Abstract
Poly(ADP-ribose) polymerase (PARP) is a major NAD-dependent modifying enzyme that mediates important steps in DNA repair, transcription, and apoptosis, but its role during development is poorly understood. We found that a single Drosophila Parp gene spans more than 150 kb of transposon-rich centromeric heterochromatin and produces several differentially spliced transcripts, including a novel isoform, PARP-e, predicted to encode a protein lacking enzymatic activity. An insertion mutation near the upstream promoter for Parp-e disrupts all Parp expression. Heterochromatic but not euchromatic sequences become hypersensitive to micrococcal nuclease, nucleoli fail to form, and transcript levels of the copia retrotransposon are elevated more than 50-fold; the variegated expression of certain transgenes is dominantly enhanced. Larval lethality can be rescued and PARP activity restored by expressing a cDNA encoding PARP-e. We propose that PARP-e autoregulates Parp transcription by influencing the chromatin structure of its heterochromatic environment. Our results indicate that Parp plays a fundamental role organizing the structure of Drosophila chromatin.
Keywords: PARP, chromatin, heterochromatin, copia, Drosophila, nucleolus
In diverse eukaryotes, protein ADP-ribosylation plays important but imperfectly understood roles in DNA repair, apoptosis, and gene transcription (for review, see de Murcia and Shall 2000; Ziegler and Oei 2001). Poly(ADP-ribose) polymerase-1 (PARP1), the major nuclear source of this activity in mice, becomes activated upon binding DNA strand breaks via its conserved amino-terminal zinc fingers. The activated protein transfers ADP-ribose moieties from NAD onto local chromatin proteins such as histones, topoisomerases, polymerases, and transcription factors (Poirier et al. 1982; Menissier-de Murcia et al. 1997). These modifications facilitate base excision repair by transiently clearing proteins from the chromosome to expose the lesioned area, by down-regulating transcription of the affected genes, and by modulating the activity of checkpoint and stress regulatory proteins. PARP1 also controls its own activity by ADP-ribosylating a conserved “automodification” domain that is required for dimerization and for target protein binding. PARP1 carries out these functions in part by forming protein complexes with transcription factors, replication proteins, and DNA damage checkpoint effectors (Dantzer et al. 1998; Pleschke et al. 2000; Ziegler and Oei 2001). Mice with defective Parp1 genes develop into fertile adults, and hence a developmental role for PARP1 has yet to be established (Wang et al. 1997). However, four other mouse genes encode distinct ADP-ribosyl transferases with related catalytic domains (Amé et al. 1999; Kickhoefer et al. 1999), including a telomere-associated form known as Tankyrase (Smith et al. 2001), and thus functional redundancy may have obscured such a role.
The model eukaryote, Drosophila melanogaster, has the potential to support detailed genetic studies of PARP function in both physiology and development. Its genome contains a single gene, Parp, related to mammalian Parp1 (Uchida et al. 1993; Hanai et al. 1998), and one homolog of tankyrase (Adams et al. 2000). The protein specified by the major Parp transcript, PARP-I, includes all the conserved domains characteristic of mammalian PARP1 except a canonical caspase cleavage site. Parp-I transcripts are expressed in nearly mature ovarian follicles and throughout embryonic development, but were not detected in larvae (Hanai et al. 1998). Parp-II transcripts lacking the automodification domain are produced via differential splicing of a single exon (Kawamura et al. 1998). However, genetic studies have been hindered because Parp is located deep within centromeric heterochromatin, and its exons are scattered among several contigs that remain unlinked to the euchromatic genome sequence (Adams et al. 2000).
Drosophila development has been studied extensively to determine how changes in chromatin structure contribute to specifying programs of tissue-specific and temporally regulated gene expression (for review, see Farkas et al. 2000; Gerasimova and Corces 2001). Zygotic transcription begins during the first 14 embryonic nuclear cycles concomitant with the establishment of heterochromatin and of nucleolus formation (Foe and Alberts 1983). During subsequent embryonic and larval stages, chromatin domains are refined under the control of multiprotein remodeling complexes (for review, see Cairns 1998; Jacobs and van Lohuizen 1999). The role of NAD-requiring enzymes in these processes is poorly understood, but in addition to Parp, Drosophila contains a gene structurally and functionally related to the NAD-dependent histone deacetylase encoded by the yeast Sir2 locus (Barlow et al. 2001; Rosenberg and Parkhurst 2002).
Here we characterize mutations in the heterochromatic Parp gene. Rather than simply functioning as a repair enzyme, Parp is necessary for viability and to organize the chromatin structure of nucleoli, heterochromatin, and other sequences during development. Reduction of Parp function causes hyperexpression of the copia retrotransposon and enhances the variegation of GAL4 transgenes. These studies show that Parp plays a fundamental role in organizing chromatin structure during Drosophila development, and suggest that ADP-ribosylation of chromosomal proteins plays an important role in chromatin remodeling.
