Abstract
Escherichia coli YaeL (EcfE) is a homolog of human site-2 protease (S2P), a membrane-bound zinc metalloprotease involved in regulated intramembrane proteolysis. We have shown previously that YaeL, having essential metalloprotease active site motifs in the cytoplasmic domain, is indispensable for viability. Here, we obtained rpoE, encoding an extracytoplasmic stress response σ factor (σE), as a multicopy suppressor against the yaeL disruption. Whereas σE is thought to be activated by regulated cleavage of RseA on the periplasmic side by the DegS protease, we found that a degradation intermediate of RseA consisting of the transmembrane and the cytoplasmic domains accumulated in the YaeL-depleted cells. This intermediate was degraded on expression of YaeL but not of its metalloprotease motif mutants. Cells depleted of YaeL were incapable of activating a σE-dependent promoter in response to an envelope stress. It is suggested that σE activation involves two successive proteolytic cleavages: first, at a periplasmic site by DegS; second, at a cytoplasmic or intramembrane site by YaeL. Thus, YaeL is positively required for the σE extracytoplasmic stress response.
Keywords: regulated intramembrane proteolysis, zinc metalloprotease, extracytoplasmic stress response, site-2 protease, cell envelope, DegS
Proteolytic reactions contribute to the maintenance of cellular integrity by eliminating abnormal or unwanted proteins. Another role of intracellular proteolysis is in regulated cleavage/degradation of functional or regulatory proteins. Targets of regulated proteolysis include membrane-integrated proteins. For example, membrane-bound forms of transcriptional factors are subject to site-specific cleavage that liberates their active forms (Hoppe et al. 2001). Thus, SREBP (sterol regulatory element binding protein) is synthesized as a 125-kD precursor that spans the endoplasmic reticulum membrane twice (Hua et al. 1995), and its two sequential cleavages liberate the N-terminal cytoplasmic domain, which is then translocated into the nucleus as a transcriptional activator for the sterol-controlled genes (Hua et al. 1996; Sakai et al. 1996). The initial cleavage of SREBP within the lumenal loop is catalyzed by a membrane-bound, subtilisin-like protease, site-1 protease (S1P; Duncan et al. 1997). The first cleavage somehow provokes the second cleavage by site-2 protease (S2P) of the N-terminal half of SREBP within the first transmembrane segment (Hua et al. 1996; Sakai et al. 1996). S2P is a membrane-embedded zinc metalloprotease, with homologs that are found in a variety of organisms, including bacteria (Brown et al. 2000). S1P and S2P are also involved in the proteolytic activation of ATF6 for the endoplasmic reticulum unfolded protein response (Ye et al. 2000b).
YaeL (EcfE) is an S2P homolog in Escherichia coli (Brown et al. 2000; Dartigalongue et al. 2001a; Kanehara et al. 2001). It spans the plasma membrane four times (Kanehara et al. 2001; Drew et al. 2002). Depletion of YaeL results in the loss of viability and cell elongation (Dartigalongue et al. 2001b; Kanehara et al. 2001). It is an essential protease, as mutations in the zinc-protease active site motif (HEXXH), the NPD motif and the periplasmic PDZ-like region inactivate the complementation activity (Dartigalongue et al. 2001a; Kanehara et al. 2001). The expression of yaeL is in part under the control of σE (Dartigalongue et al. 2001a,b). Raina and colleagues have suggested that YaeL degrades σE (see below) and σ32 (the classical heat shock σ factor) and negatively controls the heat stress responses (Dartigalongue et al. 2001a). However, our results reported in this paper do not substantiate this conclusion; YaeL positively regulates the σE pathway activation. σE is an alternative σ factor that is crucial for a pathway of extracytoplasmic stress response (Mecsas et al. 1993; De Las Peñas et al. 1997a,b). It is encoded by the rpoE gene, which forms an operon with genes for RseA, RseB, and RseC (De Las Peñas et al. 1997b). RseA is an anti-σE factor consisting of an N-terminal cytoplasmic domain, central transmembrane segment, and C-terminal periplasmic domain (Missiakas et al. 1997). It sequesters σE through a cytoplasmic domain–σE interaction (De Las Peñas et al. 1997b; Missiakas et al. 1997). RseB is a periplasmic protein with an ability to bind to the periplasmic domain of RseA, leading to enhanced RseA-σE complex formation (Missiakas et al. 1997; Collinet et al. 2000). The function of RseC is unclear.
Ades et al. (1999) showed that DegS, a plasma membrane protease with a periplasmic active site, acts against RseA. The DegS-dependent degradation of RseA is accelerated by envelope stresses (Ades et al. 1999), but its mechanism is unknown. Indispensability of DegS lies in its function to provide free (active) σE by inactivating RseA (Alba et al. 2001). However, because the isolated N-terminal domain of RseA still retains the anti-σE activity in vitro (Missiakas et al. 1997), the DegS-mediated RseA cleavage may not be sufficient for the σE activation. For instance, DegS action could well be followed by further degradation/cleavage on the cytoplasmic side.
In this paper, we show that YaeL has a positive role in σE activation by antagonizing RseA. Thus, overproduction of σE and deletion of rseA proved to circumvent the loss of viability in the absence of YaeL, and decreased YaeL content attenuated the σE-dependent stress response. Our results indicate that YaeL participates in the sequential cleavages of RseA.
