Abstract
Protein phosphatase 2C (PP2C) is a class of ubiquitous and evolutionarily conserved serine/threonine PP involved in stress responses in yeasts, mammals, and plants. Here, I present mutational analysis of two Arabidopsis thaliana PP2Cs, encoded by ABI1 and AtPP2C, involved in the plant stress hormone abscisic acid (ABA) signaling in maize mesophyll protoplasts. Consistent with the crystal structure of the human PP2C, the mutation of two conserved motifs in ABI1, predicted to be involved in metal binding and catalysis, abolished PP2C activity. Surprisingly, although the DGH177–179KLN mutant lost the ability to be a negative regulator in ABA signaling, the MED141–143IGH mutant still inhibited ABA-inducible transcription, perhaps through a dominant interfering effect. Moreover, two G to D mutations near the DGH motif eliminated PP2C activity but displayed opposite effects on ABA signaling. The G174D mutant had no effect but the G180D mutant showed strong inhibitory effect on ABA-inducible transcription. Based on the results that a constitutive PP2C blocks but constitutive Ca2+-dependent protein kinases (CDPKs) activate ABA responses, the MED141–143IGH and G180D dominant mutants are unlikely to impede the wild-type PP2C and cause hyperphosphorylation of substrates. In contrast, these dominant mutants could trap cellular targets and prevent phosphorylation by PKs required for ABA signaling. The equivalent mutations in AtPP2C showed similar effects on ABA responses. This study suggests a mechanism for the action of dominant PP2C mutants that could serve as valuable tools to understand protein–protein interactions mediating ABA signal transduction in higher plants.
The plant hormone abscisic acid (ABA) modulates a wide spectrum of responses, including gene activation and repression, guard cell closure, cell cycle blockage, and photosynthesis inhibition, under multiple environmental stress conditions such as drought, cold, and salinity (1–7). ABA also plays a pivotal role in the developmental program of seed maturation, desiccation, dormancy, and germination (3–8). Elegant physiological studies based on ABA-mediated guard cell closure (9–11) and ABA inhibition of α-amylase secretion in aleurone cells (12) have suggested a role for Ca2+ in ABA signaling. Extensive molecular analyses of ABA-inducible promoters in tissue culture, aleurone cells, and transgenic plants have identified important cis-acting sequences such as ABA-responsive DNA elements (ABRE) (13–17). Several transcription factors, EmBP-1, TAF-1, GBF, VP1, ABI3, and GF14 (the 14-3-3 protein), have been proposed to be involved in ABA-inducible transcription (4–8, 18, 19). Both cell surface (20–22) and intracellular locales (11, 23) have been suggested to be possible ABA-binding sites. However, the molecular mechanisms underlying the perception and transduction of the ABA signal to the target genes in the nucleus remain elusive.
Genetic approaches have been taken to select ABA response mutants in maize, tomato, potato, tobacco, barley, pea, and Arabidopsis thaliana (At) (3–8, 24–28). The selection of mutants in ABA signaling pathways has been especially fruitful in Arabidopsis. For example, many mutants selected for ABA-insensitive (abi) seedling growth (3–5, 24–28) or enhanced response to ABA (era) during germination (25) have been isolated. Two of the abi mutants (abi1 and abi2) exhibit pleiotropic phenotypes both in seeds and in vegetative tissues (3–5, 24–28). It is postulated that ABI1 and ABI2 are involved in the early steps of ABA signal transduction. Recent molecular cloning revealed that both ABI1 and ABI2 encode serine/threonine phosphatase 2C (PP2C) (26–28). Most interestingly, both the abi1 and abi2 mutants are dominant and have the equivalent G to D mutation in the PP2C domain (26–28). Because no null or recessive mutations are available, the precise role of the PP2C activity and the mechanism of dominant mutants in ABA signal transduction remain obscure.
Here, I show that the overexpression of two constitutively active PP2Cs block ABA signaling in a maize mesophyll protoplast transient assay. The role of PP2Cs as negative regulators in ABA signaling is supported by the identification of mutations that abolish PP2C activity and its inhibitory effect on ABA-inducible transcription. Interestingly, two types of dominant interfering mutants with greatly diminished PP2C activity can also block ABA signaling. This study reveals a novel mechanism for the action of dominant interfering mutants that does not impede the wild-type PP2C activity in the same cells. These PP2C mutations could serve as valuable tools to identify other essential components in the ABA signal transduction pathway in higher plants.
MATERIALS AND METHODS
Plasmid Constructions.