Results
Sequences encoding a novel PARP isoform are expressed in embryos
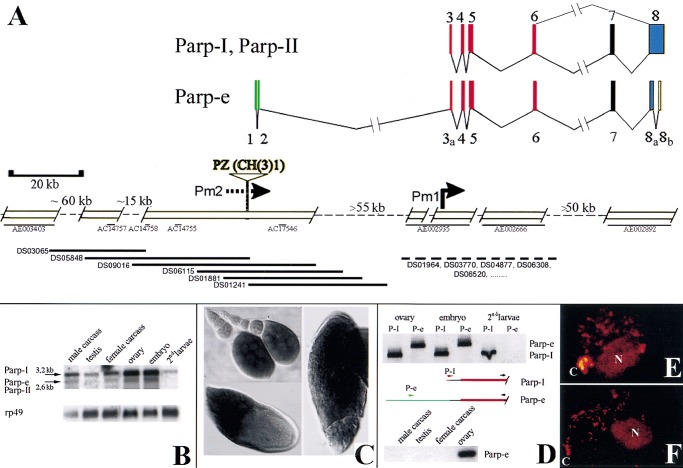
The previously determined structure of the genomic region encoding Parp-I is shown on the right in Figure 1A (Hanai et al. 1998; Adams et al. 2000). To search for additional Parp transcripts, clones corresponding to 14 Parp-related EST sequences (Rubin et al. 2000) were analyzed, and GM10715, derived from an early ovarian RNA library, was found to differ from Parp-I. The complete sequence of GM10715 was determined, revealing an additional 920 bp intron within exon 8 encoding the PARP catalytic domain as well as 287 bp of novel 5′ sequence that splice into the first Parp-I exon (exon 3) about 40 bp downstream from its 5′-end but 6 bp before the AUG codon (Fig. 1A). The PARP isoform predicted by GM10715, which we name PARP-e (embryonic), should lack enzymatic function since the new intron removes conserved amino acids essential for catalytic activity including the NAD binding site.
Figure 1.
Structure and expression of the Drosophila Parp locus. (A) Deduced genomic structure of the 300-kb Parp region; open boxes are sequenced. The arrangement of the exons encoding Parp-I is shown at top (Uchida et al. 1993; Hanai et al. 1998). (Bottom) The positions of three unlinked Drosophila genomic contigs (thin black lines: AE002935, AE002666, and AE002892) homologous to Parp-I exons are shown at right (Adams et al. 2000). Pm1 indicates the Parp-I promoter deduced from 5′ cDNA sequences (Hanai et al. 1998). A single cDNA isolated from early ovarian stages, GM10715, defines the alternatively spliced Parp-e transcript. The 5′-most 273 bp of GM10715 matches the genomic sequence flanking a P element insertion, CH(3)1 (Zhang and Spradling 1994). A map of this region (left portion of figure) was constructed by chromosome walking using a P1 genomic library (Kimmerly et al. 1996) (bottom, thick black lines). The two resulting scaffolds were sequenced and found to span four small preexisting genomic sequence contigs (thin black lines) and to link to a fifth (AE003403). The color code indicates which portion of PARP is encoded: DNA binding (red), automodification (purple), catalytic (blue), noncoding (green and yellow). (B) Multiple Parp transcripts. A Northern blot of poly(A)-containing RNA from the indicated developmental stages reveals both a 3.2-kb RNA, the size predicted for Parp-I, and a 2.6-kb RNA, the approximate size expected for Parp-II and Parp-e. Parp-homologous RNAs are abundant in both ovaries and embryos, and are reduced but still detectable in second instar larvae and adults. (C) Whole mount in situ hybridization using a 1.4-kb cDNA probe from the DNA-binding domain common to all isoforms labels Parp RNA in nurse cells and in oocytes from stage 9–14 follicles. (D) RT-PCR using isoform-specific primers (see diagrams) that distinguish between Parp-I (or Parp-II) and Parp-e demonstrate that Parp-e is produced in ovaries and embryos, but not at detectable levels in second instar larvae, or in adults outside the ovary. (E) Nuclei are shown from brains of third instar larvae expressing a Parp-I-DsRed fusion gene (see Materials and Methods). Protein is abundant in the chromocenter (C), the nucleolus (N), and at sites within euchromatin. (F) Third instar larval brain nuclei stained with anti-poly(ADP-ribose) antibody 10H show that protein-associated ADP-ribose is found in the same regions as PARP-DsRed.
