Abstract
Smad3 transduces the signals of TGF-βs, coupling transmembrane receptor kinase activation to transcriptional control. The membrane-associated molecule SARA (Smad Anchor for Receptor Activation) recruits Smad3 for phosphorylation by the receptor kinase. Upon phosphorylation, Smad3 dissociates from SARA and enters the nucleus, in which its transcriptional activity can be repressed by Ski. Here, we show that SARA and Ski recognize specifically the monomeric and trimeric forms of Smad3, respectively. Thus, trimerization of Smad3, induced by phosphorylation, simultaneously activates the TGF-β signal by driving Smad3 dissociation from SARA and sets up the negative feedback mechanism by Ski. Structural models of the Smad3/SARA/receptor kinase complex and Smad3/Ski complex provide insights into the molecular basis of regulation.
Keywords: TGF-β, Smad, phosphorylation, signaling, allosteric
The transforming growth factor beta (TGF-β) superfamily of ligands signal transcriptional control of diverse processes (Heldin et al. 1997; Derynck et al. 1998; Roberts 1999; Attisano and Wrana 2000; Blobe et al. 2000; de Caestecker et al. 2000a; Massague and Wotton 2000). Signaling from the ligand-activated transmembrane receptor kinases to the target genes are mediated by the Smad proteins (Heldin et al. 1997). The class of receptor-regulated Smad protein (R-Smad) is phosphorylated at the carboxy-terminal SXS sequence by the type I receptor kinase (Abdollah et al. 1997; Souchelnytskyi et al. 1997). Phosphorylation converts the R-Smad from the basal state monomer to the active state trimer, which exchanges a subunit with the common cofactor Smad4 to form the more stable heterotrimer (Kawabata et al. 1998; Chacko et al. 2001; Qin et al. 2001; Wu et al. 2001; Wrana 2002). The heteromeric complex enters the nucleus, binds to the promoters, and interacts with transcriptional comodulators to regulate gene expression.
Smad3 is an R-Smad that mediates the signals of TGF-βs and activins. Smad3 is recruited by SARA (Smad Anchor for Receptor Activation) to the receptor kinase for phosphorylation (Tsukazaki et al. 1998). SARA contains the FYVE domain for membrane localization, the Smad-binding domain (SBD) that are specific for Smad3 and Smad2, and a carboxy-terminal domain that interacts with the receptor kinase. Upon phosphorylation, Smad3 dissociates from SARA, forms a complex with Smad4, and enters the nucleus.
The transcriptional activity of the Smad3/Smad4 complex is modulated by coactivators and corepressors. Ski is a corepressor of the Smad complex (Akiyoshi et al. 1999; Luo et al. 1999; Sun et al. 1999; Xu et al. 2000; Liu et al. 2001). Ski interacts directly with Smad3 and recruits histone deacetylase to the transcriptional control site, resulting in chromatin remodeling and transcriptional repression. The interaction between Ski and Smad3 is enhanced on TGF-β stimulation.
Smad3 shares a common domain configuration with other R-Smads and Smad4, consisting of an amino-terminal DNA-binding domain (MH1 domain) and a carboxy-terminal effector domain (MH2 domain) separated by a linker region. The MH2 domain is a versatile protein–protein interaction module. At the receptor complex, the MH2 domain of Smad3 interacts with SARA and the receptor kinase (Macias-Silva et al. 1996; Feng et al. 1997; Chen et al. 1998; Lo et al. 1998; Persson et al. 1998; Tsukazaki et al. 1998; Huse et al. 2001). After dissociation from the receptor, the MH2 domain of Smad3 also interacts with Smad4. In the nucleus, the MH2 domain of Smad3 further interacts with Ski. However, it is unclear how various interactions of the MH2 domain are regulated.
Here, structural and biochemical evidence shows that the interaction between the MH2 domain of Smad3 and its signaling partners is conformation dependent. SARA and Ski interact preferentially with the monomeric and trimeric form of Smad3, respectively. Phosphorylation-induced Smad3 trimerization thus activates the TGF-β signals by facilitating Smad3 dissociation from SARA, and simultaneously sets up a feedback mechanism by allowing Smad3–Ski interactions. The data show that SARA and Ski bind to overlapping surfaces on the Smad3 MH2 domain, which undergo conformational changes upon subunit oligomerization. Structural models of the Smad3/SARA/receptor kinase complex and the Smad3/Ski complex shed light on the molecular basis of allostery, which serves as a paradigm for other members of the Smad protein family.
Results
Subdomain juxtaposition within the MH2 domain correlates with the activation state
Previously, we showed that the pseudophosphorylated Smad3, in which the carboxy-terminal SSVS phosphorylation sequence was mutated to EEVE, activates TGF-β signaling (Chacko et al. 2001). In an attempt to reveal the structural basis of activation, we crystallized the pseudophosphorylated Smad3 MH2 domain (residues 218–424) and determined the structure to 1.9-Å resolution (Table 1).
Table 1.
Summary of crystal analysis for Smad3/SARA complex and unliganded Smad3
| Parameter
|
Smad3/SARA
|
Smad3
|
|---|---|---|
| Crystal parameters and crystallographic data | ||
| Space group | P2(1)2(1)2 | P3(2) |
| Unit cell dimensions | a = 50.0, b = 71.7, c = 86.9 | a = b = 54.3, c = 59.4 |
| Diffraction limit (Å)a | 2.80 (2.87–2.80) | 1.90 (1.97–1.90) |
| Total reflections | 48600 | 64905 |
| Unique reflections | 7498 (556) | 14785 |
| Completeness (%) | 93.0 (99.8) | 96.4 (77.7) |
| Intensity/Sigma | 7.5 (3.6) | 20.1 (3.4) |
| Rmerge (%)b | 13.9 (46.4) | 6.1 (21.0) |
| Refinement statistics | ||
| Protein atoms | 1833 | 1398 |
| R factor (%)c | 22.4 | 19.8 |
| Rfree factor (%)d | 26.7 | 20.4 |
| Rmsd from ideal | ||
| Bond (Å) | 0.01 | 0.01 |
| Angle (°) | 1.51 | 1.2 |
Values in brackets are for the highest resolution shell.
Rmerge = Σ |Ihkl − <Ihkl>|/Σ Ihkl.
R factor = Σhkl ∥Fobs| − |Fcalc∥/Σhkl |Fobs| for all data.
R free = Σhkl ∥Fobs| − |Fcalc∥/Σhkl |Fobs| for 10% of the data not used in refinement.
The crystal packing arrangement of the Smad3 MH2 domain suggests that the structure is not in the active conformation. Biochemical study revealed that the pseudophosphorylated Smad3 forms a trimer, in which the pseudophosphorylated sequence promotes subunit assembly by interacting with the L3 loop region phosphoserine-binding site of the neighboring subunit (Chacko et al. 2001). This expected arrangement was not present. Rather, the subunits interact via a distinct interface covering ∼500-Å2 area (Fig. 1A). Mutation of the subunit packing residues, Tyr 296 and Phe 303, in the context of the pseudophosphorylated Smad3, does not affect solution trimerization (data not shown), confirming that the interactions observed are simply crystal packing effects. The high-salt and low-pH crystallization conditions could contribute to the disruption of the active trimer. Consistently, the pseudophosphorylated Smad3 behaves as a basal state monomer on a size-exclusion column under the crystallization conditions (data not shown). The carboxy-terminal tail (residues 419–424), the L3 loop (residues 381–387) and the amino-terminal β0-strand (residues 219–227) are disordered in the structure. We observed previously that the L3 loop in Smad1 exists in two conformations (Qin et al. 2001). The disordering of the L3 loop in the basal state Smad3 is consistent with the L3 loop undergoing dynamic movement between conformations.