The construction of rubisco-1,5-bisphosphate carboxylase/oxygenase small subunit-chloramphenicol acetyltransferase (RBCS-CAT), barley ABA-responsive CAT (HVA1-CAT), HVA1-luciferase (HVA1-LUC), and ubiquitin-β-glucuronidase (UBI-GUS) has been described previously (29–31). The plant expression vector containing the 35SC4PPDK (cauliflower mosaic virus 35S RNA/maize C4 pyruvate orthophosphate dikinase) promoter, the nopaline synthase (NOS) terminator, and the double influenza hemagglutinin (DHA) tag with the StuI site for cDNA fusion has been described (30, 31). The Escherichia coli expression vector pET19 and the BL21 cells were obtained from Novagen. The primers for generating PCR cDNAs of ABI1 (ABA insensitive 1), dABI1 (deletion), AtPP2C, dAtPP2 (deletion), and KAPP (kinase-associated protein phosphatase) (26–28, 32, 33) are as follows: ABI1, TAGGATCCATGGAGGAAGTATCTCCG and AAGGCCTGTTCAAGGGTTTGCTCTTGA; dABI1, TAGGATCCATGGCTATTACTAGCGAGAAGAAG and the same 3′ primer for ABI1; KAPP1, GCGGATCCATGGCTGTCCGTATCTCCTCTCAG and ACATTACAGGGAAGTATCGAAATC; KAPP2, GCGGATCCATGGCTAATCTGGAAAAGGATCGACTTA and the same 3′ primer for KAPP1. AtPP2C, GCGGATCCATGGCTGGGATTTGTTGC and AAGGCCTAGACGACGCTTGATTATTCCT; dAtPP2C, CATGCCATGGCTAGATCAGCGGTTACCAAT and the same 3′ primer for AtPP2C. The cotransfection results were the same when using truncated KAPP1 (293–582 aa) or KAPP2 (162–582 aa) (33). Two to four independent PCR clones were chosen for cotransfection experiments. The results of at least two independent clones were identical. The primers for generating ABI1 and AtPP2C mutants by PCR are: ABI1 MED141–143IGH, GGAAGAAGACCTGAGATCCATGGTGCTGTTTCGACT and AGTCGAAACAGCACCATGGATCTCAGGTCTTCT; ABI1 G174D, GCTCATTTCTTCGATGTTTACGACGGC and GCCGTCGTAAACATCGAAGAAATGAGC; ABI1 DGH177–179KLN, TTCTTCGGTGTTTACAAGCTTAACGGCGGTTCTCAGGTA and ACCTGAGAACCGCCGTTAAGCTTGTAAACACCGAAGAA; ABI1 G180D, TACGACGGCCATGACGGTTCTCAGGTA and ACCTGAGAACCGTCATGGCCGTCGTA. ABI1 D93A, ATAGTCGTCGTTGCTATCTCCGCCGG and CCGGCGGAGATAGCAACGACGACTAT AtPP2C G139D, CATCATTTCTACGATGTCTTTGACGG and GGCCGTCAAAGACATCGTAGAAATGATG AtPP2C G145D, TTGACGGCCATGACTGCTCTCATGT and CCGCAACATGAGAGCAGTCATGGCCGTCAA.
The mutations were identified by restriction enzyme digestion after cloning and confirmed by DNA sequencing. Four to six PCR clones were examined for each mutation, and two to four clones were used for cotransfection experiments. Identical results were obtained with at least two independent clones.
Protoplast Transient Expression.
Transient expression assays using greening or etiolated maize mesophyll protoplasts were described previously (29–31). ABA responses were the same in etiolated and greening maize mesophyll protoplasts. Transfection efficiency was around 30–50%. Transfected protoplasts were incubated at 0.5–1 × 105 per ml for 3–16 h under light (15 μEm−2 s−1) at 23°C. CAT and GUS assays were performed with cell extracts made from 5,000 protoplasts (31). LUC assay was performed with cell extracts made from 2,000 protoplasts by using a kit (Promega) and a luminometer, and normalized with the UBI-GUS activity for transfection control. The data were presented as cpm per 1,000 for CAT activity and lu per 10,000/10 sec for LUC activity (30, 31). The experiments were carried out with replicated samples repeated with different batches of protoplasts for 2–10 times and showed similar results.
Immunoprecipitation.
Transfected protoplasts were incubated for 4 hr to allow mRNA accumulation and then labeled with [35S]methionine (200 μCi/ml) for 2–12 h. Immunoprecipitation was as described (30), and the proteins were analyzed by 12.5% SDS/PAGE and visualized by fluorography.
PP2C Assays.
PP2C activity was measured as described with minor modifications (33–35). Protoplast cell extracts were made by lysing 105 protoplasts in 100 μl of protoplast lysis buffer (PLB) (20 mM Tris⋅HCl, pH 7.5/20 mM KCl/1 mM EDTA/10 mM DTT/0.5% Triton X-100/50% glycerol/10 μg/ml antipain/10 μg/ml leupeptin/10 μg/ml pepstatin/1 mM phenylmethylsulfonyl fluoride) on ice for 3 min. E. coli cell extracts were prepared from cells lysed in 100 μl PLB on ice for 5 min. Spermidine (5 mM) was added to allow the separation of lysates from DNA–protein complexes by centrifugation for 30 min at 4°C. The cell extracts were diluted 100-fold in PLB before using 2 μl for each 25 μl of reaction. The PP2C reaction was carried out in 25 μl of reaction (5 μg [32P]casein, 50 mM Tris⋅HCl, pH 7.5, 10 mM MgCl2, 5 mM DTT) with 2 μl of cell extracts at 30°C for 15 min (E. coli cell extracts) or 30–45 min (plant cell extracts). The reaction (10 μl) was stopped by adding 190 μl of cold 20% TCA. After centrifugation, 180 μl of the released 32P was counted in 3 ml of scintillation fluid. The PP2C activity was performed in the linear range (below 20% release of the total counts). The counts obtained in the presence of 5 mM EDTA without MgCl2 were subtracted as background. Experiments were performed with replicates. The data were presented as cpm/1,000.