Previous studies of Parp-I production showed that transcripts are abundant in late-stage ovarian follicles and embryos, but did not distinguish between transcripts encoding different isoforms (Hanai et al. 1998). We analyzed Parp expression throughout the Drosophila lifecycle using Northern blots (Fig. 1B), whole mount in situ hybridization (Fig. 1C), and RT-PCR with specific primers to distinguish Parp-e from Parp-I and II (Fig. 1D). The 3.2-kb Parp-I RNA and 2.6-kb Parp-II or Parp-e RNAs are expressed in ovaries, embryos, and adults. In contrast to previous results, low levels of the 3.2-kb Parp-I mRNA remain in second instar larvae (Fig. 1B). In the ovary, nurse cells express Parp RNA beginning as early as stage 4, whereas male germ cells strongly express Parp until the spermatid stage (data not shown). Parp-e expression is detected only in adult ovaries and embryos (Fig. 1D).
To further analyze Parp expression, we constructed and expressed epitope-tagged versions of the two major PARP isoforms in flies. When expressed using this UAS/GAL4-based system, PARP-I (Fig. 1E) and PARP-e (data not shown) are both highly enriched in nucleoli, heterochromatic chromosomal regions, and diverse euchromatic sites in the cells of most embryonic and adult tissues. The distribution of ADP(ribosyl)-modified proteins, as revealed by immunostaining with an antibody specific for ADP-ribose polymers (Kawamitsu et al. 1984), was very similar, strongly labeling these same regions within nuclei (Fig. 1F). These experiments provide a clearer picture of developmentally regulated Parp expression and show a correlation between PARP protein and protein ADP(ribosyl) moieties.
Parp spans a large region of 3R heterochromatin
The structure of GM10715 implies that some Parp transcripts originate from a novel promoter(s), which we denote Pm2, located at or upstream from the GM10715 5′ end. Using the isoform-specific 5′portion of GM10715 as a probe, six overlapping clones spanning ∼100 kb of genomic DNA flanking the 5′ region of Parp-e were isolated from a Drosophila P1 genomic library and used to map and sequence this region (Figs. 1A, 2). P1 clones were also recovered from the genomic region encoding Parp-I. At least 55 kb separates the upstream Parp-e sequence contig defined by DS09016 from the nonoverlapping Parp-I 5′ sequence contig AE002935. Sequence identity was also observed between the upstream region and the DNA flanking a previously described heterochromatic P element insertion, CH(3)1 (Zhang and Spradling 1994). To confirm that the upstream region defined by GM10715 and CH(3)1 really lies adjacent to PARP coding sequences, we showed that probes specific for Parp coding sequences and for the CH(3)1 insertion generated overlapping in situ hybridization signals on metaphase chromosomes (Fig. 2B). The CH(3)1 P element insertion was mapped previously to region h55-h56 of 3R heterochromatin (Zhang and Spradling 1994). Taken together, these studies define the structure of the Parp locus and confirm its location in the heterochromatin of chromosome 3R.
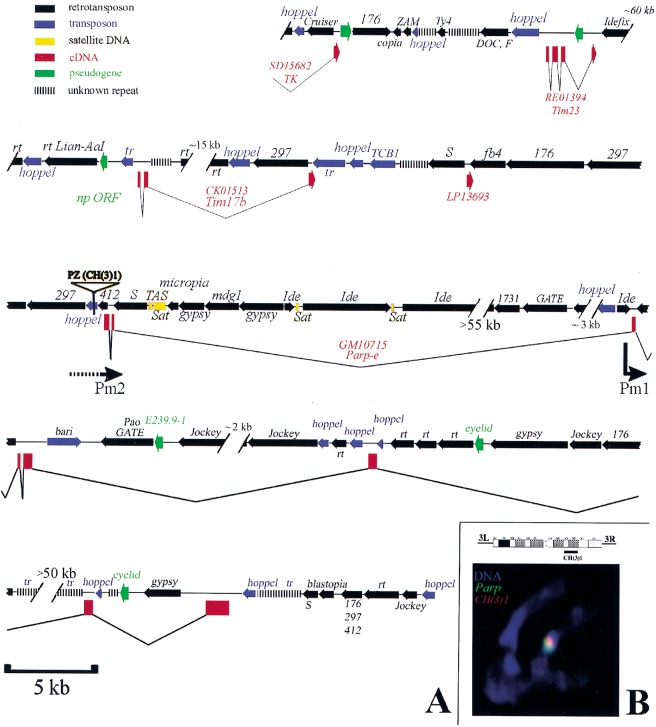
Figure 2.