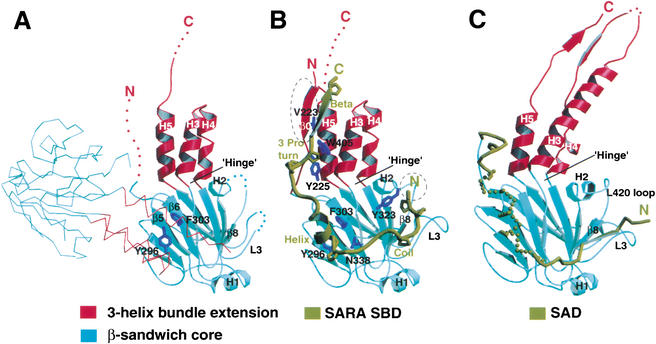
Figure 1.
Crystal structures of unliganded Smad3, Smad3/SARA complex, and S4AF. (A) Crystal structure of unliganded Smad3 shown by ribbon representation. The Cα trace of a symmetry-related subunit is shown. (B) Crystal structure of Smad3/SARA complex. The SARA SBD is in dark yellow. All proline residues are displayed. Smad3 structures that are ordered upon SARA binding are circled. (C) Crystal structure of S4AF (Qin et al. 1999). The Smad4 activation domain (SAD) is in dark yellow, in which the disordered sequence, GHYWPVHNELA, has a helical propensity. The three-helix bundle and β-sandwich subdomains are in red and cyan, respectively. The disordered regions of the structure are represented by dots and colored according to the subdomain color. The side chains are in dark blue.
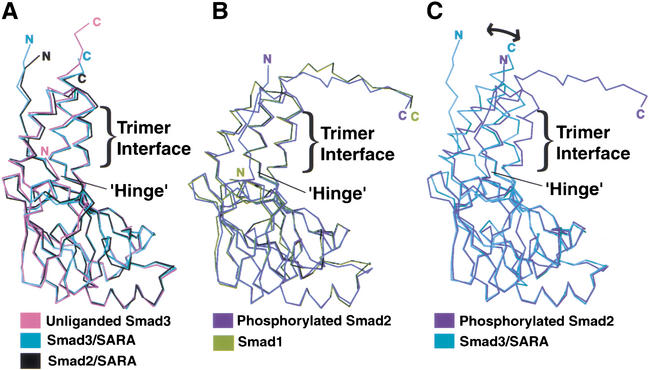
The Smad3 MH2 domain structure can be subdivided into two subdomains, the three-helix bundle extension and the β-sandwich core (Fig. 1A). Comparison between the monomeric and trimeric MH2 domain structures reveals an interesting correlation between the juxtaposition of the subdomains and the oligomerization state of the subunit (Fig. 2). Whereas the β-sandwich core of all structures superimposes well, the hinge angle between the subdomains is dependent on the oligomerization state. The MH2 domains of the Smad2/SARA complex (Wu et al. 2000) and the Smad3/SARA complex (see below), in which the MH2 domain is monomeric, have hinge angles similar to that of the unliganded Smad3. However, the MH2 domains of Smad1 (Qin et al. 2001) and phosphorylated Smad2 (Wu et al. 2001), both crystallized as a trimer, have a narrower hinge angle due to the three-helix bundle extensions tilting toward the direction of the neighboring subunit. As will be discussed, the structural transition between the two distinct states of the R-Smads serves as a critical signaling switch.
Figure 2.
Trimerization-induced tilting of the three-helix bundle subdomain relative to the β-sandwich core. (A) Superposition of the R-Smad MH2 domains that were crystallized in the monomeric form. (B) Superposition of the R-Smad MH2 domains that were crystallized in the trimeric form. (C) Superposition of the monomeric and trimeric form of the R-Smad MH2 domain. Superpositions were performed by aligning the β-sandwich subdomain. The amino termini of the unliganded Smad3 and Smad1 are shorter due to disordering of the structures in the crystal.
Stabilization of a hydrophobic surface directs Smad3 anchor to SARA
To elucidate the structural basis of Smad3 recruitment by SARA, we crystallized the Smad3/SARA complex containing the linker and MH2 domain of Smad3 (S3LC, residues 145–424) and the Smad-binding domain (SBD) of SARA (residues 665–751; Table 1). The electron density corresponding to the linker domain of Smad3 and the carboxy-terminal half of the SBD was not observed, due to disordering. The observable structure is similar to the Smad2/SARA structure (Fig. 1B; Wu et al. 2001). The SBD binds in an extended conformation to a hydrophobic groove on Smad3 burying 2700 Å2 surface. The structure of the SBD can be described from amino to carboxyl terminus as a coil, a helix, a three-proline turn, and a β-strand. Interestingly, the SARA SBD tethers the three-helix bundle and the β-sandwich subdomains of Smad3 on the opposite face of the conserved trimeric interfaces.
Comparison between the unliganded Smad3 and Smad3/SARA complex structures revealed no global change of the MH2 domain (Fig. 2A). The r.m.s. deviation of Cα trace between the MH2 domains of Smad3 and Smad3/SARA complex is 0.4 Å. Nevertheless, two structural elements in Smad3 undergo a disorder-to-order transition upon binding SARA (Fig. 1, cf. A and B). The amino-terminal β0-strand, which interacts with the β structure of the SARA SBD, is disordered in the unliganded Smad3. Also, the loop connecting helix-2 and β8, which interacts with the coil of the SBD, is disordered in the unliganded Smad3. Interestingly, the corresponding loop in Smad4 (L420 loop), interacts with the Smad4 activation domain (SAD; Fig. 1C; see also Discussion; Qin et al. 1999). These disordered regions of Smad3 contain a high percentage of hydrophobic residues that mediate direct interaction with SARA.
Dimeric model of the Smad3/SARA/receptor kinase complex
The physiological form of SARA, and, hence, the Smad3/SARA complex, is anticipated to be dimeric. First, the transmembrane receptor kinase is dimeric, which can phosphorylate a dimeric substrate (Luo and Lodish 1996; Gilboa et al. 1998; Kirsch et al. 2000; Hart et al. 2002). Second, the membrane-anchoring FYVE domain, located immediately amino-terminal of the SARA SBD, is dimeric in early endosome autoantigen (EEA1; Dumas et al. 2001). Third, sizing analysis suggested that SARA is a dimer (Jayaraman and Massague 2000). Interestingly, a dimeric Smad3/SARA complex, consisting of two copies of the complex related by a twofold symmetry, is present in the structure (Fig. 3B). Although the buried surface area between the two complexes is only 680 Å2, modeling studies suggest that the dimeric arrangement may be relevant and facilitated by the FYVE domain.
