breaks by nonhomologous end joining, terminal deoxynucleotidyl transferase, and the recently discovered Artemis protein (Grawunder and Harfst 2001; Moshous et al. 2001).
In addition to their critical roles in RSS recognition and DNA cleavage, the RAG proteins have been proposed to perform two distinct types of functions in the postcleavage phase of V(D)J recombination (for review, see Fugmann et al. 2000a; Sadofsky 2001). An architectural, or structural, function has been inferred from the finding that, after DNA cleavage in vitro, the DNA ends remain associated with the RAG proteins in a “four end” complex known as the cleaved signal complex (Hiom and Gellert 1998). Further support for this idea comes from the finding that stable association of the RAG proteins with coding and signal ends can be detected in mobility shift experiments (Bailin et al. 1999) and from the observation that a C-terminal region of RAG1 is closely associated with coding DNA (Mo et al. 2001). In addition, after release of the coding ends in vitro, and after coding joint formation in vivo (Perkins et al. 2002), the RAG proteins remain in a stable signal end complex (SEC) containing the two signal ends (Agrawal and Schatz 1997; Hiom and Gellert 1998). These postcleavage complexes may serve as essential scaffolds for the second phase of the reaction, with the RAG proteins acting to organize the DNA processing and joining events (Sadofsky 2001). This model is consistent with the finding that removing the RAG proteins prior to DNA joining drastically reduces coding joint formation in vitro (Leu et al. 1997; Ramsden et al. 1997).
The second type of RAG protein-mediated postcleavage activity is the catalysis of phosphodiester bond hydrolysis and strand transfer reactions. In the context of the postcleavage complex, the RAG proteins are capable of opening hairpin coding ends in vitro (Besmer et al. 1998; Shockett and Schatz 1999). The RAG proteins also show 3′ flap endonuclease activity that may contribute to coding end processing/joining (Santagata et al. 1999). Furthermore, the RAG proteins can utilize the 3′ OH group on the signal ends to attack hairpin coding ends (forming hybrid or open/shut joints) or virtually any DNA duplex (forming a transposition product) (Agrawal et al. 1998; Hiom et al. 1998; Melek et al. 1998).
Several RAG1 and RAG2 mutants that are active in DNA cleavage but impaired for V(D)J recombination have recently been identified, and two of these exhibited a defect in opening an artificial hairpin in Mn2+. These findings provide direct evidence of a role for the RAG proteins in the joining phase of V(D)J recombination, and suggest that their hairpin opening activity may be important in this role (Qiu et al. 2001; Schultz et al. 2001). In contrast, there is no direct evidence to indicate that the postcleavage architectural functions of the RAG proteins play an important role in V(D)J recombination, in large part because mutants defective specifically in these functions have not been identified.
In the course of characterizing a putative RAG1 GTP binding domain (C.-L. Tsai and D. Schatz, in prep.), we isolated RAG1 mutants (substitution of serine 723 with alanine or cysteine, S723A and S723C, respectively) that exhibit a phenotype similar to previously identified “SOS” mutants of IS10 transposase (Haniford et al. 1989). Like many of these IS10 mutants, the S723A and S723C RAG1 mutants are proficient in DNA cleavage but severely defective in transposition, with the defect in transposition due to a failure to form the noncovalent target capture complex. Strikingly, the RAG1 mutations also result in defects in the stability of the postcleavage complex, in the postcleavage activities of the RAG proteins in vitro, and in V(D)J recombination in vivo. The identification of such mutants provides strong evidence of the importance of a structural role for the RAG proteins in stabilizing postcleavage complexes in the joining phase of V(D)J recombination. Such RAG mutations might result in an elevated frequency of chromosomal translocations involving antigen receptor loci, raising the possibility that somatic mutation of the RAG proteins could contribute to the development of lymphoid malignancies.
Results
Isolation of serine 723 mutants
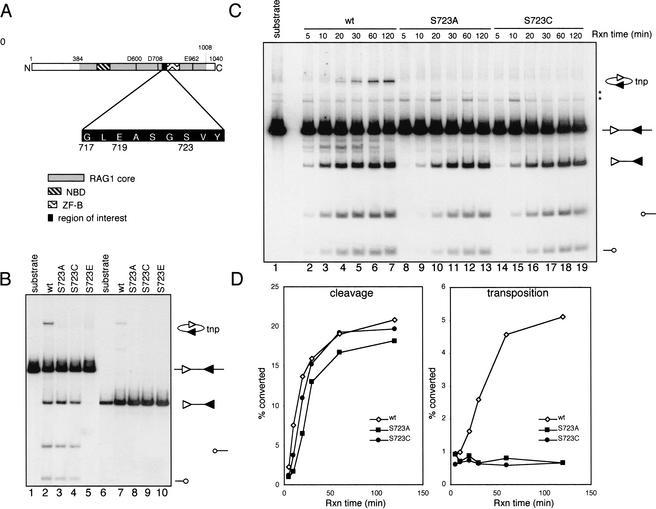
Serine 723 was chosen for mutagenesis because it resides within a putative GTP binding domain of murine RAG1 (C.-L. Tsai and D. Schatz, in prep.). It is located C-terminal to a catalytic aspartic acid residue at position 708 and at the beginning of a zinc finger domain of RAG1 (aa 723–754; Rodgers et al. 1996) thought to interact with RAG2 (Fig. 1A, Aidinis et al. 2000). Highly purified, dimeric core RAG1 protein (murine aa 384–1008), fused at its N-terminus to maltose binding protein (MBP), was prepared from bacteria, and partially purified core RAG2 protein (murine aa 1–383) was prepared from the murine cell line F2A1 (Eastman et al. 1999) (see Materials and Methods). RAG1 proteins were assayed with RAG2 and HMG2 for their cleavage and transposition activity by a standard 12/23 coupled cleavage reaction (Fig. 1B). DNA cleavage results in production of a central fragment containing two signal ends (a signal end fragment) and two coding ends. RAG-mediated intramolecular transposition can also be visualized in this assay because the inversion circle transposition products migrate to a distinct position in the gel, above that of the input substrate (Fig. 1B,C; Agrawal et al. 1998).
Figure 1.
Coupled cleavage and intramolecular transposition. (A) Schematic diagram of the murine core RAG1 protein depicting three active site residues (D600, D708, and E962), the nonamer binding domain (NBD), the ZFB zinc finger domain, and the amino acid sequence surrounding S723. (B) Cleavage and transposition activities were assessed using a body-labeled cleavage substrate containing a 12-RSS and a 23-RSS (lanes 1–5) or a precleaved signal end fragment (lanes 6–10). Structures of the substrates and reaction products are indicated at the right, with the 12-RSS and 23-RSS depicted as open and filled triangles, respectively, and coding ends as open circles. The MBP-RAG1 protein used is indicated above each lane. tnp, intramolecular transposition product. (C) Kinetic analysis of coupled cleavage. The asterisks indicate background bands present in the substrate. (D) Quantitation of cleavage and transposition activities in (C). The cleavage activity was calculated as the ratio of the intensity of the signal end fragment (including both the cleavage product and transposition product) to the total radioactivity in each reaction. Transposition activity was calculated as the ratio of the intensity of the transposition product to the sum of the signal end fragment and the transposition product.
RAG1 proteins bearing mutations at serine 723 exhibited two distinct phenotypes. Substitution with alanine or cysteine resulted in a severe defect in transposition, while leaving DNA cleavage activity comparable to that of wild-type RAG1 (Fig. 1B, lanes 3,4). In contrast, replacement of the same residue by glutamic acid (S723E) completely abolished DNA cleavage activity (lane 5). All three mutant RAG1 proteins also exhibited a transposition-deficient phenotype in an assay that bypasses DNA cleavage by using a PCR-generated precleaved signal end fragment as substrate (Fig. 1B, cf. lane 7 and lanes 8–10).