RESULTS
Constitutive PP2C Blocks ABA-Repressible Gene Expression in Leaf Cells.
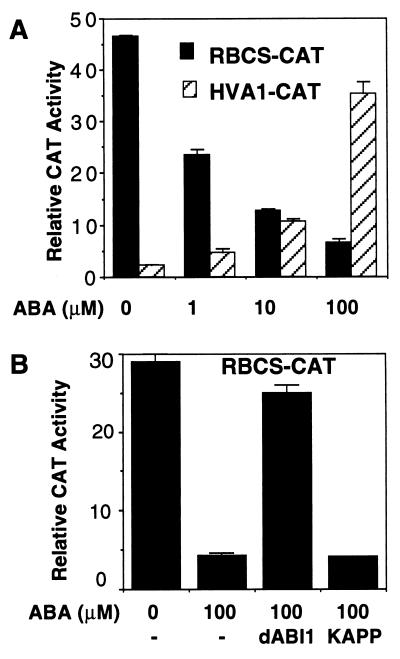
I have shown that a constitutive PP2C derived from the truncated Arabidopsis ABI1 (dABI1, 105–434 aa) can block ABA-inducible gene expression in maize mesophyll protoplasts (30). However, it was not clear whether PP2C is also involved in ABA-repressible gene expression (36–38). I first tested a maize photosynthetic gene promoter to see whether the signal transduction pathway mediating ABA repression exists in isolated maize mesophyll protoplasts. The result showed that the CAT expression controlled by the maize RBCS promoter (29) was inhibited by ABA (Fig. 1A). In contrast, the same ABA concentrations activated the HVA1 promoter (Fig. 1A). The GUS activity driven by the UBI promoter was not affected in the cotransfected protoplasts (30) and was used as an internal control to normalize transfection efficiency.
Figure 1.
Constitutive PP2C blocks ABA repression in maize mesophyll protoplasts. (A) Greening maize mesophyll protoplasts (2 × 105) were coelectroporated with 4 μg of UBI-GUS (internal control) and 40 μg of RBCS-CAT or HVA1-CAT plasmid DNA. Transfected protoplasts were divided into four parts (0.5 × 105/ml) and incubated without (0) or with 1, 10, or 100 μM ABA. (B) Greening maize mesophyll protoplasts were electroporated with the reporter plasmid RBCS-CAT and UBI-GUS or with an effector plasmid expressing dABI1 or KAPP. Transfected cells were incubated with ABA for 16 h before CAT assay.
To determine the role of PP2C in ABA-dependent gene repression, the RBCS-CAT and 35SC4PPDK-dABI1 constructs were cotransfected into maize mesophyll protoplasts by electroporation. As shown in Fig. 1B, the CAT activity was no longer repressed by ABA in the presence of PP2C activity. As a control, the overexpression of KAPP, containing a different PP2C domain (33), could not block ABA-repressible transcription. The results suggest that specific PP2C activity can negatively regulate the ABA signal transduction pathway controlling both gene activation and repression.
The N-Terminal Domain of ABI1 Provides a Regulatory Function Independent of the Putative Ca+2-Binding Motif.
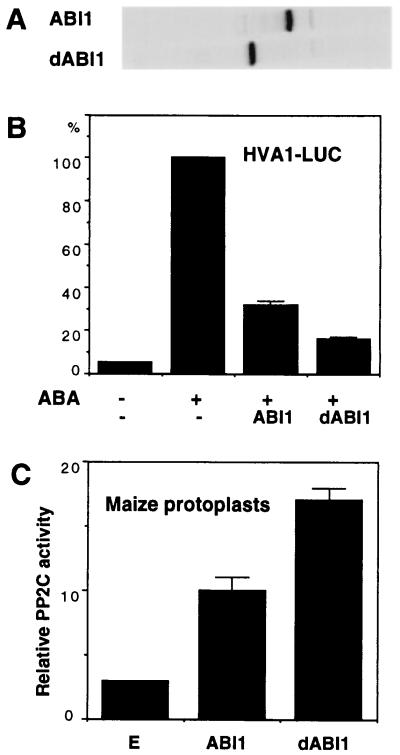
One distinct feature of three Arabidopsis PP2C genes is the presence of a short N-terminal domain with unknown function (26–28, 32). To determine whether the N-terminal domain of ABI1 plays a regulatory role in PP2C activity and ABA signaling, I compared the effect of the wild-type, full-length ABI1 and the truncated dABI1 on ABA-inducible gene expression and measured the PP2C activity from electroporated maize mesophyll protoplasts. To allow easy quantitation of the expressed ABI1 and dABI1 proteins, a double HA epitope tag (30) was fused in-frame to the C-terminus of both proteins. Similar levels of ABI1 and dABI1 were detected in electroporated protoplasts by immunoprecipitating the [35S]methionine-labeled proteins with the anti-HA antibody (Fig. 2A). Based on an equal molar ratio, dABI1 blocked ABA signaling more effectively than ABI1 (Fig. 2B). The analysis of samples from an early time point (3 h) after DNA transfection allowed clear differentiation of the effects of ABI1 and dABI1 on ABA signaling. The different effects of ABI1 and dABI1 on blocking ABA responses detected in vivo correlated with their PP2C activities measured in vitro by using cell extracts prepared from transfected protoplasts (Fig. 2C) and transformed E. coli cells (data not shown). The N-terminal domain of ABI1 is likely playing an inhibitory role in PP2C activity and ABA responses.