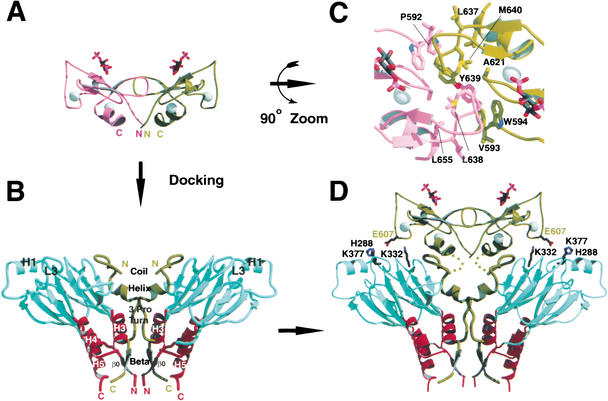
Figure 3.
Proposed structural model of the dimeric Smad3/SARA complex. (A) The dimeric SARA FYVE domain model. The model includes residues 590 to 657 of SARA. The two zinc ions within each subunit are shown in spheres. Ins(1,3)P2 is shown using stick presentation. (B) Crystal structure of the dimeric arrangement of the Smad3/SARA complex. The two copies of the complex are related by a vertical twofold crystallographic axis. The coloring is based on Fig. 1B. (C) Close-up view of the dimer interface of the SARA FYVE domain dimer. (D) Model of the Smad3/SARA complex containing the FYVE domain. The linker between the FYVE domain and the SBD contains 14 residues not included in the model (residues 658–670) and are shown by dots.
Structure of the EEA1 FYVE domain reveals a homodimeric arrangement in which the phosphoinositide-binding pocket of each subunit faces the same direction for membrane anchorage (Dumas et al. 2001). On the basis of the EEA1 FYVE domain structure, the dimeric SARA FYVE domain was constructed (Fig. 3A). The model reveals additional hydrophobic contacts within the subunit interface, which can stabilize the dimer (Fig. 3C). Furthermore, the carboxyl termini of the dimeric SARA FYVE domain can be docked closed to the amino termini of the SBD in the dimeric Smad3/SARA crystal structure with little steric hindrance (Fig. 3D). In the model, Smad3 contacts not only the SARA SBD, but also the membrane-distal face of the SARA FYVE domain, with the three-helix bundle pointing away from the membrane. The L3 loop region phosphoserine-binding residues of Smad3, Lys332, and Lys377, and the nearby His288 form potential salt bridges with the side chain of Glu 607 from the FYVE domain.
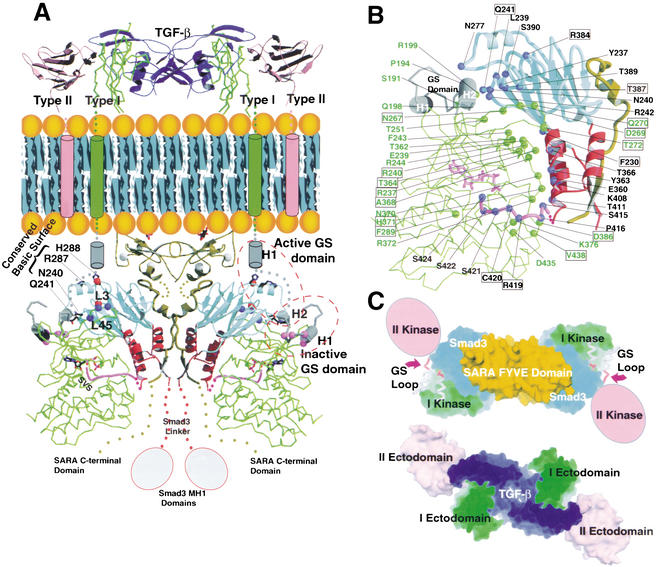
The dimeric Smad3/SARA complex can be modeled as a substrate for the type I receptor kinase (Fig. 4). The convex face of the kinase domain, located toward the back of the catalytic face, matches the concave surface of the Smad3 MH2 domain, shaped by the β-sandwich and three-helix bundle subdomains (Fig. 4A,B). The carboxy-terminal phosphorylation tail of Smad3 extends and docks in the catalytic site. The kinase domains of the dimeric receptor do not interact with each other, but form a complex through interaction with Smad3. Although the model uses the basal state kinase structure (Huse et al. 1999), the postulated consequence of activation is also drawn. The model is functionally relevant. First, the L45 loop of the receptor kinase domain and the L3 loop of Smad3, which contain matching specificity determinants of the receptor/R-Smad interaction, are adjacent in the model for interaction (Feng et al. 1997; Chen et al. 1998; Lo et al. 1998; Persson et al. 1998). Previous studies showed that the signaling specificity could be switched by engineering these determinants. Second, the GS domain within the juxtamembrane region of the type I receptor kinase, phosphorylation of which by the type II receptor kinase leads to activation, is in close proximity to the conserved basic surface of Smad3 (Wrana et al. 1994; Wieser et al. 1995; Willis et al. 1996). The GS domain forms a helix-loop-helix structure, in which the phosphorylation sites are located within the loop. In the basal state, the phosphorylation sites are partially buried, suggesting that the GS domain would undergo conformational changes upon phosphorylation (Huse et al. 1999). Recent studies revealed that phosphorylation of the GS loop creates an acidic-binding site for the conserved basic surface of Smad2 (Huse et al. 2001). Consistently, the model depicts that helix 1 and the phosphorylation loop undergo conformational changes to allow the phosphorylated GS loop to dock to the Smad3 basic surface (Fig. 4A). The GS domain in the active conformation can potentially interact with Asn 240, Gln 241, Arg 287, and His 288 of Smad3, consistent with the report that mutation of the corresponding residues in Smad2 led to defects in interaction with the receptor and phosphorylation (Huse et al. 2001). Third, the phosphorylation tail of Smad3, modeled using the Smad1 carboxy-terminal structure, extends into the kinase catalytic center. Peptide screening revealed optimal phosphorylation substrate that bears no resemblance to the conserved carboxy-terminal sequence of the R-Smads, suggesting a requirement for positioning of the phosphorylation sites in the catalytic center (Luo et al. 1995). Fourth, the kinase interacts with Smad3 via an extensive interface, burying 3000 Å2 surface. The interface contains matching hydrophilic and hydrophobic interactions, which are mediated mostly by conserved and subtype-specific residues (Fig. 4B). Lastly, the cytoplasmic organization of the receptor complex is consistent with the ectodomain organization of the type I/II receptors bound to the ligand (Fig. 4C; Hart et al. 2002). The GS loop of the type I kinase points to the direction in which the type II kinase is anticipated.
Figure 4.
Proposed structural model of Smad3/SARA/receptor kinase complex. (A) Proposed structure of the Smad3/SARA/receptor kinase complex. The type I receptor kinase model is shown in green, with the exception of the GS domain, which is in gray. The kinase active site has an ATP molecule. The type II receptor (ectodomain and transmembrane helix only) is shown in pink. The TGF-β dimer is in dark blue. Smad3 and SARA are colored based on Fig. 3D. The specificity determinants in the kinase L45 loop and Smad3 L3 loop are shown by blue spheres (left to right, N267, D269, and N270) and red spheres (top to bottom, R384 and T387), respectively. The GS loop phosphorylation sites are shown using pink spheres. The structural consequence of GS domain phosphorylation is depicted by a cartoon and is labeled active GS domain. The carboxy-terminal tail of Smad3, modeled using the Smad1 crystal structure, is shown in pink. The ligand/ectodomain structure is based on the crystal structures of the bone morphogenetic protein (BMP) in complex with the BMP type I receptor and TGF-β in complex with type II receptor (Kirsch et al. 2000; Hart et al. 2002). (B) The kinase/Smad3 interface contains residues that are mostly conserved or subtype specific in the type I receptor kinases and the R-Smads. Subtype-specific residues are boxed. Smad3 and kinase residues are labeled in black and green, respectively. (C) The cytoplasmic organization of the receptor complex is consistent with the ligand/ectodomain complex. Top views of cytoplasmic section (top) and extracellular section (bottom) are shown. The coloring is based on Fig. 4A. The type II kinase, which phosphorylates the GS loop, is shown in a cartoon.
