Abstract
The genomic architecture of protocadherin (Pcdh) gene clusters is remarkably similar to that of the immunoglobulin and T cell receptor gene clusters, and can potentially provide significant molecular diversity. Pcdh genes are abundantly expressed in the central nervous system. These molecules are primary candidates for establishing specific neuronal connectivity. Despite the extensive analyses of the genomic structure of both human and mouse Pcdh gene clusters, the definitive molecular mechanisms that control Pcdh gene expression are still unknown. Four theories have been proposed, including (1) DNA recombination followed by cis-splicing, (2) single promoter and cis-alternative splicing, (3) multiple promoters and cis-alternative splicing, and (4) multiple promoters and trans-splicing. Using a combination of molecular and genetic analyses, we evaluated the four models at the Pcdh-γ locus. Our analysis provides evidence that the transcription of individual Pcdh-γ genes is under the control of a distinct but related promoter upstream of each Pcdh-γ variable exon, and posttranscriptional processing of each Pcdh-γ transcript is predominantly mediated through cis-alternative splicing.
Keywords: Protocadherin, alternative splicing, trans-splicing, DNA recombination
Cadherin superfamily proteins are characterized by the presence of extracellular cadherin-like ectodomain repeats that are known to mediate calcium-dependent homophilic protein-protein interactions (Takeichi 1990; Suzuki 1996; Yap et al. 1997; Yagi and Takeichi 2000). This superfamily can be divided into two subfamilies: classic cadherins and protocadherins. Classic cadherins consist of five ectodomain repeats, a single transmembrane region, and a highly conserved cytoplasmic domain that interacts with actin-based cytoskeleton components through β-catenin (Gumbiner 2000). Classic cadherins are well known for their essential roles in morphogenesis of multiple tissues during development (Takeichi 1991, 1995; Marrs and Nelson 1996; Radice et al. 1997; Carmeliet et al. 1999; Garcia-Castro et al. 2000), and have also been implicated in maintenance of neuronal connectivity and formation of synaptic plasticity in the nervous system (Yamagata et al. 1995; Fannon and Colman 1996; Iwai et al. 1997; Tang et al. 1998; Bozdagi et al. 2000; Manabe et al. 2000; Tanaka et al. 2000; Lee et al. 2001).
Protocadherins include several subclasses of cadherin-like molecules (Wu and Maniatis 1999; Suzuki 2000; Yagi and Takeichi 2000). Like classic cadherins, protocadherins share the same protein structure: multiple (usually six or more) ectodomain repeats, a transmembrane region and a cytoplasmic domain. However, their cytoplasmic domains are distinct in sequence not only from classic cadherins, but also from each other among different classes of protocadherins (Kohmura et al. 1998; Yagi and Takeichi 2000; Homayouni et al. 2001). The significant molecular diversity of protocadherins was revealed by the discovery of the striking genomic organization of three protocadherin gene clusters (designated Pcdh-α, -β and -γ, which consist of nearly 60 Pcdh genes) in both the human and mouse genomes (Wu and Maniatis 1999; Sugino et al. 2000; Wu et al. 2001). These three gene clusters are tandemly arrayed on the same chromosome (human chromosome 5 and mouse chromosome 18) and transcribed in the same direction. For Pcdh-α and -γ, each gene cluster contains a variable region with multiple variable exons, and a constant region with three constant exons. Each of the variable exons is separately spliced to constant exons to generate diverse Pcdh mRNAs. The variable exons encode a similar but distinct extracellular cadherin-like domain, a transmembrane region, and part of the cytoplasmic domain, and the three constant exons encode the rest of the cytoplasmic domain shared by all members of the same cluster. This unique genomic organization of Pcdh gene clusters and its resemblance to Ig and TCR gene clusters suggested that a novel mechanism may be involved in the generation of protocadherin diversity and their cell-specific expression pattern in the brain (Wu and Maniatis 1999).
Pcdh genes are abundantly expressed in the central nervous system during embryonic development and in adulthood (Sano et al. 1993; Kohmura et al. 1998; Obata et al. 1998; Hirano et al. 1999; Wu and Maniatis 1999). Pcdh-γ proteins have been shown to mediate homophilic cell adhesion in transfected L1 cells (Sano et al. 1993; Obata et al. 1995). Some Pcdh-α proteins (CNR1) have been localized at synapses (Kohmura et al. 1998). Pcdh-γ proteins have also been identified by mass spectrometry in a “presynaptic web” preparation (Phillips et al. 2001). The unique genomic structure of Pcdh gene clusters can generate a significant amount of molecular diversity that is required for establishment and maintenance of complex neural networks in the brain (Wu and Maniatis 1999). All these observations have led to the hypothesis that Pcdh proteins are primary candidates for establishing specific neuronal connectivity and synapse formation (Serafini 1999; Shapiro and Colman 1999; Bruses 2000; Yagi and Takeichi 2000; Benson et al. 2001).
Expression studies using in situ hybridization have demonstrated that individual neurons can express an overlapping but distinct subset of Pcdh genes (Kohmura et al. 1998). Therefore, the subset of protocadherins which individual neurons express may provide specific molecular codes for neuron–neuron connections. If Pcdh proteins function in establishing the specificity of neural circuitry, a key question is: How is the cell-specific expression pattern of Pcdh genes achieved? Maniatis and his colleagues proposed four models (Wu and Maniatis 1999; Wu et al. 2001) for the cell-specific Pcdh gene expression, including (1) DNA recombination, (2) single promoter and cis-alternative splicing, (3) multiple promoters and cis-alternative splicing, and (4) multiple promoters and trans-splicing. These models have provided a framework to investigate the mechanism of the cell-specific expression of Pcdh genes.
Here, we used a combination of genetically modified alleles at the mouse Pcdh-γ locus to distinguish the four possible mechanisms of Pcdh gene expression. Although Pcdh gene clusters share striking similarity with Ig and TCR gene clusters, somatic DNA recombination does not appear to be involved in the regulation of Pcdh gene expression. Trans-splicing between putative RNA precursors only contributes a minor portion of functional Pcdh mRNAs. The transcription of each Pcdh-γ variable exon initiates at sequences immediately upstream of each translational start site without a common 5′UTR sequence, arguing against a single promoter scenario. Furthermore, a common DNA motif upstream of each variable exon is essential for the transcription of individual Pcdh-γ members. Thus, our data are most consistent with the multiple promoters and cis-alternative splicing model for the cell-specific Pcdh gene expression.
Results
Multiple genetically modified alleles at the mouse Pcdh-γ locus
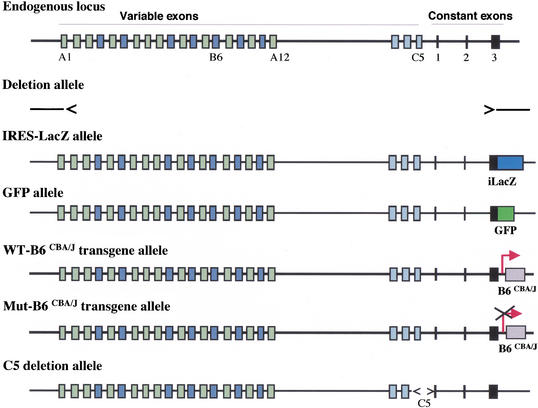
To investigate the molecular mechanisms of cell-specific expression of Pcdh genes, we focused on the mouse Pcdh-γ locus and generated multiple genetically modified alleles in mice or in mouse ES cells. These alleles are summarized in Figure 1: (1) A deletion allele that removed all Pcdh-γ coding sequences from variable exon A1 to constant exon 3; (2) An IRES-GFPLacZ fusion reporter was targeted into constant exon 3 to generate a gene expression reporter allele; (3) A GFP cDNA was fused in-frame with the constant exon 3 to encode Pcdh-γ-GFP fusion proteins; (4) A transgene carrying the B6 variable exon from the CBA/J strain was inserted at the end of Pcdh-γ locus; (5) A mutant transgene similar to the fourth allele, but with a deletion of the putative promoter sequence was inserted at the same location as the wild-type transgene; and (6) A 5-kb sequence upstream of constant exon 1 including the fragment encoding the last variable exon (C5) was deleted by gene targeting. We used combinations of these alleles to distinguish the four possible models for cell-specific Pcdh gene expression.
Figure 1.
Mouse Pcdh-γ locus and genetically modified alleles. Multiple modified Pcdh-γ alleles used in this study are shown in relation to the endogenous locus. Deletion allele, IRES-LacZ reporter allele, GFP fusion allele, WT-B6CBA/J transgene allele, Mut-B6CBA/J transgene allele, and C5 deletion allele.
Multiple Pcdh-γ members are expressed from a single Pcdh-γ allele in a single neuron
The genomic structure of Pcdh gene clusters is strikingly similar to that of the immunoglobulin genes; therefore, it has been postulated that similar somatic DNA recombination may occur at the Pcdh locus and control Pcdh gene expression in a cell-specific manner (Wu and Maniatis 1999). Using a combination of these alleles, we first addressed the question of whether DNA recombination similar to Ig genes occurs at the Pcdh-γ locus. The DNA recombination model makes two predictions (Fig. 2A). First, cell-specific somatic recombination brings a distal variable exon to the proximity of a hypothetical enhancer and activates the transcription of the closest variable exon, implying that only one specific form of variable exon is transcribed from each functional allele. Second, since both variable and constant exons are transcribed in the same orientation, somatic recombination should lead to the deletion of intervening sequences.
