Abstract
Saccharomyces mating-type switching results from replacement by gene conversion of the MAT locus with sequences copied from one of two unexpressed donor loci, HML or HMR. MATa cells recombine with HMLα ∼90% of the time, whereas MATα cells choose HMRa 80%–90% of the time. HML preference in MATa is controlled by the cis-acting recombination enhancer (RE) that regulates recombination along the entire left arm of chromosome III. Comparison of RE sequences between S. cerevisiae, S. carlsbergensis, and S. bayanus defines four highly conserved regions (A, B, C, and D) within a 270-bp minimum RE. An adjacent E region enhances RE activity. Multimers of region A, D, or E are sufficient to promote selective use of HML. Regions A, D, and E each bind in vivo the transcription activator forkhead proteins Fkh1p and Fkh2p and their associated Ndd1p, although there are no adjacent open reading frames (ORFs). Deletion of FKH1 significantly reduces MATa's use of HML, as does mutation of the Fkh1/Fkh2-binding sites in a multimer of region A. We conclude that Fkh1p regulates MATa donor preference through direct interaction with RE.
Keywords: Saccharomyces cerevisiae, donor preference, recombination enhancer, FKH1, FKH2, mating-type switching
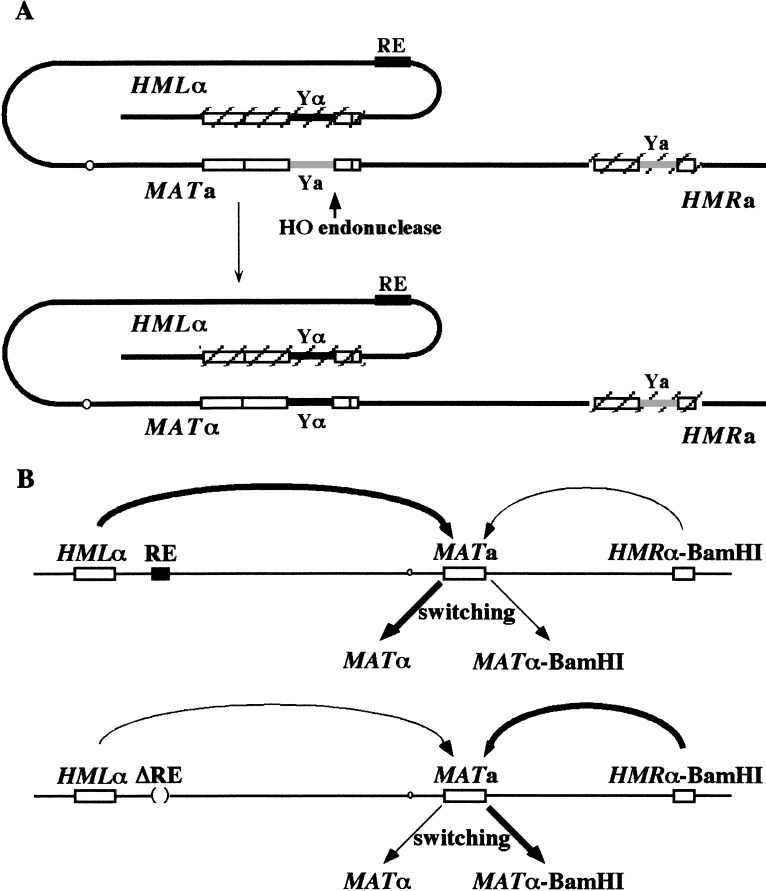
Mating-type gene switching in Saccharomyces cerevisiae has provided one of the best-studied examples of DNA double-strand break (DSB)-induced recombination in mitotic cells. Recombination is initiated by cleavage of the MAT locus with the sequence-specific HO endonuclease. The ends of the DSB can recombine with one of two silenced and heterochromatic donor loci, HML and HMR, located near the extremities of the same chromosome (Fig. 1A). Gene conversion occurs without crossing-over, often resulting in the replacement of MAT-Ya or -Yα sequences that regulate whether cells will be a- or α-mating type (Strathern 1989; Haber 1992, 1998a,b; 2002). In most strains of S. cerevisiae, HML carries Yα sequences (HMLα), whereas HMR carries Ya (HMRa). One of the remarkable aspects of this process is the phenomenon of donor preference, in which MATa cells preferentially recombine with HMLα, ensuring that recombination will usually result in a change of mating type; similarly, MATα cells preferentially select HMRa (Klar et al. 1982; Weiler and Broach 1992; Wu and Haber 1995; Wu et al. 1996). Donor selection depends on the location of the sequences but not their content, as MATa cells will preferentially recombine with HMLa even versus HMRα (Weiler and Broach 1992). Moreover, MATa's choice of a left-arm donor occurs even if the entire HML region is replaced by HMR (Weiler and Broach 1992) or if the donor is placed at other locations along the left arm (Wu and Haber 1995). MATα cells continue to choose a right-arm donor even if HMR is replaced by HML sequences, again regardless of whether the donor carries Ya or Yα (Wu et al. 1996).
Figure 1.
Mating-type switching at the MATa locus. (A) Expression of HO endonuclease creates a double-strand break (DSB) at the Y/Z1 border of the MAT locus and initiates repair by using the silent donor HML and leads to gene conversion. The two donor loci (HML and HMR) are maintained in a transcriptionally inactive chromatin structure. Other shared regions of homology are indicated. The preferential use of HML donor in MATa cells is regulated by a cis-acting element, recombination enhancer (RE), located ∼17 kb centromere-proximal to HML. The positions of HML, RE, centromere, MAT and HMR are indicated. (B) HMRa is replaced by HMRα modified to carry a point mutation that creates a BamHI site, so that it is possible to distinguish which sequences have been gene converted into the MAT locus. Donor preference is assayed in populations of cells induced to switch from MATa to either MATα or MATα-B (Wu and Haber 1995). MATa cells recombine with HML ∼90% of the time in the presence of RE; when RE is deleted, MATa cells prefer to choose HMR 80%–90% of the time.
The control of donor preference depends on a small cis-acting sequence, the recombination enhancer (RE), which acts as a locus-control region to control recombination along the entire left arm of chromosome III (Wu and Haber 1996). RE lies in a 2.5-kb intergenic region. It is located ∼29 kb from the left end of chromosome III (i.e., 17 kb centromere-proximal to HML) between KAR4 and SPB1 genes, but it retains its ability to influence donor choice even if it is inserted 43 kb further toward the centromere (G.-F. Richard and J.E. Haber, unpubl.). Deletion of the entire RE region causes a profound change in MATa donor preference, so that HML is used only 10% of the time, compared with 85%–90% when RE is present (Wu and Haber 1996). These studies indicated that RE acted to make the left arm of chromosome III “hot” for recombination in MATa cells, whereas in MATα cells the left arm was made unusually inaccessible for recombination (thus allowing HMR to be the preferred donor). These changes in recombination are not accompanied by any change in the mRNA levels of genes along the left arm (Galitski et al. 1999; A.P. Gasch, M.B. Vaze, D. Botstein, P.O. Brown, and J.E. Haber, unpubl.).