SARA allosterically inhibits Smad3 trimerization
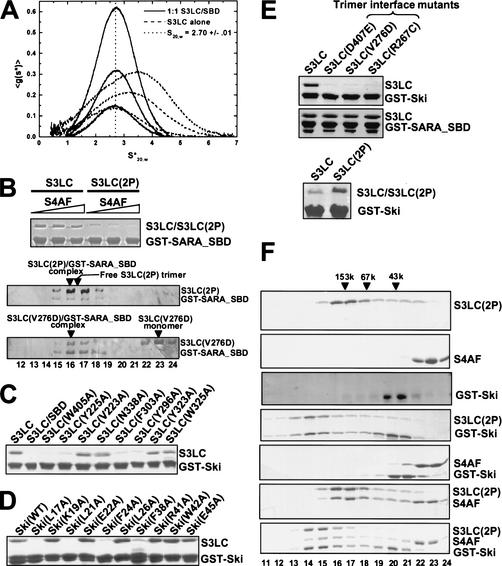
Previous analysis revealed that Smad3 has a propensity to trimerize, which is further enhanced by carboxy-terminal phosphorylation (Correia et al. 2001). The structure of the Smad3/SARA complex does not reveal a trimeric arrangement of Smad3, raising the possibility that SARA may inhibit Smad3 oligomerization. Using analytical ultracentrifugation, we show that in contrast to the unliganded Smad3, which undergoes concentration-dependent trimerization, the Smad3/SARA complex exhibits little propensity to oligomerize (Fig. 5A). When the Smad3/SARA complex was modeled onto the trimeric scaffold of phosphorylated Smad2, SARA is located at the peripheral of the trimer and does not physically block the trimer interface (data not shown). However, in the trimeric model, the Smad3/SARA complex makes poor intersubunit contacts, due to the three-helix bundle subdomain not tilting enough toward the neighboring subunit (Fig. 2C). We rationalize that because SARA tethers the three-helix bundle and the β-sandwich core of Smad3 on the opposite face of the trimer interface, the function of SARA may be to inhibit Smad3 trimerization by restricting the three-helix bundle from tilting. Consistently, the SARA SBD is proline rich (25%), which provides rigidity to the structure. The three-proline turn packs against the hinge between the three-helix bundle and the β-sandwich subdomains of Smad3, at a perfect position to hinder the hinge motion (Fig. 1B). The dimeric arrangement of SARA could further reinforce the SBD (Fig. 3D), providing a mechanism for Smad3 dissociation upon phosphorylation-induced conformational change. Consistently, the phosphorylated Smad3 exhibits a lower affinity to the SARA SBD in vitro, as shown by a lower level of binding in the GST pull-down assay (Fig. 5B). Furthermore, once bound to the SBD, the phosphorylated Smad3 becomes monomeric, as the complex elutes at the same position as the complex formed by the trimer interface mutant of Smad3 (V276D) on the size-exclusion column (Fig. 5B). These results are consistent with the following: SARA binds only the Smad3 monomers, and that SARA–Smad3 interaction and Smad3 homotrmeric interaction are competing and mutually exclusive. Although Smad4 only has a small effect on Smad3/SARA interaction in the binding assay, it is possible that Smad4 or other chaperones may more significantly influence the kinetics of Smad3 dissociation in vivo.
Figure 5.
Biochemical characterization of Smad3–SARA and Smad3–Ski interactions. (A) DCDT+ analysis of a sedimentation velocity experiment conducted on a 1:1 mixture of S3LC and SARA SBD conducted at 42K rpm, 24.7°C and total protein concentrations of 4.0, 8.6, and 17.5 μM complex. Fitting with DCDT+ (and SVEDBERG) gives an average S20,w = 2.698 ± 0.012 (2.700 ± 0.037) and an average MW = 42,183 ± 1264 (39,407 ± 1287). The absence of a shift in the peak position, especially relative to the obvious shift upon trimerization by S3LC alone (reproduced from Correia et al. 2001), and the similarity of the sedimentation coefficient to that of monomeric Smad4 (2.46 S20,w), corrected for the size of SBD, are consistent with this being a 1:1 complex. (B) Phosphorylated Smad3 has a weaker affinity for SARA SBD. (Top) GST–SARA_SBD was used to detect interaction with either the unphosphorylated or phosphorylated S3LC by use of the GST pull-down assay. The effect of S4AF was performed by including S4AF in the first two washes, followed by regular washes. (first lane) No S4AF; (second lane) 0.5 mg/mL S4AF; (third lane) 2 mg/mL S4AF. The bound S3LC in the absence of S4AF reflects ∼20% to 30% of the input. (Middle and bottom gels) Size exclusion chromatography of S3LC(2P)/SARA complex and S3LC(V276D)/SARA complex. The loading sample contains 0.5 mg/mL of GST–SARA–SBD and 1 mg/mL of S3LC. The positions of the eluted species are indicated above the SDS–polyacrylamide gels. The fraction numbers are marked below the bottom gel. (C) Ski binds to subsite of the SARA-binding site. The GST–Ski(17–45) was used to detect interaction with the S3LC mutants. The bound S3LC in the first lane reflects ∼20% to 30% of the input. (D) Hydrophobic residues in Ski mediate direct interaction with Smad3. The GST–Ski(17–45) mutants were used to detect interaction with S3LC. The bound Ski(WT) reflects ∼20% to 30% of the input. (E) Smad3–Ski interaction requires trimerization of Smad3. (Top) The GST–Ski(17–45) and GST–SARA_SBD were used to detect interaction with the trimer interface mutants of S3LC. The bound S3LC in the first lane reflects ∼20% to 30% of the input. (Bottom) S3LC(2P) binds Ski better than the unphosphorylated S3LC after extensive washing. In addition to the standard conditions, the beads were equilibrated with the washing buffer for 60 min between washes. (F) Ski interacts with the Smad3/Smad4 heterotrimeric complex. Protein compositions as indicated to the right of each gel panel were injected into the size exclusion column. The eluted fractions were analyzed by SDS-PAGE. The molecular weight standards are shown at top. The fraction numbers are shown at bottom. All gels in this figure were stained with Coomassie blue.
Ski interacts with Smad3 via the SARA-binding surface
Ski is a proto-oncogene that suppresses the transcriptional activity of Smad3 through direct interactions. To elucidate the structural basis of Ski–Smad3 interaction, we mapped the segment in Ski that contacts Smad3. By use of limited proteolysis of the complex, followed by mass spectrometry and systematic deletional analysis, the Smad3 interaction domain of Ski was mapped to residues 17–45. This 29-residue segment, Ski(17–45), is highly conserved in SnoN, a Ski homolog that also mediates transcriptional repression of Smad3. The GST fusion protein of Ski(17–45) mediates strong interaction with Smad3 (Fig. 5C).