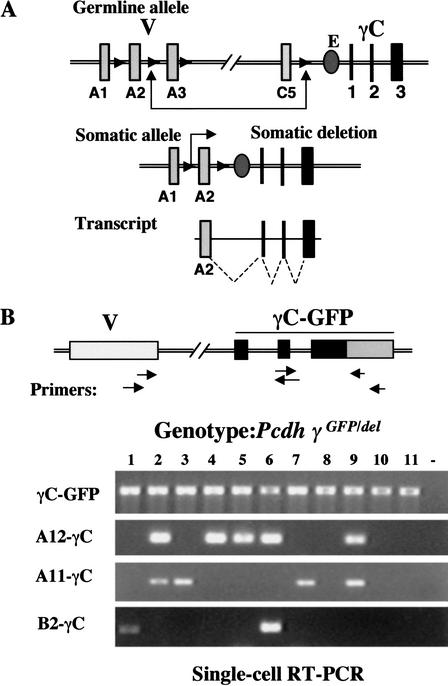
Figure 2.
A single Pcdh-γ allele expresses multiple variable exons. (A) DNA recombination model for Pcdh gene expression (Wu and Maniatis 1999). (B) Detection of multiple different Pcdh-γ genes using single-cell RT-PCR. Single neurons from Pcdh-γdel/GFP cerebellum (postnatal P5) were used to detect γC-GFP, A12-γC, A11-γC, and B2-γC.
To evaluate the recombination model, we performed a single-cell RT-PCR analysis from Pcdh-γdel/GFP mice carrying only one functional Pcdh-γ-GFP allele. Mouse cerebellum was dissociated into a single-cell suspension. Single cells were isolated and the RT-PCR analysis was initiated using a GFP-specific primer. The PCR primer pairs and subsequent nested primers were designed to span exon-exon junctions, excluding possible contamination from genomic DNA. As shown in Figure 2B, all cells (neurons) expressed Pcdh-γ (γC-GFP panel). Individual Pcdh variable exons (A12, A11, and B2) were only expressed in a subpopulation of neurons, and some of the neurons expressed multiple variable exons. Because the cells were obtained from mice carrying only one allele of Pcdh-γ, this result demonstrated that more than one variable exon can be transcribed from a single allele, providing evidence that is inconsistent with the first prediction of the DNA recombination model.
Deletion of common intervening sequences at the Pcdh-γ cluster is not detected
Pcdh genes are primarily expressed in the central nervous system. Neurons are postmitotic cells that cannot be propagated as clones; therefore, we searched for a population of cells that express Pcdh-γ. Expression studies using both the LacZ allele (Fig. 3A) and the GFP allele (Fig. 3B,C) showed that in adult mouse brains the majority of cells in the cortex region expressed Pcdh-γ. We reasoned that if somatic DNA recombination is required for the expression of Pcdh-γ in neurons, a significant portion of cells would share a common deletion of intervening sequence in a population of cells that almost all express Pcdh-γ. Therefore, we dissected a region of the cortex from adult mouse brains and compared the gene dosage around the Pcdh-γ locus by Southern blot analyses. The analysis focused on the sequences upstream of the constant region that are more likely to be deleted and shared by different individual cells. In this experiment, we used the mice that carry only one Pcdh-γ allele to emphasize any dosage difference, should such differences exist. As shown in Figure 3D, the comparison of hybridization signals between liver DNA (nonPcdh-γ-expressing tissue) and cortex DNA (Pcdh-γ-expressing tissue) showed no significant change in gene dosage with three probes that lie immediately upstream from constant exon 1 (C5-1), further upstream from variable exon C3 (i), and within the A12 variable exon (A12). This line of investigation suggested that the deletion of common intervening sequences upstream of the constant region does not occur in a population of neurons that express Pcdh-γ, providing evidence against the second prediction of the DNA recombination model.
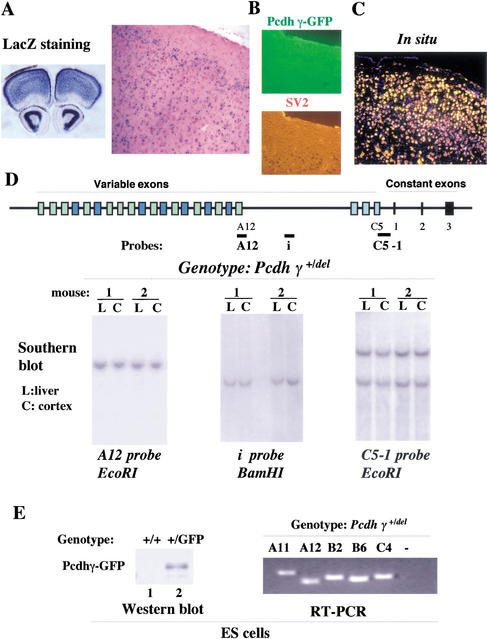
Figure 3.
No deletion of common intervening sequences upstream of the constant region. The majority of cells express Pcdh-γ in the cortex of adult mouse brain. (A) LacZ staining of a vibratome section (left) and a frozen section (right) from adult mouse brain (Pcdh-γ+/LacZ); (B) Immunostaining of a frozen section of Pcdh-γ+/GFP mouse brain with rabbit anti-GFP antibodies and mouse anti-SV2 antibody. (C) In situ hybridization of a frozen section from adult Pcdh-γ+/GFP mouse brain using a fluorescein-labeled GFP riboprobe (in yellow) and stained with DAPI (in blue); (D) No deletion of common intervening sequences upstream from the constant region. Genomic DNAs were obtained from liver (L) and cortex (C ) of Pcdh-γ+/del adult mice. Gene dosage comparison by Southern blot analysis using the three probes indicated. (E) Expression of multiple Pcdh-γ genes in heterozygous (Pcdh-γ+/del) ES cells. Western blot analysis using anti-GFP antibodies detected Pcdh-γ-GFP fusion proteins in Pcdh-γ+/GFP ES cells. RT-PCR analysis detected multiple spliced forms of Pcdh-γ cDNAs in Pcdh-γ+/del ES cells.
We also detected a low level of Pcdh-γ-GFP fusion proteins in undifferentiated ES cells with a targeted GFP fusion allele (Fig. 3E, Western blot, left). RT-PCR analysis showed that multiple forms of spliced Pcdh-γ mRNAs were expressed in mouse ES cells carrying only one Pcdh-γ allele (Fig. 3E, right). LacZ staining of ES cells with a targeted IRES-LacZ allele showed that a significant portion of cells expressed a low level of Pcdh-γ, suggesting that the detected Pcdh-γ expression was not due to a minor contribution from differentiated ES cells (data not shown). Pcdh-γ mRNAs were also detected in different cell types of testis, including the mature germ cells (Johnson et al. 2000). These observations suggested that at least in germ cells, an irreversible genetic change (DNA recombination) is not required for the expression of multiple Pcdh-γ genes. Although we cannot completely rule out the possibility that DNA recombination, different from that which occurs at the Ig and TCR loci, occurs at the Pcdh-γ locus, we obtained no evidence suggesting that DNA recombination is involved in the control of Pcdh-γ gene expression.
Individual Pcdh-γ transcripts do not contain a shared 5′ noncoding leader exon
We next examined the single promoter hypothesis. In this model, a common promoter activates transcription of the whole locus and functional mRNAs are assembled by alternative splicing (Fig. 4A; Wu and Maniatis 1999). The prediction of this model is that individual Pcdh transcripts should share a common 5′ UTR sequence. To test this hypothesis, we performed a 5′ RACE analysis on two variable exons, B2 and A12. Sequencing of multiple 5′RACE products and the comparison of these sequences with genomic sequences revealed that transcripts encoding both B2 and A12 variable exons initiated immediately upstream of each variable exon (Fig. 4B). Multiple transcription start sites for each variable exon have been identified (Fig. 4B). More importantly, these transcripts share no common 5′ UTR sequences, arguing against the common single promoter model.
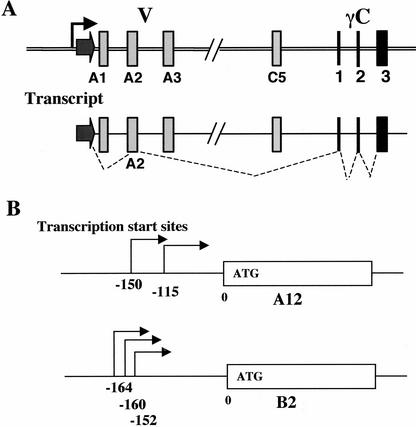
Figure 4.