Kinetics of coupled cleavage
The cleavage defect of S723E raised the possibility that S723A and S723C might also have a subtle defect in DNA cleavage. We performed a kinetic analysis of the coupled cleavage reaction in an attempt to detect such a defect. Cleavage products generated by wild-type RAG1 were readily detectable after 5 min and accumulated rapidly for almost 1 h (Figs. 1C,D, left panel). S723A and S723C exhibited approximately a twofold reduction in the initial rate of cleavage, but generated similar levels of cleavage products as wild-type RAG1 at later time points of the reaction. No transposition products were detected with the mutants at any time point (Figs. 1C,D, right panel). Together with the fact that S723E is completely defective in cleavage, these results indicate that serine 723 plays a role in DNA cleavage. In light of this, we also measured cleavage activity on oligonucleotide substrates. S723A and S723C exhibited nicking and hairpin formation activities equal to that of wild-type RAG1 with substrates containing high efficiency coding flanks, but were less active than wild-type RAG1 with substrates in which the first two coding flank nucleotides 5′ of the heptamer were TT (resembling previously described low-efficiency coding flanks; data not shown; Ezekiel et al. 1997; Yu and Lieber 1999). The substrates used for the in vitro and in vivo experiments reported here did not contain such low-efficiency coding flanks, and could be cleaved efficiently by S723A and S723C.
Precleavage complexes
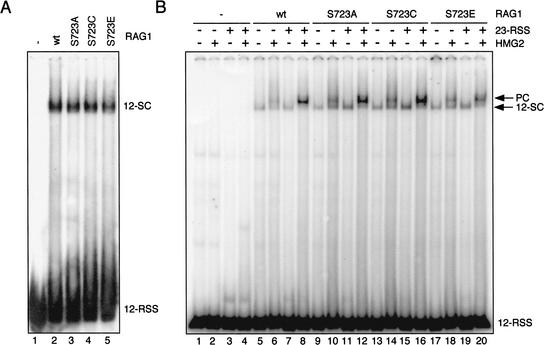
The RAG proteins form specific protein–DNA complexes with RSSs prior to DNA cleavage, and a defect in DNA binding might explain the cleavage defect of the S723 mutant RAG1 proteins. To address this, the ability of mutant RAG1 proteins to form various precleavage complexes was assessed using electrophoretic mobility shift assays (EMSAs) with Ca++ as the sole divalent cation (Ca++ supports DNA binding but not cleavage by the RAG proteins). Incubation of labeled 12-RSS substrate with wild-type MBP-core RAG1, GST-core RAG2, and HMG2 resulted in the formation of a specific RSS-RAG complex (the 12-SC; Fig. 2A, lane 2) (Hiom and Gellert 1997). Reactions using S723A, S723C, or S723E mutant proteins also generated a complex of the same mobility although with somewhat reduced efficiency (Fig. 2A, lanes 3–5), indicating that the mutant RAG1 proteins were able to interact with RAG2 and the 12-RSS with the same stoichiometry as wild-type RAG1. In addition, in the presence of an unlabeled 23-RSS substrate, S723A and S723C were able to form what is thought to be the paired complex, the immediate precursor of 12/23 coupled cleavage, at least as efficiently as wild-type RAG1, whereas S723E showed only mildly reduced activity in this assay (Fig. 2B). Hence, S723A and S723C have essentially normal DNA binding properties, whereas S723E shows reduced binding and no cleavage activity, indicative of a defect in multiple steps in the process leading to double strand breaks (C.-L. Tsai and D. Schatz, in prep.).
Figure 2.
Formation of the 12-SC and the paired complex. (A) EMSA for detection of the 12-SC was performed using MBP-RAG1, GST-RAG2, and HMG2 proteins in 5 mM Ca++ with a 5′ end-labeled double stranded 12-RSS oligonucleotide substrate. Complexes were resolved on a native 4% polyacrylamide gel. (B) The assay for paired complex formation was performed with labeled 12-RSS in a buffer containing Ca++ in the presence or absence of unlabeled 23-RSS and HMG2 as indicated. Complexes were analyzed on a native 6% (80:1) polyacrylamide gel. The positions of the bands corresponding to the 12-SC as well as the paired complex are indicated at the right.
The signal end complex, target capture complex, and intermolecular transposition
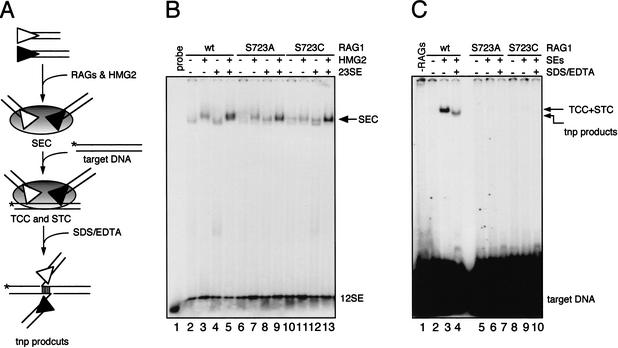
After DNA cleavage, the resulting four DNA ends are retained in a postcleavage complex termed the cleaved signal complex (Hiom and Gellert 1998). Release of the coding ends from this complex results in the RAG proteins bound to the two signal ends in a signal end complex (SEC) (Agrawal and Schatz 1997; Hiom and Gellert 1998). In vitro, the SEC can capture target DNA to form a target capture complex (TCC), within which the RAG proteins can catalyze the integration of signal ends into target DNA (Agrawal et al. 1998; Hiom et al. 1998). The protein–DNA complex containing signal ends covalently linked to target DNA is termed the strand transfer complex (STC). The transposition defect of S723A and S723C could result from destabilization of the SEC, a defect in target capture, or a failure to perform strand transfer. To examine these possibilities, we tested the ability of these mutants to form the complexes leading to transposition. Incubating an equimolar mixture of a labeled 12-signal end substrate and an unlabeled 23-signal end substrate with wild-type MBP-RAG1, GST-RAG2, and HMG2 led to the generation of the SEC (Fig. 3B, lane 5). Consistent with previous results, formation of the SEC was stimulated by the presence of both types of signal ends and HMG2 (cf. lanes 2–4 and lane 5) (Hiom and Gellert 1998; Fugmann et al. 2000b). Reactions containing mutant RAG1 proteins yielded virtually identical results (lanes 6–13), indicating that they were able to interact with signal ends and proceed to SEC formation.
Figure 3.
Formation of the SEC, the TCC, and intermolecular transposition products. (A) Schematic diagram of the complexes leading to transposition. The SEC is formed from a pair of signal ends, whereas in the TCC and STC, the signal ends are associated noncovalently or covalently, respectively, with target DNA (which can be revealed by denaturation with SDS/EDTA). (B) SEC formation reactions were carried out using 5′ end-labeled 12-signal end oligonucleotide in a reaction buffer containing 4 mM Mg++ in the presence or absence of HMG2 and unlabeled 23-signal end as indicated above the lanes. Reactions were analyzed on a native 6% (80:1) polyacrylamide gel. (C) TCC formation and intermolecular transposition reactions were carried out in a two-stage fashion. First, the SEC was formed as in (B) except that both the 12-signal end and 23-signal end were unlabeled. Formation of the TCC and STC were initiated by adding 5′ end-labeled oligonucleotide target DNA. Deproteinized transposition products were visualized by treating the reaction with SDS/EDTA prior to loading on a native 6% (80:1) polyacrylamide gel (lanes 4,7,11). Treatment with higher concentrations of EDTA (25 mM) together with 0.5% SDS yielded identical results (data not shown). The transposition product band seen in lane 4 is temperature-labile (55°C, 5′), as predicted from the structure shown at the bottom of panel A.