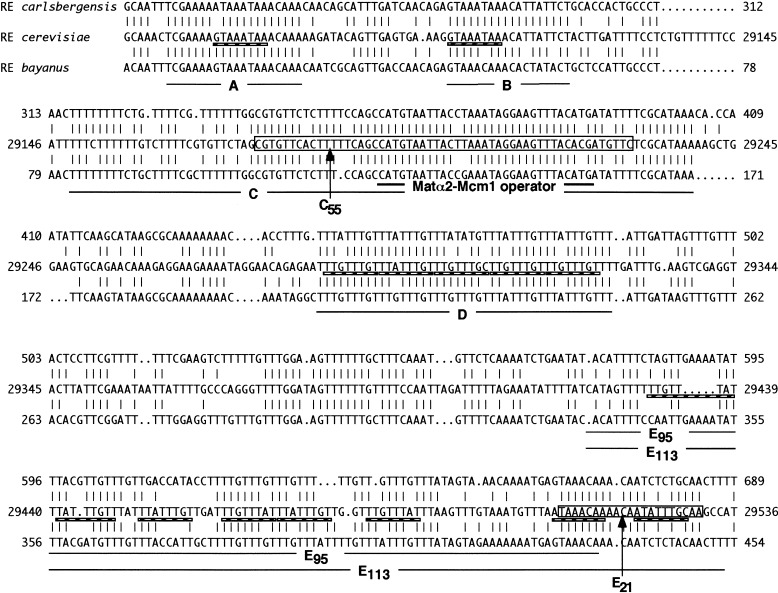
Comparison of the RE regions of S. cerevisiae and S. carlsbergensis allowed us to identify four highly conserved sequences (A, B, C, and D) within a 270-bp region (Wu et al. 1998). Deletion analysis revealed that the A, C, and D segments were required for activity, so that a minimum enhancer consisting of these three regions increased HML usage from 10% without RE to 45% (Wu et al. 1998). It is likely that an additional region, E (Fig. 2), which shares strong sequence homology with D, especially in S. cerevisiae, is also important. That some of these regions function in a redundant fashion is indicated by the finding that a 700-bp region lacking A, but containing B, C, D, and E, was nearly as functional as a 753-bp region containing all five elements (Wu and Haber 1996). Both regions, D and E, contain repeats of TTT(A/G), the only sites in the yeast genome to have such similar-length arrays of this sequence.
Figure 2.
Comparison of RE among three species: Saccharomyces carlsbergensis, S. cerevisiae, and S. bayanus (analyzed in “LASERGENE navigator”). Identical residues in the alignment of the DNA sequences of RE carlsbergensis (top), RE cerevisiae (middle), and RE bayanus (bottom) are indicated by vertical lines. The four most conserved subdomains (designated as A, B, C, and D) and a second near-perfect array of TTT(G/A) repeats (designated as E113) are underlined. Region C contains the Matα2-Mcm1 consensus binding site (double underlined). Regions A, B, D, and E contain a total of 15 Fkh1p and Fkh2p consensus binding sites (hatched line). The fragments used to replace the wild-type RE in donor preference assay—designated as C55 (boxed), E21 (boxed), E95 (solid line), and E113 (solid line)—are indicated.
How RE works to promote MATa donor preference is not yet understood; to date only one trans-acting mutation, a deletion of CHL1, has been found that alters MATa donor preference, and in this case the effect is modest (Weiler et al. 1995). We have a better idea how RE is inactivated in MATα cells, causing a phenotype of MATa strain lacking RE; region C contains a binding site for the Mcm1-Matα2 repressor complex that is known to turn off transcription of a set of a-specific genes when the MATα genes are expressed. In MATα cells, the RE region is covered with highly positioned nucleosomes that apparently inactivate RE (Weiss and Simpson 1997; Wu et al. 1998). This inactivation is similar to that seen at the promoter of the a-specific STE6 gene (Shimizu et al. 1991; Murphy et al. 1993), although no open reading frame (ORF) is found adjacent to RE. The activation of RE in MATa cells depends on binding of Mcm1p. In MATa cells, mutation of the Mcm1-binding site in RE also leads to the formation of a positioned array of nucleosomes and the loss of RE activity (Wu et al. 1998). At STE6, Mcm1p appears to be a trans-activating transcription factor. The RE does not contain an ORF, but there are several noncoding RNAs (Szeto et al. 1997). Whether Mcm1p is needed for transcription of these noncoding transcripts has not been established; moreover, it should be noted that the minimum RE and synthetic REs that lack these transcribed regions are not seriously impaired in activating HML in MATa cells (Wu et al. 1998) (results below).
In the present study, we have further characterized the sequences that are critical for RE activity in MATa cells. We have refined the identity of these sequences by sequencing the RE region from S. bayanus. We show that there is substantial redundancy in the S. cerevisiae RE region, so that even multimers of A, D, or E segment have strong RE activity. Second, we show that Fkh1 and Fkh2 transcription activators bind to region A in vivo, as well as to regions D and E, and that a fkh1Δ strain has a marked reduction in MATa donor preference. We conclude that, in addition to their key role in regulating expression of a class of cell cycle-regulated genes, forkhead proteins play an important role in the cell cycle-independent activation of the RE.
Results
In combination with the 270-bp minimum RE, a second region containing TTT(G/A) repeats fully restores RE activation
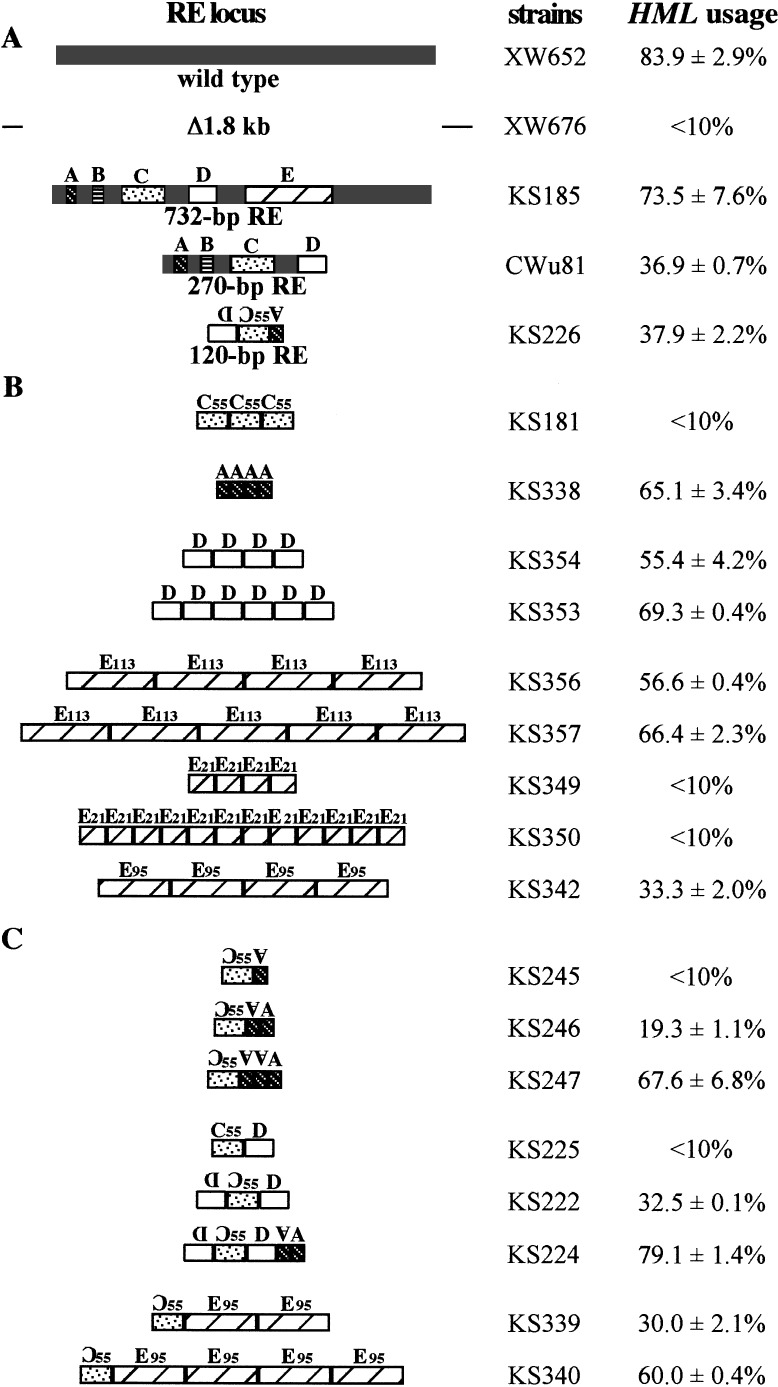
By comparing the sequences from originally identified 700-bp RE between S. carlsbergensis and S. cerevisiae, a 270-bp minimum RE was defined, containing four highly conserved segments designated A, B, C, and D (Wu et al. 1998) (Fig. 2). When this 270-bp region was then inserted into a chromosome in place of a 1.8-kb sequence containing RE and other intergenic sequences between KAR4 and SPB1, this minimum enhancer only retained part of the donor preference activity. A strain carrying a 753-bp RE region selected HML ∼75% of the time, whereas a strain with 270-bp RE used HML ∼45% of the time, which is still significantly above the 10% level seen when RE is absent (Wu et al. 1998). Further analysis showed that region B is dispensable for MATa's use of HML, but the other three regions were required (Wu et al. 1998).