Individual variable exon transcripts share no common 5′UTR sequence. (A) The single promoter and cis-alternative splicing model (Wu and Maniatis 1999). (B) 5′RACE analysis on two Pcdh-γ variable exon transcripts identified multiple transcription start sites immediately upstream of translation start ATG codon. These transcripts have a different 5′ UTR sequence.
Individual Pcdh-γ variable exons are expressed under the control of their own promoter
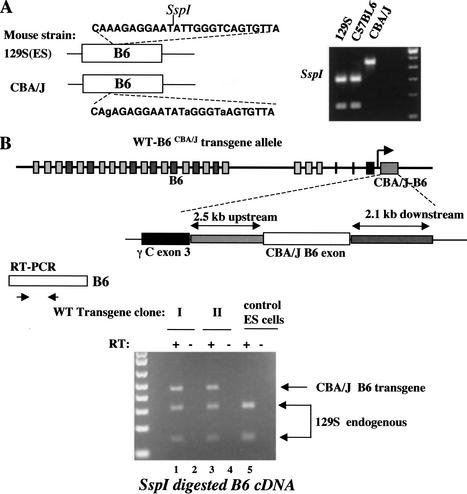
Having obtained evidence that no common promoter exists for the Pcdh-γ gene cluster, we explored the existence of promoters for individual variable exons. We identified Pcdh-γ cDNAs from a mouse hypothalamic cDNA library that was derived from C57BL6/CBA/J F1 hybrid mice. From sequence analysis of 50 Pcdh-γ cDNA clones, we identified three single-nucleotide polymorphisms (SNPs) and one restriction fragment-length polymorphism (RFLP) in one Pcdh-γ variable exon, B6 variable exon (Fig. 5A). Analysis of genomic DNAs amplified from different mouse strains confirmed that these RFLP and SNP markers are also informative between 129S and CBA/J strains (Fig. 5A, inset).
Figure 5.
Individual Pcdh-γ variable exons have their own promoters. (A) Identification of SNPs and RFLPs in variable exon B6 between 129S and CBA/J mouse strains. The relevant sequences from different mouse strains are shown as well as SspI RFLP analysis of genomic DNA from different mouse strains. (B) Targeting of a B6 transgene from the CBA/J strain into the Pcdh-γ locus in mouse ES cells (129S strain). The CBA/J B6 transgene consists of 2.5-kb 5′ flanking sequence, the coding exon and 2.1 kb of intronic sequence. This transgene is transcribed in the same orientation as the endogenous Pcdh-γ gene cluster. cDNA RFLP analysis revealed that the CBA/J-B6 exon was expressed at a level similar to the endogenous 129S-B6 exon (lanes 1,3). Wild-type ES cells serve as a control (lane 5). The RT (minus) control excludes the possibility of genomic DNA contamination.
Taking advantage of the RFLP marker in the B6 exon, we generated a transgene from the CBA/J strain that contains 2.5 kb of 5′ upstream sequence, the B6 coding exon, and 2.1 kb of 3′ intronic sequence. We inserted this transgene at the end of constant exon 3 of the Pcdh-γ gene cluster by gene targeting in mouse ES cells (129S strain) (Fig. 5B). This transgene is transcribed in the same direction as the endogenous locus. An SspI RFLP was used to distinguish the B6 exon encoded by the transgene from the endogenous B6 exon encoded by the 129S allele. RT-PCR was used to recover B6 exon cDNAs from the transgene-targeted ES clones (undifferentiated). RFLP analysis on these cDNAs revealed that the expression of the CBA/J B6 exon was at a similar level compared to the 129S B6 exon from the endogenous locus (Fig. 5B). Thus the 2.5-kb upstream sequences used for this transgene include the promoter cis-elements necessary for transcription of the CBA/J B6 exon.
Interspecies comparative sequence analysis also suggests that individual exons have their own promoters (Wu et al. 2001). This analysis identified a correlation between the distribution of CpG islands and the locations of Pcdh variable exons, significant sequence identity within the 5′ flanking sequences of orthologous variable exons and the existence of a common DNA sequence motif upstream of each individual Pcdh gene. We noticed that this sequence motif is located immediately upstream of the transcription start sites we identified in the 5′ RACE analysis (Fig. 4B).
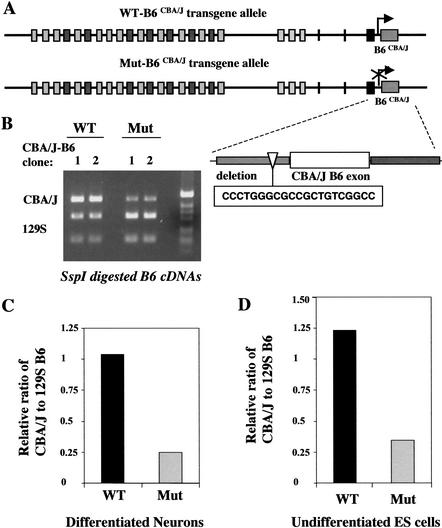
To evaluate the role of the conserved DNA motif, we generated a mutant CBA/J B6 transgene with a deletion of this DNA motif (20 bp) (Fig. 6A). The mutant transgene was targeted to the same location as the wild-type transgene (Fig. 6A) so that we were able to compare the expression levels of the B6 exon independent of any positional effect commonly associated with the integration sites of transgenes. ES cells carrying different transgenes were differentiated into neurons in vitro, and RT-PCR was used to recover Pcdh-γ cDNAs. RFLP analysis on these cDNAs showed that the representation of the CBA/J B6 exon encoded by the mutant transgene was significantly decreased (Fig. 6B). The relative ratio of the expressed CBA/J B6 exon to the 129S B6 exon was decreased to approximately 25% of the levels observed from the wild-type transgene (Fig. 6C). A similar result was also obtained in the undifferentiated ES clones (Fig. 6D). Thus, this conserved DNA motif appears to be important for the transcription of the individual variable exons in both ES cells and neurons. Since a similar conserved DNA motif exists for each variable exon, it is most likely that each variable exon has its own promoter.
Figure 6.
A conserved DNA motif is required for the transcription of each variable exon. (A) Targeting of a mutant CBA/J B6 transgene. This mutant transgene includes a deletion of the 20-bp DNA motif present upstream of each variable exon. The mutant transgene was targeted to the same location as the wild-type (WT) transgene (Fig. 5A). (B) Both the WT transgene- and mutant transgene-targeted ES clones were differentiated into neurons in vitro. RT-PCR and RFLP analysis showed a significant decrease of the CBA/J B6 exon encoded by the mutant transgene. (C,D) Analysis of the relative ratio of the expressed CBA/J-B6 and 129S-B6 transcripts in the differentiated neurons (C) and undifferentiated ES cells (D).
The diversity of Pcdh-γ transcripts
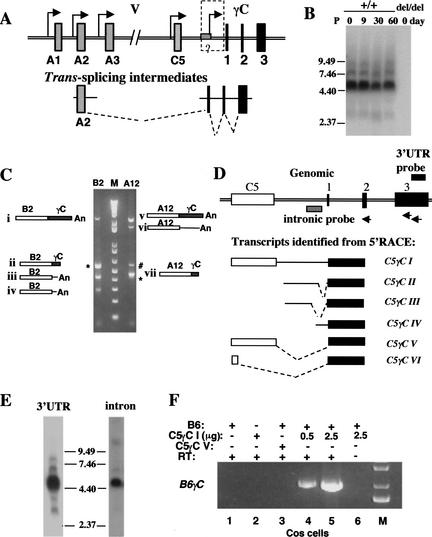
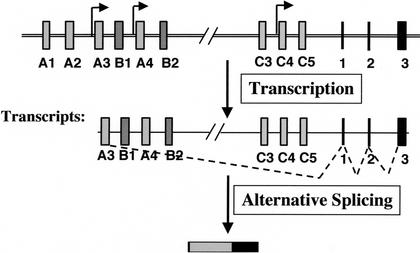
Having demonstrated that multiple promoters exist for individual Pcdh variable exons, we evaluated the possibility of trans-splicing at the Pcdh locus. The trans- splicing model predicts the existence of variable exon-containing 5′ splicing intermediate transcripts and constant exon-containing 3′ splicing intermediate transcripts (Fig. 7A; Wu and Maniatis 1999). Northern blot analysis using a 3′γUTR probe common to all Pcdh-γ members showed that at different stages of brain development, Pcdh-γ transcripts range from 2.5 to 9.0 kb (Fig. 7B). To understand the complexity of these transcripts, we performed a 3′RACE analysis on Pcdh-γ B2 and A12 transcripts using newborn mouse brain RNA (Fig. 7C). Spliced transcripts (Fig. 7C, i, ii, v, and vii) and nonspliced transcripts with intronic sequence downstream of each variable exon (Fig. 7C, iii, iv, and vi) were identified. Similar transcripts are commonly found in EST databases for individual Pcdh variable exons, suggesting that nonspliced transcripts are most likely common to all Pcdh-γ members (Wu and Maniatis 1999). Furthermore, from a 5′RACE analysis using primers in the constant exons, we identified a number of alternatively spliced transcripts between the last variable exon C5 and the constant exons (Fig. 7D, I–VI). Northern blot analysis using the intronic probe confirmed that the intron-containing transcripts are present in the mouse brain (Fig. 7E, right). To exclude the possibility that these alternatively spliced transcripts originated from a larger intermediate RNA, we identified representative cDNA clones by screening duplicate filters of a mouse hypothalamic cDNA library with both intronic and constant exon 3 probes. Sequence analysis of double-positive cDNA clones confirmed that the constant exons of these transcripts were spliced and terminated at an authentic poly A site. The existence of both nonspliced variable exon transcripts and alternatively spliced C5-constant exon transcripts is consistent with the hypothesis that trans-splicing occurs at the Pcdh locus.