Binding studies showed that Ski(17–45) interacts with Smad3 at the SARA-binding surface (Fig. 5C). In contrast to the unliganded Smad3, the Smad3/SARA complex has a dramatically decreased ability to interact with GST–Ski(17–45), suggesting that the Ski-binding site is rendered inaccessible in the presence of SARA. Consistently, site-specific mutations of several SBD-binding residues on Smad3 abolish Ski interaction. These residues include Trp 405 and Tyr 225, which bind the β structure of SBD, as well as Phe 303 and Tyr 296, which bind the helical structure of SBD. However, the binding determinants for SARA and Ski are not identical, as mutation of Tyr 323, Asn 338 and Val 223, which participate in SBD binding, has no effect on Ski interaction. Smad3 interaction with Ski(17–45) probably does not involve the surface that contacts the SARA coil region, as mutations in this area do not affect Ski interaction.
Hydrophobic residues in Ski are involved in direct contact with Smad3 (Fig. 5D). Point mutation of Leu 17, Leu 21, Phe 24, or Phe 38 in Ski dramatically reduces interaction with Smad3. However, point mutation of Lys 19, Glu 22, Leu 26, Arg 41, Trp 42, or Glu 45 in Ski has no significant effect on the interaction.
Ski interacts specifically with the trimeric form of Smad3
The interaction between Ski and Smad3 is dramatically reduced when the conserved trimer interface residues of Smad3 are mutated (Fig. 5E). These mutations stabilize Smad3 in the monomeric form (Chacko et al. 2001). In contrast, trimer interface mutations have no effect on SARA binding, consistent with the analytical ultracentrifugation studies that SARA stabilizes the monomeric form of Smad3. The phosphorylated Smad3 also binds Ski better than the unphosphorylated Smad3, but only becomes apparent after a longer period of washing than the standard condition, presumably due to different rates of dissociation. Thus, despite sharing a common surface of Smad3 for interaction, Ski and SARA interact specifically with the trimeric and monomeric form of Smad3, respectively. Because Smad3 trimerization is promoted upon TGF-β stimulation, these data explain the in vivo observation that Ski–Smad3 interaction is enhanced upon TGF-β stimulation (Akiyoshi et al. 1999; Luo et al. 1999; Sun et al. 1999; Xu et al. 2000). The 29-residue Ski fragment therefore mimics the cellular response and constitutes a sensor for Smad3 trimerization.
Ski(17–45) also interacts with the Smad3/Smad4 heterotrimer (Fig. 5F). The phosphorylated form of S3LC, S3LC(2P), and the transcriptionally active Smad4 fragment (S4AF, residues 273–552 of Smad4) were used in these experiments. On a size-exclusion column, S3LC(2P) and S4AF independently elute as a trimer and monomer, respectively, consistent with the previous findings (Fig. 5F, first and second panels). GST–Ski(17–45) alone elutes as a dimer, presumably through GST dimerization (Fig. 5F, third panel). GST–Ski(17–45) interacts with S3LC(2P), as shown by their coelution with an apparent molecular weight larger than that of S3LC(2P) alone (Fig. 5F, cf. fourth panel and first panel). GST–Ski(17–45) also interacts with the S3LC(2P)/S4AF heterotrimer, as shown by the coelution of all three species with an apparent molecular weight larger than that of the S3LC(2P)/S4AF trimeric complex (Fig. 5F, cf. seventh panel and sixth panel). Two observations suggest that Smad4 alone or within the heterotrimeric Smad complex does not interact with Ski(17–45). First, S4AF and Ski do not form a complex on the size-exclusion column (Fig. 5F, cf. second, third panels and fifth panel). Second, the S3LC(2P)/Ski complex elutes with an apparent molecular weight larger than that of the S3LC(2P)/S4AF/Ski complex (Fig. 5F, cf. fourth panel, peak fraction 14 and seventh panel, peak fraction 15), suggesting that S4AF within the heteromeric Smad complex does not bind Ski. The result is consistent with the previous reports that Smad4–Ski interaction involves a different region of Ski.
Trimeric model of the Smad3/Ski complex
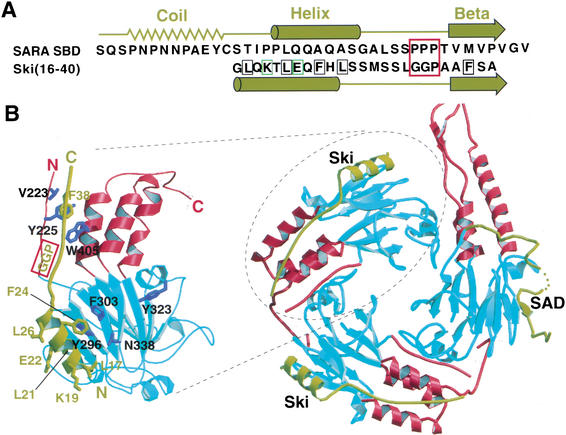
The analysis described above shows that SARA and Ski bind to an overlapping surface on Smad3 but recognize specifically the monomeric and trimeric form of Smad3, respectively. Although the Smad3 interacting domains of SARA and Ski do not share sequence homology, they likely have structural counterparts over the helical and β-strand region (Fig. 6A). Further truncation of Ski showed that residues 16–40 are sufficient for Smad3 interaction (data not shown). This minimal Smad3 interaction domain of Ski was modeled approximately to the binding site on Smad3 on the basis of putative secondary structure homology to SARA and the mutagenesis results (Fig. 6B). In the model, residues 16–24 of Ski form an amphipathic helix, in which the hydrophobic side chains important for Smad3 interaction face the hydrophobic surface of Smad3. At the position corresponding to the three-proline turn in SARA contains the more flexible Gly–Gly–Pro sequence in Ski. Residues 37–40 of Ski form a putative β-strand, which interacts hydrophobically with the β0 strand and helix 5 of Smad3. The model is consistent with the mutagenesis data and provides insights as to why Ski interacts with the trimeric Smad3. The more flexible Gly–Gly–Pro sequence in Ski, which packs against the Smad3 hinge, may be adaptable to the tilting of the three-helix bundle structure in the trimeric form of Smad3. The flexible glycine turn and the unique flanking interactions mediated by the helical and β structures may jointly recognize the more curved binding surface in the trimeric form of Smad3. The detailed mechanism awaits a high-resolution crystal structure of the Smad3/Ski complex.
Figure 6.
Proposed structural model of the Smad3/Ski complex. (A) Sequence and secondary structural comparison between SARA SBD and Ski(16–40). The secondary structures of SARA SBD from the crystal structure are shown above the SARA sequence. The predicted secondary structures of Ski(16–40) based on the Garnier-Osguthorpe-Robson method in the GCG program are shown below the Ski sequence. Ski mutations that weaken Smad3–Ski interaction are boxed in black. Ski mutations that have no effect on Smad3–Ski interaction are boxed in green. The three-proline turn in SARA SBD and the corresponding sequence in Ski are boxed in red. (B, right) Proposed structure of the Smad3/Smad4/Ski complex. The Smad4 subunit contains the SAD domain. (Left) Close-up view of the Smad3/Ski subunit model. The coloring is based on Fig. 1.