We then further refined our examination of RE by including sequences obtained by sequencing the intergenic region between highly homologous KAR4 and SPB1 genes in S. bayanus. Three of the four subregions we had previously identified (A, C, and D) were strongly conserved; however, region B was less conserved among the three species (Fig. 2). We showed that this RE sequence was functional by inserting a 3.1-kb sequence containing the S. bayanus RE in place of the S. cerevisiae RE and showing that HML usage in MATa cells was >80% (data not shown).
A notable feature of RE region is that it contains many TTT(G/A) repeats. Conserved region D has 10 TTT(G/A) repeats. Region A and B also have two or three of such repeats. Among the sequences outside of the 270-bp minimum RE, a less well-conserved segment (E113) also contains TTT(G/A) repeats (Fig. 2). The E113 region has been shown as a unique nuclease hypersensitive region to micrococcal nuclease digestion flanked by two footprinted regions in S. cerevisiae (Weiss and Simpson 1997).
In the alignment of the DNA sequences from S. carlsbergensis, S. cerevisiae, and S. bayanus (Fig. 2), the overall region E is not conserved among all three species. Interestingly, included in the E113 region is an 18/21 match between S. carlsbergensis and S. cerevisiae and a 17/21 match between S. bayanus and S. cerevisiae, centromere-proximal to the less-conserved TTT(G/A) repeats (bp29512–bp29532, designated E21, boxed in Fig. 2).
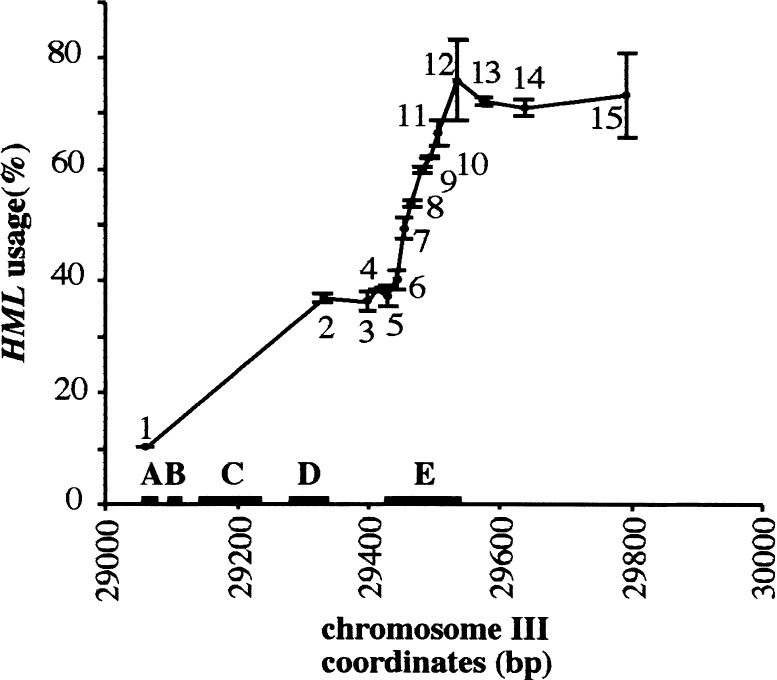
To investigate whether the other important cis-acting elements other than A, C, and D contribute to the function of RE, a series of deletions were made in a 732-bp RE fragment that has the same activity as the 753-bp RE shown previously (Wu et al. 1998; Fig. 3). To assess donor preference, MATa cells carrying a galactose-inducible HO gene were transferred from YEP-lactate medium to YEP-galactose for 1.5 h to induce gene conversion of MATa, which can use either HMLα or HMRα-B as a donor (Wu and Haber 1995). HMRα-B carries a single base-pair mutation that creates a BamHI site and allows us to distinguish which sequences have been gene converted into the MAT locus (Fig. 1B). As shown in Figure 3, deletions removing the second TTT(G/A) repeat-rich region (E) caused a significant reduction in RE activity (Fig. 3, cf. lanes 12 and 5). A small deletion (bp29505–bp29535) including the highly conserved E21 region in a 732-bp RE decreased MATa's use of HML from ∼75% to ∼65% (Fig. 3, #12 and #11). Further deletions in TTT(G/A) region diminished the activity from ∼65% to ∼55% when at least seven TTTG/A repeats remained (Fig. 3, #8). Activity fell to 40% when only three TTT(G/A) repeats were retained (Fig. 3, #6). This result argues that region E is an important contributor to overall RE activity.
Figure 3.
Identification of region E, which, in combination with A, B, C, and D, can provide full RE activity in MATa cells through a series of deletions from centromere-proximal end of RE region. The different RE fragments were introduced into the chromosome III to replace a 1.8-kb sequence containing RE. The strains used here (XW676, CWu81, KS210, KS209, KS208, KS207, KS206, KS205, KS204, KS203, KS202, KS198, KS199, KS184, and KS185) are indicated as 1–15, respectively. Number 1 is the strain that has no RE fragment inserted into a 1.8-kb deletion. Number 2 carries the chromosome III region from 29059 bp to 29330 bp, which was reported previously as the 270-bp RE (Wu et al. 1998). Numbers 3–14 each represent strains that carry a fragment starting from 29059 bp of chromosome III to centromere-proximal end. The numbers of TTT(G/A) repeats are #3 (0), #4 (0), #5 (0), #6 (3), #7 (5), #8 (7), #9 (11), #10 (14), #11 (16), #12 (20), #13 (20), and #14 (20). Number 15 carries the 732-bp RE starting from 29059 bp to 29790 bp. Donor preference is shown as the percentage of HML usage in MATa cells. Important region E is from ∼29423 to ∼29535 bp as indicated above.
Linker sequences between regions A, C, and D are dispensable
To define the active elements within RE more precisely, we ligated together various combinations of the conserved regions, A, C, D, and E. In all cases, the constructs were introduced into a 1.8-kb deletion that removed the native RE. A construct containing a 22-bp A region, a 55-bp segment of the C region including the Mcm1p-binding site (designated C55, in Fig. 2), and a 43-bp D region had approximately the same activity as the entire 270-bp minimum RE (Fig. 4A, KS226). This construct was inserted in the opposite orientation as the normal RE, but previous studies had shown that RE is orientation independent (Wu and Haber 1996). Thus the regions in between A, C, and D are not important for minimum RE function, which therefore resides in these 120 bp. This result also means that the exact positioning of these elements on the chromosome is not crucial.
Figure 4.
Effect of different combinations of regions A, C, D, and E on the percentage of HML usage in MATa cells. The different synthetic RE fragments were introduced into the chromosome III to replace the 1.8-kb sequence containing RE. C55, E21, E95, and E113 are indicated in Fig. 2. All other regions, A, C, and D, used here contain full length of consensus sequences.