Figure 7.
The diversity of Pcdh-γ transcripts. (A) Multiple promoters and trans-splicing model (Wu and Maniatis 1999). (B) Northern blot analysis shows that a variety of Pcdh-γ transcripts are present at different stages of mouse brain development. (C) 3′RACE analysis of Pcdh-γ B2 and A12 transcripts identified both spliced (i, ii, v, and vii) and nonspliced transcripts (iii, iv, and vi). B2-ii and A12-vii are 3′RACE products generated by annealing of an oligo-dT primer in the A-rich region of constant exon 3. The # indicates a background band resulting from single oligo-dT primer amplification. The * indicates PCR fragments containing the chimeric transcripts B2αC or A12αC described in Fig. 9. (D) 5′RACE analysis identified a variety of constant exon-containing transcripts. The schematic drawing illustrates the genomic region containing the Pcdhγ C5 variable exon and three constant exons (not to scale) and alternatively spliced C5γC transcripts (I–VI). The primers used in the 5′RACE are depicted as arrows. (E) Northern blot analysis shows that the intron-containing Pcdh-γ transcripts are present in newborn mouse brain (intronic probe, right panel). (F) In vitro trans-splicing of the Pcdh-γ B6 variable exon with a C5γC intermediate. COS-7 cells were transfected with combinations of the indicated expression vectors. RT-PCR analysis of the RNA from transfected cells shows B6γC trans-spliced transcripts (lanes 4,5) between B6 and C5γC I. Note that trans-splicing did not occur in the cells coexpressing the B6 gene and the C5γC VI cDNA (lane 3).
To test whether trans-splicing can occur between these candidate Pcdh-γ intermediate RNAs, we coexpressed the nonspliced variable exon B6 and the C5γC I transcript in COS cells. RT-PCR analysis detected the spliced B6γC transcript (Fig. 7F, lanes 4,5). Although this result was positive, this is not conclusive because trans-splicing can occur between separate transcripts harboring 5′ and 3′ splicing sites in this type of in vitro assay (Bruzik and Maniatis 1995; Chiara and Reed 1995).
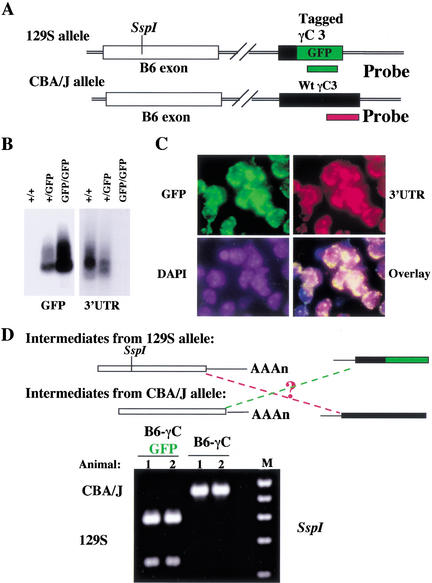
Interallelic trans-splicing is infrequent
In search of in vivo evidence for trans-splicing of Pcdh-γ, we utilized allelic differences in both variable and constant exons. As shown in Figure 8, in a 129S allele, the SspI RFLP marks the B6 variable exon and the constant exon 3 is tagged with GFP by gene targeting (Fig. 1). Mice with the 129S Pcdh-γ-GFP reporter allele were crossed to CBA/J mice to generate heterozygous mice carrying both the reporter allele and the CBA/J-B6 variant. Both Pcdh-γ alleles are actively transcribed in mouse brain, as shown by Northern blot analysis (Fig. 8B) and in single cells as shown by in situ hybridization using allele-specific probes (Fig. 8C). Consequently, intermediate transcripts encoded from each allele coexist in a single cell, allowing us to test whether trans-splicing between intermediates from different alleles can occur (Fig. 8D). From brain RNA, we amplified spliced B6γC cDNAs by RT-PCR, using a common primer in the B6 exon and an allele-specific primer in the constant region. For each type of spliced B6γC cDNA identified, RFLP analysis revealed that the vast majority originated from the same allele (Fig. 8D). To quantify this result, we subcloned the PCR products by TA cloning. Clonal PCR and SspI digestion followed by sequence analysis identified just two interallelic transcripts out of 392 inserts. Thus, interallelic trans-splicing does not occur frequently.
Figure 8.
In vivo assessment of trans-splicing between different Pcdh-γ alleles. (A) Allelic differences in both variable and constant exons. An SspI RFLP detected the allelic difference in the variable B6 exon. The second allelic difference in the constant region was introduced by tagging constant exon 3 with a GFP cDNA in the 129S allele. (B) Northern blot analysis using allele-specific probes shows that both Pcdh-γ alleles are active in transcription (comparing the hybridization signals between heterozygous and homozygous mice). (C) Both Pcdh-γ alleles are actively transcribed in single neurons. Shown is double-fluorescence-labelled in situ hybridization of frozen sections of adult mouse brain (Pcdh-γ+/GFP), using allele-specific probes. (D) Trans-splicing between different alleles is not abundant. Two heterozygous mice were analyzed (1 and 2), each carrying both the targeted allele and CBA/J allele. RFLP analysis of two types of B6γC cDNAs demonstrated that the spliced B6γC transcripts predominantly originated from the same allele. B6γC-GFP cDNA was from the 129S allele; and B6γC cDNA was from the CBA/J allele. Interallelic B6γC transcripts were not abundant.
Low level of Pcdh-α/γ chimeric transcripts
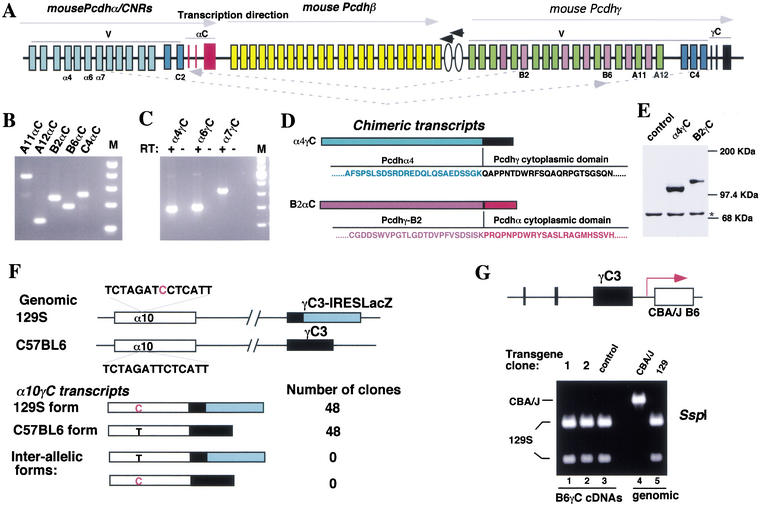
In addition to the Pcdh-γ transcripts containing a γ variable exon and γ constant exons, we identified chimerictranscripts containing Pcdh-γ variable exons (B2 or A12) and Pcdh-α constant exons from the 3′RACE analysis (Fig. 7C). The Pcdh-α, Pcdh-β, and Pcdh-γ gene clusters are located on the same chromosome and are transcribed in the same direction (Fig. 9A). These transcripts can be explained by two possible mechanisms: intermolecular trans-splicing and intramolecular exon scrambling (Nigro et al. 1991). We confirmed the existence of these products by RT-PCR on mouse brain RNA and demonstrated the existence of trans-splicing products between Pcdh-γ variable exons and Pcdh-α constant exons for all the members we tested (Fig. 9B). Furthermore, we identified transcripts containing a Pcdh-α variable exon and Pcdh-γ constant exons (Fig. 9C).
Figure 9.