DNA sequence of the heterochromatic region containing Parp. (A) A diagram summarizing the sequence organization of the region as determined from this study (see Materials and Methods) and from Adams et al. (2000) is shown. Genes defined by cDNAs sequenced as part of this study are shown in red (boxes are exons). The names of retrotransposons (black) and of transposons (blue) are given above the region of homology represented by an arrow (arrowhead: 3′ end). Regions containing only small sequence blocks related to a particular transposon are indicated by parallel bars. The position of the CH(3)1 insertion and the location of the putative Parp promoters Pm1 and Pm2 are indicated. Gaps in the sequence of known or estimated size are represented by hash marks. (B) An ideogram of chromosome 3 heterochromatin shows the cytological region of CH(3)1 insertion (Zhang and Spradling 1994). (Bottom) A chromosome set from a CH(3)1/TM3 third instar larval neuroblast is shown that has been hybridized in situ with Parp cDNA (green) and transposon-specific sequences (red). The partial overlap of the Parp and CH(3)1 sequences indicates that Parp and CH(3)1 are located near each other in 3R heterochromatin. (Note: the TOTO-3 used for this confocal micrograph does not reveal full morphological detail, but chromosomes were also scored using DAPI; CH(3)1 was localized previously to h55 (Zhang and Spradling 1994).
Analyzing the genomic DNA sequence surrounding the Parp transcription unit revealed several striking features of this 300-kb heterochromatic chromosome region (Fig. 2A). About 210 kb (70%) lies within Parp and four other genes, including a thiamine kinase ortholog (TK) and two apparent mitochondrial translocase subunits (Tim17b and Tim23). Most of the DNA within and surrounding the genes consists of transposons (Fig. 2A, blue) and retrotransposons (black) that are strikingly organized over the entire region studied. Nearly all are oriented in the same direction on the chromosome and opposite to the genes. These transposons have lost LTR homologies, and the gypsy elements lack insulator sequences that can disrupt enhancer-promoter interactions. Because unselected genomic sequences diverge rapidly during Drosophila evolution, our observations suggest that in recent evolutionary time the Parp region underwent extensive transposon invasion subject to some large-scale selective or mechanistic constraint on insertional orientation.
CH(3)1 defines a complementation group that disrupts Parp expression
We characterized the CH(3)1 strain to learn whether its recessive lethality (Zhang and Spradling 1994) is caused by disrupting Parp gene function. A second allele of the CH(3)1 locus was found within another P element insertion strain, CH(3)4, but the CH(3)4 P element cannot be responsible for the allelism as it is located on the opposite chromosome arm (3L) and mutates a different gene. However, both CH(3)1 and CH(3)1/CH(3)4 animals display a similar phenotype. Mutant CH(3)1 homozygotes develop slowly and usually die during the second larval instar after 6–9 d (Fig. 3A). Evidence of mitotic cell cycle defects was not seen; predominantly diploid larval tissues such as the brain are of normal size. However, examination of larval mouth hooks shows that up to 50% of the mutant larvae are arrested at the onset of ecdysis II (Fig. 3B). When the CH(3)1 element was imprecisely excised, about 7% of the derived chromosomes were homozygous viable and complemented CH(3)4, arguing strongly that the CH(3)1 P element was responsible for the original lethality.
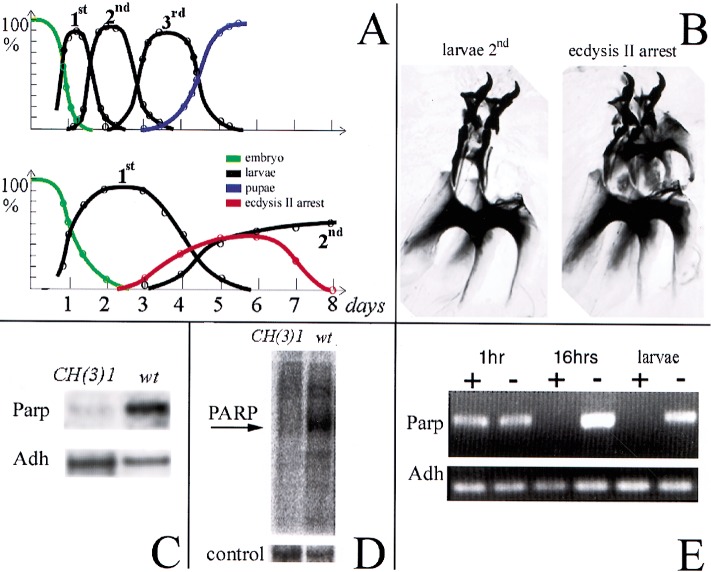
Figure 3.
The CH(3)1 complementation group disrupts Parp expression and activity. (A) Timelines of development of wild-type (top) and CH(3)1 homozygotes (bottom) are shown. The fraction of animals at each developmental stage are plotted as a function of time, revealing the strong developmental delay caused by CH(3)1. (B) Preparation of larval mouth hooks, which distinguish larval instars, are illustrated showing the characteristic appearance of the normal l2 mouthhooks (left) and of mouthhooks from CH(3)1 mutants arrested at the onset of ecdysis 2 (right). (C) Northern blot of poly(A)-containing RNA from wild-type larvae and 4-d-old CH(3)1 larvae showing reduced levels of Parp 3.2-kb mRNA. (D) Proteins labeled by ADP-ribosylation in wild-type (wt) and CH(3)1 mutant larvae. An autoradiogram of a gel of 32P-labeled protein is shown (see Materials and Methods). The prominent band at 117 kD in wild-type has the expected molecular weight of Parp itself. Stained protein in a segment of the same gel is shown as a loading control. (E) RNAi treatment of embryos eliminates detectable Parp mRNA in 16-h embryos and larvae. An RT-PCR assay recognizing all forms of the Parp transcripts is shown; primers specific for the alcohol dehydrogenase gene (Adh) gene serve as a loading control.