To assess the ability of the SEC to perform target capture and strand transfer, we used a modification of the assay described by Roth and colleagues (Neiditch et al. 2001), and assembled the TCC and STC from unlabeled signal ends and a labeled target oligonucleotide (Fig. 3A). Incubation of the preformed SEC with target DNA resulted in the generation of a slowly migrating protein–DNA complex (Fig. 3C, lane 3). Formation of this complex was absolutely dependent on the presence of RAG1 and RAG2, signal ends, and HMG2 (data not shown). Since strand transfer can take place under the reaction conditions used and is the rate-limiting step of the transposition reaction (Neiditch et al. 2001), this complex is a mixture of the TCC and the STC at the early time point (15 min) used in this experiment. To estimate the relative contribution of each to this complex, we treated the reaction with SDS and EDTA to disrupt DNA–protein interactions and to separate noncovalently linked target DNA from signal ends. This resulted in the generation of a faster migrating band containing only 40% of the label seen in the shifted complex in the absence of denaturants. Therefore, at least 40% of the signal seen in lane 3 derives from the STC (Fig. 3B, cf. lanes 3 and 4). To investigate this further, we purified the slowly migrating bands from large-scale reactions identical to those of lanes 3 and 4, and found that intact target DNA was more abundant in the sample that had not been exposed to SDS/EDTA (data not shown). We infer that a substantial fraction of the target DNA found in the slow mobility complex in lane 3 is present in a noncovalent TCC. In striking contrast, in reactions containing S723A and S723C, essentially no shifted complexes were detectable (Fig. 3B, lanes 6,9). These results demonstrate that S723A and S723C are defective in transposition due to a defect in formation of a stable TCC.
The stability of postcleavage complexes
Coding ends and target DNA interact with the RAG proteins primarily in a nonsequence-specific fashion, and both can be the target of nucleophilic attack by the 3′ hydroxyl groups of the signal ends. Thus, it is plausible that the RAG proteins utilize the same binding site to interact with the coding end and the target DNA, and that mutations affecting target capture might also impair the RAG protein-coding end interaction in postcleavage complexes. To measure the stability of these complexes, a biotin molecule was added to the 5′ end of the 12-RSS coding flank, allowing various postcleavage complexes to be purified by virtue of biotin-streptavidin bead interactions (Fig. 4A). The DNA substrate was bound to beads, unbound material was removed by washing, and the bound substrate was then subject to DNA cleavage. One-quarter of each reaction was removed prior to the purification procedure to allow assessment of the amount of cleavage performed by each protein (input; Fig. 4B, lanes 2–4). Beads were washed twice to remove nonspecifically associated DNA, and elution was performed with SDS/EDTA under conditions that disrupt postcleavage complexes while maintaining most of the biotin-streptavidin interactions. The stability of postcleavage complexes should be reflected by the amount of the signal end fragment or the nonbiotinylated coding end retained on the beads and eluted by SDS/EDTA.
Figure 4.
Stability of the postcleavage complex. (A) Schematic diagram of a biotin-labeled postcleavage complex bound to a streptavidin magnetic bead. (B) Large-scale coupled cleavage reactions were carried out with streptavidin bead-bound DNA substrate at 37°C for 1 h. Samples are indicated as: input, one-quarter of each reaction prior to purification; unbound, DNA in the reaction supernatant, not retained on the beads; wash, DNA in the supernatant of the second wash; elution, DNA eluted by treatment with SDS/EDTA; beads, DNA eluted by subsequent harsh treatment of the beads with proteinase K for 60 min at 55°C followed by phenol/chloroform extraction (this removes only ∼50% of the bound, biotinylated substrate from the beads; data not shown). wt, wild-type RAG1; SA, S723A RAG1; SC, S723C RAG1. Arrowheads indicate the signal end fragment resulting from coupled cleavage. Shown here is a representative gel from three independent experiments. (C) Quantitation of the stability of the postcleavage complex as measured by the ratio of intensity of the signal end fragment in the elution fraction to that in the input fraction (corrected for loading).
As expected, wild-type RAG1, S723A, and S723C performed equivalent amounts of cleavage (lanes 2–4), and the majority of the nonbiotinylated reaction products, as well as a small fraction of biotinylated DNA, were released from the beads during the 1-h cleavage reaction (lanes 5–7). In the second wash with reaction buffer, virtually no DNA was eluted from the beads (lanes 8–10). Two major nonbiotinylated DNA fragments were recovered upon elution with SDS/EDTA, the signal end fragment (arrowhead), and the product of single-site cleavage at the 12-RSS (lanes 11–13). In the reaction containing wild-type RAG1, approximately 36% of the signal end fragment in the reaction was eluted (Fig. 4B, lane 11, and 4C), whereas with S723A and S723C, the value was 11% and 13%, respectively. This indicates that these mutations of serine 723 destabilize association of the signal end fragment with the biotinylated coding end. Because steps leading up to elution are carried out in the cleavage buffer, some cleavage likely occurs during the washes and some weakly bound signal end fragments may not have had an opportunity to be released from the beads. Hence, the defect in postcleavage complex stability of the mutant RAG1 proteins may be greater than is suggested by these results. The presence of single site cleavage products in the elution fraction was not expected, and may arise from cleavage within alternative synaptic complexes (e.g., between two 12-RSSs) whose formation is favored by alignment of substrate molecules on the beads prior to cleavage. We conclude that S723 is likely involved in stabilizing the postcleavage association of signal ends with coding ends. This conclusion is further supported by the finding that while the SEC formed by wild-type RAG1 is able to capture hairpin target molecules (Lee et al. 2002), the SEC formed by S723A and S723C cannot (data not shown).
Postcleavage activities
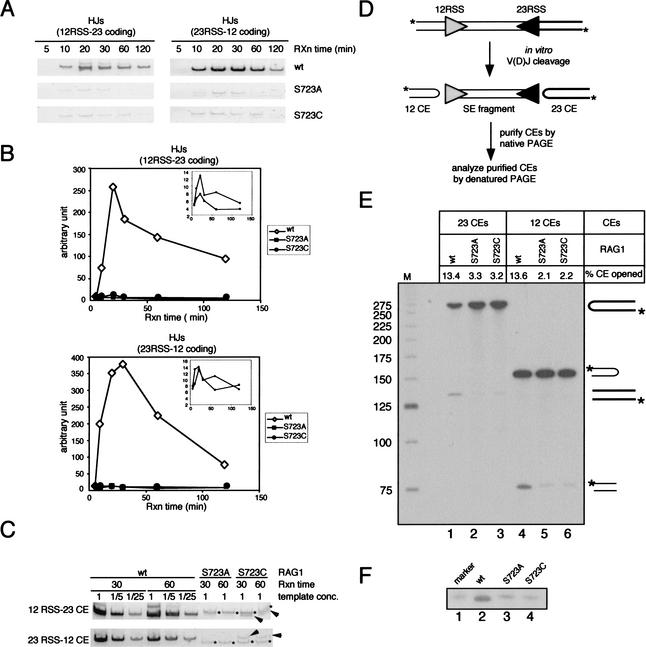
Formation of a stable postcleavage complex should be essential for postcleavage activities of the RAG proteins such as hybrid joint formation and hairpin opening, leading us to predict that S723A and S723C should be compromised for these activities. To measure hybrid joint formation, we used the products of 12/23-coupled cleavage reactions from Figure 1B as templates for PCR amplification of the two expected hybrid joint products. In the wild-type RAG1 reaction, hybrid joints are readily detectable after 10 min, accumulate for 20 min, and then decline (Fig. 5A,B). The decline at later time points may be due to the reverse reaction, DNA cleavage, followed by release of coding ends from the postcleavage complex. Hybrid joints were barely detectable at any time point of the reactions containing S723A or S723C (Fig. 5A), but the kinetics of hybrid joint formation by these mutants paralleled that of wild-type RAG1 (Fig. 5B, insets). We estimate that hybrid joint formation by the mutant RAG1 proteins is reduced by at least 25-fold compared to wild-type RAG1 (Fig. 5C). Mutation of Ser 723 also results in a severe defect in hybrid joint formation in vivo (data not shown), as assessed by PCR in a transient transfection recombination assay (see below).
Figure 5.