Figure 2.
The N-terminal domain of ABI1 has an inhibitory function. (A) The [35S]methionine-labeled ABI1 and dABI1 proteins were shown from transfected etiolated maize mesophyll protoplasts after immunoprecipitation and SDS/PAGE. (B) Protoplasts were cotransfected with HVA1-LUC and ABI1 or dABI1 and analyzed at 3 h to determine LUC activity without or with ABA (100 μM) treatment. The level of LUC activity in the presence of ABA without any effector was set as 100% for easy comparison. (C) PP2C activity (cpm/1,000) from transfected maize mesophyll protoplasts incubated for 3 h was determined. Endogenous (E) PP2C activity was detectable in protoplasts electroporated with control plasmid DNA.
To determine whether the putative EF-hand Ca2+ binding site in the N-terminal domain (26, 28) plays a role in the regulation of ABI1, site-directed mutagenesis was used to change the D residue (D93A), which is essential for Ca2+ binding in authentic EF hand domains (39). The mutant was analyzed in transfected maize mesophyll protoplasts and did not alter the effect of ABI1 on ABA-inducible transcription and PP2C activity (data not shown). Thus, the putative EF-hand motif in ABI1 does not seem to have a true physiological function. This conclusion is in agreement with the previous finding that ABI1 does not show high-affinity binding to Ca2+ (40).
PP2C Activity Is Required as a Negative Regulator in ABA Signal Transduction.
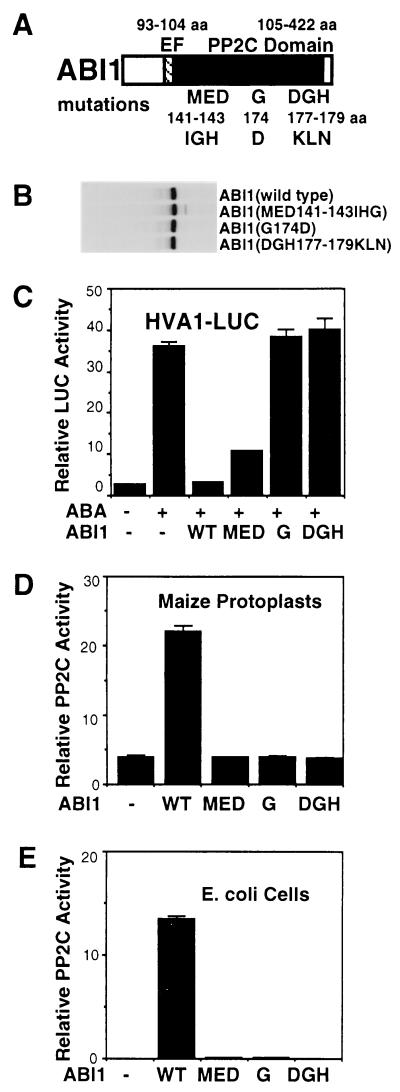
To further support the notion that PP2C activity is required to block ABA responses, I attempted to create null PP2C mutants by site-directed mutagenesis. Three ABI1 mutants (MED141–143IHG, G174D, and DGH177–179KLN) were generated, tagged with DHA, and inserted into the plant expression vector (30, 31) (Fig. 3A). The MED and DGH sequences were chosen because of their high conservation in all PP2Cs and their involvement in the PP2C active sites recently revealed by the crystal structure (41). The mutated sequences carried new restriction sites for convenient identification and encoded unrelated amino acids. The G174 residue was mutated because it is conserved in three Arabidopsis PP2C genes and is adjacent to the conserved DGH177–179 motif and the G180D mutation in abi1. None of these mutations affected the expression level of the proteins in electroporated maize mesophyll protoplasts (Fig. 3B). However, two mutations (G174D and DGH177–179KLN) abolished the ability of ABI1 to block ABA-inducible transcription (Fig. 3C). PP2C activity assays using cell extracts from transfected maize mesophyll protoplasts (Fig. 3D) or transformed E. coli cells (Fig. 3E) confirmed that PP2C activity was crucial for ABI1 to act as a negative regulator. G174D and DGH177–179KLN are true null mutations perhaps owing to dramatic changes in amino acid charges near or at the PP2C active site. It is interesting to note that the MED141–143IGH mutant could still partially block ABA induction despite null PP2C activity (Figs. 3 C–E). Because E142 and D143 are involved in metal binding at the active site of PP2C (41), the MED141–143IGH mutation probably hinders the catalytic process but retains or gains protein-binding ability and impedes ABA signaling by a dominant effect independent of PP2C activity.
Figure 3.