Regions A, D, and E function in a redundant fashion and the requirement of Mcm1 binding is bypassed in synthetic REs
To determine if there were redundancies among regions A, C, D, and E, we tested the function of multimers of individual subdomains. We found that trimers of C55 alone were unable to activate the use of HML as a donor; in this case, HML usage was <10% (KS181 in Fig. 4B). Similarly dimers of A, D, or E alone had no activity (data not shown). However, dimers of A, D, or E combined with one C55 domain increased the use of HML (Fig. 4C, 19.3% in KS246, 32.5% in KS222, and 30.0% in KS339). Again, the orientation and precise position of these additional elements did not seem to be important. A strain with a synthetic RE consisting of A-A-C55-D-D used HML at nearly wild-type levels (Fig. 4C, 79% in KS224).
Surprisingly, tetramers of A (KS338), D (KS354), and E (KS356) were able to substantially activate HML as a donor (50%–65% HML usage). Adding more copies of D (KS353) or E (KS357) gave still further increases (66%–69%), approaching the level of activity seen with the native RE (Fig. 4B).
By a similar approach, we found that the conserved E21 region lacking TTT(A/G) repeats cannot by itself activate the use of HML as a donor (Fig. 4B, KS350). In contrast, a tetramer of the TTT(A/G)-containing part of region E (E95) can provide an activation of HML (Fig. 4B, 33% in KS342), although not as strongly as the more well-preserved TTT(A/G)-containing D region (60%). We conclude that region A, D, or E each is sufficient to provide at least some RE function.
In MATa cells, the binding of Mcm1p in the intact RE region is required for the activation of RE because of its ability to remove highly positioned nucleosomes that lead to the repression of RE activity (Wu et al. 1998). Here we show that multimers of A, D, or E are each active by themselves and that the requirement for binding of Mcm1 in the wild-type RE can be bypassed. It is possible that the role of Mcm1p is to facilitate chromatin reorganization to provide access to the A, D, and E sites.
In MATα cells, the binding of Matα2/Mcm1p at the RE region was shown to be responsible for the repression of RE because of its ability to organize highly positioned nucleosomes that led to the repression of RE activity (Tanaka et al. 1984; Szeto and Broach 1997; Szeto et al. 1997; Weiss and Simpson 1997; Wu et al. 1998). Previously, we showed that mutations of the Matα2-binding site removed most of this repression (15% of HML usage in wild-type MATα vs. 55% in Matα2-binding site mutant) (Wu et al. 1998). In this study, we replaced the wild-type RE with tetramers of region A in MATα cells. Here, too, there was no repression caused by Matα2/Mcm1p. The use of HML increased from ≦10% without RE to 50% (data not shown). Thus, the repression normally seen in RE does not occur when there is no Matα2p-Mcm1p binding.
Region A contains a consensus binding site for forkhead transcription factors
We focused our attention on region A, where four tandem copies gave 65% HML usage in MATa cells. To determine the binding site for any potential trans-acting factors in this region, a series of deletions was created and then two such truncated region As were joined to C55 and D to assess donor preference. As shown in Figure 5A, region A activity was apparently contained within a 15-bp region, which in this assay had as much activity as the intact A region. This region contains the 13-bp sequence 5′-GTAAATAAACAAA-3′ conserved in S. carlsbergensis, S. bayanus, and S. cerevisiae. Deletions of either the left 8 bp or the right 5 bp of this 13-bp region abolished RE activity from ∼50% to ∼10% HML usage in MATa cells. We tested one TA to GC mutation within this sequence (5′-GTAAAGCAACAAA-3′) and found that it also severely impaired MATa's use of HML when two copies of the mutated A region (Am) were combined with single copies of C55 and D (Fig. 5A, KS251). In contrast, three other 2-bp mutations did not affect MATa's use of HML (Fig. 5A, see strains KS325, KS326, and KS327).
Figure 5.
Identification of functional region A-binding site. (A) Identification of essential sequences within region A by introduction of the A + A + C55 + D synthetic REs carrying different deletions or 2 bp mutations on each A into the chromosome III to replace a 1.8-kb sequence containing RE. The 2-bp changes are boxed. The conserved region is underlined. Donor preference is shown as the percentage of HML usage in MATa cells. (B) Comparison of consensus Fkh1p- and Fkh2p-binding sites reported previously (Zhu et al. 2000) with region A sequence. R: A or G; Y: T or C; W: A or T.
Having identified a mutation within region A that appears to abolish its function, we used MatInspector V2.2 (Quandt et al. 1995) to search the region A and the entire RE region for the binding sites of transcription factors. The 13-bp consensus A region was shown to contain a potential binding site (5′-GTAAATAAA-3′) for the forkhead family of transcription factors (Fig. 5B). Similar putative forkhead-binding sites were found in regions B, D, and E (Fig. 2).
The fkh1 fkh2 double mutant reduces donor preference in MATa cells
In S. cerevisiae, there are four forkhead homologs, FKH1, FKH2, HCM1, and FHL1 (Zhu et al. 1993; Hermann-Le Denmat et al. 1994; Hollenhorst et al. 2000). Among the four members of the forkhead protein family, only Fkh1p and Fkh2p share significant sequence similarity (47% identity overall) between each other outside of the highly conserved forkhead DNA-binding domain. Fkh1p and Fkh2p were shown to activate transcription of the cell-cycle-dependent CLB2 cluster and to affect HMR transcriptional silencing (Hollenhorst et al. 2000, 2001; Koranda et al. 2000; Kumar et al. 2000; Zhu et al. 2000). In addition, FKH1 and FKH2 also have redundant functions in preventing pseudohyphal growth (Hollenhorst et al. 2000). On the basis of these observations, we made fkh1Δ and fkh2Δ single mutations and the fkhΔ1 fkh2Δ double mutant in different strains containing either wild-type RE or synthetic REs and tested them for donor preference.
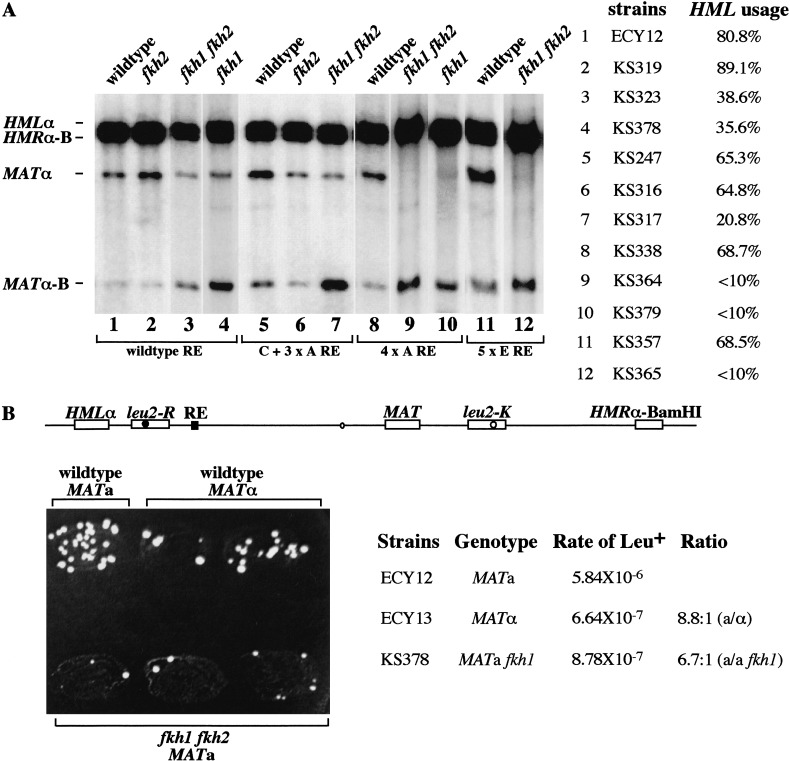
In a strain carrying wild-type RE, HML was used ∼81% of the time (Fig. 6A, lane 1). A fkh2Δ derivative did not affect donor preference (Fig. 6A, lane 2). However a fkh1Δ fkh2Δ double or a fkh1Δ single mutant reduced HML usage to <40% (Fig. 6A, lanes 3,4).