Pcdh-α/γ chimeric transcripts. (A) Schematic diagram of the mouse Pcdh gene clusters. Note that all three clusters are transcribed in the same direction, and two additional genes illustrated by ovals are transcribed in the opposite direction. (B) RT-PCR analysis identified chimeric transcripts between the Pcdh-γ variable exons A11, A12, B2, B6, and C4 and the Pcdh-α constant exons (αC) from mouse brain RNA. (C) RT-PCR analysis identified chimeric transcripts between the Pcdhα variable exons α4, α6, and α7 and the Pcdh-γ constant exons (γC). (D) The chimeric proteins (α4γC and B2αC) encoded by the chimeric transcripts. (E) Western blot analysis confirmed the expression of an in-frame α4γC protein in transfected cells. The * indicates a background band serving as the loading control. (F) Interallelic trans-splicing assay of α10γC chimeric transcripts (similar to Fig. 8). An SNP (red C on 129S strain α10 exon) and the targeted IRES-LacZ cassette define allelic differences in both α10 variable exon and γ constant exon 3. Sequence analysis of two different types of α10γC cDNAs showed that each originated from its original allele, suggesting that trans-splicing might have occurred on the same chromosome. (G) Trans-splicing is infrequent on the same chromosome within the Pcdh-γ cluster. The CBA/J-B6 transgene was targeted to the end of the Pcdh-γ locus and is transcribed in the same orientation as the endogenous locus. This mimics the configuration of the γ variable exon spliced to the α constant exons (in B). RT-PCR was used to amplify spliced B6γC cDNA from the undifferentiated ES cells using a specific primer for the constant exon. RFLP analysis of the RT-PCR products showed that splicing predominantly occurred from the endogenous 129S-B6 exon to the γ constant exons but not from the CBA/J-B6 exon (lanes 1 and 2, cf. lane 3 of control ES cells).
To assess the relative abundance of these transcripts, we screened both unamplified and amplified mouse hypothalamic cDNA libraries. From two million recombinants, we obtained over 500 positive clones with a Pcdh-γ 3′UTR probe and over 100 positive clones with a Pcdh-α 3′UTR probe. Screening duplicate filters with probes to α variable exons (α4, α6, α7, and α10) and γ constant exons identified only one double-positive clone. Thus, the α/γ chimeric transcripts are expressed at a much lower level (0.2%) compared to the regular Pcdh transcripts. Sequence analysis confirmed that all chimeric transcripts have an in-frame fusion, for instance, between the Pcdh-α4 exon and Pcdh-γ constant exons, or between the Pcdh-γ B2 exon and Pcdh-α constant exons (Fig. 9D). Overexpression of the α4γC cDNA in COS cells gave rise to a protein of the expected size (Fig. 9E). To determine whether the α/γ chimeric transcripts originated from the different alleles or the same allele, we performed an interallelic trans-splicing assay using an α10 variable exon SNP between 129S and C57BL6 strains (Fig. 9F). Sequence analysis confirmed that α10γC transcripts predominantly originated from the same allele.
Chimeric transcripts between different Pcdh gene clusters occurred at a frequency similar to those generated by interallelic trans-splicing, favoring the interpretation that these transcripts are generated through trans-splicing rather than exon scrambling. However, analysis of the chimeric α10γC transcripts suggested that trans-splicing must have occurred from intermediates from the same chromosome, presumably through a transcription-coupled mechanism if these transcripts were indeed generated by trans-splicing.
To test the idea of trans-splicing on the same chromosome within the same gene cluster, we again used the CBA/J B6 transgene-targeted ES cells. The transgene mimics the configuration of γ variable exons spliced to α constant exons. Although the CBA/J-B6 exon was expressed at a similar level to the endogenous B6 exon mRNA in the targeted ES cells (Fig. 5B), most of the spliced B6γC transcripts were still generated from the endogenous 129S-B6 exon (Fig. 9G). RFLP and sequence analyses of spliced B6γC cDNAs showed that only 0.4% were generated by trans-splicing from the CBA/J B6 exon. Since the targeted CBA/J-B6 transgene was actively transcribed, it should contribute 50% of the spliced mRNA species from the same allele. This result does not support the idea that trans-splicing is a major mechanism in generating spliced Pcdh-γ mRNA. However, this assay could not rule out the possibility that trans-splicing occurs in a highly transcription-coupled fashion and can only proceed efficiently in its native configuration.
Cis-splicing of Pcdh-γ in the absence of possible promoter(s) for the constant region
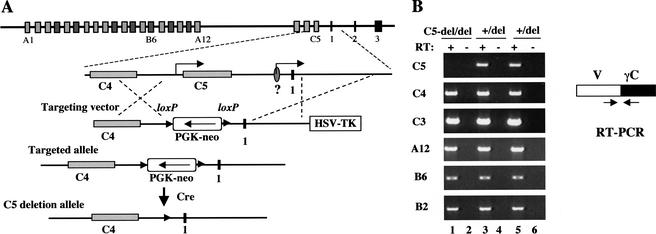
The interallelic trans-splicing and CBA/J B6 transgene assays suggested that trans-splicing is rare, but did not completely rule out this possibility. Therefore, we designed another allele to disprove the trans-splicing model. An essential component of the trans-splicing hypothesis is that a putative promoter somewhere upstream of the constant region drives the expression of constant exon-containing intermediate transcripts (Fig. 7A). These intermediates are essential for the trans-splicing to occur. The likely locations of the promoter include the region upstream of the constant exon 1, and the promoter for the last variable exon. Highly conserved DNA sequences between mouse and human exist a few hundred basepairs upstream of constant exon 1 of Pcdh-α and Pcdh-γ (Wu et al. 2001), suggesting the possibility that these sequences may function as a transcription regulatory element. From 5′RACE analysis, we observed that some constant exon-containing cDNAs initiated downstream of the conserved sequences (Fig. 7D, IV). However, we do not know whether these cDNAs were derived from incomplete cDNA synthesis of a larger RNA species. Other cDNAs identified from 5′RACE contained the conserved DNA sequences or extended further 5′ to the C5 variable exon (Fig. 7D, I–III). This led to the speculation that the promoter for the C5 variable exon may serve as a promoter to generate constant exon-containing intermediates.
We constructed a targeting vector that was designed to generate a 5-kb deletion, which removed both the conserved DNA sequence and the whole C5 gene including the promoter (Fig. 10A). This modification was generated in the ES cells in which one allele of the Pcdh-γ gene cluster had been deleted. We were able to directly measure the effect of this mutation on the splicing of Pcdh-γ mRNAs. In the feeder-free C5-deleted ES cells, the C5γC cDNA was absent as expected (Fig. 10B, C5 panel, lane 1). However, other individual variable exons were still spliced to constant exons (Fig. 10B), suggesting that the expression of constant exon containing intermediates is not necessary for the generation of spliced Pcdh-γ mRNAs. One possible argument against this interpretation is that the upstream C4 promoter might substitute for the function of the original prooter(s) for the constant region in the C5-deleted cells. However, this did not appear to be the case. Although the C4 expression was significantly decreased in the C5-deleted clones prior to Cre excision of the PGK-Neo cassette (possibly due to the transcription of Neo cassette in the opposite orientation), other distal variable exons were still efficiently spliced to the constant exons (data not shown). These results provided strong evidence against trans-splicing as a major mechanism for generating spliced mRNAs. Therefore, functional Pcdh-γ mRNA is most likely generated through cis- alternative splicing in vivo.
Figure 10.
Cis-splicing in the absence of upstream sequence from constant exon 1. (A) Deletion of C5 and conserved upstream DNA sequences from constant exon 1. A 5-kb sequence containing the entire C5 gene and the conserved DNA sequences upstream of constant exon 1 was deleted by gene targeting in ES cells carrying only one Pcdh-γ allele. (B) RT-PCR analysis showed that individual variable exons (C3, C4, B2, B6, and A12) were efficiently spliced to γ constant exons in the absence of the 5-kb upstream sequence from constant exon 1.
Discussion
A model for cell-specific expression of individual Pcdh-γ genes
A combination of molecular and genetic analyses led us to conclude that each Pcdh-γ variable exon is under the control of its own promoter. Trans-splicing beween variable exon-containing and constant exon-containing intermediates exists, but only occurs at a very low frequency compared to cis-splicing events. Therefore, the major mechanism for the generation of functional transcripts is through cis-alternative splicng. In this model (Fig. 11), cell-specific expression of protocadherins is determined by a combination of differential promoter activation and cis-alternative splicing.
Figure 11.
Cell-specific expression of a subset of Pcdh genes: Differential promoter activation and cis-alternative splicing. In this model, individual cells express a distinct subset of protocadherins. The expression of the specific combination of protocadherins is achieved by differential promoter activation, followed by cis-alternative splicing.
Alternative splicing, a mechanism generating molecular diversity in the nervous system
It is a general notion that the large neuronal diversity and staggering complexity of neuronal processes and connections in the nervous system require significant diversity of cell surface molecules to specify a neuronal type and its connectivity. The understanding of molecular diversity and the mechanisms generating it may provide a guideline to unveil sophisticated neural networks. The discovery of Drosophila DSCAM, an axon guidance molecule, is a remarkable demonstration of the enormous number of distinct mRNAs and proteins that can be generated by alternative splicing (Schmucker et al. 2000). In this case, alternative splicing could potentially produce a total of 38,016 different DSCAM proteins if all combinations of these exons were used. Protocadherin gene clusters (Pcdh) represent another example of a genomic structure that could generate significant diversity in the nervous system. Of the possible mechanisms, our present analysis demonstrated that cis-alternative splicing is the one utilized to generate functional Pcdh mRNAs. The number of protocadherins that can be generated from the Pcdh gene cluster is greatly reduced compared to the Drosophila DSCAM gene. However, from earlier studies of Pcdh-α (CNR) (Kohmura et al. 1998) and our study of Pcdh-γ gene products, it is clear that single neurons express an overlapping but distinct subset of Pcdh molecules. This raises the possibility that a significant number of combinatorial protocadherin codes could impart connection specificity. It appears that alternative splicing is widely used to generate large mRNA and protein diversity in the nervous system. However, it remains a challenge to explain what might control DSCAM or Pcdh splicing, since the mechanisms of regulating alternative splicing are poorly understood.