PP2C activity is required for ABI1 function as a negative regulator in ABA signaling. (A) The sequences and locations of three ABI1 mutations in the PP2C domain are shown. (B) ABI1 mutations did not affect protein expression as shown after [35S]methionine labeling, immunoprecipitation, and SDS/PAGE. (C) Etiolated maize mesophyll protoplasts were electroporated with the reporter gene HVA1-LUC with an inert construct or with different ABI1 constructs: wild-type ABI1 (WT), MED141–143IGH mutation (MED), G174D mutation (G), and DGH177–179KLN mutation (DGH). Transfected cells were incubated without or with ABA (100 μM) for 16 h to observe the maximal effect on ABA-inducible transcription. PP2C activity was measured from transfected maize mesophyll protoplast (D) and transformed E. coli cells (E).
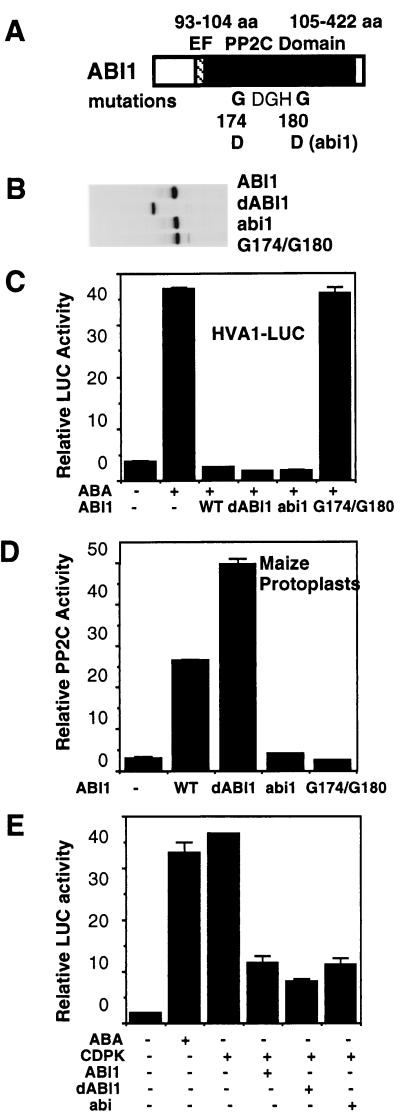
abi1 Blocks ABA Signal Transduction with Diminished PP2C Activity in Maize Mesophyll Protoplasts.
Although I have shown that PP2C activity is required as a negative regulator in ABA signaling, the analysis of abi1 and abi2 proteins in E. coli extracts suggested that both mutants block ABA responses with diminished PP2C activity (27, 40). To understand the mechanism of abi1 and abi2 action, it is important to determine the PP2C activity and its effect on ABA signaling in the same plant cells. An abi1 mutant (G180D) was created by site-directed mutagenesis (Fig. 4A), tagged by DHA, and cloned into the plant expression vector with the 35SC4PPDK promoter (30). The expression of the protein from abi1 was not affected by the mutation (Fig. 4B). Interestingly, in transfected maize mesophyll protoplasts, the abi1 protein was as effective as ABI1 and dABI1 in eliminating ABA-inducible transcription (Fig. 4C). The measurement of PP2C activity by using both transfected protoplasts (Fig. 4D) and E. coli cells (data not shown) revealed that the mutation in abi1 virtually abolished PP2C activity. Thus, the data support the model that abi1 blocks ABA responses through a dominant interfering effect without PP2C activity. Together with the MED141–143IGH mutant, this class of dominant mutants might facilitate the identification of new protein components essential for ABA signaling.
Figure 4.
The abi1 mutant abolishes ABA signaling by a dominant interfering effect. (A) The locations of the ABI1 mutations in the PP2C domain are shown. (B) Protein expression of the ABI1 mutants is shown by [35S]methionine labeling, immunoprecipitation, and SDS/PAGE. (C) Etiolated maize mesophyll protoplasts were electroporated with the reporter gene HVA1-LUC with an inert plasmid or with an ABI1 construct: wild-type ABI1 (WT), truncated ABI1 (dABI1), G180D mutation (abi1), and the double mutation G174D and G180D (G174/G180). Transfected cells were incubated without or with ABA (100 μM) for 16 h, and the LUC activity was measured. (D) PP2C activity of ABI1 mutants in transfected maize mesophyll protoplasts. (E) Etiolated maize mesophyll protoplasts were electroporated with the reporter gene HVA1-LUC and an inert plasmid or with CDPK or/and various ABI1 constructs.
To test whether the dominant property of abi1 could be abrogated by introducing the mutation G174D that eliminates normal ABI1 functions (Fig. 3), a double mutant (G174D/G180D) was created (Fig. 4A). The double mutant does not affect protein expression (Fig. 4B). Remarkably, the dominant interfering effect of the abi1 protein was completely abolished in the double mutant (Fig. 4C). Although the dominant effect of the protein may not be related to the normal function of ABI1, the analysis of the double mutant (G174D/G180D) indicates that the functions of ABI1 and abi1 require the same G174 residue, perhaps essential for binding the same targets. No PP2C activity was detected from the double mutant in the maize (Fig. 4D) and E. coli cell extracts (data not shown).