Figure 6.
The effects of Fkh1p and Fkh2p on donor preference and leu2 heteroallelic recombination in MATa cells. (A) Analysis of HMLα usage in different strains carrying single or double forkhead deletions in either wild-type RE or synthetic RE with C + 3 × A, 4 × A, and 5 × E. HO-induced switching of MATa to MATα (from HMLα) or MATα-Bam (from HMRα-Bam) can be assessed densitometrically on a Southern blot of DNA digested with BamHI and HindIII restriction endonucleases. Yα-specific probe was used to give rise to four specific bands indicated above. Donor preference is shown as the percentage of HML usage in MATa cells. (B) Spontaneous recombination between leu2-R inserted at kb 22 of chromosome III and leu2-K located at kb 233 of the right arm of MAT produces Leu+ prototrophs. The wild-type MATa and MATα and mutant fkh1 fkh2 MATa were patched on a YEPD plate and then replica-plated to synthetic medium lacking leucine. The rate of LEU2 prototroph formation was determined by a fluctuation test (Lea and Coulson 1949).
In strains containing a synthetic RE, the wild-type RE was replaced with different fragments containing either C55 + 3 × A (KS247 in Fig. 4B), 4 × A (KS338 in Fig. 4B), or 5 × E (KS357 in Fig. 4B). As shown in Figure 6, the strains carrying these synthetic REs used HML ∼65% to 68% of the time (Fig. 6, lanes 4,7,9). There was no effect on HML donor preference in the fkh2Δ derivative (Fig. 6, lane 5), but there was a significant decrease of HML donor preference in the fkh1Δ or the fkh1Δ fkh2Δ double mutant strain, so that HML was only used <20% of the time instead of ∼65% (Fig. 6, lanes 7,9,10,12). To make sure that the fkh mutations do not affect the efficiency of switching, we used the same genomic DNA made from fkh1Δ fkh2Δ (Fig. 6, lane 7) and confirmed that the efficiency of switching (i.e., the repair of the DSB) was very high (94.6%), and that (in this independent measurement of donor preference) the use of HML was 19.9% (data not shown), in excellent agreement with the measurement of 20.8% in Figure 6A. Thus, especially in the synthetic REs in which regions A, D, and E are not redundant with each other, the effect of deleting FKH1 is profound. The effect of the double fkh1Δ fkh2Δ mutant is similar to that of the single fkh1Δ mutant, indicating that Fkh1p plays a more important role in regulating donor preference through the RE.
The fkh1Δ fkh2Δ mutant eliminates mating-type activation of heteroallelic leu2 recombination
It has been shown that fkh1Δ weakly increases the expression of a1 mRNA from the HMR locus when the adjacent I silencing region is deleted and the E silencer is simplified; however, this change was not seen in a fkh1Δ fkh2Δ double mutant (Hollenhorst et al. 2000). To be sure that the changes in donor preference we saw in fkh1Δ or fkh1Δ fkh2Δ strains were not attributable to some change in the heterochromatic structure of HML or HMR, we assayed the effect of the forkhead proteins on spontaneous intrachromosomal recombination between two leu2 alleles to produce Leu2+ recombinants. In this assay, neither leu2 allele is silenced nor is recombination dependent on HO endonuclease cleavage. Previously, we showed that a leu2-R allele, replacing HML, recombined with a leu2-K allele, located near MAT, 25–30-fold more frequently in MATa cells than in MATα (Wu and Haber 1995). In the present assay, HML is present and leu2-R is inserted at kb 22 of chromosome III, between HML and RE (Fig. 6B). In this strain, there is a >10-fold increase in the rate of spontaneous Leu+ recombinants in MATa cells versus MATα. In the leu2 recombination assay, we found a very similar effect as we did in the direct monitoring of the use of HML and HMR in the fkh1Δ or fkh1Δ fkh2Δ mutant. As shown in Figure 6B, numbers of Leu+ papillae were much lower in the fkh1Δ fkh2Δ double mutant than in the wild-type MATa cells, and all the patches had the lower number of Leu+ papillae as seen in MATα cells. We also measured the rate of spontaneous Leu+ recombinants by a fluctuation test based on a minimum of seven independent cultures of each strain, initiated from 100 cells and grown to saturation (Lea and Coulson 1949). As shown in Figure 6B, there is approximately a nine-fold difference in the rate of spontaneous Leu+ prototroph formation in MATa versus MATα cells and approximately a seven-fold difference between wild-type MATa cells and fkh1Δ MATa cells. Thus, we concluded that the changes in donor preference we saw in fkh1Δ or fkh1Δ fkh2Δ strains were not attributable to some change in the heterochromatic structure of HML or HMR. The fkh1Δ mutant has the same effect on both donor preference in HO-induced MAT switching and the mating-type dependent difference in spontaneous heteroallelic leu2 recombination.
Ndd1p binds to region A
As an alternative way to screen for proteins that bind to region A, we did one-hybrid screening in which one assays the activation of transcription by a GAL4 activation domain fused to another protein domain that can bind to a specific sequence upstream of two reporter genes. Four tandem copies of domain A were ligated together and subcloned upstream of the minimal promoter of both the pHISi-1 and pLacZi reporter plasmids and then integrated into the yeast genome of YM4271 (Materials and Methods). We screened 5 × 106 transformants and isolated 21 positive clones that were His+ on 3-AT containing plates lacking histidine and also showed high β-galactosidase activity. Among these 21 positive clones, 18 encoded a known transcription factor, Gln3p, which recognizes an AT-rich binding site (GATAAG) (Stanbrough and Magasanik 1996). Two were from the uncharacterized ORF YDR317W. However, neither the deletions of GLN3 and YDR317W nor a double mutant had an effect on donor preference in mating switching (data not shown).
The last candidate was NDD1. NDD1 is an essential gene whose function is required during mitosis (Loy et al. 1999). Recently, it has been shown that Ndd1p is associated with the CLB2 and SWI5 promoters in chromatin immunoprecipitation (ChIP) assays (Koranda et al. 2000). Its recruitment at these sites depends on Mcm1p and on both Fkh2p and Fkh1p. Because NDD1 is essential, we could not assay directly its role in donor preference, but we could obtain evidence of its participation by ChIP assays.
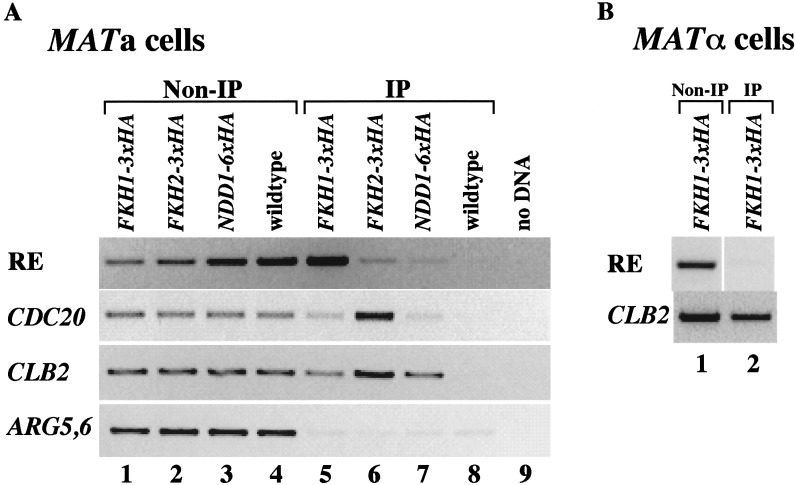
Fkh1p, Fkh2p, and Ndd1p bind to the RE region in vivo
If the consensus Fkh1p- and Fkh2p-binding sites represented bona fide Fkh-binding sites and Ndd1p can interact with region A in a one-hybrid assay, then the RE region should be enriched in a protein-DNA fraction prepared by ChIP with antibodies against Fkh1p, Fkh2p, or Ndd1p. Previous studies of enrichment of the CLB2-cluster promoter by ChIP experiments showed that both Fkh1p and Fkh2p bound to CLB2-cluster promoters, although generally Fkh2p bound more efficiently than Fkh1p (Koranda et al. 2000; Hollenhorst et al. 2001). Interestingly, some CLB2-promoters without Mcm1-binding sites or with a mutant Mcm1p-binding site were occupied equally well or more efficiently by Fkh1 (Hollenhorst et al. 2001). Moreover, Ndd1p was also associated with CLB2 and SWI5 promoters in ChIP assays (Koranda et al. 2000).