If CH(3)1 alleles mutate Parp, then its gene transcripts should be reduced in the affected larvae. As predicted, we found that Parp expression is severely affected in both CH(3)1 and CH(3)1/CH(3)4 animals. We observed that 3.2-kb Parp-I mRNA levels are strongly reduced on Northern blots of RNA from mutant larvae (Fig. 3C) and by using quantitative RT-PCR (data not shown). ADP-ribosylation of proteins is also dramatically decreased in mutants (Fig. 3D). However, the effects observed on all forms of Parp were surprising because mutation of Pm2-initiated transcripts by the CH(3)1 insertion might have been expected to only disrupt production of the 2.6-kb Parp-e mRNA and the enzymatically inactive PARP-e protein isoform. The small amount of Parp mRNA and enzymatic activity that does remain in the mutant larvae might come from transcripts initiated at Pm1 or from remaining maternal stores of Parp transcripts. To try and remove all Parp mRNA, we injected a 587-bp dsRNA specific for Parp into early embryos and observed that all traces of Parp mRNA detectable by RT-PCR were lost after 16 h of embryonic development (Fig. 3E). More than 70% of the animals receiving Parp RNAi injections, unlike buffer-injected controls, arrested after hatching into first instar larvae, that is, at an earlier point than in the mutants that retain low levels of residual Parp RNA. These observations further strengthen the connection between the CH(3)1 locus and Parp.
Loss of PARP derepresses the copia retrotransposon
Because Parp is located in transposon-rich heterochromatin, we looked for effects of the mutation on transposon activity. We found that CH(3)1, CH(3)4, and CH(3)1/CH(3)4 animals dramatically overproduce the 5.5-kb transcript of the copia retrotransposon (Fig. 4A). A similar large accumulation of copia-specific RNA was observed in embryos and larvae following injection of Parp-specific RNAi (Fig. 4B), providing further support that the CH(3)1 mutation acts directly on Parp. Tests using several other retrotransposable elements showed no increase in transcripts in CH(3)1 mutant animals, so the increased expression appeared to be specific for copia. Copia hyperexpression in CH(3)1 mutants and normal copia expression in their wild-type sibs could be abolished by injecting copia-specific dsRNA into preblastoderm embryos (Fig. 4C), but CH(3)1 lethality was not rescued. Thus, disrupting Parp expression causes copia hyperexpression, but this effect is not responsible for the lethal effects of CH(3)1.
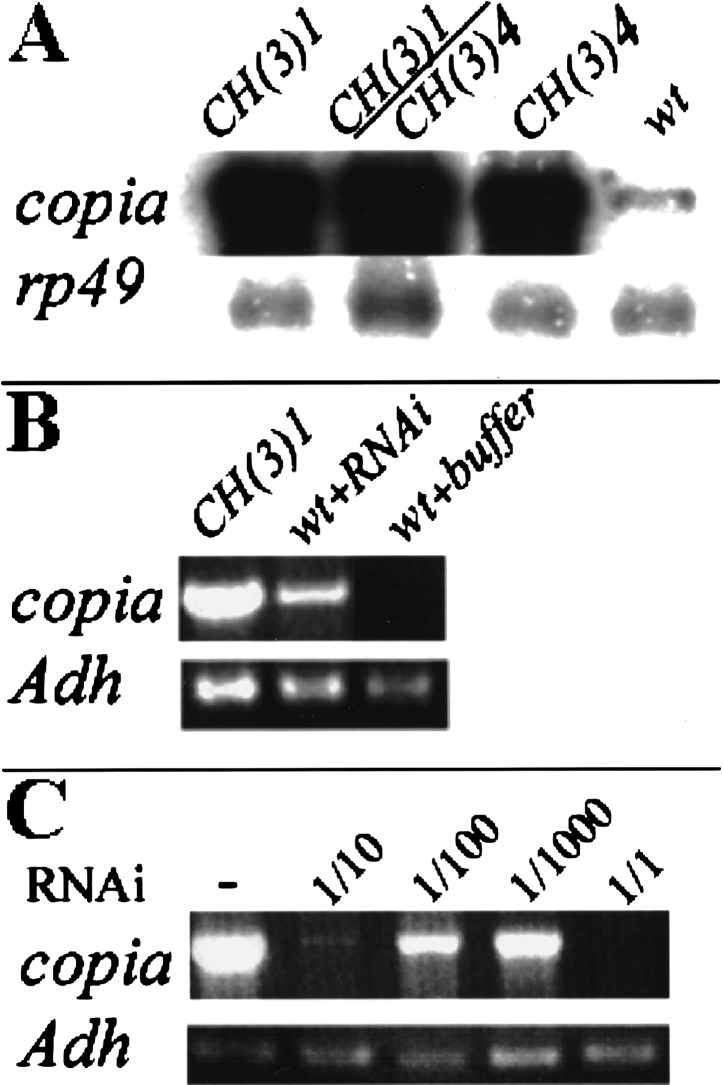
Figure 4.