Results
Identification of rpoE, encoding σE, as a multicopy suppressor against the yaeL disruption
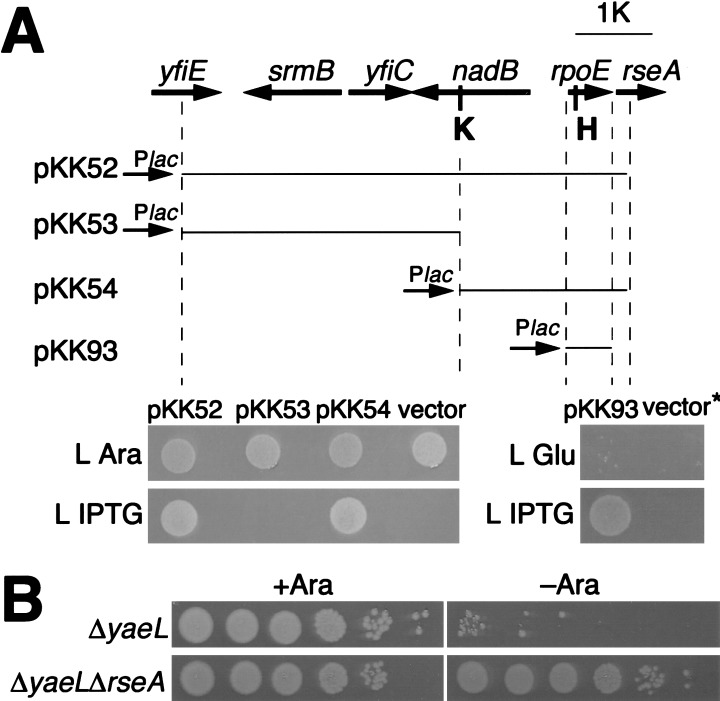
As a means to investigate into the cellular role of YaeL, we isolated multicopy suppressors against a yaeL null mutation. KK31 has a chromosomal yaeL-disruption (yaeL∷kan) and a complementing plasmid (pKK6) in which yaeL+ has been placed under the control of the arabinose-promoter. Growth of KK31 is strictly dependent on arabinose (Fig. 1A, lower panel). A chromosomal DNA library on plasmid was constructed from wild-type strain (W3110) and introduced into KK31. Transformants were selected in the absence of arabinose. Among 66 plasmid clones thus isolated, 65 carried yaeL itself and one (named pKK52) carried a fragment corresponding to a 58-min region of the chromosome (Fig. 1A). Subcloning experiments established that rpoE in this fragment is responsible for the suppression of ΔyaeL. Thus, pKK54 carrying rpoE as the sole uninterrupted open reading frame supported arabinose-independent growth of KK31 (Fig. 1A). In a yaeL+ strain, pKK52 and pKK54 elevated the expression of rpoHP3-lacZ, a reporter with expression that depends on σE, more than threefold. Thus, these plasmids indeed directed σE oversynthesis (data not shown). We also constructed pKK93 that carried only the rpoE gene under the lac promoter. This plasmid, which isopropyl-β-d-thiogalactopyranoside (IPTG) dependently elevated the expression of rpoHP3-lacZ 3.5-fold, enabled the yaeL-depeleted cells to grow only in the presence of IPTG (Fig. 1A). A frame shift by a 4-bp insertion at the HindIII site in rpoE on pKK93 (see Fig. 1A) abolished the multicopy suppressor activity (data not shown). These results indicate that the suppression was caused by an increased σE abundance but not to an increased number of some regulatory nucleotide sequence on the plasmids.
Figure 1.
A multicopy suppressor, rpoE, and a loss-of-function suppressor, ΔrseA, against the yaeL disruption. (A, top) Chromosomal regions carried on pKK52 and its derivatives. Thick arrows indicate genes carried on pKK52 isolated as a multicopy suppressor against ΔyaeL, with arrowheads showing directions of transcription. yfiE and rseA are not totally included, as shown by broken lines. A segment carried in each plasmid is shown by a thin line. Plac, K, and H represent the lac promoter, a KpnI site, and a HindIII site. (bottom) ΔyaeL suppression abilities of plasmids. Plasmids pKK52, pKK53, pKK54, pTWV229 (vector), pKK93, and pTWV228H (vector*) were introduced into strain KK31 (yaeL∷kan/pKK6 [Para-yaeL]) in the presence of arabinose. Cultures with arabinose (0.02%) were diluted 104-fold with 0.9% NaCl, and 4 μL each was spotted on L-agar plate containing 0.2% arabinose (L Ara), 0.4% glucose (L Glu), or 1 mM isopropyl-β-d-thiogalactopyranoside (L IPTG). The plates were incubated for 12 h at 37°C. (B) Effect of the rseA disruption on growth under the YaeL-depleted condition. Fully grown cultures of KK326 (yaeL∷kan/pKK64 [yaeL+]) and KK334 (yaeL∷kan rseA∷cat/pKK64) in the presence of arabinose (0.02%) were serially diluted 10-fold with 0.9% NaCl, and 4 μL each of the samples (102 to 107 dilution) was spotted on L-agar plates with (Ara+) or without (Ara−) 0.2% arabinose as indicated. The plates were incubated for 24 h at 30°C.
The yaeL∷kan marker was introduced successfully by P1 transduction into cells without any yaeL plasmid but carrying pKK54 (data not shown), indicating that yaeL is totally dispensable in the presence of excess σE. We repeated isolation of multicopy suppressors using another genomic library from the yaeL rseA double-disruptant strain (see below) and isolated five independent clones carrying rpoE (data not shown).
The above results raised a question of whether enhanced σE activity was responsible for the YaeL dispensability. As another means to activate σE, we then used the mutation ΔrseA that knocks out RseA, the membrane-bound anti-σE factor. The ΔrseA mutation was introduced into a YaeL-depletable strain KK326 (yaeL∷kan/pKK64 [Para-yaeL]) by P1 transduction in the presence of arabinose. However, the resulting strain (KK334) proved to grow even in the absence of arabinose (Fig. 1B). Moreover, the ΔyaeL∷kan mutation was introduced into a ΔrseA strain but not into an isogenic rseA+ strain (without a complementing yaeL plasmid). Taken together, these results establish that the elevated σE activity fully alleviates the YaeL requirement of E. coli.
Degradation of RseA by sequential actions of DegS and YaeL
One possible explanation for the YaeL-σE connection is that YaeL proteolytically inactivates RseA. In this case, the loss of YaeL function would stabilize RseA, leading to oversequestration of σE and eventual cell death; σE is essential even under nonstressed conditions (De Las Peñas et al. 1997a). We thus investigated the effects of the yaeL disruption on stability of RseA.
We constructed an RseA derivative with an N-terminal hemagglutinin (HA) tag (HA-RseA; Fig. 2). Expression of HA-RseA in cells carrying fkAp-lacZ, a σE reporter gene (SP887; Danese and Silhavy 1997), significantly lowered the LacZ activity, indicating that HA-RseA retained at least partial anti-σE activity. In the following experiments, HA-RseA was induced in the ΔrseA strain for 2–3 h. During this time span, cells continued to grow, although they should have been eventually dying. Two factors might have contributed to the continued growth. First, the positive autoregulation of σE allowed its overaccumulation under the rseA-disrupted conditions. Second, HA-RseA might have lower anti-σE activity owing to the attached HA sequence.