Postcleavage activities of the RAG proteins in vitro. (A) Hybrid joint formation. Products of the coupled cleavage reactions described in Fig. 1B were used as templates for PCR reactions to amplify hybrid joints. PCR products were resolved on 6% polyacrylamide gels and visualized by staining with cybergreen I. (B) Quantitation of hybrid joints in (A) by FluorImager (Molecular Dynamics). (C) Two time points (30 and 60 min) of the coupled cleavage reaction products were further analyzed by a semiquantitative PCR assay. The reactions containing the wild-type RAG1 protein were subjected to successive fivefold dilutions prior to PCR amplification, whereas the reactions containing the mutant RAG1 proteins were used as templates for PCR without dilution. Asterisks indicate nonspecific PCR products, and arrows indicate specific HJ products. (D) Schematic diagram showing the assay to detect hairpin opening by the RAG proteins in the context of coupled cleavage. Large-scale coupled cleavage reactions were carried out using a 5′ end-labeled PCR substrate, and the resulting coding ends were purified from a 4% native polyacrylamide gel. Purified coding ends were then analyzed on a 10% denaturing gel containing 40% (v/v) formamide, shown in (E). Unprocessed coding ends give rise to bands at 272 nt and 154 nt, whereas the processed coding ends yield products of ∼136 nt and 77 nt. The hairpin opening activity (% CE opened) is indicated above each lane and was determined by calculating the ratio of the processed coding end to the total coding end and multiplying by 100. (F) Purified 12-coding ends were further analyzed by comparing to a synthetic oligonucleotide identical in length and sequence to the top strand of the 12-coding end that has been nicked at the hairpin tip (lane 1). The processed coding ends in the wild-type RAG1 reaction migrated slightly faster than the marker and the processed coding ends from the reactions containing mutant RAG1 proteins (cf. lane 2 and lanes 1,3,4).
To measure hairpin opening activity, we synthesized a 5′ end-labeled cleavage substrate by PCR using labeled primers (Fig. 5D). Cleavage products from large-scale coupled cleavage reactions were separated on a native polyacrylamide gel, from which coding ends were excised, purified, and then subjected to denaturing PAGE. Unprocessed coding ends have a hairpin structure and migrate under denaturing conditions as 154-nt (12 coding end) or 272-nt (23 coding end) molecules, whereas the labeled strand resulting from nicking near the tip of the hairpins should be approximately 77 nt (12 coding end) or 136 nt (23 coding end) (Fig. 5E). About 13.5% of coding ends formed by wild-type RAG1 together with RAG2 were nicked (lanes 1,4), whereas only 2%–3% of coding ends formed in the reactions using mutant RAG1 proteins were nicked (lanes 2,3,5,6). We also noticed that the major product of hairpin opening with wild-type RAG1 migrated slightly faster than the products with S723A or S723C, and faster than a synthetic oligonucleotide representing the product of nicking at the tip of the hairpin (Fig. 5F, cf. lane 2 and lanes 1,3,4). This suggests that coding ends are opened 1 to 2 nt 5′ of the tip by wild-type RAG proteins, consistent with previous findings (Shockett and Schatz 1999). It also rules out the possibility that the coding ends purified from the wild-type reaction were substantially contaminated with the products of initial nicking adjacent to the 12-RSS. In summary, these results demonstrate that both hybrid joint formation and hairpin opening by the S723 mutant proteins are severely impaired.
In vivo V(D)J recombination
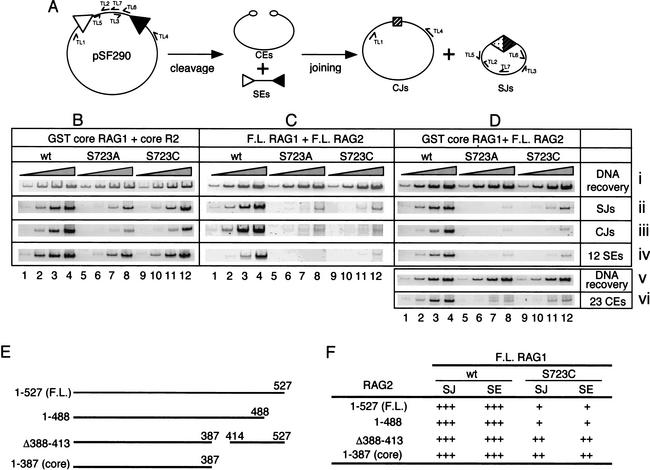
The S723A and S723C mutants perform DNA binding and cleavage relatively normally, but are defective in many postcleavage functions of the RAG proteins, in part as a result of a defect in the stability of postcleavage complexes. This unique phenotype allowed us to address the importance of RAG-mediated postcleavage complex stabilization for V(D)J recombination in vivo. We carried out a standard transient transfection assay in 293T cells to measure the recombination activity of S723A and S723C in vivo (Fig. 6A). V(D)J recombination products, signal joints (SJs) and coding joints (CJs), as well as intermediates (signal ends and coding ends), were analyzed by PCR and ligation-mediated PCR (LM-PCR), respectively, using samples harvested 48 h posttransfection. We first studied the combination of GST-core RAG1 and core RAG2, which best mimics the RAG proteins used in the in vitro analyses (Fig. 6B). S723A and S723C exhibited >10-fold and 3–5-fold reductions, respectively, in the formation of SJ and CJ products compared to wild-type RAG1 (Fig. 6B, panels ii and iii; serial 10-fold dilutions of the input substrate for PCR are shown throughout). Strikingly, the levels of signal end recombination intermediates generated by the mutant RAG1 proteins were reduced to the same extent (Fig. 6B, panel iv), despite the nearly normal cleavage activity of these mutants in vitro. We then performed the same assays using full-length, untagged RAG1 and RAG2 proteins. Interestingly, in this case the S723 mutants exhibited a much more severe impairment in the accumulation of recombination intermediates and products (about 100-fold; Fig. 6C). A similar 100-fold reduction in signal ends, SJs, and CJs was observed for the mutants using the combination of GST-core RAG1 and full-length RAG2 (Fig. 6D, panels ii, iii, and iv), indicating that the more severe phenotype is attributable to the full-length RAG2 protein. The S723 mutants exhibited an equally severe defect in signal end and SJ formation, relative to wild-type RAG1, when the activity of the full-length RAG proteins was assessed at 12 and 24 h posttransfection (data not shown), suggesting that intermediates do not accumulate transiently at an early time point in the assay.
Figure 6.
In vivo V(D)J recombination. (A) Schematic diagram of recombination of the pSF290 recombination substrate. Specific PCR primers used to amplify recombination intermediates and products are indicated. In vivo recombination assays were carried out with the combinations of (B) GST-core RAG1 and his-myc tagged core RAG2; (C) untagged full-length RAG1 and RAG2; and (D) GST-core RAG1 and untagged full-length RAG2. Each group of four lanes represents three successive 10-fold dilutions of the input PCR substrate, as indicated above the lanes by the shaded triangles. Panels i and v, DNA recovery (primers TL5 and TL6); panel ii, SJ detection (primers TL2 and TL3); panel iii, CJ detection (primers TL1 and TL4); panel iv, 12-signal end detection (LM-PCR with primers DR20 and TL7); panel vi, 12-coding end detection (LM-PCR with primers DR20 and TL4). SJ formation for all of the combinations of RAG proteins was assayed at least twice and representative results are shown. For the combination of GST-core RAG1 and full-length RAG2, signal end accumulation was measured in two independent experiments, and the deficit in CJ formation by the S723 mutant RAG1 proteins (for the combination of full-length RAG1 and RAG2) was confirmed by a standard bacterial transformation assay (Fugmann and Schatz 2001). (E) Schematic diagram of RAG2 proteins used in the in vivo recombination assays. (F) Summary of recombination activities of wild-type and S723C RAG1 (full-length) together with various RAG2 proteins. “+++” indicates activity comparable to that of full-length RAG1/2 (as in C, lanes 1–4). “++” and “+” indicate a 5–10-fold and a 30–100-fold reduction, respectively, of recombination activity relative to this.
Using the combination of GST-core RAG1 and full-length RAG2, we also measured the accumulation of coding ends. DNA samples were treated with mung bean nuclease to nick open hairpin coding ends, and were then subjected to a standard LM-PCR analysis. The S723A and S723C mutants exhibited a severe defect in coding end accumulation (Fig. 6D, panel vi), which paralleled that in signal end, SJ, and CJ accumulation.