Parp mutations or Parp (RNAi) elevate copia transcript levels. (A) A Northern blot of total RNA from second instar larvae of the indicated genotypes was probed with copia sequences. The 5.5-kb copia transcript is overproduced up to 50-fold in CH(3)1 or CH(3)4 homozygotes, and in CH(3)1/CH(3)4 trans-heterozygotes compared to wild-type. An rp49 probe was used as a loading control. (B) Quantitative RT-PCR shows that injection of Parp-specific RNAi, but not buffer, causes copia RNA to be overproduced. Primers specific to Adh served as a loading control. (C) Copia RNA accumulation does not cause lethality. Injection of mutant CH(3)1 embryos with RNAi specific to copia suppressed the accumulation of excess copia RNA and resulted in the elimination of all copia transcripts detectable by RT-PCR within 16 h. Sequential dilutions of the RNAi gave a graded response. However, the treatment did not rescue larval lethality.
Disrupting Parp expression alters heterochromatin structure
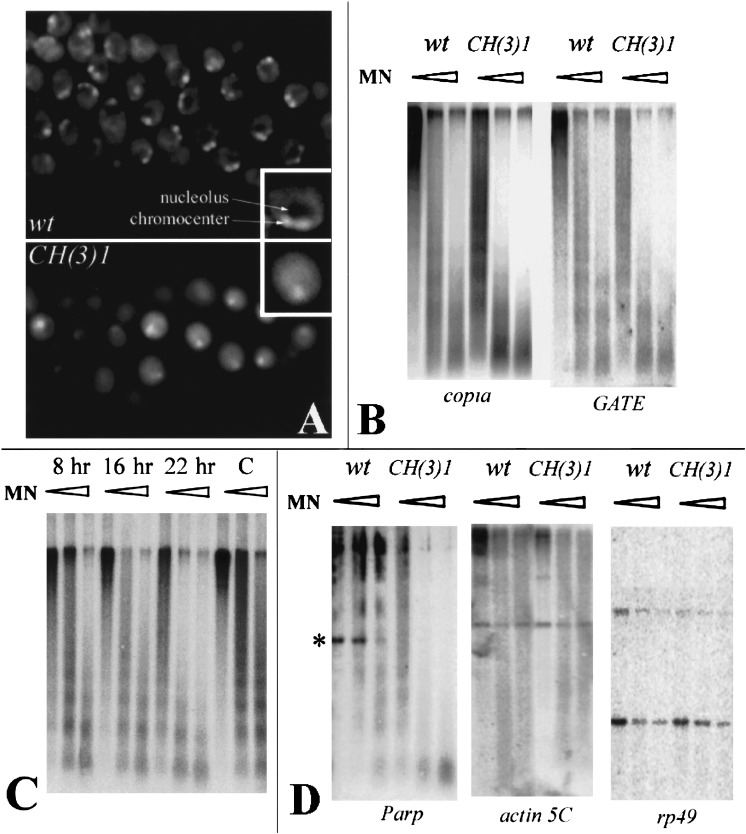
The CH(3)1 mutation might affect a transcription factor that negatively regulates copia transcription or it might disrupt a protein that acts at the level of chromatin. We looked for global effects on chromatin by examining DAPI-stained nuclei from various tissues of CH(3)1 homozygotes, and by carrying out nuclease sensitivity experiments. CH(3)1 alleles dramatically alter nuclear morphology (Fig. 5A). DAPI-stained nuclei from all mutant tissues examined appear more uniform than wild-type, show a less distinct chromocenter, and lack a nucleolar region of low DNA density. Copia chromatin is specifically affected, because copia-homologous sequences are much more sensitive to micrococcal nuclease digestion within CH(3)1 mutant compared to wild-type nuclei (Fig. 5B). Even the lowest levels of nuclease, which digested very little copia-specific DNA in wild-type, cleaved it extensively in CH(3)1 homozygotes. Elevated sensitivity could also be induced by injecting Parp-specific RNAi (Fig. 5C).
Figure 5.