DNA recombination in the nervous system?
Of all the proposed mechanisms for generating protocadherin diversity, the most intriguing one is somatic DNA recombination. The possible involvement of DNA recombination for neural development came from the observation that mice lacking functional XRCC4 and LigIV genes, which are required for double-stranded DNA break repair, die during embryogenesis, largely due to massive apoptosis of postmitotic neurons (Gao et al. 1998; Chun and Schatz 1999). The resemblance of the Pcdh gene clusters to the Ig and TCR genes made this possibility very intriguing (Wu and Maniatis 1999). However, it was clear at the outset that RAG1/2 recognition sequences are not present in the Pcdh locus (Wu and Maniatis 1999). Our analysis revealed that the Pcdh locus is different from the immunoglobulin locus in many ways. Both Pcdh-γ alleles are active in a single cell, clearly different from allelic exclusion in the immune system. A single allele can encode multiple forms of Pcdh genes. Somatic recombination has not been detected. Moreover, mice deficient for the entire Pcdh-γ locus exhibit relatively late and regional onset of neuronal cell death and degeneration (X. Wang, J.A. Weiner, S. Levi, A. Craig, A. Bradley, and J.R. Sanes, in prep.), clearly different from early extensive postmitotic neuronal death observed in mice deficient in XRCC4 and LigIV genes. In summary, we did not obtain any evidence suggesting that DNA recombination occurs at the Pcdh locus.
The significance of trans-splicing and Pcdh-chimeric transcripts
Trans-splicing, a process of joining two RNA precursors (Konarska et al. 1985; Solnick 1985; Agabian 1990), is used to generate mRNA and protein diversity in many organisms, including trypanosoma, nematodes, plants, and fruit flies (Bonen 1993; Dorn et al. 2001). In mammalian cells, trans-splicing has been described mostly as a way to duplicate exons, and often occurs at a low frequency (Chapdelaine and Bonen 1991; Eul et al. 1995; Akopian et al. 1999; Frantz et al. 1999; Chatterjee and Fisher 2000; Takahara et al. 2000). Our present findings show that trans-splicing between two distinct Pcdh gene clusters does generate a novel set of chimeric transcripts but at a low frequency. The functional significance if any of this low level of trans-spliced protocadherins remains to be determined. Nevertheless, it is worth mentioning that a trans-spliced protocadherin shares the same homophilic cadherin-like domain as its cis-spliced counterpart, but the cytoplasmic domains encoded by Pcdh-α and Pcdh-γ constant exons respectively are distinct in their sequences and mostly likely in their functions. For example, the cytoplasmic domain of Pcdh-α (CNR) has been shown to bind to Fyn tryosine kinase, but Pcdh-γ's does not (Kohmura et al. 1998). Therefore, homophilic interactions between cis-and trans-spliced protocadherins may provide a basis for asymmetric signaling at presynaptic and postsynaptic terminals of synapses (Sudhof 2001). Alternatively, such molecules may provide a crosslink between two distinct intracellular signaling pathways.
The unresolved questions
In this study, we obtained evidence that is most consistent with a model of multiple promoters and cis-alternative splicing for achieving cell-specific Pcdh gene expression. Several issues remain to be resolved. At the transcriptional level, how does each cell selectively express one particular subset of Pcdh genes? Two possible nonexclusive mechanisms might be involved. First, the context of the proximal promoter for each variable exon may determine the transcription activity in a cell-specific manner. Second, a distal enhancer (e.g., locus control region, LCR) may contribute to the specific expression of individual variable exons. A rational step to advance our understanding of the transcriptional regulation of Pcdh gene expression is to identify transcription factors that bind to the common DNA motif upstream of each variable exon. The identification of such factors and cell-specific proteins that interact with the factors may eventually shed light on our understanding of how the cell-specific Pcdh expression pattern is achieved. At the posttranscriptional level, the mechanism of alternative splicing that enables individual transcripts to be unresponsive to downstream splicing sites and efficiently assemble functional Pcdh mRNAs remains unknown.
Materials and methods
Gene targeting
To target the Pcdh-γ constant exon 3, we constructed a targeting vector. The 5′ homology arm is a NotI/SalI-digested 6.4-kb fragment (corresponding to genomic sequence between constant exons 2 and 3 including part of constant exon 3) generated by long-range PCR with two primers (3774, 5′-CTTATGCGGC CGCAGAGTGATCACGTGATCCACTGAG-3′; 3775, 5′-CAT ATGTCGACATCGAGCGCTCAGACCCATGGTGC-3′). An IRES-GFPLacZ fusion gene reporter and floxed PGK-neo positive selection marker was placed downstream of the 5′ homology arm. The 3′ homology arm is a NheI/EcoRI-digested 1.6-kb PCR fragment amplified using two primers (3776: 5′-ACTAT GCTAGCCTTGAACCACAAGGCAGC-3′; 3777, 5′-CGATGA ATTCAGGAGCTACGTGACTATGC-3′). Downstream of the 3′ arm is a PolII TK negative selection marker. We identified two targeting events by long-range PCR using the primer pairs: 3769 (5′-CGATGAATTCAGGAGCTACGTGACTATGC-3′) and 3984 (5′-CGAGATCAGCAGCCTCTGTTCCACATACA C-3′) to detect recombination on the 3′ side, and the primer pairs 3755 (5′-GATACAGAGATGCTGCAAGCCATGATCT TGGCCTCT-3′) and 3744 (5′-GTTTACGTCGCCGTCCAGCT CGAC-3′) to detect 5′ recombination.
To target the CBA/J-B6 transgene to the 3′ end of the Pcdh-γ constant region, we constructed an insertion targeting vector. The homology region is a HindIII/SfiI-digested 5-kb fragment generated by PCR with the primer pairs (5466, 5′-CGACCCCTC CCTGTACTGACTTCTCTA-3′; 5472, 5′-GCATTGGCCATCA TGGCCAGGATCTCATAGCTGAGG-3′). This contains the last 138 bp of Pcdh-γ constant exon 3 and the downstream genomic sequences. Downstream of the homology region is a 6.3-kb fragment containing the CBA/J-B6 genomic sequences amplified with PCR primer pair 5473 (5′-GCATTGGCCATGATGGCCTTTA ACTCCAGCACTCA-3′) and 5475 (5′-CTCTAGGCGCGCCAC TATAATATTTCAGCAAATGC-3′). A PGK-Neo selection cassette was placed in the opposite orientation to the transgene. The vector was linearized by Asp718/MfeI double digestion to generate the gap (also used as a probe to detect the targeted insertion). The targeted clones were identified by PCR with primer set 1 (GAP primer, 5′-GTTTAGCCAGAAGCCATGCAATTGTGGC T-3′; and vector primer, 5′-CCTGAGTGCTGGAGTTAAAGGC CATCAT-3′) and primer set 2 (5466, 5′-CGACCCCTCCCTGTA CTGACTTCTCTA-3′; and 3984, 5′-CGAGATCAGCAGCCTC TGTTCCACATACAC-3′). The PCR positive clones were confirmed by Southern blot analysis with the gap probe (Asp718/MfeI fragment). The deletion of the conserved motif in the CBA/J-B6 transgene was carried out using overlapping PCR.
To delete the upstream sequences of constant exon 1, we constructed a targeting vector that contained an Asp817/Nhe I-digested 3.9-kb genomic fragment generated by PCR with the primers (5616, 5′-CGGTAGGTACCAGTCTCAGATAGCCA C-3′; and 5617, 5′-GCGTTGCTAGCCACAATGGCCTCTAG GTT-3′), a floxed PGK-Neo selection marker, and a BamHI/NotI-digested 3.1-kb fragment amplified by PCR with the primers (5619, 5′-CGTTATGCGGCCGCCTTGAACTTCACCCT GGTT-3′; and 5221, 5′-CCTGTCAGATCACCATCCCATAT CGTCACGT-3′), followed by an HSV-TK negative selection marker. The targeting events were identified by long-range PCR. Transient expression of Cre in ES cells removed the PGK-Neo cassette. The Pcdh-γ-GFP allele and Pcdh-γ deletion allele were generated as described elsewhere (X. Wang, J.A. Weiner, S. Levi, A. Craig, A. Bradley, and J.R. Sanes, in prep.). Gene targeting, blastocyst injection, and chimeric mouse production were carried out as described (Ramirez-Solis et al. 1993). The targeted allele was generated in AB2.2 129S7 ES cells, and the germline-transmitted allele has been maintained in a C57BL6J/129S7 hybrid background. In vitro neuronal differentiation of ES cells was carried out as described (Bain et al. 1995; Gajovic et al. 1997).