Yeast cells harboring integrated FKH1–3xHA, FKH2–3xHA or NDD1–6xHA genes (Materials and Methods) and wild-type cells containing untagged version of these genes were used in ChIP experiments with an antibody specific for the hemagglutinin epitope (α-HA). To test whether Ndd1, Fkh1p, and/or Fkh2p bound to the RE region in vivo, a 500-bp wild-type RE region was analyzed by ChIP. As shown in Figure 7A, Fkh1p, Fkh2p, and Ndd1p bound to the RE region, although to different extents. Unlike the CLB2-cluster promoter region (Hollenhorst et al. 2001), the signal generated by FKH1–3xHA was much stronger than the signal generated by FKH2–3xHA, although there is a Mcm1p-binding site inside the RE region. The assay did not allow a precise determination of the binding stoichiometry, but it indicated that Fkh1p may have a more important role than Fkh2p in controlling RE activity.
Figure 7.
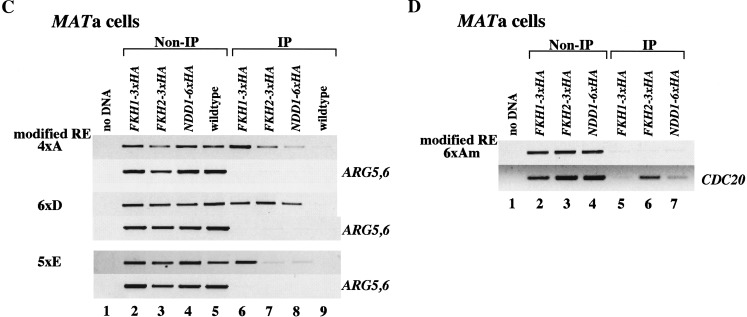
Fkh1p, Fkh2p, and Ndd1p are recruited to the RE in vivo. (A,B) Chromatin immunoprecipitation (ChIP) assays with a different pair of primers that can specifically amplify the wild-type RE region, the promoter regions of CDC20 and CLB2 (positive controls), and the coding region of ARG5,6 (negative control). Strains used in this assay are CFY480 (A, lanes 1,5) and CFY854 (A, lanes 2,6) (Hollenhorst et al. 2000), 155 (A, lanes 3,7) (Koranda et al. 2000), W303 (A, lanes 4,8), and KS369 (MATα; B, lanes 1,2). (C,D) ChIP assays with pairs of primers that can specifically amplify either the modified RE region, the promoter of CDC20 (positive controls), or the coding region of ARG5,6 (negative control). The wild-type RE is replaced by either four copies of region A, six copies of D, five copies of E, or six copies of mutant region A, indicated as 4 × A, 6 × D, 5 × E, or 6 × Am at left. Controls that have fragments amplified from ARG5,6coding region and CDC20 promoter are indicated at right. All strains used here are MATa. Strains used in this assay are KS358 (4 × A), KS359 (6 × D), and KS360 (5 × E) (C, lanes 2,6); KS370 (4 × A), KS371 (6 × D), and KS372 (5 × E) (C, lanes 3,7); KS374 (4 × A), KS375 (6 × D), and KS376 (5 × E) (C, lanes 4,8); KS361 (4 × A), 362 (6 × D), and 363 (5 × E) (C, lanes 5,9); KS365 (6 × Am) (D, lanes 2,5); KS373 (6 × Am) (D, lanes 3,6); and KS377 (6 × Am) (D, lanes 4,7).
In MATα cells, RE is covered with highly positioned nucleosomes that lead to the repression of RE activity; whereas the RE chromatin is less organized in MATa cells (Weiss and Simpson 1997). Our results clearly indicated that Fkh1p can be recruited to RE region in MATa cells. To ask whether Fkh1p can also be recruited to the RE in MATα cells, MATa cells carrying FKH1–3xHA were switched to MATα by introducing a plasmid containing GAL-HO. As shown in Figure 7B, unlike what we saw in MATa cells, Fkh1p was not recruited to the RE region in the wild-type MATα cells. This result indicates that Fkh1p can only be recruited to the less-organized RE chromatin found in MATa cells.
To show that the recruitment of Fkh1p and Fkh2p to the RE region are the result of their direct interactions with the putative forkhead-binding sites and Ndd1p is recruited to the same regions as forkhead proteins are, we replaced wild-type RE with synthetic REs carrying 4 × A, 6 × D, or 5 × E and then performed ChIP using the same antibody. As shown in Figure 7C and D, Fkh1p, Fkh2p, and Ndd1p can be recruited to the 4 × A, 6 × D, and 5 × E, where the consensus forkhead protein-binding sites are present. In contrast, when we inserted six copies of mutant region Am (a TA to GC mutation within forkhead protein-binding site, see KS251 in Fig. 5A) into a 1.8-kb deletion that removed the native RE, Fkh1, Fkh2, and Ndd1 were not recruited to the 6 × Am region. Thus, the consensus Fkh1p- and Fkh2p-binding sites identified within the RE region represents bona fide Fkh-binding sites. Ndd1p can also be recruited to the same region as are forkhead proteins.
Discussion
We have investigated the manner in which the RE acts to control donor preference during mating-type switching in MATa cells and made several important findings. First, the RE consists of several redundant domains whose exact spacing and orientation is surprisingly flexible. Second, multimers of region A, D, or E are sufficient to promote selective use of HML. Third, activation of HML donor preference does not require the presence of a Mcm1p-binding site. Finally, we have showed that the transcription regulator Fkh1p plays an important role in the activation of RE in MATa cells, in a region where there are no transcribed genes.
Our previous characterization of RE focused on the C domain in which there is a Mcm1p-Matα2p-binding site. It has been shown that Mcm1p-Matα2p is responsible for the repression of RE because of its ability to organize highly positioned nucleosomes that lead to the repression of RE activity in MATα cells (Weiss and Simpson 1997; Wu et al. 1998). In MATa cells, the binding of Mcm1p, in the context of the complete RE, is required to activate donor preference (Wu et al. 1998). However, the C region is not essential for the activation of HML usage in synthetic REs, because multimers of 4 × A regions, as well as 4 × D or 4 × E regions, lacking a Mcm1p-binding site are nearly as active as the entire RE region. We believe that the role of Mcm1 is to facilitate the removal of the highly positioned nucleosomes so that other factors can then be recruited to the RE and activate the RE. However, the possibility that Mcm1p could be recruited to the synthetic REs directly by binding to the Fkh2 protein (Hollenhorst et al. 2001) cannot be excluded, although fhk2Δ did not affect donor preference.
The C region and its Mcm1p-Matα2p-binding site do seem to be essential for the repression of RE in MATα cells. The absence of a Mcm1p-Matα2p site in the 4 × A construct leads to constitutive RE activity in MATα cells, although the lower usage of HML in MATα cells (50%) versus MATa (66%) indicates the possibility of a MATa-specific gene product that may contribute to the full activation of RE in MATa cells. This difference between MATa and MATα was previously seen in an intact RE in which the two Matα2-binding sites were mutated (Wu et al. 1998).