To explore further the role of the C-terminal region of RAG2 in the recombination activity of S723 mutant RAG1 proteins, we measured the recombination activities (signal joint formation and accumulation of signal ends) of S723C in the context of two additional RAG2 mutants (depicted in Fig. 6E). Interestingly, S723C exhibited only two distinct phenotypes: a severe (30-fold to 100-fold) impairment of recombination activity in the context of full-length RAG2 or truncation mutant 1-488, and a mild (3-fold to 10-fold) impairment in the context of core RAG2 or deletion mutant Δ388-413 (data not shown; summarized in Fig. 6F).
Taken together, these results demonstrate that S723A and S723C are defective in V(D)J recombination and that a C-terminal region of RAG2 (aa 388–488) plays an important role in the recombination activity of S723 RAG1 mutants.
Mammalian cell-expressed S723A and S723C are active
The steady-state levels of signal and coding ends detected by LM-PCR reflect the rate at which the ends are produced (DNA cleavage by the RAG proteins) and the rate at which they are consumed (end joining and DNA degradation). We considered two possibilities to account for decreased signal end and coding end accumulation in cells expressing S723A or S723C. First, the cleavage activity of the mutant RAG1 proteins in vivo may be similar to that of wild-type RAG1, as suggested by the in vitro analyses, but the signal ends and coding ends might be quickly degraded due to instability of postcleavage complexes. Second, the mutant RAG1 proteins may not be active when expressed in 293T mammalian cells (the in vitro analyses of Figs. 1–5 were performed with RAG1 proteins purified from bacteria). To explore the latter possibility, we used a coimmunoprecipitation assay to examine the interaction of full-length wild-type or mutant RAG1 proteins with full-length RAG2 protein in vivo. We found that wild-type and mutant RAG1 proteins interact equally well with RAG2 (Fig. 7A, cf. lane 6 and lanes 7,8). In addition, we coexpressed GST-core RAG1 and untagged full-length RAG2 in 293T cells, partially purified the RAG complexes on glutathione beads, and measured their cleavage activity in vitro. All RAG1 proteins copurified with equal amounts of untagged RAG2, indicating that the mutant RAG1 proteins interact with RAG2 as efficiently as wild-type RAG1 (data not shown). In a standard coupled cleavage assay, wild-type and mutant RAG1 proteins were able to perform coupled cleavage, with the mutants exhibiting a mild twofold reduction in cleavage at all concentrations tested (Fig. 7B). This contrasts with their ∼100-fold defect in signal end and coding end accumulation in vivo (Fig. 6D).
Figure 7.
Coimmunoprecipitation, DNA cleavage activity ex vivo, and RSS binding activity in vivo, of RAG proteins expressed in mammalian cells. (A) Full-length untagged RAG1 protein (wild-type or mutants) and polyhistidine/myc tagged full-length RAG2 proteins were coexpressed in 293T cells. The lysate was subjected to immunoprecipitation using anti-myc antibodies. Five microliters of lysate (input, lanes 1–4) and half of the precipitated proteins (IP, lanes 5–8) were analyzed on a 7.5% SDS–polyacrylamide gel, and after transfer to a membrane, proteins were detected with anti-RAG1 (R1P1) and anti-RAG2 antibodies. Asterisk indicates breakdown products of the RAG1 proteins. (B) GST-core RAG1 and full-length untagged RAG2 were coexpressed in 293T cells and purified by glutathione affinity chromatography. Yields of RAG1 and RAG2 were equal for wild-type and mutant RAG1 proteins (not shown). Cleavage activity was assessed in a standard coupled cleavage reaction using serial twofold dilutions of the partially purified RAG proteins. (C) One-hybrid in vivo DNA binding assay. The ability of the mutant RAG1 proteins to interact with RAG2 and the 12-RSS was determined using a mammalian one-hybrid system. Cells were transfected with the p(12)8 reporter plasmid (expressing firefly luciferase), pRL-CMV (expressing renilla luciferase), and plasmids expressing GST-core RAG1 and core RAG2-VP16. Firefly luciferase values were normalized by dividing them by the renilla luciferase values, which corrects for variations in transfection efficiency. The normalized value obtained in transfections of RAG2 alone was arbitrarily set to one, and all other values are expressed relative to this. The values represent the mean of results from three independent transfections, and error bars indicate the standard error of the mean.
Finally, we asked whether the S723 mutant RAG1 proteins were able to bind the RSS in vivo using a one-hybrid assay previously developed by our laboratory (Difilippantonio et al. 1996). This assay uses a reporter vector in which a luciferase gene is under the control of a minimal promoter containing eight copies of a 12-RSS. This vector was cotransfected with plasmids expressing GST-core RAG1 (wild-type or mutants) and core RAG2 fused to the transcriptional activation domain of the VP16 protein of herpes simplex virus (RAG2-VP16), and luciferase levels were measured after 2 d (see Materials and Methods). We found that S723A, S723C, and wild-type RAG1 stimulated luciferase expression approximately equally well (Fig. 7C).
Together, these experiments indicate that the S723 mutant RAG1 proteins bind well to the RSS in vivo, and are catalytically active when expressed in mammalian cells. These findings support the hypothesis that much of the V(D)J recombination defect we observe with these proteins is due to their severe defect in postcleavage steps of the reaction.
Discussion
The role of serine 723 in DNA cleavage by the RAG proteins
S723A and S723C exhibit subtle defects in coupled cleavage (Fig. 1C,D), which may arise from a slight reduction in their ability to form the RAG1-RAG2-RSS complex (Fig. 2A). The defect in precleavage complex formation is more evident with S723E, which also exhibits a much more severe defect in cleavage. How does serine 723 contribute to RSS recognition and DNA cleavage? We found that the cleavage defect of S723A and S723C is more pronounced with oligonucleotide cleavage substrates with low-efficiency coding flanks than with those with high-efficiency flanks (data not shown). In addition, the nicking activity of S723E can be partially rescued by introducing a mismatch at the second position of the heptamer of the RSS (C.-L. Tsai and D. Schatz, in prep.). Based on these observations, we propose that upon interaction of the RAG proteins with the heptamer of the RSS, serine 723 is involved in creating a DNA structure favorable for stable precleavage complex formation and cleavage. It is worth noting that serine 723 is located within the region of RAG1 thought to interact with the heptamer of the RSS and coding flank (Arbuckle et al. 2001; Mo et al. 2001).
Mutation of serine 723 impairs target capture complex formation and destabilizes the postcleavage complex
S723A and S723C are severely defective in both intramolecular and intermolecular transposition (Figs. 1B, 3C), and this defect is due primarily to their failure to perform target capture (Fig. 3C). This parallels the observation that the postcleavage complex formed by the mutant RAG1 proteins is less stable than that formed by the wild-type protein (Fig. 4), supporting the possibility that the binding sites for coding ends and for target DNA are overlapping or identical. We propose that a binding pocket for target DNA/coding ends may be created as a result of conformational changes that occur upon RSS recognition and DNA cleavage, and that serine 723 is involved (directly or indirectly) in the formation of this pocket. Thus, the SEC formed by the S723 mutant proteins may be different structurally from that formed by the wild-type protein (see below). Recent UV crosslinking experiments support a direct role for RAG1 in binding to coding DNA (Mo et al. 2001). We could find no evidence for the possibility that serine 723 achieves these functions by directly coordinating metal ions, since Mn++ did not appreciably rescue S723C-mediated transposition (data not shown). In addition to their defect in transposition, S723A and S723C are also defective in hybrid joint formation and hairpin opening. It is likely that these defects result at least in part from the instability of the postcleavage complex formed by the mutant RAG1 proteins, but we cannot rule out the possibility that a catalytic defect also underlies the large deficiency in these postcleavage activities.
The idea that target DNA and coding ends occupy a common binding pocket formed as a result of DNA cleavage must be reconciled with recent findings that target commitment by the RAG proteins can occur prior to DNA cleavage (Neiditch et al. 2001). There may be two modes of interaction with target DNA that involve distinct binding surfaces. In this regard, it is interesting to note that competitor-resistant target commitment only occurs under conditions that support DNA cleavage (i.e., in Mg++, not Ca++; Neiditch et al. 2001).