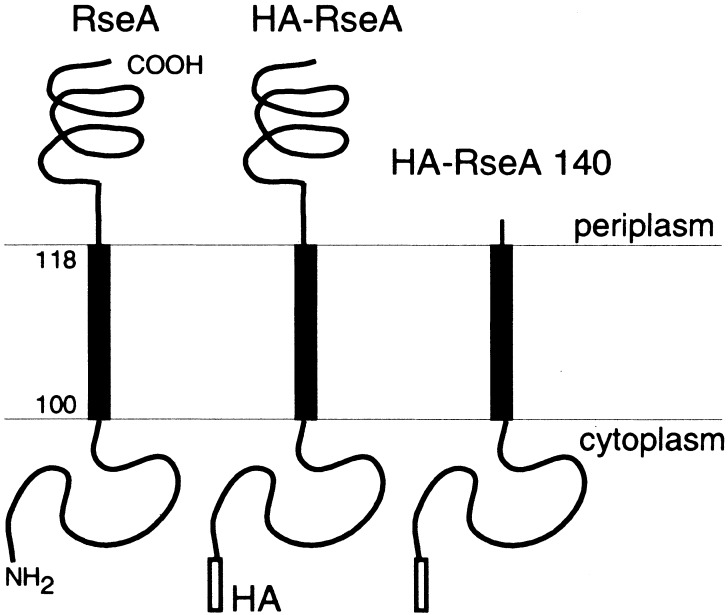
Figure 2.
Schematic diagrams of RseA and its derivatives used in this study. HA indicates the hemagglutinin tag. HA-RseA140 lacks residue 141 to the C terminus, most of the C-terminal periplasmic region.
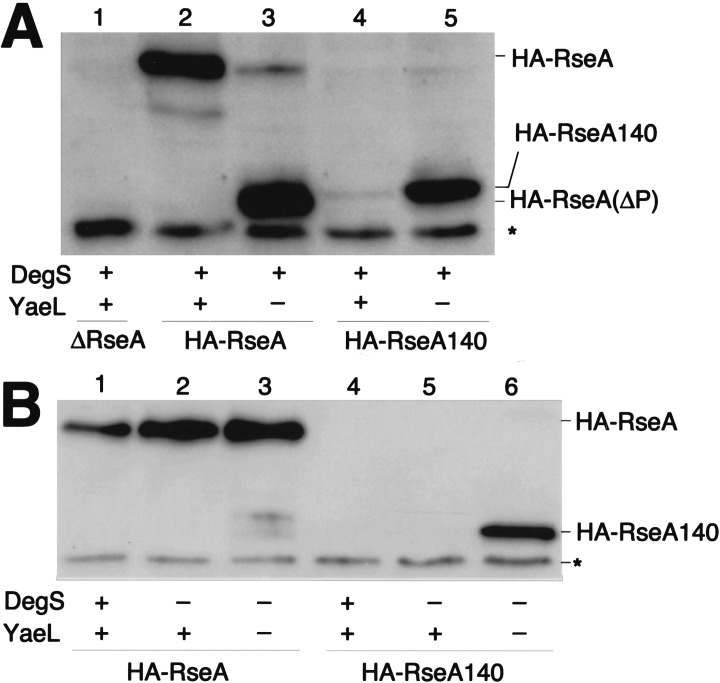
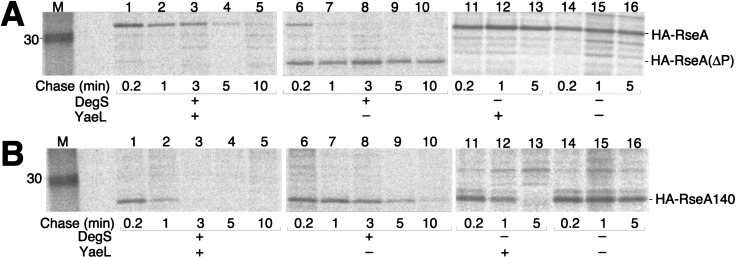
HA-RseA was expressed in cells with the ΔyaeL ΔrseA genotype and in cells with the yaeL+ ΔrseA genotype. Its accumulation was examined by anti-HA immunoblotting (Fig. 3A). Whereas the ΔrseA yaeL+ cells contained only the intact HA-RseA molecule (Fig. 3A, lane 2), the ΔrseA ΔyaeL cells accumulated a smaller species (HA-RseA[ΔP]) in addition to a small amount of the full-length protein (lane 3). The ratio of full-length HA-RseA to HA-RseA(ΔP) varied considerably in different experiments (see Fig. 4B). Although the exact reason for this variability is unknown, HA-RseA seems to be destabilized in the absence of YaeL (see Fig. 5) and after prolonged induction of itself (data not shown). HA-RseA(ΔP) was a C-terminally truncated form, as it retained the N-terminal HA tag. Its molecular size was similar to that of HA-RseA140 (Fig. 2), a construct in which a C-terminal periplasmic region (residue 141 to the C terminus) was deleted from HA-RseA (Fig. 3A, cf. lanes 3 and 5). Ades et al. (1999) suggested that RseA is degraded by DegS, a membrane-bound protease with a periplasmic active site. The striking accumulation of HA-RseA(ΔP) in the ΔyaeL cells raised the possibility that DegS proteolysis produces HA-RseA(ΔP), which is subsequently degraded by YaeL. In this respect, HA-RseA140 can be regarded as mimicking the putative product that is generated after the DegS cleavage. Indeed, HA-RseA140 only accumulated under the YaeL-deficient conditions (Fig. 3A, cf lanes 4 and 5).
Figure 3.
Cellular accumulation of hemagglutinin (HA)-RseA and HA-RseA140 in the presence and absence of YaeL and/or DegS. (A) Cells of AD1811 (yaeL+ ΔrseA)/pTWV229 (vector; lane 1), AD1811/pKK55 (ha-rseA; lane 2), KK211 (ΔyaeL ΔrseA/pKK55; lane 3), AD1811/pKK58 (ha-rseA140; lane 4), and KK211/pKK58 (lane 5) were grown in L-broth containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 1 mM cAMP at 30°C. Portions (containing about 1.2 × 108 cells) were withdrawn and mixed with an equal volume of 10% trichloroacetic acid for subsequent SDS-PAGE and anti-HA immunoblotting. HA-RseA(ΔP) indicates the C-terminally truncated intermediate form of HA-RseA. Asterisk indicates a nonspecific background. (B) Cells of AD1811 (degS+ yaeL+ ΔrseA)/pKK55 (ha-rseA; lane 1), AD1839 (ΔdegS yaeL+ ΔrseA)/pKK55 (lane 2), AD1840 (ΔdegS ΔyaeL ΔrseA)/pKK55 (lane 3), AD1811/pKK58 (ha-rseA140; lane 4), AD1839/pKK58 (lane 5), and AD1840/pKK58 (lane 6) were grown, and proteins were analyzed by SDS-PAGE and anti-HA immunoblotting as above. Asterisk indicates a nonspecific background.