Parp mutations alter nuclear morphology and chromatin accessibility to nuclease. (A) DAPI-stained nuclei from second instar larval salivary glands of wild-type (top) or CH(3)1 mutants (bottom). A single nucleus is presented at higher magnification in the insets. Nuclei in the mutant appear more diffuse, have a less distinct chromocenter, and lack the region of low DNA density caused by the presence of a normal nucleolus. (B) Nuclei from CH(3)1 mutant larvae were treated with increasing concentrations of micrococcal nuclease (triangles) prior to DNA extraction, digestion with PstI, and analysis on Southern blots probed with a copia or GATE probe. Pst digestion produces no small internal fragment of copia or GATE resolvable within the molecular weight range of the gel. At all concentrations, retrotransposon-specific sequences were far more sensitive to digestion in the mutant. (C) The same analysis as in B was carried out using nuclei at the indicated times after injection of Parp-specific RNAi. Copia sequences from RNAi-injected animals become increasingly sensitive to micrococcal nuclease digestion with increasing time after RNAi injection, compared to buffer-injected controls (C). (D) Micrococcal nuclease assays were carried out as in B and analyzed with a probe from the Parp gene region encoding exons 3, 4, and 5, and with probes specific for the single-copy euchromatic genes actin 5C and rp49. Parp sequences are much more accessible to digestion in the mutant, including a band containing exon 3 and Pm1 (asterisk). To ensure that experiments with heterochromatic and single-copy probes were comparable, the same blot was used for copia, GATE, actin 5C, and rp49. The blot assayed with Parp in D was reprobed with copia as a control and showed the same differential digestion as in B.
Many additional nuclease sensitivity tests were carried out to develop a picture of which genomic sequences and regions become nuclease-sensitive in the mutant. All the repetitive sequences tested were strongly affected (Fig. 5B; data not shown). These include the transposons GATE, gypsy, mdg1, hoppel, the S element, 297, Idefix, the rDNA-specific R1 element, and the Stellate repeats. In contrast, no changes in micrococcal nuclease sensitivity of the unique euchromatic genes actin 5C and rp49 were observed (Fig, 5D). The single-copy Parp gene resides within a region of highly repetitive sequences including many of the transposons shown to be affected in deficient animals. We tested three Parp exons, including exon 3 which lies adjacent to Pm1, and found that they became much more accessible to nuclease digestion in CH(3)1 homozygotes (Fig. 5C). Taken together, these observations suggest that reducing PARP activity selectively alters the chromatin structure of heterochromatic and repetitive sequences but not of euchromatic, single-copy DNAs.
Expression of PARP-e but not PARP-I rescues CH(3)1 mutations
Despite strongly reduced Parp expression in CH(3)1 mutant animals and the correlation between the mutant phenotype and the effects of removing PARP activity using RNAi, we sought to verify that CH(3)1 mutates Parp by rescue. Because of its size and unclonable structure, it is impractical to attempt rescuing CH(3)1 using genomic Parp DNA. Consequently, we generated constructs that express Parp-I or Parp-e cDNAs under the control of a UAS promoter. Following transformation, we studied the effects of expressing these cDNAs throughout many tissues using the Armadillo-GAL4 driver. Ectopic expression of Parp-I cDNA, but not Parp-e cDNA, in wild-type flies causes rough eyes and abdominal cuticle defects (data not shown). Parp-I-expressing animals arrest at the pupal stage if two doses of the driver are present. Thus, as in mammals, excess PARP-I levels cause deleterious effects.
Expressing Parp-e cDNA in a CH(3)1 mutant background revealed that the PARP-e isoform can completely suppress larval lethality and give rise to a small number of viable, fertile adults. Mutant flies bearing two copies of the Parp-e cDNA and driver can be readily maintained as a homozygous stock. In contrast, mutant animals expressing Parp-I cDNA die as third instar larvae but still develop significantly farther than in the absence of the construct. The ability of Parp cDNAs to partially or wholly rescue CH(3)1 animals demonstrates that the CH(3)1 lesion directly disrupts Parp gene expression. We have therefore renamed the CH(3)1 and CH(3)4 alleles ParpCH1 and ParpCH4, respectively.
Expressing Parp transcripts dramatically restores the nuclear morphology and the Parp expression of the mutant larvae. Parp-I expression causes a nucleolus to form that can be visualized with the specific antibody AJ1 in many but not all nuclei (Fig. 6A). The mosaic nature of the response, which may result from cell-to-cell variation in either the production or effects of ectopic PARP-I, is likely to explain the failure of this construct to rescue fully. All nuclei in the Parp-e-expressing animals appear wild-type in morphology. Surprisingly, larvae rescued by Parp-e contain higher than wild-type levels of both the 2.6-kb Parp-e and the 3.2-kb Parp-I mRNA species (Fig. 6B). Thus, the enzymatically inactive Parp-e isoform may rescue CH(3)1 by inducing production of Parp-I mRNA. Consistent with this model, ADP-ribosyl transferase enzymatic activity is also restored, because the amount of poly(ADP-ribose)-containing protein detectable by anti-poly(ADP-ribose)-specific antibody increases to well above wild-type levels (Fig. 6C, 117 kD PARP-I band). PARP-e may induce a more physiological pattern of Parp-I expression, leading to fewer deleterious effects than when Parp-I is misexpressed globally. How expression of an enzymatically inactive protein rescues Parp-I expression and ADP-ribosyl transferase activity is discussed below.