Histology
For LacZ staining, anesthetized mice were perfused with ice-cold 4% paraformaldehyde in phosphate-buffered solution. Vibratome- or cryostat-cut brain sections were collected and stained with X-gal overnight at room temperature to visualize the β-galactosidase activity. For immunofluorescent staining, frozen sections were stained using rabbit anti-GFP (Molecular Probes) and mouse anti-SV2 (Developmental Studies Hybridoma Bank). For in situ hybridization, allelic specific riboprobes were synthesized using T3/T7 RNA polymerase (Invitrogen) labeled with digoxigenin or fluorescein, and hybridized to frozen sections. Both riboprobes were hybridized together and detected sequentially using the TSA Plus system (Perkin-Elmer Life Sciences).
RNA isolation
Total RNAs from mouse tissues or cultured cells were prepared with an RNAgents Total RNA Isolation System (Promega). To eliminate DNA contamination, some RNA samples were treated with RNase-free DNase (Roche) followed by second purification using the QIAGEN RNeasy Mini Kit.
Cell transfection and RNA, DNA, and protein analysis
COS-7 cells were purchased from ATCC. The DEAE-Dextran transfection, Northern blot, Southern blot, and Western blot analyses were performed as described (Wang et al. 1998).
3′, 5′ RACE and RT-PCR analyses
3′ RACE analysis of Pcdhγ RNAs was performed essentially according to the Clontech SMART RACE cDNA Amplification protocol. Three μg of total RNA was primed with oligo-dT in cDNA synthesis. The oligo dT primer (5′-ATTCTAGAGGC CTGATCGGCCGACATGT30VN-3′), B2-specific primers (1S, 5′-CTACCAGAAGATTCTCCACTAGGAACAGTCAT-3′; 3S, 5′-AGGCGATACCGGAATACAACGTCACCAT-3′; 5S, 5′-GT ATTGCCTCACAGTCAGCTAAGACAGAGT-3′), and A12-specific primers (1S, 5′-CATACTGGGCCTACATCCCAGA GAATAAC-3′; 3S, 5′-CACCTCCGAAACTTCAGGGCTCAC T-3′; 5S, 5′-GGGTGATTCGGTTTTTTCTAAAGACAATCAT GC-5′) were used in three rounds of 3′RACE PCR amplification.
The 5′ RACE analysis was carried out according to GIBCO BRL's 5′RACE protocol with modifications. The modification included the tailing of cDNA with dATP (750 nM) and the subsequent PCR amplification of the tailed cDNA with the same oligo-dT primer used in the 3′RACE analysis. In the 5′RACE reaction, 3 μg of total RNA was used for cDNA synthesis with a gene-specific primer (γC2AS, 5′-CAAGTAAGGACGAGCTG GACT-3′). Primers used in three rounds of PCR amplifications were: γC4AS (5′-TCCGTATCGAGCGCTCAGACCCATGGT3′), γC6AS (5′-GAGATGCTGCAAGCCATGATCTTGGCCT C-3′), and γC8AS (5′-GGCCAGGTGCCAGTTTCATCACCA TT-3′). To identify transcription start sites for A12 and B2 variable exons, the following primers were used in the 5′ RACE analysis. A12 5′RACE 2AS (5′-GAGCTGGTAGCTCTGAAGC GAGT-3′), 4AS (5′-GAAGTAGGGAGCGTTGTCGTTAATG TC-3′) and 6AS (5′-CAGCTCCTCGGGAACCGAATAGC-3′); B2 5′RACE 2AS (5′-CTGGTCTGGTGGACTCTCCAATG-3′), 4AS (5′-GTCGTTCACATCCTGAAGAATTACAGC-3′), and 6AS (5′-GCGAGGTTTCCTACTATCGAGTTCTTG-3′).
In the RT-PCR analysis, 2 μg of total RNA was used in cDNA synthesis with oligo-dT or a gene-specific primer. To detect the RFLP in the B6 exon, B6.1S (5′-GTTAGTGCTGCAAGTGAC AGCTTC-3′) and B6.2AS (5′-CACTAACTTGTAATAATTCT TGGAGGTGG-3′) were used to amplify the B6 sequence from either genomic DNA or cDNA. To detect the B6γC transcripts shown in Figure 7F, cDNAs were synthesized with the primer (pcDNA.6AS, 5′-GCGAGCTCTAGCATTTAGGTGACAC-3′) and amplified with primer pairs (B6.3S, 5′-GCTCCTCAATTC GAGAAGAAAGAAACG-3′; and γC8AS, see above). To detect B6γC-GFP transcripts (Fig. 8D), cDNA was synthesized with GFP.2AS (5′-GGTGCGCTCCTGGACGTAG-3′) and amplified with B6.3S and GFP.4AS (5′-GTTTACGTCGCCGTCCAGCT CGAC-3′). For B6γCWt transcripts, cDNAs were synthesized with the primer 5232.AS (5′-GCCCTGGCTCCTACCAGTAT CCT-3′) and amplified with B6.3S and 5227.AS (5′-GGTACA AGTAAGGACGAGCTGGACTGGCTG-3′). To detect α10γC-IRESLacZ transcripts (Fig. 9F), cDNA was synthesized with the primer (IRES.2AS, 5′-CCTAGGAATGCTCGTCAAGAAGAC AG-3′) and amplified with 5541 (5′-CTGGTGCCTGCTGCT CTCACTTTTG-3′) and IRES.4AS (5′-CAAGCGGCTTCGGC CAGTAACGTTAG-3′). To detect α10γC transcripts, cDNA was synthesized with the primer (C.2AS, 5′-GTGGTTCAAG GCCTGGCCTCCAT-3′) and amplified with 5541 and C.4AS (5′-GAGCCTTGCCATCTCGTTTGCCAGC-3′). To detect B6γC transcripts (Fig. 9G), cDNAs were synthesized and amplified with the primers (5232.AS, B6.3S and γC8AS, see above). To detect trans-splicing products between Pcdh-γ and Pcdh-α from mouse brain RNAs, the cDNAs were synthesized with 5232.AS (see above) or 5457.αAS (5′-GTTAGAGGATGGCCATCAT-3′). The following primer pairs were used in RT-PCR to identify the trans-splicing products: A11.1S (5′-TGGTACTGCTGGCTCT CAAGCTTAG-3′) and αC8AS (5′-GAATGCCAGCCTCCTC CAGGTG-3′), A12.5S (5′-GGGTGATTCGGTTTTTTCTAAA GACAATCATGC-3′) and αC8AS, B2.5S (5′-GTATTGCCTCA CAGTCAGCTAAGACAGAGT-3′) and αC8AS, B6.5360 (5′-CA CCTTTGCCTTCTTCTCAAGAGATCGT-3′) and αC8AS, C4.7S (5′-CCTACTCGGAGTGATAGCTTCATGATG-3′) and αC6AS (5′-TCTGGTGTTGCACTGGATACTGTTGG-3′), α4.1S (5′-CA CTGTCCCCAGTGAGAGCGTGT-3′) and γC8AS, α6.1S (5′-CG GTGCTCAGTTCCACCTACAGAG-3′), and γC8AS, α7.1S (5′-G CCAAACACAGAAGGTCTCATCTAAGGT-3′) and γC8AS. To detect the multiple forms of Pcdh-γ described in Figures 3 and 10, A11.1S, A12.3S, A12.5S, B2.5S, B6.5360, C4.7S, γC8AS (see above), C3.1S (5′-AGGCAGATTCTCACAGTCTTGATCTC AGA-3′), C4.1S (5′-GTTATGCAGGGGAAGTACGGACTG CT-3′), C5.1S (5′-CTCTGTACTGTGGCTGCCTCTGATC-3′), and B2.5177 (5′-AGGCGATACCGGAATACAACGTCACCAT-3′) were used in RT-PCR. Single-cell RT-PCR analysis was carried out according to the instruction manual (Cell-to-cDNA, Ambion) using the primer GFP.2AS for RT. A12.3S, A11.1S, B2.1S, 5437 (5′-CAGAGATGCTGCAAGCCATGATCTTG-3′), and GFP.4AS were used for the first-round PCR, and A12.5S, A11.3S, B2.5177, and γC8AS were used for the nested PCR.
cDNA, genomic DNA cloning, and expression vector construction
To identify mouse Pcdh-γ cDNAs, we screened a mouse hypothalamic/thalamic cDNA ZAPII phage library with a variety of probes. The PCR-amplified cDNA or genomic DNA fragments were cloned using the TOPO TA cloning kit (Invitrogen). The identities of all RACE and RT-PCR products in the studies were confirmed by sequence analysis. The expression vectors for a variety of Pcdh cDNAs were constructed by cloning restriction enzyme-digested or PCR-amplified DNA fragments into either pcDNA3 or pcDNA3.1 V5/His TOPO TA vector (Invitrogen).
GenBank sequence submission
All sequences reported here have been submitted to GenBank; the accession nos. are AF464151–AF464181.