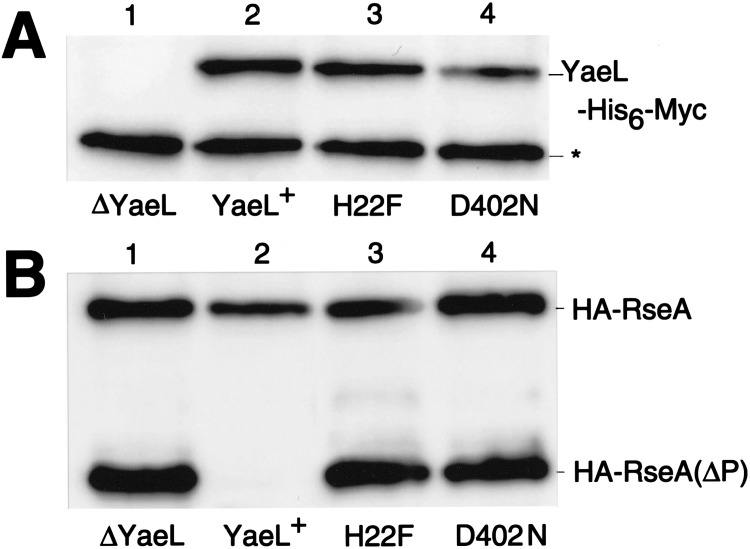
Figure 4.
Effects of conserved motif mutations of yaeL on cellular accumulation of the C-terminally truncated intermediate form (HA-RseA[ΔP]) of RseA. Strain KK211 (ΔyaeL ΔrseA)/pKK55 (ha-rseA) was transformed further with either pMPM-T3 (vector; lane 1), pSTD630 (yaeL+-his6-myc; lane 2), pSTD631 (yaeL[H22F]-his6-myc; lane 3), or pSTD632 (yaeL[D402N]-his6-myc; lane 4). Plasmid-bearing cells were grown at 30°C in L-broth containing 1 mM isopropyl-β-d-thiogalactopyranoside and 1 mM cAMP. Total cellular proteins were subjected to SDS-PAGE and anti-Myc (A) or anti-HA (B) immunoblotting. Asterisk indicates a nonspecific background.
Figure 5.
Stability of hemagglutinin (HA)-RseA and HA-RseA140 in the presence and absence of YaeL and/or DegS. Cells of AD1811 (degS+ yaeL+ ΔrseA; lanes 1–5), KK211 (degS+ ΔyaeL ΔrseA; lanes 6–10), AD1839 (ΔdegS yaeL+ ΔrseA; lanes 11–13), and AD1840 (ΔdegS ΔyaeL ΔrseA; lanes 14–16), each containing pKK55 (ha-rseA; A) or pKK58 (ha-rseA140; B), were grown in M9 medium supplemented with 18 amino acids (other than Met and Cys), 2 μg/mL thiamine, and 0.4% glucose at 30°C and induced with 1 mM isopropyl-β-d-thiogalactopyranoside and 1 mM cAMP. Cells were then pulse-labeled with [35S]methionine for 1 min, followed by chase with unlabeled methionine for the indicated periods. Samples were processed for anti-HA immunoprecipitation, SDS-PAGE, and phosphor imaging. Lane M was for molecular mass markers.
The above interpretation was further supported by examining the effects of the ΔdegS mutation on the RseA metabolism. As shown in Fig. 3B (lanes 2,3), accumulation levels of HA-RseA were similar and no HA-RseA(ΔP) was produced in the absence of DegS, whether or not YaeL had additionally been disrupted. Thus, YaeL may not be able to degrade intact RseA. In the presence of YaeL, HA-RseA140 did not accumulate irrespective of the presence or absence of DegS (Fig. 3B, lanes 4,5). These results are consistent with the YaeL-dependent degradation of HA-RseA(ΔP), a natural product after DegS action, as well as of HA-RseA140, which was genetically designed to mimic HA-RseA(ΔP). The above conclusion was further supported by the observation that coexpression of the wild-type YaeL abolished the accumulation of HA-RseA(ΔP) (Fig. 4B, lane 2), but that of YaeL derivatives with mutations His22Phe or Asp402Asn in the putative protease active site regions failed to do so (Fig. 4B, lanes 3,4).
Stability of RseA derivatives was studied directly by pulse-chase experiments (Fig. 5). In the yaeL+ ΔrseA cells, HA-RseA was degraded with an approximate half-life of 3 min without generating any detectable degradation products (Fig. 5A, lanes 1–5). In contrast, intact HA-RseA was degraded within 1 min in the ΔyaeL ΔrseA cells, which also produced HA-RseA(ΔP) as a major product (Fig. 5A, lanes 6–10). HA-RseA(ΔP) was slowly degraded even in the absence of YaeL, whereas it was undetectable in the yaeL+ cells. HA-RseA140 was degraded very rapidly (half-life, <1 min) in the yaeL+ ΔrseA cells (Fig. 5B, lanes 1–5) and much slowly (half-life, ∼3 min) in ΔyaeL ΔrseA cells (Fig. 5B, lanes 6–10).
In the ΔdegS background, HA-RseA was stabilized markedly both in the presence and absence of YaeL (Fig. 5A, lanes 11–16). In contrast, the DegS states did not further affect the stability of HA-RseA140, which was degraded solely in YaeL-dependent manners (Fig. 5B, lanes 11–16). These results strongly suggest that degradation of HA-RseA includes at least two successive proteolytic events: the initial DegS-dependent cleavage on the periplasmic side and the subsequent YaeL-dependent degradation on the transmembrane-cytoplasmic portion.