Discussion
An efficient signaling switch
This work reveals an allosteric mechanism through which receptor-mediated Smad3 phosphorylation is coupled directly to transcriptional regulation. Phosphorylation-induced Smad3 trimerization serves as a single switch for two functions, promoting Smad3 dissociation from the receptor complex and facilitating Smad3 interaction with the nuclear repressor Ski. Although the two activities may seem contradicting, the transcriptional activity of Smad3 is likely balanced by the steady-state level of Ski versus the coactivators in the nucleus. One possible scenario is that the coactivators may compete with Ski for similar binding sites, also requiring the trimeric form of Smad3 for interaction. This view is supported by the observation that many Smad coactivators interact with the Smad proteins only after ligand stimulation. Another possibility is that the coactivators and Ski may bind simultaneously to distinct sites on Smad3, and the overall transcriptional activity is regulated by higher order protein–protein interactions mediated by Ski and the coactivators. Further studies are required to dissect the complexity of Smad transcriptional regulation.
The Smad active site
The SARA-binding surface on Smad3 may be a common active site in Smad proteins. In the S4AF structure, the corresponding surface interacts with the SAD, a 50-residue segment in the linker region of Smad4 indispensable for Smad4 transcriptional and signaling function (Fig. 1C; de Caestecker et al. 1997, 2000b). Besides a remarkable similarity in the disposition of the SAD and SARA SBD relative to their MH2 domains (Fig. 1, cf. B and C), the same secondary structural elements on the MH2 domains are involved in interactions, the loop between β5 and β6, the loop between helix 2 and β8, β8, and helix 5. Although a segment of the SAD corresponding to the helix in SBD is disordered in the crystal structure, secondary structure prediction suggests that the disordered sequences have a helical propensity. Nevertheless, <5% of the MH2 domain residues involved in the interactions are conserved. Also, the SBD-binding surface is considerably more hydrophobic than the SAD-binding surface. The interaction between SBD and Smad3 is predictably much tighter than that between the SAD and Smad4. The SAD may undergo conformational changes upon Smad3/Smad4 complex formation to free the binding surface for other transcriptional regulators and reposition the MH1 domain for DNA binding. Because Ski binds to a subsite of the SBD-binding site on Smad3, the analysis also reveals a common transcriptional regulatory surface on Smad3 and Smad4. Recently, characterization of the Smad2 transcriptional regulator Mixer revealed a conserved Smad interaction motif (SIM) in the Fast-1 or the Mix family proteins, which binds Smad2 in the same hydrophobic pocket as does the coil region of the SARA SBD (Randall et al. 2002). Ski and Mixer appear to occupy nonoverlapping subsites of the SARA SBD site, suggesting that they may coexist on Smad2/3 to regulate gene expression.
Structural basis of signaling
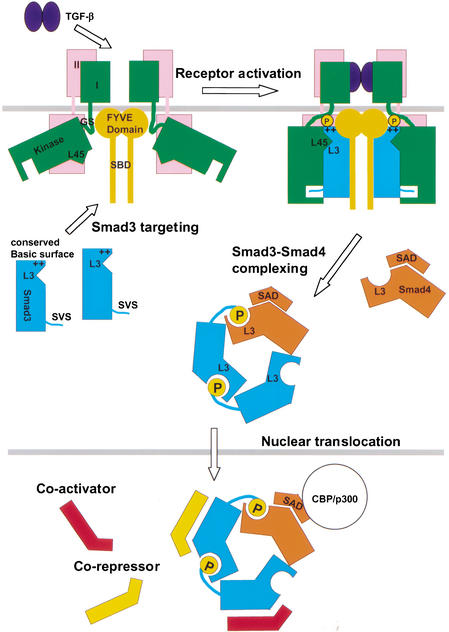
The present work suggests a TGF-β-signaling mechanism that is critically dependent on Smad protein trimerization (Fig. 7). The common observation of ligand-induced association between Smad proteins and transcriptional comodulators suggests that trimerization-dependent interaction between Smad3 and Ski may represent a more general paradigm of Smad nuclear interactions.
Figure 7.
Trimerization-dependent Smad signaling in the TGF-β pathway.
Comparison between the unliganded Smad3 and Smad3/SARA complex structures reveals no global structural change, suggesting that Smad3 is monomeric before recruitment to SARA. Structural modeling suggests that SARA promote Smad3-kinase recognition by the following mechanisms. First, SARA stabilizes the monomeric form of Smad3 through the proline-rich structure of the SBD. The receptor model precludes a trimeric Smad3 to the receptor due to steric hindrance. Second, SARA presents Smad3 for kinase recognition by tethering the three-helix bundle and the β-sandwich subdomains of Smad3 on the opposite face of the Smad3/kinase interface. Finally, SARA precisely positions the phosphorylation site of Smad3 in the kinase calatytic center. Purified receptor kinase domain phosphorylates Smad3 at nonspecific sites (data not reported), suggesting that stereo-specific docking of the Smad3 carboxy-terminal tail is required. The model further suggests that the GS domain undergoes a conformational change upon phosphorylation to dock to the conserved basic surface of the Smad protein. Because the GS domain is linked to the membrane-spanning helix, the conformational change is likely manifest through movement of the kinase domain.
The receptor model suggests that competition of protein–protein interactions and protein conformational changes are responsible for phosphorylation-induced dissociation of Smad3 from the receptor complex. The conserved basic surface and the three-helix bundle subdomain of Smad3, which contact the kinase, are also involved in the subunit interface of the phosphorylated Smad3 trimer (Chacko et al. 2001). The Smad3 L3 loop, which interacts with the kinase L45 loop and the SARA FYVE domain, also interacts with the phosphorylated carboxy-terminal tail of the neighboring subunit in the Smad3 trimer. The key conformational change in Smad3 upon phosphorylation is the trimerization-induced tilting of the three-helix bundle subdomain relative to the structural core. This global change likely disturbs interactions of Smad3 with both the SARA and the receptor kinase, which sandwich Smad3. In addition, the conformational change of the Smad3 L3 loop upon trimerization, as shown previously in Smad1, will perturb its interaction with the kinase (Qin et al. 2001). Although the receptor model will require verification by further analysis, the model fulfills multiple biochemical constraints and is consistent with the current biological data.
Once the phosphorylated Smad3 dissociates from the receptor, it forms a 2:1 heterotrimeric complex with Smad4 (Chacko et al. 2001). In the nucleus, Ski binds to the SARA-binding surface of Smad3 in the trimeric complex. The selectivity of Ski binding for the trimeric Smad3 over the basal, monomeric state of Smad3 appears to be due to Ski being able to recognize the global structural change of Smad3 upon trimerization. Biochemical and modeling analyses indicate that the trimerization-induced tilting of the three-helix bundle subdomain, which weakens Smad3/receptor interaction, shapes a unique recognition surface for Ski to bind. It should be noted that a Smad2–Smad4 heterodimeric model has been proposed, which assumes the same subunit arrangement as the Smad3–Smad4 heterotrimeric model, but with one Smad3 subunit missing (Wu et al. 2001). The discrepancy between Smad3 and Smad2 should be investigated in the future.