Because ABI1 and abi1 inhibit ABA signaling by different mechanisms, they may act at different steps in the ABA signal transduction pathway. It was also speculated that abi1 might impede endogenous PP2C and cause hyperphosphorylation of substrates (40). I have previously shown that a constitutively active Ca2+-dependent protein kinase (CDPK) can activate an ABA-inducible promoter in the absence of ABA (30). I tested the effect of ABI1, dABI1, and abi1 on CDPK-activated gene expression. If abi1 blocked the ABA signaling pathway at a step upstream of CDPK, then the gene expression activated by a constitutively active CDPK would not be repressed by abi1. As shown in Fig. 4E, all three versions of ABI1 significantly blocked CDPK-activated gene expression in a similar fashion. Thus, it is likely that abi1 and ABI1 act at the same level or downstream of the positive regulator CDPK in the ABA signal transduction pathway. Instead of inhibiting the function of wild-type PP2C, abi1 prevents the action of CDPK as a positive regulator in ABA signaling. Because all three versions of PP2C can block ABA-inducible transcription completely (Fig. 4C), the remaining LUC activity enhanced by CDPK (Fig. 4E) is likely ABA-independent, perhaps mediated through distinct transcription factors in response to other stress signals (16, 17, 30).
A Third Arabidopsis PP2C Provides Redundant Functions in ABA Signaling.
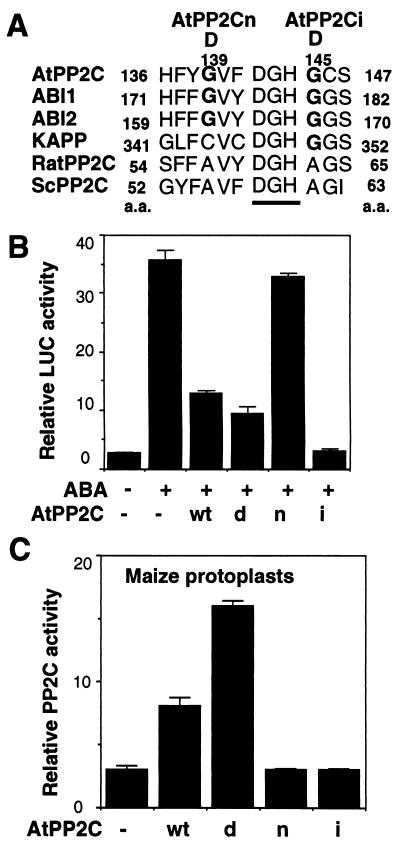
Because PP2C functions are provided by three partially redundant genes in fission (35) and budding yeasts (34), it is interesting to determine whether the third Arabidopsis gene encoding PP2C (AtPP2C) (32) is important for ABA signaling. AtPP2C shares high sequence similarity to ABI1 and ABI2 especially around the conserved active site with two unique G residues (Fig. 5A). To test its role in ABA signaling, four AtPP2C constructs were made. They carry the cDNAs for the wild-type AtPP2C, an N-terminal deletion dAtPP2C, a null G139D mutation AtPP2Cn, and a dominant ABA-insensitive G145D mutation AtPP2Ci similar to abi1 and abi2 (Fig. 5A). These constructs were cotransfected with the reporter HVA1-LUC into maize mesophyll protoplasts and evaluated for their ability to inhibit ABA-inducible transcription 3 h after electroporation. The overexpression of AtPP2C blocked ABA-inducible transcription like ABI1. Similar to dABI1, the N-terminal deletion dAtPP2C was more effective than AtPP2C. The null mutation AtPP2Cn had little effect. Finally, the dominant interfering mutant AtPP2Ci showed the highest efficacy in repressing ABA responses at the early time point (Fig. 5B). The measurement of PP2C activity in protoplasts extracts supported the involvement of PP2C activity, except for the dominant interfering mutant AtPP2Ci (Fig. 5C). Thus, redundant functions are clearly provided by at least three related Arabidopsis PP2C genes to modulate ABA signal transduction.
Figure 5.
AtPP2C provides redundant functions in ABA signaling. (A) AtPP2C, ABI1, and ABI2 possess two uniquely conserved G residues around the DGH (underlined) active site. Cotransfection was performed with HVA1-LUC and an inert plasmid or with four different plasmids expressing the wild-type AtPP2C (wt), the N-terminal deletion dAtPP2C (d), the null mutant AtPP2Cn (n), and the dominant mutant AtPP2Ci (i). LUC activity (B) and PP2C activity (C) were determined from the extracts of maize mesophyll protoplast harvested 3 h after DNA transfection.
DISCUSSION
PP2C represents a unique class of serine/threonine PP found ubiquitously in eukaryotes. The involvement of multiple PP2Cs in stress responses is well documented in yeasts. For example, genetic analyses have identified three PP2C genes with redundant functions in osmotic and heat shock stress responses in fission and budding yeasts (34, 35). I show here that the PP2Cs derived from Arabidopsis ABI1 and AtPP2C are involved in ABA signal transduction. Expression of three other serine/threonine PP1, PP2A, or PP2B did not block ABA signaling in maize leaf cells (data not shown). Recently, the surprising discovery of the Arabidopsis ABI2 as another PP2C closely related to ABI1 (27) supports the notion that redundant PP2C activities are involved in stress responses mediated by ABA. Interestingly, ABI2 also has the two characteristic G162 and G168 flanking the DGH165–167 active site uniquely conserved in ABI1 and AtPP2C (Fig. 5A). Similar to the G180D mutation of abi1 and G145D mutation of AtPP2C, the G168D mutation found in abi2 confers a dominant ABA-insensitive effect (27). It is highly possible that all three PP2Cs provide redundant functions as negative regulators in the ABA signal transduction pathway in higher plants. The functional redundancy also explains the lack of recessive null mutants in any PP2C genes in Arabidopsis as in budding and fission yeasts (24, 26–28, 34, 35). However, not all Arabidopsis genes carrying a PP2C domain are involved in ABA responses. For instance, the Arabidopsis KAPP was isolated based on its interaction with an Arabidopsis receptor-like kinase RLK5, and its PP domain showed authentic PP2C activity when expressed and purified from E. coli (33). However, the two truncated forms of KAPP carrying the PP2C catalytic domain do not block ABA-dependent gene expression in maize leaf cells (Fig. 1B; data not shown).