S723 mutations result in a severe impairment of V(D)J recombination in vivo
S723A and S723C exhibit reduced V(D)J recombination activity in vivo, and strikingly, the levels of V(D)J recombination products (SJs and CJs) and V(D)J recombination intermediates (signal ends and coding ends) are reduced to the same extent (Fig. 6). Our data argue against the possibility that this is due solely to an in vivo cleavage defect of the mutant RAG1 proteins. The S723 mutants are able to interact with RAG2 and the RSS in vivo (Fig. 7A,C) and exhibit only mildly reduced cleavage activity in vitro when isolated from mammalian cells (Fig. 7B) or when purified from bacteria (Fig. 1). A defect exclusively in the cleavage phase of the reaction is also hard to reconcile with the more severe deficiency in recombination observed when the mutant RAG1 proteins are expressed with full-length versus core RAG2 (Fig. 6). We note that in previous studies, full-length RAG2 was at least as active as core RAG2 in generating recombination products (Cuomo and Oettinger 1994; Sadofsky et al. 1994; Steen et al. 1996). Importantly, the C-terminal region of RAG2 missing from the core protein (aa 384–527) has been implicated in postcleavage activities in V(D)J recombination (Steen et al. 1999; Sekiguchi et al. 2001), and has been suggested to contribute to the destabilization/disassembly of postcleavage complexes (Steen et al. 1999). Consistent with this notion, we have mapped a C-terminal region of RAG2 (aa 388–488) that exacerbates the recombination defects of the S723 mutants, perhaps by further destabilizing the postcleavage complexes. We note that wild-type RAG1 exhibited similar recombination activities with the different RAG2 proteins, in contrast to a previous study (Steen et al. 1999). The difference may be due to either the RAG1 proteins used in defining the function of RAG2 (full-length RAG1 in our study vs. core RAG1 in Steen et al.) or the use of different expression systems.
Overall, the deficiency in recombination observed for the S723 RAG1 mutants likely results both from a mild cleavage defect and from the severe postcleavage defects identified by our biochemical analyses.
It is unclear why signal ends fail to accumulate in cells expressing the S723 mutant RAG1 proteins. The mutants form the SEC in vitro as efficiently as does wild-type RAG1 (Fig. 3B), indicating that signal ends can remain associated with the mutants after cleavage in vitro. That signal ends and SJs do not accumulate in vivo may reflect abnormal features of the disassembly/structure of postcleavage complexes containing the mutant RAG1 proteins. The SEC formed by S723A or S723C (together with RAG2 and HMG2) exhibits defects in its interactions with coding ends and with target DNA, and it is plausible that it differs from the SEC formed by wild-type RAG1 in ways that lead to its destabilization in vivo. It is also possible that inappropriate release of coding ends from postcleavage complexes containing mutant RAG1 (as would be predicted by our biochemical data) is detected by damage sensors and results in destabilization of the SEC and degradation of both coding and signal ends.
Based on this model, it was possible that the CJ and SJ products produced by S723A and S723C would exhibit excessive deletions, but this was not the case: the joints generated by the mutant RAG1 proteins had a normal structure (data not shown). It is worth noting that the assays used here to detect CJs and SJs (PCR and bacterial transformation) are not suitable for detecting joining products containing large deletions.
Postcleavage defects, end release errors, and chromosomal translocations
The Roth laboratory recently reported mutations of RAG1 or RAG2 that result in a postcleavage defect in V(D)J recombination (Qiu et al. 2001; Schultz et al. 2001). The recombination phenotype of these mutants—defective coding and/or signal joint formation accompanied by the accumulation of significant levels of signal and coding ends in vivo—is distinct from that of the S723 RAG1 mutants. When a subset of these mutants was analyzed biochemically, all were able to catalyze the formation of nearly wild-type levels of hybrid joints and transposition products, and two were defective in opening of synthetic hairpins in Mn2+. While direct DNA binding studies were not performed, these results suggest that the Roth mutants form stable postcleavage complexes (capable of mediating hybrid joint formation and transposition) and that their joining deficiency arises from defects in other postcleavage activities. The biochemical phenotype of the S723A/C mutants is different in several important regards, most notably in exhibiting a defect in association of the SEC with coding ends and target DNA, with a resulting severe defect in hybrid joint formation and transposition. Thus, distinct biochemical phenotypes have distinct in vivo correlates: RAG proteins that form stable postcleavage complexes support accumulation of cleaved intermediates in vivo, while those that form unstable postcleavage complexes do not.
It is noteworthy that mutants of IS10 transposase with a phenotype similar to that of S723A and S723C have been identified (Kennedy and Haniford 1996). These mutants exhibit rather minor cleavage defects in vitro, but fail to accumulate transposition products or cleaved intermediates in vivo. Importantly, these mutants, like the S723 mutant RAG1 proteins, generate unstable postcleavage complexes in vitro, leading to the proposal that cleaved intermediates fail to accumulate in vivo because of rapid breakdown of postcleavage complexes with attendant degradation of the cleaved DNA (Kennedy and Haniford 1996). Very similar processes may occur in vivo with the S723 mutant RAG1 proteins.
It is becoming increasingly clear that errors in the process of V(D)J recombination can lead to chromosomal translocations and thereby contribute to the malignant transformation of lymphocytes (Kuppers and Dalla-Favera 2001). Two general types of errors have been suggested to be involved: recognition errors, in which incorrect target sequences are bound and cleaved by the RAG complex, and end release errors, in which cleavage is followed by premature release of the DNA ends (Lewis 1994). The S723A and S723C mutant RAG1 proteins appear to be prone to end release errors, and expression of such a mutant in developing lymphocytes might result in an elevated incidence of chromosomal translocations and lymphomas, an idea that is currently being tested.
Our study of serine 723 of RAG1 provides evidence that the stability/structure of postcleavage complexes is critical for the postcleavage activities of the RAG proteins in vitro and for V(D)J recombination in vivo. In light of this, we propose that the RAG proteins play an important postcleavage, “scaffold” role in preparing broken DNA ends for proper and efficient end processing and joining; and that, in addition, they serve a protective function, helping to shield DNA ends in the postcleavage complex from nuclease digestion. The importance of this scaffold function for efficient V(D)J recombination suggests that the RAG proteins are key players in the joining phase of V(D)J recombination and therefore contribute to the maintenance of genome integrity in developing lymphocytes.
Materials and methods
Mutagenesis and protein expression
Serine 723 of RAG1 was mutated to alanine, cysteine, and glutamic acid using the QuiKChange kit (Stratagene) according to the manufacturer's instructions. Mutations were made initially in the bacterial expression vector pCJM233 (Rodgers et al. 1999) and were confirmed by DNA sequencing. To construct vectors that express mutant proteins in mammalian cells, a 0.7-kb BsrGI fragment from pCJM233 containing amino acid residue 723 was substituted for the corresponding fragment of pEBG-core RAG1 (expressing GST-core RAG1) or pEBB-RAG1 (expressing untagged full-length RAG1). Recombinant MBP-tagged RAG1 proteins were expressed in bacteria and purified by a three-step purification. Partially purified proteins from amylose affinity chromatography (Rodgers et al. 1996) were subjected to Ni-NTA Superflow (QIAGEN) affinity chromatography and subsequently to gel filtration chromatography (Superdex 200, Pharmacia). This procedure yields dimeric RAG1 that is >95% pure. GST-tagged core RAG1 and untagged full-length RAG2 were coexpressed in 293T cells and purified by glutathione-agarose affinity chromatography (Spanopoulou et al. 1996).