Conclusion and perspective
Phosphorylation-induced Smad protein trimerization functions as a master allosteric switch of the TGF-β pathway, converting Smad-receptor interactions to Smad-nuclear interactions. The conformational transition allows Smads to use common structural determinants to interact with multiple signaling molecules in a spatio and temporal manner. This study provides a molecular framework for further mechanistic investigation of the signaling pathway and possibly therapeutic strategies to regulate TGF-β signaling.
Materials and methods
Construction of expression plasmids and mutagenesis
The cDNA fragments were generated by PCR and subcloned into the pGEX vectors (Amersham). Site-directed mutants were generated using the Quik-Change kit (Strategene) and confirmed by sequencing.
Protein expression and purification
All proteins, except the phosphorylated S3LC, were expressed with a glutathione S-transferase tag, extracted by glutathione sepharose and released by thrombin. The proteins were loaded to a DEAE column equilibrated with 20 mM Tris (pH 7.4), 10 mM NaCl, 0.1 mM EDTA, and 1 mM DTT. Proteins were eluted by a sodium chloride gradient from 10 to 300 mM. The S3LC/SBD complex was made by incubating purified S3LC with GST–SBD on glutathione sepharose, followed by thrombin cleavage. The S3LC/SBD complex was further purified using a Superdex 200 HR column.
Protein structure determination by X-ray crystallography
Crystals of the unliganded Smad3 and the Smad3/SARA complex were obtained by the hanging drop vapor diffusion technique. For the unliganded Smad3, the well solution contains 700 mM ammonium dihydrogen phosphate, 20% ethylene glycol (pH 4.8). For the Smad3/SARA complex, the well solution contains 1.8 M sodium chloride and 100 mM sodium acetate (pH 5.0). Crystals were transferred to cryosolvents consisting of 25% (v/v) glycerol and 75% of the well solution, and were flash frozen in liquid nitrogen. The data for the unliganded Smad3 were obtained from beamline X-25 of NSLS at Brookhaven National Laboratory. The data for the Smad3/SARA complex were obtained from the ALS at Berkeley lab. Data were integrated and reduced using DENZO and Scalepack (Otwinowski and Minor 1997).
The structure of the unliganded Smad3 and the Smad3/SARA complex were determined by molecular replacement using the CNS package (Brunger et al. 1998). In both cases, the crystal structure of the Smad4 MH2 domain was used as a model for the rotation and translation searches (Qin et al. 1999). The initial solutions were further refined by use of the rigid body, conjugated gradient, and simulated annealing refinement protocols implicated in the CNS package. Structures were rebuilt using CHAIN (Sack 1988). For the unliganded Smad3 structure, the final model includes residues 228–322, 328–379, and 387–417. For the Smad3/SARA complex, the final model includes residues 219–415 from Smad3 and residues 671–708 from SARA.
Construction of the Smad3/SARA/receptor kinase model and the Smad3/Ski model
The dimeric SARA FYVE domain model was constructed by substituting the side chains of the EEA1 FYVE domain with those of the SARA FYVE domain on the basis of sequence alignment (Dumas et al. 2001). The docking of the SARA FYVE domain and the kinase domain to the Smad3/SARA complex was performed using 3D-Dock (G. Moont, G.R. Smith, and M.J.E. Sternberg 2001). The program scans all translational and rotational space for potential complexes and ranks the solutions on the basis of the surface complementarity and electrostatic interaction scores. Biological constraints were applied to filter out solutions inconsistent with the biochemical information. The Smad3/Ski model was constructed using the crystal structure of phosphorylated Smad2 and the SARA SBD structure as the starting models. The Smad3 side chains were introduced into the Smad2 structure. The Ski side chains were built onto the backbone of the SARA SBD on the basis of predicted secondary structure homology. The Ski model was then adjusted to fit the mutagenesis data and secondary structure prediction.
Analytical ultracentrifugation
Experiments were conducted in a Beckman Optima XLA analytical ultracentrifuge equipped with absorbance optics and an An60Ti rotor as described previously (Correia et al. 2001). Buffer conditions were 20 mM Hepes, 100 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT (pH 7.4), plus 2 mM TCEP at 24.7 °C. Sedimentation velocity experiments were performed at 42 K rpm in charcoal-filled Epon double-sector centerpieces. Velocity data were analyzed using DCDT+, version 1.14 (Philo 2000) and SVEDBERG, version 6.39 (Philo 1997) and corrected to S20,w values using measured density and viscosity values and partial specific volume values estimated with Sednterp, version 1.06 (0.7229 for S3LC, 0.7262 for SBD, and 0.7236 for the S3LC/SBD complex) (Laue et al. 1992).
Sedimentation equilibrium experiments were performed at 24K rpm for 5, 10, and 15 μM S3LC/SBD complex, in charcoal-filled Epon 6 channel centerpieces. Data from the three channels were globally fit with Nonlin to a single species model and gave results consistent with the presence of a 1:1 complex (S3LC/SBD MW = 37,489 ± 709, expected MW = 40,575). The slightly lower MW than expected could be due to slight dissociation or <1:1 stoichiometry.
Size-exclusion chromatography
Size-exclusion chromatography was performed using the Superdex 200 HR column on the Akta Explore 10 FPLC (Amersham). The column was equilibrated at room temperature with 20 mM HEPES (pH 7.4), 0.1 mM EDTA, 100 mM NaCl, and 1 mM DTT. Protein samples were incubated in 1 mM TCEP, a reducing agent, for 60 min at room temperature. Sample injection, elution, and data analysis were performed using the UNICORN software (Amersham). The flow rate was 0.7 mL/min and the fraction volume was 0.5 mL. The column was calibrated with molecular weight standards blue dextran, ovalbumin (43 k), albumin (67 k), aldolase (158 k), catalase (232 k), and ferritin (440 k).
GST pull-down assay
The GST fusion form of one protein was immobilized onto the glutathione-sepharose beads followed by washing with a buffer containing 20 mM Tris (pH 7.4), 0.1 mM EDTA, 10 mM NaCl, and 1 mM DTT. The second protein, purified without the GST tag, was then added to the beads and incubated at 4°C for 1 h. The beads were then washed and analyzed by SDS-PAGE and the gels were stained with Coomassie blue. The standard condition uses 100 μL of the beads, 100 ug of the GST-fusion proteins and 200 μg of the binding partners. Unless otherwise stated, the beads were washed six times, each time by adding 1 mL of washing buffer, followed by rapid mixing, centrifugation, and removal of the buffer.
Generation of phosphorylated Smad3
S3LC without the carboxy-terminal CSSVS sequence (S3LCΔC5) was cloned into the pTXB1 bacterial expression vector (New England Biolab) upstream of GyrA intein and the chitin-binding domain (CBD). The protein was expressed at room temperature for 24 h, and the cells were harvested in a harvesting buffer containing 20 mM Tris, 500 mM NaCl, 0.1 mM EDTA, and 1 mM DTT. After cell lysis, the lysate was loaded onto a chitin column equilibrated with the harvesting buffer. After extensive washing of the column, the column was incubated with 50 mM MESNA in the harvesting buffer at 4°C overnight to induce cleavage of S3LCΔC5 from intein-CBD. The eluted S3LCΔC5 protein was concentrated to 10 mg/mL and fourfold excess of the phosphorylated synthetic peptide Cys–Ser–pSer–Val–Ser was added for chemical ligation to the carboxyl terminus of S3LCΔC5.