It is clear that the function of ABI1 and AtPP2C as negative regulators in ABA signaling requires PP2C activity, which may directly inactivate PKs and/or counteract the effect of PKs mediating ABA responses (30). It remains to be shown whether CDPK, mitogen-activated PK (MAPK) (42, 43), or other PKs (44, 45) are involved in ABA signaling in diverse cell types of different plant species. It will be interesting to determine whether different PKs act sequentially in the same pathway or independently in distinct pathways. The identification of substrates or regulators of PP2Cs and CDPKs will be critical for the future effort in unraveling the ABA signal transduction pathways. The ability of ABI1 to interact with components in the ABA signaling pathway is strongly supported by the fact that the abi1 mutant, with greatly diminished PP2C activity, can potently block ABA responses by a dominant interfering effect. It is possible that all three dominant mutants, abi1, abi2, and AtPPCi, interact with and block the function of the same protein essential for ABA signaling. The double mutant that abolished the effect of abi1 could be a valuable tool to study protein–protein interactions involved in the ABA responses. The determination of the subcellular localization of ABI1 will also be important in elucidating its function in controlling ABA-regulated gene expression.
In summary, I have conclusively demonstrated the role of PP2C in ABA signaling by examining the effects of distinct ABI1 mutants. The specific role of PP2C as a negative regulator in ABA signal transduction is further supported by the discovery of functionally redundant AtPP2C and ABI2, its ability to diminish the effect of a specific CDPK as a positive regulator, and the lack of involvement of other PPs (data not shown). Based on the results that the constitutive PP2Cs block but constitutive CDPKs activate ABA responses, it is unlikely that these PP2C dominant mutants compete with the wild-type PP2C and cause hyperphosphorylation of the substrates (40, 46) to inhibit ABA responses in maize leaf cells. In contrast, these dominant interfering mutants could prevent the action of PKs required for ABA signaling. It is most likely that multiple PP2Cs play a crucial role in maintaining physiological homeostasis of plant cells before and after the disturbance caused by stress signals. The system used here provides powerful tools to complement the classical genetic approach in dissecting plant signal transduction pathways, especially when redundant functions are involved.
Acknowledgments
I thank R. Kincaid, F. Katagiri, and H. Saito for their generous gifts of PP2B and ABI1 cDNAs and SSK2 and SSK22 genes, respectively; P. Leon and V. Walbot for the LUC vector; P. Quail for the maize UBI promoter; G. Lazar and L. Zhou for pET19 and BL21 E. coli cells; W. Zheng for the immunoprecipitation protocol; L. Zhou for the DHA tag; and W.-L. Chiu, J.-C. Jang, T. Jones, Lena Kovtun, and J. Nardone for critical comments on the manuscript. J.S. is supported by the U.S. Department of Agriculture (NRICGP 94-37306-0784 and 97-35306-4581), National Science Foundation (IBN-9723610), and Hoechst A.G.
ABBREVIATIONS
- PP2C
protein phosphatase 2C
- ABA
abscisic acid
- ABI
ABA insensitive
- KAPP
kinase-associated protein phosphatase
- HVA
barley ABA responsive
- CAT
chloramphenicol acetyltransferase
- LUC
luciferase
- GUS
β-glucuronidase
- CDPK
calcium-dependent protein kinase
- UBI
ubiquitin
- RBCS
rubisco-1,5-bisphosphate carboxylase/oxygenase small subunit
References
- 1.Assmann S M. Annu Rev Cell Biol. 1993;9:345–375. doi: 10.1146/annurev.cb.09.110193.002021. [DOI] [PubMed] [Google Scholar]
- 2.Chandler P M, Robertson M. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:113–141. [Google Scholar]
- 3.Finkelstein R R, Zeevaart J A D. In: Arabidopsis. Meyerowitz E M, Somerville C R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 523–553. [Google Scholar]
- 4.Giraudat J, Parcy F, Bertauche N, Gosti F, Leung J, Morris P-C, Bouvier-Durand M, Vartanian N. Plant Mol Biol. 1994;26:1557–1577. doi: 10.1007/BF00016490. [DOI] [PubMed] [Google Scholar]
- 5.Rock C D, Quatrano R S. Curr Biol. 1994;4:1013–1015. doi: 10.1016/s0960-9822(00)00229-3. [DOI] [PubMed] [Google Scholar]
- 6.Ingram J, Bartels D. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- 7.Quatrano R S, Bartels D, Ho T-H D, Pages M. Plant Cell. 1997;9:470–475. [Google Scholar]
- 8.McCarty D R. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:71–93. [Google Scholar]