From the various arrangements of A, C, D, and E domains that activate HML usage, it is clear that the spacing and orientation of these regions are not critical for RE activity. What seems to be common to all of the different active arrangements of sequences is the presence of multiple sites where either Fkh1p or Fkh2p can bind (Fig. 2). Consistent with this observation is the fact that the fkh1Δ single mutant or the fkh1Δ fkh2Δ double mutant show reduced HML usage in MATa cells. This is especially evident in the 4 × A or 5 × E constructs in which donor preference is reduced from ∼65% to <10%. However, donor preference is not completely abolished when FKH1and FKH2 are deleted in a strain carrying an unmodified RE (compare KS323 in Fig. 6A to the complete absence of RE, XW676, in Fig. 3: HML was used ∼39% vs. 10%). The residual RE activity indicates that additional proteins contribute to RE function, probably interacting with other sequences.
The Fkh1p and Fkh2p proteins have been best understood in their role in activating transcription of the CLB2-cluster of cell cycle-regulated genes including CLB2 and SWI5 in the G2 phase of the cell cycle (Hollenhorst et al. 2000, 2001; Kumar et al. 2000; Pic et al. 2000; Zhu et al. 2000). A third protein, Ndd1p, is apparently recruited to these promoters by the Fkh proteins, but Ndd1p must have additional functions, as it is an essential protein, whereas even fkh1Δ fkh2Δ is viable.
We have shown by ChIP that RE and even simple multimers of A, D, or E regions bind Fkh1p, Fkh2p, and Ndd1p. This can occur even in the absence of a Mcm1p-binding site, as in the case of 4 × A, whereas in CLB2 cluster promoters, the recruitment of Fkh2p appears to require the binding of Mcm1p (Hollenhorst et al. 2001). We cannot rule out that Fkh2p is able to recruit Mcm1p to promoters that lack Mcm1p-binding sites (Hollenhorst et al. 2001). In many other respects, the action of the Fkh proteins appears to be quite different from their role in CLB2-cluster promoters. First, RE activation does not seem to depend on transcription. In the region including region E, Szeto et al. did find a weak transcript that does not appear to encode a protein (Szeto et al. 1997); however, RE activity does not depend on these sequences because they were absent in the minimum enhancer (Wu et al. 1998). This is most evident in the case of the multimers such as 4 × A, each unit of which is only 22 bp.
It should be noted that the role of Fkh proteins, in combination with Mcm1p and Ndd1p, is to activate the transcripts of CLB2-cluster in G2. Donor preference is not significantly different at different phases of the cell cycle (Wu et al. 1997); indeed, normal MAT switching occurs in G1 cells. Recently, it has been shown that Fkh1 and Fkh2 are also associated with genes expressed in G1 and S phase, whereas the binding of Mcm1 could not be detected (Simon et al. 2001). It seems clear that the way the Fkh proteins and Ndd1p act in donor preference is significantly different from their role at G2-regulated promoters.
Despite our progress in delineating the cis-acting domains and some of the key proteins that combine to create RE activity, we still do not know how RE acts to alter the ability of the entire left arm of chromosome III to recombine with the HO-cut MAT locus. Our previous results indicated that in MATα cells, the entire left arm is inaccessible for recombination (Wu et al. 1996). We imagine that there must be additional sites along this chromosome arm that serve to alter (perhaps immobilize) the left arm to prevent its participation in recombination, but without impairing transcription, both in MATα cells in which RE is repressed and in MATa cells when RE is deleted. The forkhead proteins and Ndd1p bound to RE may then interact with these repressive sites, thus liberating the left arm for recombination.
Materials and methods
Strains
All strains used for donor preference experiments were derivatives of DBY745 (ho MATa ade1–100 ura3–53 leu2–3,112). Strains used to monitor MATa donor preference carry HMRα-B, containing a single base-pair mutation that creates the BamHI site (Wu and Haber 1995). They also carry a galactose-inducible HO endonuclease gene integrated at the ADE3 locus (Sandell and Zakian 1993).
All strains used in ChIP were derivatives of W303 provided by C. Fox and G. Ammerer.
All yeast transformations were performed by one-step transformation (Chen et al. 1992) or high-efficiency method (Gietz et al. 1995, 1997). All the disruptions were verified by Southern blot analysis. To disrupt FKH1, a PCR fragment was amplified from the strain that has Δfkh1∷KAN (Record #32290) obtained from ResGen. To introduce subfragments into a deletion of the RE, a plasmid pKS58 was constructed from pCWU155 (described previously; Wu et al. 1998) except two more unique sites, BamHI and PmlI, were introduced adjacent to SalI. Details of other specific strain constructions are available on request. The selections of introduction of subfragments into a deletion of the RE were performed as described (Wu and Haber 1996).
Identification of the RE in S. bayanus
An S. bayanus clone containing genes homologous to KAR4 and SPB1 of S. cerevisiae was obtained as part of the study of the evolution of hemiascomycetes (Bon et al. 2000). The S. bayanus DNA was inserted into plasmid pBAM3 in Escherichia coli strain DH10B (as described in Bon et al. 2000). The intervening region was sequenced and aligned with RE regions from S. cerevisiae and S. carlsbergensis (Wu et al. 1998). A 3.1-kb fragment containing the conserved S. bayanus sequences was inserted in place of the 1.8-kb deletion of S. cerevisiae RE, as described previously for testing the S. carlsbergensis RE (Wu et al. 1998). The S. bayanus sequence has the GenBank accession no. AY123283.
Analysis of donor preference
The measurement of donor preference was described previously (Wu and Haber 1995, 1996). The bands (MATα and MATα-BamHI) corresponding to the use of HML and HMR were quantified using ImageQuant V1.2 (Molecular Dynamics). In some experiments, YEP-raffinose was used instead of YEP-lactate medium.
One-hybrid screen
The yeast MATCHMAKER one-hybrid system was purchased from CLONTECH. The screening was essentially performed as recommended by the manufacturer. The S. cerevisiae genomic library is provided by Philip James (James et al. 1996). Four tandem copies of 22 bp of wild-type region A were inserted into the polylinker of two plasmids, pHISi-1 and placZi, just upstream of the minimal promoter of HIS3 and lacZ genes to generate pKS133b and pKS132–2. Four tandem copies of 22 bp of nonbinding mutant region A (TA-GC, see KS251 in Fig. 5A) were also inserted into the polylinker of pHISi-1 at the same location to generate pKS165. A wild-type dual reporter yeast strain (KS257) was constructed by integrating pKS133b and pKS132–2 into the genome, and a nonbinding mutant reporter strain (KS259) was also constructed by integrating pKS165 into the genome. Both strains grew poorly on SD (-histidine) + 15 mM 3-aminotriazole (3-AT) and not at all on SD (-histidine) + 45 mM 3-AT plates. The yeast transformation was performed as described by Gietz et al. (1997).
ChIP and analysis of immunoprecipitated DNA
ChIPs were performed as described previously (Strahl-Bolsinger et al. 1997) with some modifications. The cross-linking time was 10 min instead of 15 min. Breakage was performed in 500 μL of lysis buffer that contained 1 mM PMSF, 1 mM Benzamidine, and 1 mg/mL of Bacitracin. The extract was sonicated for a total of 60 sec (resulting in an average DNA size of 500 bp–1 kb), clarified by centrifugation, and subjected to immunoprecipitation with anti-HA monoclonal antibody (12CA5, Roche) and protein G-Agarose beads (Roche).