Acknowledgments
We thank S. Vaishnav, L. Arango, G. Schuster, S. Rivera, and J. Wesley for technical assistance. This work was supported by grants from the NIH to A.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL abradley@sanger.ac.uk; FAX 44-1223-494714.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1004802.
References
- Agabian N. Trans splicing of nuclear pre-mRNAs. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- Akopian AN, Okuse K, Souslova V, England S, Ogata N, Wood JN. Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett. 1999;445:177–182. doi: 10.1016/s0014-5793(99)00126-x. [DOI] [PubMed] [Google Scholar]
- Bain G, Kitchens D, Yao M, Huettner JE, Gottlieb DI. Embryonic stem cells express neuronal properties in vitro. Dev Biol. 1995;168:342–357. doi: 10.1006/dbio.1995.1085. [DOI] [PubMed] [Google Scholar]
- Benson DL, Colman DR, Huntley GW. Molecules, maps and synapse specificity. Nat Rev Neurosci. 2001;2:899–909. doi: 10.1038/35104078. [DOI] [PubMed] [Google Scholar]
- Bonen L. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 1993;7:40–46. doi: 10.1096/fasebj.7.1.8422973. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron. 2000;28:245–259. doi: 10.1016/s0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- Bruses JL. Cadherin-mediated adhesion at the interneuronal synapse. Curr Opin Cell Biol. 2000;12:593–597. doi: 10.1016/s0955-0674(00)00137-x. [DOI] [PubMed] [Google Scholar]
- Bruzik JP, Maniatis T. Enhancer-dependent interaction between 5′ and 3′ splice sites in trans. Proc Natl Acad Sci. 1995;92:7056–7059. doi: 10.1073/pnas.92.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Lampugnani MG, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oostuyse B, Dewerchin M, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Chapdelaine Y, Bonen L. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: A trans-splicing model for this gene-in-pieces. Cell. 1991;65:465–472. doi: 10.1016/0092-8674(91)90464-a. [DOI] [PubMed] [Google Scholar]
- Chatterjee TK, Fisher RA. Novel alternative splicing and nuclear localization of human RGS12 gene products. J Biol Chem. 2000;275:29660–29671. doi: 10.1074/jbc.M000330200. [DOI] [PubMed] [Google Scholar]
- Chiara MD, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- Chun J, Schatz DG. Rearranging views on neurogenesis: Neuronal death in the absence of DNA end-joining proteins. Neuron. 1999;22:7–10. doi: 10.1016/s0896-6273(00)80671-6. [DOI] [PubMed] [Google Scholar]
- Dorn R, Reuter G, Loewendorf A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc Natl Acad Sci. 2001;98:9724–9729. doi: 10.1073/pnas.151268698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eul J, Graessmann M, Graessmann A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J. 1995;14:3226–3235. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- Frantz SA, Thiara AS, Lodwick D, Ng LL, Eperon IC, Samani NJ. Exon repetition in mRNA. Proc Natl Acad Sci. 1999;96:5400–5405. doi: 10.1073/pnas.96.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajovic S, St-Onge L, Yokota Y, Gruss P. Retinoic acid mediates Pax6 expression during in vitro differentiation of embryonic stem cells. Differentiation. 1997;62:187–192. doi: 10.1046/j.1432-0436.1998.6240187.x. [DOI] [PubMed] [Google Scholar]
- Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Vielmetter E, Bronner-Fraser M. N-Cadherin, a cell adhesion molecule involved in establishment of embryonic left-right asymmetry. Science. 2000;288:1047–1051. doi: 10.1126/science.288.5468.1047. [DOI] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin adhesive activity. J Cell Biol. 2000;148:399–404. doi: 10.1083/jcb.148.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Ono T, Yan Q, Wang X, Sonta S, Suzuki ST. Protocadherin 2C: A new member of the protocadherin 2 subfamily expressed in a redundant manner with OL-protocadherin in the developing brain. Biochem Biophys Res Commun. 1999;260:641–645. doi: 10.1006/bbrc.1999.0950. [DOI] [PubMed] [Google Scholar]
- Homayouni R, Rice DS, Curran T. Disabled-1 interacts with a novel developmentally regulated protocadherin. Biochem Biophys Res Commun. 2001;289:539–547. doi: 10.1006/bbrc.2001.5998. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Johnson KJ, Patel SR, Boekelheide K. Multiple cadherin superfamily members with unique expression profiles are produced in rat testis. Endocrinology. 2000;141:675–683. doi: 10.1210/endo.141.2.7334. [DOI] [PubMed] [Google Scholar]
- Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Konarska MM, Padgett RA, Sharp PA. Trans splicing of mRNA precursors in vitro. Cell. 1985;42:165–171. doi: 10.1016/s0092-8674(85)80112-4. [DOI] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, Zipursky SL. N-cadherin regulates target specificity in the Drosophila visual system. Neuron. 2001;30:437–450. doi: 10.1016/s0896-6273(01)00291-4. [DOI] [PubMed] [Google Scholar]
- Manabe T, Togashi H, Uchida N, Suzuki SC, Hayakawa Y, Yamamoto M, Yoda H, Miyakawa T, Takeichi M, Chisaka O. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. Mol Cell Neurosci. 2000;15:534–546. doi: 10.1006/mcne.2000.0849. [DOI] [PubMed] [Google Scholar]
- Marrs JA, Nelson WJ. Cadherin cell adhesion molecules in differentiation and embryogenesis. Int Rev Cytol. 1996;165:159–205. doi: 10.1016/s0074-7696(08)62222-6. [DOI] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Obata S, Sago H, Mori N, Davidson M, St John T, Suzuki ST. A common protocadherin tail: Multiple protocadherins share the same sequence in their cytoplasmic domains and are expressed in different regions of brain. Cell Adhes Commun. 1998;6:323–333. doi: 10.3109/15419069809010791. [DOI] [PubMed] [Google Scholar]
- Obata S, Sago H, Mori N, Rochelle JM, Seldin MF, Davidson M, St John T, Taketani S, Suzuki ST. Protocadherin Pcdh2 shows properties similar to, but distinct from, those of classical cadherins. J Cell Sci. 1995;108:3765–3773. doi: 10.1242/jcs.108.12.3765. [DOI] [PubMed] [Google Scholar]
- Phillips GR, Huang JK, Wang Y, Tanaka H, Shapiro L, Zhang W, Shan WS, Arndt K, Frank M, Gordon RE, et al. The presynaptic particle web: Ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32:63–77. doi: 10.1016/s0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Ramirez-Solis R, Davis AC, Bradley A. Gene targeting in embryonic stem cells. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- Sano K, Tanihara H, Heimark RL, Obata S, Davidson M, St John T, Taketani S, Suzuki S. Protocadherins: A large family of cadherin-related molecules in central nervous system. EMBO J. 1993;12:2249–2256. doi: 10.1002/j.1460-2075.1993.tb05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- Serafini T. Finding a partner in a crowd: Neuronal diversity and synaptogenesis. Cell. 1999;98:133–136. doi: 10.1016/s0092-8674(00)81008-9. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Solnick D. Trans splicing of mRNA precursors. Cell. 1985;42:157–164. doi: 10.1016/s0092-8674(85)80111-2. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. In: The synaptic cleft and synaptic cell adhension. Cowan M, Sudhof TC, Stevens CF, editors. Baltimore, MD: The Johns Hopkins University Press; 2001. [Google Scholar]
- Sugino H, Hamada S, Yasuda R, Tuji A, Matsuda Y, Fujita M, Yagi T. Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics. 2000;63:75–87. doi: 10.1006/geno.1999.6066. [DOI] [PubMed] [Google Scholar]
- Suzuki ST. Protocadherins and diversity of the cadherin superfamily. J Cell Sci. 1996;109:2609–2611. doi: 10.1242/jcs.109.11.2609. [DOI] [PubMed] [Google Scholar]
- Suzuki ST. Recent progress in protocadherin research. Exp Cell Res. 2000;261:13–18. doi: 10.1006/excr.2000.5039. [DOI] [PubMed] [Google Scholar]
- Takahara T, Kanazu SI, Yanagisawa S, Akanuma H. Heterogeneous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J Biol Chem. 2000;275:38067–38072. doi: 10.1074/jbc.M002010200. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: A molecular family important in selective cell–cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Shan W, Phillips GR, Arndt K, Bozdagi O, Shapiro L, Huntley GW, Benson DL, Colman DR. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang T, Cheng JF, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan JP, Zhang MQ, Myers RM, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Takeichi M. Cadherin superfamily genes: Functions, genomic organization, and neurologic diversity. Genes & Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- Yamagata M, Herman JP, Sanes JR. Lamina-specific expression of adhesion molecules in developing chick optic tectum. J Neurosci. 1995;15:4556–4571. doi: 10.1523/JNEUROSCI.15-06-04556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap AS, Brieher WM, Gumbiner BM. Molecular and functional analysis of cadherin-based adherens junctions. Annu Rev Cell Dev Biol. 1997;13:119–146. doi: 10.1146/annurev.cellbio.13.1.119. [DOI] [PubMed] [Google Scholar]