Acknowledgments
We thank K. Luo, R. Derynck, and C. Sagerstrom for constructive criticism of the manuscript, the staffs of the ALS at Berkeley laboratory and beamline X-25 of NSLS at Brookhaven National Laboratory for assistance with x-ray data collection, D.G. Lambright for sharing the coordinates of EEA1 before publication; R. Derynck for Smad3 and Smad4 cDNA, J. Wrana for SARA cDNA, K. Luo and S.L. Stroschein for Ski cDNA, and B.M. Chacko for phosphorylated S3LC. This work is supported by grants from the Juvenile Diabetes Foundation and the National Institutes of Health to K.L. The analytical ultracentrifugation work was performed at the UMMC Analytical Ultracentrifuge Facility (J.J.C)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kai.lin@umassmed.edu; FAX (508) 856-2398.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1002002.
References
- Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana JL. TbetaRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J Biol Chem. 1999;274:35269–35277. doi: 10.1074/jbc.274.49.35269. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. New Eng J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol. 2001;8:248–253. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massague J. Determinants of specificity in TGF-β signal transduction. Genes & Dev. 1998;12:2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia JJ, Chacko BM, Lam SS, Lin K. Sedimentation studies reals a direct role of phosphorylation in Smad3:Smad4 homo- and hetero-trimerization. Biochemistry. 2001;40:1475–1482. doi: 10.1021/bi0019343. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Hemmati P, Larisch-Bloch S, Ajmera R, Roberts AB, Lechleider RJ. Characterization of functional domains within Smad4/DPC4. J Biol Chem. 1997;272:13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-β signaling in Cancer. J Natl Cancer Inst. 2000a;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, Shioda T, Roberts AB, Lechleider R J. The smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem. 2000b;275:2115–2122. doi: 10.1074/jbc.275.3.2115. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH. Smads: Transcriptional activators of TGF-β responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG. Multivalent endosome targeting by homodimeric EEA1. Mol Cell. 2001;8:947–958. doi: 10.1016/s1097-2765(01)00385-9. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling specificity. EMBO J. 1997;16:3912–3923. doi: 10.1093/emboj/16.13.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa L, Wells RG, Lodish HF, Henis YI. Oligomeric structure of type I and type II transforming growth factor β receptors: Homodimers form in the ER and persist at the plasma membrane. J Cell Biol. 1998;140:767–777. doi: 10.1083/jcb.140.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PJ, Deep S, Taylor AB, Shu Z, Hinck CS, Hinck AP. Crystal structure of the human TβR2 ectodomain–TGF-β3 complex. Nat Struct Biol. 2002;9:203–208. doi: 10.1038/nsb766. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Huse M, Chen YG, Massague J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF β receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- Huse M, Muir TW, Xu L, Chen Y, Kuriyan J, Massague J. The TGF β receptor activation process. An inhibitor- to substrate-binding switch. Mol Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Massague J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-β pathway. J Biol Chem. 2000;275:40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. In: Analytical ultracentrifugation in biochemistry and polymer science. Harding S, Rowe A, Horton J, editors. Cambridge, U.K: Royer Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- Liu X, Sun Y, Weinberg RA, Lodish HF. Ski/Sno and TGF-β signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Lo RS, Chen Y-G, Shi Y, Pavletich NP, Massague J. The L3 loop: A structural motif determining specific interactions between SMAD proteins and TGF-β receptors. EMBO J. 1998;17:996–1005. doi: 10.1093/emboj/17.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Lodish HF. Signaling by chimeric erythropoietin-TGF-β receptors: Homodimerization of the cytoplasmic domain of the type I TGF-β receptor and heterodimerization with the type II receptor are both required for intracellular signal transduction. EMBO J. 1996;15:4485–4496. [PMC free article] [PubMed] [Google Scholar]
- Luo K, Zhou P, Lodish HF. The specificity of the transforming growth factor β receptor kinases determined by a spatially addressable peptide library. Proc Natl Acad Sci. 1995;92:11761–11765. doi: 10.1073/pnas.92.25.11761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q. The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes & Dev. 1999;13:196–206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Silva M, Abdollah S, Hoodless P, Pirone R, Attisano L, Wrana JL. MADR-2 is a substrate of the TGF-β receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- Massague J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation model. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engstrom U, Heldin CH, Funa K, ten Dijke P. The L45 loop in type I receptors for TGF-β family members is a critical determinant in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- Philo JS. An improved function for fitting sedimentation velocity data for low-molecular-weight solutes. Biophys J. 1997;72:435–444. doi: 10.1016/S0006-3495(97)78684-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem. 2000;279:151–163. doi: 10.1006/abio.2000.4480. [DOI] [PubMed] [Google Scholar]
- Qin B, Lam SSW, Lin K. Crystal structure of a transcriptionally active Smad4 fragment. Structure. 1999;7:1493–1503. doi: 10.1016/s0969-2126(00)88340-9. [DOI] [PubMed] [Google Scholar]
- Qin BY, Chacko BM, Lam SS, de Caestecker MP, Correia JJ, Lin K. Structural basis of Smad1 activation by receptor kinase phosphorylation. Mol Cell. 2001;8:1303–1312. doi: 10.1016/s1097-2765(01)00417-8. [DOI] [PubMed] [Google Scholar]
- Randall RA, Germain S, Inman GJ, Bates PA, Hill CS. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J. 2002;21:145–156. doi: 10.1093/emboj/21.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB. TGF-β signaling from receptors to the nucleus. Microbes Infect. 1999;1:1265–1273. doi: 10.1016/s1286-4579(99)00258-0. [DOI] [PubMed] [Google Scholar]
- Sack JS. CHAINS. J Mol Graph. 1988;6:244. [Google Scholar]
- Shi Y, Hata A, Lo RS, Massague J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, Dijke P, Heldin C-H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liu X, Eaton EN, Lane WS, Lodish HF, Weinberg RA. Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol Cell. 1999;4:499–509. doi: 10.1016/s1097-2765(00)80201-4. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGF-β receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Wieser R, Wrana JL, Massague J. GS domain mutations that constitutively activate TβR-1, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SA, Zimmerman CM, Li LI, Mathews LS. Formation and activation by phosphorylation of activin receptor complexes. Mol Endocrinol. 1996;10:367–379. doi: 10.1210/mend.10.4.8721982. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Wieser R, Ventura F, Massague J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- Wrana JL. Phosphoserine-dependent regulation of protein-protein interactions in the Smad pathway. Structure. 2002;10:5–7. doi: 10.1016/s0969-2126(01)00702-x. [DOI] [PubMed] [Google Scholar]
- Wu G, Chen Y-G, Ozdamar B, Gyuricza CA, Chong PA, Wrana JL, Massague J, Shi Y. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science. 2000;287:92–97. doi: 10.1126/science.287.5450.92. [DOI] [PubMed] [Google Scholar]
- Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, et al. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-β signaling. Mol Cell. 2001;8:1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- Xu W, Angelis K, Danielpour D, Haddad MM, Bischof O, Campisi J, Stavnezer E, Medrano EE. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type β transforming growth factor. Proc Natl Acad Sci. 2000;97:5924–5929. doi: 10.1073/pnas.090097797. [DOI] [PMC free article] [PubMed] [Google Scholar]