PCR reactions were performed in 50 μL volume with one fifth of the immunoprecipitated material or one fiftieth of the nonimmunoprecipitated material and serial two-fold dilutions thereof as templates. Hotstar Taq polymerase (Qiagen) was used in 1 X manufacturer's buffer supplemented with 2.5 mM MgCl2, 200 μM each dNTP, and 500 nM or 1 μM of each primer. Typically, 25–30 cycles of amplification were performed. The following primers were used to amplify the RE region: SUN575, 5′-CGCGGATCCAAACTCGAAAAGTAAATA-3′, and Wu027, 5′-ACGCTCGAGCCCGGGCTTGCAAATATTGT-3′; SUN540, 5′-CTTGTCGACATAAATCCTTGCTTAGC-3′, and P-MAB09, 5′-CAATACCTTCTTGAACCATTTCCC-3′; CDC20 promoter region: SUN844, 5′-ACGTTAGTTGAACTTGAATTCCG-3′, and SUN845, 5′-AAGGTGATAAATTCTTTGCCTGC-3′; CLB2 promoter region: SUN842, 5′-GACAGATTTTATTCCAAAT GCGG-3′, and SUN 843, 5′-CGCTTTTCAGAAGTATCAA TTCG-3′; ARG5,6 coding region: ARG5,6 p1, 5′-CAAGGATC CAGCAAAGTTGGGTGAAGTATGGTA-3′, and ARG5,6 p2, 5′-GAAGGATCCAAATTTGTCTAGTGTGGGAACG-3′.
Acknowledgments
We are grateful to Catherine Fox and Gustav Ammerer for providing us strains, Philip James for providing the yeast genomic library, Ranjan Sen for advice and critically reading the manuscript, Cherry Wu for her early work on mutations of region A, and Neal Sugawara for his invaluable technical advice. This work was supported by the National Institutes of Health (grant GM20056). E.C. was supported in part by a grant from l'Association pour la Recherche sur le Cancer.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL haber@brandeis.edu; FAX (781) 736-2405.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.994902.
References
- Bon E, Neuveglise C, Casaregola S, Artiguenave F, Wincker P, Aigle M, Durrens P. Genomic exploration of the hemiascomycetous yeasts: 5. Saccharomyces bayanus var. uvarum. FEBS Lett. 2000;487:37–41. doi: 10.1016/s0014-5793(00)02276-6. [DOI] [PubMed] [Google Scholar]
- Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Triggs-Raine B, Robbins A, Graham KC, Woods RA. Identification of proteins that interact with a protein of interest: Applications of the yeast two-hybrid system. Mol Cell Biochem. 1997;172:67–79. [PubMed] [Google Scholar]
- Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Trends Genet. 1992;8:446–452. doi: 10.1016/0168-9525(92)90329-3. [DOI] [PubMed] [Google Scholar]
- ————— A locus control region regulates yeast recombination. Trends Genet. 1998a;14:317–321. doi: 10.1016/s0168-9525(98)01501-7. [DOI] [PubMed] [Google Scholar]
- ————— Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998b;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- ————— . Mobile DNA II. 2002. Switching of Saccharomyces cerevisiaemating-type genes. Craig, N.L., Craigie, R., and Gellert, M., eds. pp. 927–951. ASM Press, Washington, D.C. [Google Scholar]
- Hermann-Le Denmat S, Werner M, Sentenac A, Thuriaux P. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol Cell Biol. 1994;14:2905–2913. doi: 10.1128/mcb.14.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Bose ME, Mielke MR, Muller U, Fox CA. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics. 2000;154:1533–1548. doi: 10.1093/genetics/154.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Pietz G, Fox CA. Mechanisms controlling differential promoter-occupancy by the yeast forkhead proteins Fkh1p and Fkh2p: Implications for regulating the cell cycle and differentiation. Genes & Dev. 2001;15:2445–2456. doi: 10.1101/gad.906201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two- hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar AJ, Hicks JB, Strathern JN. Directionality of yeast mating-type interconversion. Cell. 1982;28:551–561. doi: 10.1016/0092-8674(82)90210-0. [DOI] [PubMed] [Google Scholar]
- Koranda M, Schleiffer A, Endler L, Ammerer G. Forkhead-like transcription factors recruit Ndd1 to the chromatin of G2/M-specific promoters. Nature. 2000;406:94–98. doi: 10.1038/35017589. [DOI] [PubMed] [Google Scholar]
- Kumar R, Reynolds DM, Shevchenko A, Goldstone SD, Dalton S. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol. 2000;10:896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Loy CJ, Lydall D, Surana U. NDD1, a high-dosage suppressor of cdc28–1N, is essential for expression of a subset of late-S-phase-specific genes in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3312–3327. doi: 10.1128/mcb.19.5.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MR, Shimizu M, Roth SY, Dranginis AM, Simpson RT. DNA-protein interactions at the S. cerevisiaealpha 2 operator in vivo. Nucleic Acids Res. 1993;21:3295–3300. doi: 10.1093/nar/21.14.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, West AG, Johnston LH, Sharrocks AD, Morgan BA. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. Embo J. 2000;19:3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT. Nucleosomes are positioned with base pair precision adjacent to the alpha 2 operator in Saccharomyces cerevisiae. Embo J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, Wyrick JJ, Zeitlinger J, Gifford DK, Jaakkola TS, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Magasanik B. Two transcription factors, Gln3p and Nil1p, use the same GATAAG sites to activate the expression of GAP1 of Saccharomyces cerevisiae. J Bacteriol. 1996;178:2465–2468. doi: 10.1128/jb.178.8.2465-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Strathern JN. Control and execution of mating type switching in Saccharomyces cerevisiae. Washington, D.C.: American Society for Microbiology; 1989. [Google Scholar]
- Szeto L, Broach JR. Role of alpha2 protein in donor locus selection during mating type interconversion. Mol Cell Biol. 1997;17:751–759. doi: 10.1128/mcb.17.2.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto L, Fafalios MK, Zhong H, Vershon AK, Broach JR. Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes & Dev. 1997;11:1899–1911. doi: 10.1101/gad.11.15.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Oshima T, Araki H, Harashima S, Oshima Y. Mating type control in Saccharomyces cerevisiae: A frameshift mutation at the common DNA sequence, X, of the HMLalpha locus. Mol Cell Biol. 1984;4:203–211. doi: 10.1128/mcb.4.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Broach JR. Donor locus selection during Saccharomyces cerevisiaemating type interconversion responds to distant regulatory signals. Genetics. 1992;132:929–942. doi: 10.1093/genetics/132.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Szeto L, Broach JR. Mutations affecting donor preference during mating type interconversion in Saccharomyces cerevisiae. Genetics. 1995;139:1495–1510. doi: 10.1093/genetics/139.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K, Simpson RT. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. Embo J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Weiss K, Yang C, Harris MA, Tye BK, Newlon CS, Simpson RT, Haber JE. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomycesmating-type switching. Genes & Dev. 1998;12:1726–1737. doi: 10.1101/gad.12.11.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Haber JE. MATa donor preference in yeast mating-type switching: Activation of a large chromosomal region for recombination. Genes & Dev. 1995;9:1922–1932. doi: 10.1101/gad.9.15.1922. [DOI] [PubMed] [Google Scholar]
- ————— A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- Wu X, Moore JK, Haber JE. Mechanism of MAT alpha donor preference during mating-type switching of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:657–668. doi: 10.1128/mcb.16.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu C, Haber JE. Rules of donor preference in Saccharomycesmating-type gene switching revealed by a competition assay involving two types of recombination. Genetics. 1997;147:399–407. doi: 10.1093/genetics/147.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Muller EG, Amacher SL, Northrop JL, Davis TN. A dosage-dependent suppressor of a temperature-sensitive calmodulin mutant encodes a protein related to the fork head family of DNA-binding proteins. Mol Cell Biol. 1993;13:1779–1787. doi: 10.1128/mcb.13.3.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]