- 9.Schroeder J I, Hagiwara S. Proc Natl Acad Sci USA. 1990;87:9305–9309. doi: 10.1073/pnas.87.23.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAinsh M R, Brownlee C, Hetherington A M. Plant Cell. 1992;4:1113–1122. doi: 10.1105/tpc.4.9.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allan A C, Fricker M D, Ward J L, Beale M H, Trewavas A J. Plant Cell. 1994;6:1319–1328. doi: 10.1105/tpc.6.9.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilroy S, Jones R L. Proc Natl Acad Sci USA. 1992;89:3591–3595. doi: 10.1073/pnas.89.8.3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcotte W R, Russell S H, Quatrano R S. Plant Cell. 1989;1:969–976. doi: 10.1105/tpc.1.10.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mundy J, Yamaguchi-Shinozaki K, Chua N-H. Proc Natl Acad Sci USA. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pla M, Vilardell J, Guiltinan M J, Marcotte W R, Niogret M F, Quatrano R S, Pages M. Plant Mol Biol. 1993;21:259–266. doi: 10.1007/BF00019942. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi-Shinozaki K, Shinozaki K. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Q, Zang P, Ho T-H D. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiltinan M J, Marcotte W R, Jr, Quatrano R S. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 19.Lu G, Paul A-L, McCarty D R, Ferl R J. Plant Cell. 1996;8:847–857. doi: 10.1105/tpc.8.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson B E, Ward J M, Schroeder J I. Plant Physiol. 1994;104:1177–1183. doi: 10.1104/pp.104.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilroy S, Jones R L. Plant Physiol. 1994;104:1185–1192. doi: 10.1104/pp.104.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knox J P, Peart J, Neil S J. Planta. 1995;196:266–270. [Google Scholar]
- 23.Schwartz A, Wu W-H, Tucker E B, Assmann S M. Proc Natl Acad Sci USA. 1994;91:4019–4023. doi: 10.1073/pnas.91.9.4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koornneef M, Reuling G, Karssen C M. Plant Physiol. 1984;61:377–383. [Google Scholar]
- 25.Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- 26.Leung J, Bouvier-Durand M, Morris P-C, Guerrier D, Chefdor F, Giraudat J. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 27.Leung J, Merlot S, Giraudat J. Plant Cell. 1997;9:759–771. doi: 10.1105/tpc.9.5.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer K, Leube M P, Grill E. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 29.Schäffner A R, Sheen J. Plant Cell. 1991;3:997–1012. doi: 10.1105/tpc.3.9.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheen J. Science. 1996;274:1900–1902. doi: 10.1126/science.274.5294.1900. [DOI] [PubMed] [Google Scholar]
- 31.Sheen J. EMBO J. 1993;12:3497–3505. doi: 10.1002/j.1460-2075.1993.tb06024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuromori T, Yamamoto M. Nucleic Acids Res. 1994;22:5296–5301. doi: 10.1093/nar/22.24.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stone J M, Collinge M A, Smith R D, Horn M A, Walker J C. Science. 1994;266:793–795. doi: 10.1126/science.7973632. [DOI] [PubMed] [Google Scholar]
- 34.Maeda T, Rsai A M, Saito H. Mol Cell Biol. 1993;13:5408–5417. doi: 10.1128/mcb.13.9.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiozaki K, Russell P. EMBO J. 1995;14:492–502. doi: 10.1002/j.1460-2075.1995.tb07025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartholomew D M, Bartley G E, Scolnik P A. Plant Physiol. 1990;96:291–296. doi: 10.1104/pp.96.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y C, Walling L L. Plant Physiol. 1991;97:1260–1264. doi: 10.1104/pp.97.3.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weatherwax S C, Ong M S, Degenhardt J, Bray E A, Tobin E M. Plant Physiol. 1996;111:363–370. doi: 10.1104/pp.111.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiser J R, van Tuinen D, Brockerhoff S E, Neff M M, Davis T. Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- 40.Bertauche N, Leung J, Giraudat J. Eur J Biochem. 1996;241:193–200. doi: 10.1111/j.1432-1033.1996.0193t.x. [DOI] [PubMed] [Google Scholar]
- 41.Das A K, Helps N R, Cohen P T W, Bardford D. EMBO J. 1996;15:6798–6809. [PMC free article] [PubMed] [Google Scholar]
- 42.Knetsch M L W, Wang M, Snaar-Jagalska B E, Heimovaara-Dijkstra S. Plant Cell. 1996;8:1061–1067. doi: 10.1105/tpc.8.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori I C, Muto S. Plant Physiol. 1997;113:833–839. doi: 10.1104/pp.113.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderberg R J, Walker-Simmons M K. Proc Natl Acad Sci USA. 1992;92:9520–9524. [Google Scholar]
- 45.Li J, Assmann S M. Plant Cell. 1996;8:2359–2368. doi: 10.1105/tpc.8.12.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armstrong F, Leung J, Grabov A, Brearley J, Giraudat J, Blatt M. Proc Natl Acad Sci USA. 1995;92:9520–9524. doi: 10.1073/pnas.92.21.9520. [DOI] [PMC free article] [PubMed] [Google Scholar]