YaeL is positively required for the σE-dependent stress response
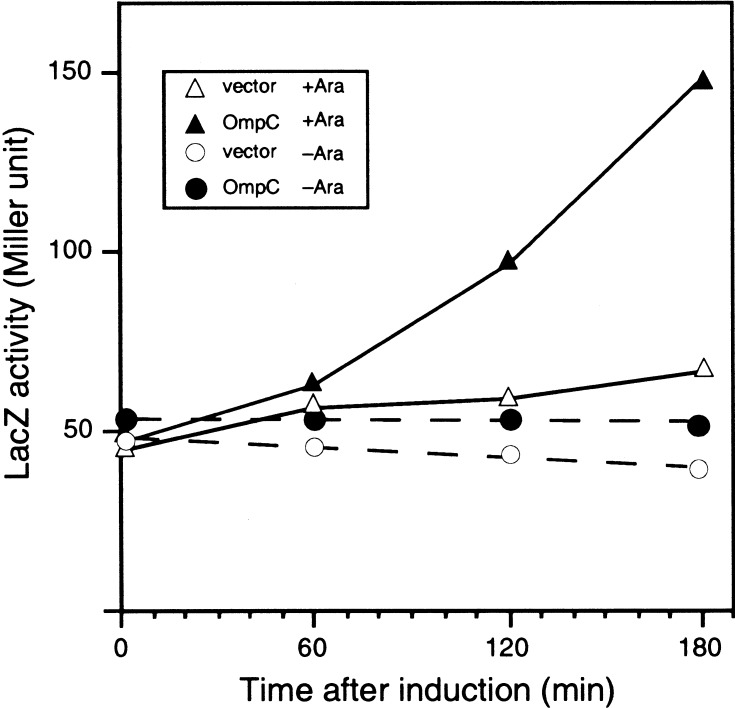
Our results so far presented suggest, contrary to the conclusion of Dartigalongue et al. (2001b), that YaeL participates in activation of σE by degrading the negative factor, RseA. To know whether YaeL-dependent degradation of RseA plays a role in regulation of the σE activity, effects of the YaeL depletion on the expression of rpoHP3-lacZ (Fig. 6) were studied. As an extracytoplasmic stress, we overproduced an outer membrane protein, OmpC (Mecsas et al. 1993). Thus, OmpC was overproduced in the ΔyaeL strain that harbored pKK6 having Para-controlled yaeL. In the presence of arabinose, cellular LacZ activity was increased evidently after induction of OmpC synthesis (Fig. 6, cf. solid and open triangles). In contrast, YaeL-depleted cells grown for 2 h in the absence of arabinose did not show any significant increase in the LacZ activity, even when OmpC was overproduced (Fig. 6, cf. solid and open circles). Under the present experimental conditions, cell growth continued at almost normal rate up to 6 h after arabinose removal and then ceased at 8 h. These results show that YaeL is required for the activation of σE in response to envelope stresses.
Figure 6.
YaeL depletion prevents activation of σE-pathway on OmpC overexpression. Plasmid pKK74 (ompC; solid symbols) and pMPM-T1 (vector; open symbols) were introduced into KK360 (λ[rpoHP3-lacZ] yaeL∷kan/pKK6 [yaeL+]). Cells were grown at 30°C in L − 0.02% arabinose medium. Cells at a mid log phase were harvested, washed five times by repeated centrifugation/suspension with arabinose-free L medium, and inoculated into L-arabinose medium (triangles) or arabinose-free L medium (circles). After further incubation for 2 h at 30°C, isopropyl-β-d-thiogalactopyranoside (final concentration, 1 mM) and cyclic AMP (final concentration, 1 mM) were added to induce the OmpC overexpression. At the indicated time points, samples were withdrawn and assayed for β-galactosidase activity.
Discussion
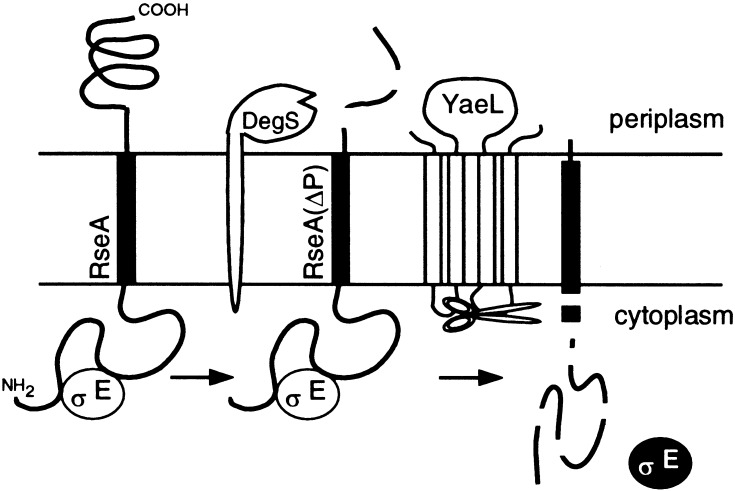
Alba et al. (1999) showed that proteolytic cleavage of RseA by DegS leads to activation of the σE stress response. We have shown here that YaeL also contributes to the σE activation process. A contrasting feature of these two membrane-bound proteases lies in the cellular locations of their proteolytic active sites. Although DegS has the periplasmic active site (Alba et al. 2001), the metalloprotease active site motifs in YaeL all reside in the cytosolic side of the membrane (Kanehara et al. 2001). Our results indicate that the membrane-bound anti-σE factor RseA is subject to proteolysis at least at two sites that are topologically separated by the plasma membrane, as obligatory processes of the stress response (Fig. 7). It is suggsted that DegS introduces the first cleavage/degradation at a periplasmic side. We were able to identify the resulting degradation product, HA-RseA(ΔP), but only under the YaeL-depleted conditions or when YaeL contained a mutational alteration in the putative protease active sites. The intact HA-RseA molecule was stable in the absence of DegS, even when functional YaeL was present. This suggests that YaeL cannot degrade intact RseA. On the other hand, DegS is not involved in the YaeL-dependent second proteolysis because HA-RseA140, with the genetically truncated periplasmic region, was degraded rapidly both in the degS+ yaeL+ and ΔdegS yaeL+ strains. The simplest interpretation of our results is that RseA is inactivated by sequential proteolytic actions of DegS and YaeL. Thus, our findings support the idea that YaeL functions as a “site-2” protease in E. coli.
Figure 7.
Two-step proteolytic processing of RseA. RseA first receives a cleavage by DegS on the periplasmic side, which somehow triggers the second cleavage by YaeL on the cytoplasmic side or within the membrane. The end result will be elimination of RseA and release of active σE.
Our results show that not only the DegS-mediated first cleavage but also the YaeL-dependent proteolysis of RseA is relevant in terms of cellular ability to elicit the σE pathway of extracytoplasmic stress response. Thus, overproduction of OmpC resulted in up-regulation of a σE-dependent promoter in the yaeL+ cells but not in the absence of YaeL. The fact that yaeL itself is σE-regulated (Dartigalongue et al. 2001a) indicates that activation of σE is under the positive feedback control. This positive control will contribute to the rapid response of cells to envelope stresses.
The two-step processing mechanism of σE activation is analogous to that of the ATF6 and SREBP activation in mammalian cells. Whereas the end result of the proteolysis is inactivation of a membrane-bound anti-transcription factor (RseA) in the former case, the cleavages of the membrane-bound precursors by S1P and the S2P directly produces active transcription factors in the latter (Sakai et al. 1996; Ye et al. 2000b). It is interesting to note that in both cases, the sequential events of proteolysis near or within the membrane activate transcription factors, and that RseA and ATF6 are both involved in responses against stresses in an extracytosolic compartment. The SREBP pathway equivalent in Drosophila has also been suggested to function to maintain membrane integrity (Seegmiller et al. 2002). Sequential proteolysis of a membrane protein at both sides of the membrane appears to be a universal mechanism of regulated transmembrane signal transduction across the membrane.