Core RAG2 protein with polyhistidine and myc epitope tags was purified from the murine cell line F2A1 (Eastman et al. 1996) expressing both his-myc-core-RAG1 and his-myc-core RAG2 by a three-step purification procedure that separates fractions containing the RAG1-RAG2 complex from those containing only RAG2. Briefly, 30 L of cell pellets was extracted with 150 mL of extraction buffer (300 mM NaCl, 3 mM MgCl2, 25 mM Tris-Cl at pH 7.5; 0.2 mM EDTA, 0.2 mM EGTA, 20% glycerol, 0.1% Nonidet P-40, 5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride). After centrifugation at 30,000 rpm for 1 h, the supernatant was loaded onto a 6-mL Ni-NTA Superflow column (QIAGEN). The column was washed with 60 mL of lysis buffer and 60 mL of lysis buffer containing 20 mM imidizole (pH 7.4) and then eluted with 60 mL of lysis buffer containing 100 mM imidizole. The fractions containing the RAG proteins were collected and dialyzed against 1 L of buffer Q (20 mM Tris-HCl at pH 7.4, 100 mM NaCl, 3 mM MgCl2, 10% glycerol, and 2 mM DTT) at 4°C overnight. Proteins were loaded on a 1-mL MonoQ column (Pharmacia) and eluted with a 20-mL gradient of NaCl (from 100 to 600 mM). The fractions containing the RAG proteins were collected and diluted with buffer S (20 mM HEPES-Na at pH 7.4, 3 mM MgCl2, 10% glycerol) to lower the NaCl concentration to 100 mM. The diluted proteins were then loaded on a 1-mL MonoS column and eluted with a 20-mL gradient of NaCl (100 to 600 mM). The RAG1-RAG2 fractions and the RAG2-only fractions were pooled and assayed for their cleavage activity. The purified RAG2 protein is devoid of detectable RAG1 and alone does not exhibit any cleavage activity (data not shown).
DNA substrates
The body-labeled coupled cleavage substrate was generated by PCR from pC317 (Agrawal et al. 1998). The 12-RSS and 23-RSS oligonucleotide substrates for 12-SC and paired complex formation have been described (Fugmann et al. 2000b). The 12-signal end and 23-signal end oligonucleotide substrates for signal end complex formation, target capture complex formation, and intermolecular transposition were identical to the 12-RSS and 23-RSS, but lack coding flank sequences. The double-stranded oligonucleotide target DNA for intermolecular transposition was made by annealing 5′-end-labeled CLT011 (5′-CGCTCGGTT GCCGCCGGGCGTACTATATTGA-3′) with CLT012 (5′-TCA ATATAGTACGCCCGGCGGCAACCGAGCG-3′).
DNA cleavage, intramolecular transposition, and DNA binding
Coupled cleavage and intramolecular transposition reactions (25 μL final volume) contained 10 ng of body-labeled substrate, 50 ng of each RAG protein, and 15 ng of HMG2. Reactions were incubated for 2 h at 37°C in a buffer containing 20 mM HEPES-Na, pH 7.4, 75 mM NaCl, 4 mM MgCl2, 1 μM ZnSO4, 0.1 mg/mL BSA, and 2 mM DTT. Binding reactions to detect the 12-SC contained 10 fmol of 5′ end-labeled 12-RSS substrate, 50 ng of each RAG protein, and 15 ng of HMG2. They were performed in the buffer described previously (Fugmann et al. 2000b) for 15 min at 30°C in a final volume of 12 μL. Next, 3 μL of a 50% glycerol solution was added, and the samples were immediately loaded onto a 4% native polyacrylamide gel. Gels were electrophoresed in 1× TBE buffer at 175V for 3 h. Binding reactions to detect the paired complex were performed as described for the 12-SC except that they contained 10 fmol of unlabeled 23-RSS substrate and were performed for 30 min at 37°C in the buffer described previously (Hiom and Gellert 1998), then electrophoresed on 6% (acrylamide:bis = 80:1) native polyacrylamide gels at 175V in 1× TBE for 3.5 h. All quantitation of bands was performed with a PhosphorImager (Molecular Dynamics).
Assays for formation of the SEC, TCC, and intermolecular transposition products
Reactions for SEC formation contained 0.05 pmol-labeled 12-signal end oligonucleotide substrate, 50 ng of MBP-core RAG1, and 50 ng of GST-core RAG2 in 25 mM MOPS-NaOH (pH 7.0), 75 mM potassium acetate, 100 μg/mL BSA, 4 mM DTT, and 5.4 mM CaCl2 at 37°C for 15 min in a final volume of 12 μL; then 3 μL of a 50% glycerol solution was added, and the samples were immediately loaded onto a 6% (80:1) native polyacrylamide gel. HMG2 (15 ng) and/or equimolar unlabeled 23-signal end substrate were added to some reactions. The assay for target capture complex formation was based upon that described by Roth and colleagues (Neiditch et al. 2001). Briefly, the signal end complex was formed using equimolar unlabeled 12- and 23-signal end DNA substrates (0.1 pmol) under the same reaction conditions as the assay for formation of the SEC. Three-microliter mixtures of 0.6 pmol of labeled target DNA and 25 mM MgCl2 were added to the reaction mixtures and incubated at 37°C for 15 min. Three microliters of 50% glycerol was added to all reaction mixtures in the presence or absence of additional 1 μL of 20 mM EDTA/10% SDS. Entire reaction mixtures were loaded onto a 6% (80:1) native polyacrylamide gel and run at 175V for 3.5 h.
Assay for the stability of the postcleavage complex
Biotinylated coupled cleavage substrate used in the assay was generated by using a biotinylated primer Cit4a (5′-GCAACT GACTGAAATGCCTC-3′) and nonbiotinylated primer LE1 (5′-GGAATTGTGAGCGGATAAC-3′). To ensure maximal recovery of the postcleavage complex, the biotinylated coupled cleavage substrate was prebound to streptavidin beads for 10 min at room temperature. To remove unbound substrate, the beads were wash twice with 200 μL of cleavage buffer prior to the cleavage reaction. Large-scale cleavage reactions (100 μL) were carried out in a rotating chamber at 37°C to avoid precipitation of DNA substrates. After 1 h, one-quarter of each reaction was removed and treated with proteinase K, phenol/chloroform-extracted, and served as the input fraction. The remainder of each reaction was then placed on a magnet stand for 5 min, and the DNA free in solution was collected (unbound fraction; Fig. 4B, lanes 5–7). The beads were washed with the reaction buffer twice to remove DNA associated nonspecifically with the beads, and nonbiotinylated DNA fragments remaining bound to the beads were eluted with the reaction buffer containing 2% SDS and 10 mM EDTA. All samples were subject to proteinase K treatment (55°C for 1 h), phenol/chloroform extraction, and ethanol precipitation, followed by analysis on a 4% native polyacrylamide gel.
Hybrid joint formation
For hybrid joint formation, 1/20 of the products from the reactions in Figure 1B were used for PCR amplification of hybrid joints (24 cycles). The primers for amplifying 12-RSS/23-coding HJs were TL6 and Cit4a. The primers for amplifying 23-RSS/12-coding HJs were TL3 and LE1 (Leu et al. 1997).
In vivo V(D)J recombination
RAG proteins were expressed in pEBB vector (Roman et al. 1997). Vectors for the expression of full-length and GST-core RAG1, and full-length and core RAG2 have been described (Roman et al. 1997; Fugmann et al. 2000b), and the RAG2 mutants 1-488 and Δ388-413 were generous gifts from M. Sadofsky (Sadofsky et al. 1994) and were subcloned into pEBB vector.
The transient recombination assay was performed as described (Hesse et al. 1987). Briefly, 30% confluent 10-cm dishes of 293T cells were transfected with 5 μg RAG1 expression vector, 5 μg RAG2 expression vector, and 5 μg recombination substrate pSF290 (Fugmann and Schatz 2001). After 48 h, plasmids were recovered from the cells by the rapid alkaline method (Hesse et al. 1987) and then subjected to a serial 10-fold dilution prior to PCR amplification for signal joints (primers TL2 and TL3; 30 cycles) and for the measurement of DNA recovery (primer TL5 and TL6; 21 cycles). To detect coding joints, recovered DNA was subject to ClaI digestion, which reduces PCR amplification of unrearranged pSF290, prior to PCR amplification (primers TL1 and TL4; 26 cycles). To detect 12 RSS signal ends, the recovered plasmids were ligated (16°C, overnight) to the double stranded anchor primers made by annealing DR19 with DR20 (Zhu and Roth 1995). The ligation products were then subjected to PCR amplification using primers DR20 and TL7 (5′-CAGAACGCTCGGTTGCCGCC-3′) (25 cycles). To detect coding ends, plasmids were recovered from transfected cells by the Hirt method (Hirt 1967). One-fifth of the recovered DNA was treated with 20 units of mung bean nuclease (New England Biolabs) in a buffer containing 10 mM Tris-HCl (pH 7.9), 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, and 1 mM ZnSO4 at 30°C for 2 h. The reactions were terminated by phenol/chloroform extraction. Nuclease-treated samples were then subjected to ligation and PCR as described above, using primers DR20 and TL4 (29 cycles). Mung bean nuclease treatment dramatically stimulated detection of coding ends (data not shown).