Although the site-2 proteolysis of RseA and ATF6/SREBP is catalyzed by homologous proteases (YaeL and S2P), the enzymes for the site-1 cleavage share little homology. SpoIVFB, a S2P homolog in Bacillus subtilis, is involved in the processing of pro-σK, the precursor form of an alternative σ factor dedicated for spore formation (Rudner et al. 1999). Pro-σK, having an N-terminal transmembrane segment, seems to be cleaved by SpoIVFB without prior cleavage by another protease. Thus, S2P homologs are well conserved in primary sequences, but they differ in their modes of dependence on the site-1 cleavage.
In vitro, the isolated cytoplasmic domain of RseA can associate with and antagonize against σE (Missiakas et al. 1997). Thus, a single cleavage of RseA by YaeL within or just after the transmembrane region may not be sufficient for activation of σE. In fact, HA-RseA(ΔP) in yaeL+ cells was degraded such that its N-terminal region was immunologically undetectable. Although this degradation clearly depends on YaeL, other protease(s) might also participate in the effective elimination of the anti-σE domain of RseA.
A key biological question is the mechanism by which the successive proteolytic events against RseA are regulated. The S1P action against SREBP and ATF6 appears to be regulated at the level of transport from the ER to the compartment of the S1P action (DeBose-Boyd et al. 1999). Because YaeL could not act on intact RseA, DegS-dependent removal of the periplasmic domain of RseA is a prerequisite for YaeL to act on the cytoplasmic side. Thus, a primary regulatory target will be the DegS action. Indeed, it has been suggested that RseB down-regulates the DegS-dependent cleavage of RseA (Ades et al. 1999). We noted that degradation of HA-RseA was much faster (Fig. 5) than that reported for the chromosomally encoded RseA (Ades et al. 1999). It is conceivable that RseB contributed to the degradation retardation of the latter but only insufficiently of the former, because HA-RseA was overproduced from plasmid. Possible mechanisms for the envelope stress–induced destabilization of RseA have been discussed by Ades et al. (1999).
It was proposed that the S1P cleavage of SREBP induces its structural transition such that the otherwise membrane-embedded S2P cleavage site is exposed to the cytosol (Ye et al. 2000a). By analogy, RseA could undergo a similar conformational change on cleavage by DegS. Alternatively, YaeL could interact with a periplasmic region of RseA and directly sense its cleavage by DegS. The periplasmic loop of YaeL contains a region of homology with the PDZ domain (Dartigalongue et al. 2001b; Kanehara et al. 2001) involved in a variety of protein–protein interactions (Harrison 1996; Pallen and Ponting 1997). YaeL and DegS might interact with each other, and this interaction could play a role in the regulated degradation of RseA. In addition, roles of RseB and RseC in the first and/or the second proteolytic events should be clarified for our full understanding of regulatory mechanisms of the σE pathway stress response.
YaeL, an essential E. coli protein, is dispensable in the presence of excess σE or in the absence of RseA. This is very similar to the nature of essentiality observed for DegS (Alba et al. 2001) and understandable in terms of their common function of inactivating RseA, the antagonist against σE, another essential protein. Thus, the essentiality of YaeL should lie at least in part in its σE-activating function. It is still possible that YaeL has a more direct role for the viability, and the lack of this function is compensated for by some σE-inducible factor of an overlapping function. This possibility should be considered, because we found that the ΔompR mutation, which alleviates the σE requirement of the cell and suppresses ΔdegS, is unable to suppress ΔyaeL (K. Kanehara, K. Ito, and Y. Akiyama, unpubl.). Thus, YaeL could perform some essential functions, presumably proteolytic, in addition to the up-regulation of the σE pathway.
Dartigalongue et al. (2001a) reported that the loss of YaeL function stabilizes σE, leading to increased transcription from σE-dependent promoters, whereas overproduction of YaeL lowered the cellular content and activity of σE. These investigators claimed further that YaeL showed in vitro activities to degrade σE. Their conclusion that YaeL has a negative regulatory role in the σE pathway is in sharp contrast to our results presented in this paper. Our results that YaeL acts on a membrane-integrated protein after a periplasmic cleavage seem to be more consistent with its being a S2P homolog. Although Dartigalongue et al. (2001a) documented that purified YaeL did not degrade RseA, we believe that such in vitro reproduction will require the use of a physiologically relevant substrate. In this case, the DegS-processed form of RseA should be used instead of the intact molecule.
We have been unsuccessful in obtaining in vivo results that support the notion that YaeL overproduction down-regulates the σE pathway. Specifically, we used strains that carried reporters degP-lacZ (Danese et al. 1995) and rpoHP3-lacZ (Danese and Silhavy 1997) for the same promoters as Dartigalongue et al. (2001a) used. Media and temperature were also the same. Then YaeL was overproduced using two different plasmids of different extents of overproduction. In all of these experiments, the LacZ activities were unaltered or slightly increased on induction of YaeL (K. Kanehara, K. Ito, and Y. Akiyama, unpubl.). Although the exact cause of the discrepancy has not been solved, our experimental results strongly suggest that the YaeL action is positive with regard to the σE stress response.