RAG1-RAG2 coimmunoprecipitation
To coexpress the RAG1 and RAG2 proteins, 293T cells were transfected with 5 μg of the vector expressing untagged full-length RAG1 proteins (wild-type or mutants) and 5 μg of the vector expressing full-length RAG2 protein with polyhistidine and myc epitope tags using a calcium phosphate method. Forty-eight h after transfection, cells were harvested and the cell pellet was resuspended in 500 μL of RSB (10 mM Tris at pH 7.4, 10 mM NaCl, 5 mM MgCl2, and 0.5% NP-40). Cells were lysed in 750 μL of LSB (20 mM Tris at pH 7.4, 1M NaCl, 0.2 mM MgCl2, and 0.2% NP-40) followed by incubation at 4°C for 2 h. Debris was removed by ultracentrifugation at 32,000 rpm for 1 h at 4°C, and 200 μL of the cleared cell lysate was used for coimmunoprecipitation. The lysate was precleared for 30 min at 4°C with 20 μL of protein-G agarose beads (GIBCO-BRL), followed by addition of 20 μg of anti-myc antibodies (9E10). Reactions were rotated overnight at 4°C and then centrifuged and washed twice with a 1:1.5 ratio mixture of RSB and LSB. Thereafter, the beads were transferred to a new tube and subjected to two additional washes. Precipitated proteins were resuspended in 2× protein loading buffer and separated on a 7.5% SDS-PAGE, and Western blotted with anti-R1P1 and anti-RAG2 antibodies (Leu and Schatz 1995).
One-hybrid system
One-hybrid experiments were performed as described (Difilippantonio et al. 1996). Briefly, 293T cells were cotransfected with the core RAG2-VP16 fusion protein expression vector pCJM170, RAG1 expression vector pEBG-core RAG1 (or mutant versions), the firefly luciferase reporter construct pMJD112, and the renilla luciferase expression vector pRL-CMV (Promega), which allows measurement of transfection efficiency. After 48 h, cells were harvested and the luciferase activities were determined using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Firefly luciferase values were normalized relative to the renilla luciferase values, and the normalized value obtained in the absence of RAG1 was set to one.
Acknowledgments
We thank M.A. Oettinger and M.J. Sadofsky for providing plasmids, J. Repasky for her contributions to the development of the assay for the stability of the postcleavage complex, N.D. Grindley, S. Ghosh, J.J. Yeh, and all members of the Schatz lab for helpful suggestions, and S.D. Fugmann, D.G.T. Hesslein, and A.I. Lee for critical reading of and comments on the manuscript. This work was supported by grant R01 AI-32524 to D.G.S. from the National Institutes of Health. D.G.S. is an investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL david.schatz@yale.edu; FAX (203) 737-1764.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.984502.
References
- Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Schatz DG. RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- Aidinis V, Dias DC, Gomez CA, Bhattacharyya D, Spanopoulou E, Santagata S. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J Immunol. 2000;164:5826–5832. doi: 10.4049/jimmunol.164.11.5826. [DOI] [PubMed] [Google Scholar]
- Arbuckle JL, Fauss LJ, Simpson R, Ptaszek LM, Rodgers KK. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J Biol Chem. 2001;276:37093–37101. doi: 10.1074/jbc.M105988200. [DOI] [PubMed] [Google Scholar]
- Bailin T, Mo X, Sadofsky MJ. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol Cell Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besmer E, Mansilla-Soto J, Cassard S, Sawchuk DJ, Brown G, Sadofsky M, Lewis SM, Nussenzweig MC, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- Eastman QM, Leu TMJ, Schatz DG. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- Eastman QM, Villey IJ, Schatz DG. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezekiel UR, Sun TH, Bozek G, Storb U. The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol Cell Biol. 1997;17:4191–4197. doi: 10.1128/mcb.17.7.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: Complexes, ends, and transposition. Annu Rev Immunol. 2000a;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000b;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Harfst E. How to make ends meet in V(D)J recombination. Curr Opin Immunol. 2001;13:186–194. doi: 10.1016/s0952-7915(00)00203-x. [DOI] [PubMed] [Google Scholar]
- Haniford DB, Chelouche AR, Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989;59:385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- ————— Assembly of a 12/23 paired signal complex: A critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: A possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kennedy AK, Haniford DB. Isolation and characterization of IS10 transposase separation of function mutants: Identification of amino acid residues in Transposase that are important for active site function and the stability of transposition intermediates. J Mol Biol. 1996;256:533–547. doi: 10.1006/jmbi.1996.0106. [DOI] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Sinden RR, Roth DB. Targeted transposition by the V(D)J recombinase. Mol Cell Biol. 2002;22:2068–2077. doi: 10.1128/MCB.22.7.2068-2077.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu TMJ, Eastman QM, Schatz DG. Coding joint formation in a cell free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- Leu TMJ, Schatz DG. Rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol Cell Biol. 1995;15:5657–5670. doi: 10.1128/mcb.15.10.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SM. The mechanism of V(D)J joining: Lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Melek M, Gellert M, van Gent DC. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- Mo X, Bailin T, Sadofsky M. A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol Cell Biol. 2001;21:2038–2047. doi: 10.1128/MCB.21.6.2038-2047.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- Mundy C, Patenge N, Matthews A, Oettinger M. Assembly of the RAG1/RAG2 synaptic complex. Mol Cell Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Lee GS, Landree MA, Roth DB. RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol Cell Biol. 2001;21:4302–4310. doi: 10.1128/MCB.21.13.4302-4310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Perkins EJ, Nair A, Cowley DO, Van Dyke T, Chang Y, Ramsden DA. Sensing of intermediates in V(D)J recombination by ATM. Genes & Dev. 2002;16:159–164. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JX, Kale SB, Schultz HY, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Paull TT, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- Rodgers KK, Bu Z, Fleming KG, Schatz DG, Engelman DM, Coleman JE. A unique zinc-binding dimerization motif domain in RAG-1 includes the C3HC4 motif. J Mol Biol. 1996;260:70–84. doi: 10.1006/jmbi.1996.0382. [DOI] [PubMed] [Google Scholar]
- Rodgers KK, Villey IJ, Ptaszek L, Corbett E, Schatz DG, Coleman JE. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 1999;27:2938–2946. doi: 10.1093/nar/27.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman CAJ, Cherry SR, Baltimore D. Complementation of V(D)J recombination deficiency in RAG-1(−/−) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity. 1997;7:13–24. doi: 10.1016/s1074-7613(00)80506-3. [DOI] [PubMed] [Google Scholar]
- Sadofsky M. The RAG proteins in V(D)J recombination: More than just a nuclease. Nucleic Acids Res. 2001;29:1399–1409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadofsky M, Hesse J, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Besmer E, Villa A, Bozzi F, Allingham JS, Sobacchi C, Haniford DB, Vezzoni P, Nussenzweig MC, Pan Z-Q, et al. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: Implications for junctional diversity in V(D)J and transpositional recombination. Mol Cell. 1999;4:935–947. doi: 10.1016/s1097-2765(00)80223-3. [DOI] [PubMed] [Google Scholar]
- Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene (RAG-1) Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- Schultz HY, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Whitlow S, Alt F. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- Shockett PE, Schatz DG. DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol Cell Biol. 1999;19:4159–4166. doi: 10.1128/mcb.19.6.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- Steen SB, Gomelsky L, Roth DB. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Yu KF, Lieber MR. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol Cell Biol. 1999;19:8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CM, Roth DB. Characterization of coding ends in thymocytes of scid mice - implications for the mechanism of V(D)J recombination. Immunity. 1995;2:101–112. doi: 10.1016/1074-7613(95)90082-9. [DOI] [PubMed] [Google Scholar]