Materials and methods
Bacterial strains
E. coli K-12 strains used in this study were derivatives of AD16 (Δpro-lac thi/F‘lacIq ZM15 Y+ pro+; Kihara et al. 1995), W3110 (wild type), and MC4100 (araD139 Δ[argF-lac]U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR; Casadaban 1976). KK9 (AD16 yaeL∷kan/pSTD479 [Plac-yaeL]) and KK31 (AD16 yaeL∷kan Δ[srl-recA]306∷Tn10/pKK6 [Para-yaeL]) were described previously (Kanehara et al. 2001). AD1811 (AD16 rseA∷cat) was constructed as follows: First, the rseA∷cat marker was introduced into AB1157 (thr-1, leu-6, thi-1, lacY1, galK2, ara-14, xyl-5, mtl-1, proA2, his-4, argE3, str-31, tsx-33, sup-37; Murphy 1998) carrying pKN201 [Ptac-red-gam; a gift of K. Murphy (Department of Molecular Genetics and Microbiology, University of Massachusetts Medical School, Worcester, MA)] by linear transformation (Murphy 1998) using a DNA fragment amplified from the chromosomal DNA of AK2168 (dsbA∷cat; Inaba and Ito 2002) with a pair of primers (5′-TAT CAGGCGTTGACGATAGCGGGATACTGGATAAGGGTAT TAGGCTATCTTCCTGGCATCTTCCAG-3′, and 5′-ATAACAG GCTACCTGTCACTAATGACATGGCAAACCAAAGTTGCTC ATCTGTATTAACGAAGCGC-3′). The rseA∷cat marker was then P1-transduced into AD16. For construction of AD1839 (AD16, rseA∷cat degS∷tet) and AD1840 (AD16, rseA∷cat yaeL∷kan degS∷tet), a degS∷tet derivative of AB1157/pKN201 was first constructed similarly using a degS∷tet fragment amplified from AD1735 (zad-220∷Tn10; primers used were 5′-GGT TAACTCGTGGTATGCTGCTGCCGTTCCCTTTTTTAATGA CGCCCGACCTCATTAAGCAGCTC-3′, and 5′-GTTAACTGC TTATCATCACGCATCACTACTACAGGGATCACCGAAAA CTAAGCACTTGTCTCCTG-3′). The degS∷tet marker was finally transferred into AD1811 and KK211 (AD16, rseA∷cat, yaeL∷kan), respectively. KK211 and KK326 (AD16, yaeL∷kan/pKK64 [Para-yaeL]) were constructed by introducing the yaeL∷kan marker into AD1811 and AD16/pKK64, respectively, by P1 transduction. KK334 (AD16, yaeL∷kan rseA∷cat/pKK64) was an rseA∷cat transductant of KK326. SP887 (MC4100, λRS88[fkpA-lacZ]; Danese and Silhavy 1997) and SP556 (same as PND2000; MC4100, λRS88[degP-lacZ]; Danese et al. 1995; Shimohata et al. 2002) were described previously. TR71 (MC4100, λRS45[rpoHP3-lacZ]) was provided by T. Silhavy (Department of Molecular Biology, Princeton University, Princeton, NJ). KK360 (TR71, ara+ yaeL∷kan/pKK6) was constructed by two-step P1 transduction, first eliminating the araD139 marker (donor, AD16) and then introducing the yaeL∷kan marker.
Plasmids
pKK6 (Kanehara et al. 2001) carried the ara promoter-controlled yaeL on pBAD33 (a pACYC184-based ara promoter vector; Guzman et al. 1995). pKK52 carried a Sau3AI-partially digested chromosomal fragment from the rpoE region of the W3110 chromosome cloned into pTWV229 (pBR322-based vector; Takara Shuzo) and obtained as a multicopy suppressor against ΔyaeL. pKK53 and pKK54 were subcloning products of pKK52, the former lacking a 2.3-kb KpnI fragment, and the latter having only this chromosomal fragment. pKK93 (encoding σE under the lac promoter control) was constructed as follows. A rpoE fragment was amplified from the AK2168 chromosome using primers 5′-ACTTGAGCTCTTTGGGGAGACTTTACCTCG-3′, and 5′-GCAGTCTAGATCCCGCTATCGTCAACGCCT-3′ (SacI and XbaI recognition sequences underlined) and cloned between these sites of pTWV228H, a derivative of pTWV228 (pBR322-based vector; Takara Shuzo) in which the HindIII site in the multicloning region had been eliminated by sequential treatments with HindIII, T4 polymerase, and T4 ligase. pKK97 was a derivative of pKK93, carrying a similarly introduced 4-bp insertion at the HindIII site within the rpoE coding region. pKK64 carried a 1.4-Kb KpnI–HindIII yaeL fragment of pKK6 cloned into pBAD30 (another pACYC184-based ara promoter vector; Guzman et al. 1995). pCH85 (a pTWV228-based vector carrying a sequence for the N-terminal HA-tag) was a gift of S. Chiba (Institute for Virus Research, Kyoto University, Kyoto, Japan). pKK55 (encoding HA-RseA) was constructed as follows. An rseA fragment was amplified from the AK2168 chromosome using primers 5′-CTAACGGTACCTCAGAAAGAACAACTT TCCGCTTTA-3′, and 5′-GTACGAAGCTTCTACTGCGATT GCGTTCCTAAAGT-3′ (KpnI and HindIII recognition sequences are underlined) and cloned into pCH85 after digestions with these enzymes. pKK58 (encoding HA-RseA140, a C-terminal truncated form) was constructed similarly: A DNA fragment was amplified from the chromosome using primers 5′-CTAAC GGTACCTCAGAAAGAACAACTTTCCGCTTTA-3′ and 5′GTACGAAGCTTCTACGGCAGTGTATTAAATACCGGCGT3′ (KpnI and HindIII recognition sequences are underlined, and a stop codon to be introduced is italicized) and cloned into pCH85 after digestions with these enzymes. pKK55 and pKK58 encoded RseA and RseA140 derivatives, respectively, with an N-terminal MYPYDVPDYASVP (HA epitope region is boldfaced, and linker region is italicized) sequence. pSTD630 (yaeL+-his6-myc), pSTD631 (yaeL[H22F]-his6-myc) and pSTD632 (yaeL[D402N]-his6-myc) were constructed by cloning 1.5-kb KpnI fragments of pKK14 (Kanehara et al. 2001), pKK28 (Kanehara et al. 2001), and pKK35 (Kanehara et al. 2001), respectively, into pMPM-T3 (a pACYC177-derived lac promoter vector; Mayer 1995). pKK74 carried ompC and lacIq and constructed by successive cloning of a blunt-ended 1.4-kb AseI fragment (containing ompC) of pMAN002 (Matsuyama et al. 1984) and a blunt-ended 1.2-kb SalI fragment (containing lacIq) from pREP4 (QIAGEN) into SmaI and blunt-ended XbaI sites, respectively, of pMPM-T1 (a pBlueScript-based lac promoter vector; Mayer 1995), with consistent directionality of ompC, lacIq, and the lac promoter.
Media
L-medium (Davis 1980) and M9 medium (Miller 1972) were used. Ampicillin (50 μg/mL), chloramphenicol (10 or 20 μg/mL), kanamycin (12.5 or 25 μg/mL), and/or tetracycline (6.25, 12.5, or 25 μg/mL) was added for selecting transformants and transductants and for growing plasmid-bearing strains.
Isolation of multicopy suppressors againstt he yaeL disruption
Genomic DNA libraries were constructed on the BamHI site of pTWV229 by cloning 5 to 15 kb Sau3AI-partially digested fragments of the chromosomal DNA from the wild-type (W3110) or the ΔyaeL ΔrseA mutant (KK211) cells. Pools of plasmids thus constructed were selected as KK31 transformants that grew at 37°C on arabinose-free L-agar supplemented with ampicillin, chloramphenicol, and 1 mM IPTG.
Pulse-chase, immunoprecipitation, and immunoblotting experiments
[35S]methionine pulse-chase experiments were performed essentially as described by Taura et al. (1993). Labeled proteins were immunoprecipitated using anti-HA (Y11; Santa Cruz Biotechnology, Inc.), separated by 12.5% SDS-PAGE, and visualized by BAS1800 phosphor image analyzer.
Immunoblotting with anti-Myc (A-14; Santa Cruz Biotechnology, Inc.) and anti-HA was performed as described previously (Shimoike et al. 1995). Antibody-decorated proteins were visualized using ECL detection kit (Amersham Biosciences) and Fuji LAS1000 lumino-image analyzer.
β-Galactosidase assays
The σE activity was assayed by monitoring β-galactosidase activity expressed from a chromosomal σE-dependent lacZ reporter gene (fkpA-lacZ, degP-lacZ, or rpoHP3-lacZ). The enzymatic activity was measured as described by Miller (1972) and expressed in Miller units.
Acknowledgments
We thank T. Silhavy and K. Murphy for strains; S. Chiba and S. Matsuyama for plasmids; H. Mori for stimulating discussion; and K. Mochizuki, M. Sano, and Y. Yoshioka for technical support. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and from CREST, Japan Science and Technology Corporation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
After completion of this work, we learned that Carol Gross and her co-workers (Alba et al. 2002) have reached a conclusion very similar to ours regarding the role of YaeL in the σE stress response.
Footnotes
E-MAIL yakiyama@virus.kyoto-u.ac.jp; FAX 81-75-771-5699.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1002302.
References
- Ades SE, Connolly LE, Alba BM, Gross CA. The Escherichia coli σE-dependent extracytoplasmic response is controlled by the regulated proteolysis of an anti-σ factor. Genes & Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba BM, Zhong HJ, Pelayo JC, Gross CA. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σEactivity. Mol Microbiol. 2001;40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- Alba, B.M., Leeds, J.A., Onufryk, C., Zen Lu, C., and Gross, C.A. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac operon to selected promoters in Escherichia coliusing bacteriophages λ and μ. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Collinet B, Yuzawa H, Chen T, Herrera C, Missiakas D. RseB binding to the periplasmic domain of RseA modulates the RseA: σE interaction in the cytoplasm and the availability of σERNA polymerase. J Biol Chem. 2000;275:33898–33904. doi: 10.1074/jbc.m006214200. [DOI] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. The σE and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes & Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- Danese PN, Synder WB, Cosma CL, Davis LJB, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coliregulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes & Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- Dartigalongue C, Loferer H, Raina S. EcfE, a new essential inner membrane protease: Its role in the regulation of heat shock response in Escherichia coli. EMBO J. 2001a;20:5908–5918. doi: 10.1093/emboj/20.21.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli σEregulon. J Biol Chem. 2001b;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- Davis RW, Botstein D, Roth JR. Advanced bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1980. [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li W-P, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: Relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- De Las Peñas A, Connolly L, Gross CA. σE is an essential sigma factor in Escherichia coli. J Bacteriol. 1997a;179:6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol Microbiol. 1997b;24:373–385. doi: 10.1046/j.1365-2958.1997.3611718.x. [DOI] [PubMed] [Google Scholar]
- Drew D, Sjöstrand D, Nilsson J, Urbig T, Chin C, de Gier J-W, von Heijne G. Rapid topology mapping of Escherichia coliinner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc Natl Acad Sci. 2002;99:2690–2695. doi: 10.1073/pnas.052018199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan EA, Brown MS, Goldstein JL, Sakai J. Cleavage site for sterol-regulated protease localized to a Leu-Ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J Biol Chem. 1997;272:12778–12785. doi: 10.1074/jbc.272.19.12778. [DOI] [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J, Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SC. Peptide-surface association: The case of PDZ and PTB Domains. Cell. 1996;86:341–343. doi: 10.1016/s0092-8674(00)80105-1. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Rape M, Jentsch S. Membrane-bound transcription factors: regulated release by RIP or RUP. Curr Opin Cell Biol. 2001;13:344–348. doi: 10.1016/s0955-0674(00)00218-0. [DOI] [PubMed] [Google Scholar]
- Hua X, Sakai J, Ho YK, Goldstein JL, Brown MS. Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J Biol Chem. 1995;270:29532–29540. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- Hua X, Sakai J, Brown MS, Goldstein JL. Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane. J Biol Chem. 1996;271:10379–10384. doi: 10.1074/jbc.271.17.10379. [DOI] [PubMed] [Google Scholar]
- Inaba K, Ito K. Paradoxical redox properties of DsbB and DsbA in the protein disulfide-introducing reaction cascade. EMBO J. 2002;21:2646–2654. doi: 10.1093/emboj/21.11.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehara K, Akiyama Y, Ito K. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene. 2001;281:71–79. doi: 10.1016/s0378-1119(01)00823-x. [DOI] [PubMed] [Google Scholar]
- Kihara A, Akiyama Y, Ito K. FtsH is required for proteolytic elimination of uncomplexed forms of SecY, an essential protein translocase subunit. Proc Natl Acad Sci. 1995;92:4532–4536. doi: 10.1073/pnas.92.10.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Inokuchi K, Mizushima S. Promoter exchange between ompF and ompC, genes for osmoregulated major outer membrane proteins of Escherichia coliK-12. J Bacteriol. 1984;158:1041–1047. doi: 10.1128/jb.158.3.1041-1047.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. The activity of σE, an Esherichia coliheat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes & Dev. 1993;7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. Modulation of the Escherichia coli σE(RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol. 1997;24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- Murphy KC. Use of bacteriophage recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Ponting CP. PDZ domains in bacterial proteins. Mol Microbiol. 1997;26:411–415. doi: 10.1046/j.1365-2958.1997.5591911.x. [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci. 1999;26:14765–14770. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- Seegmiller AC, Dobrosotskaya I, Goldstein JL, Ho YK, Brown MS, Rawson RB. The SREBP pathway in Drosophila: Regulation by palmitate, not sterols. Dev Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- Shimohata N, Chiba S, Saikawa N, Ito K, Akiyama Y. The Cpx stress response system of Escherichia colisenses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site. Genes Cells. 2002;7:653–662. doi: 10.1046/j.1365-2443.2002.00554.x. [DOI] [PubMed] [Google Scholar]
- Shimoike T, Taura T, Kihara A, Yoshihisa T, Akiyama Y, Cannon K, Ito K. Product of a new gene, syd, functionally interacts with SecY when overproduced in Escherichia coli. J Biol Chem. 1995;270:5519–5526. doi: 10.1074/jbc.270.10.5519. [DOI] [PubMed] [Google Scholar]
- Taura T, Baba T, Akiyama Y, Ito K. Determinants of the quantity of the stable SecY complex in the Escherichia colicell. J Bacteriol. 1993;175:7771–7775. doi: 10.1128/jb.175.24.7771-7775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease. Proc Natl Acad Sci. 2000a;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000b;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]