Figure 6.
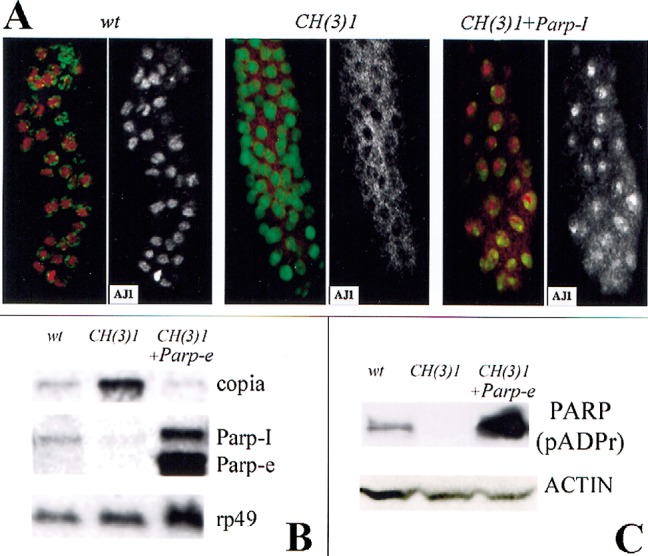
Expression of Parp-I or Parp-e cDNA rescues defects in CH(3)1 mutants. (A) Partial restoration of normal nuclear morphology by expression of Parp-I. Immunofluorescent detection of the nucleolar antigen AJ1 (red) and DNA (green) is shown in larval salivary glands of the indicated genotypes. AJ1 staining alone is shown on the right. In CH(3)1 mutants (middle), AJ1 is cytoplasmic rather than in nucleoli as in wild-type (left). Expression of Parp-I cDNA (right) restores nucleoli and nuclear AJ1 staining in a mosaic manner; note cells at the top of the figure with normal localization, but cells near the bottom still show a mostly cytoplasmic distribution of AJ1 reactivity. (B) A Northern blot of RNA from larvae of the indicated genotypes shows that Parp-e cDNA expression greatly elevates the level of 2.6-kb Parp-e mRNA and also of the 3.2-kb Parp-I mRNA. Note that copia-specific RNA accumulation is greatly reduced in CH(3)1 mutant larvae that express Parp-e cDNA. rp49 hybridization serves as a loading control. (C) A Western blot of proteins isolated from larvae of the same genotypes as in C, and probed with an antibody specific for poly(ADP-ribosyl) moieties. Expression of Parp-e cDNA in a CH(3)1 homozygous background increases the amount of poly(ADP)-ribose-modified proteins to levels greater than in wild-type. As in wild-type, diverse proteins are affected, the most prominent of which is the size of PARP-I itself (shown). An actin antibody was used as a loading control.
PARP and SIR2 modify GAL4/UAS variegation
Because the Parp gene is located in heterochromatin and acts on the chromatin structure of repetitive DNA sequences, we investigated whether it functions as an enhancer or suppressor of variegated position effects. Neither ParpCH1 nor ParpCH4 altered the level of wm4 variegation, a standard test for modifiers of classical position-effect variegation (data not shown). However, we did notice that Parp strongly affects the variegated expression commonly exhibited by many UAS/GAL4 constructs (Brand and Perrimon 1993). For example, in the presence of only one wild-type dose of Parp+, the variegation of an epitope-tagged mitochondrial protein (Tim17B-DsRed) driven from a UAS promoter is strongly enhanced (Fig. 7A,B). At six different tested sites of integration, DsRed expression is virtually silenced in a ParpCH1/+ background, while at the seventh site, nonvariegated expression becomes variegated. Altering the dose of Sir2 was found to have the opposite effect and dominantly suppress UAS-Tim17B-DsRed variegation (Fig. 7C). Similar reciprocal effects were observed with two other tested constructs, UAS-Parp-I-DsRed (data not shown) and UAS-Sir2-DsRed (Fig. 7D–F), suggesting that the dosage of Parp and Sir2 may affect the expression of UAS/GAL4 constructs generally. PARP and SIR2 levels may alter the chromatin structure of sequences such as those in UAS and GAL4 constructs that are prone to silencing during development.
Figure 7.
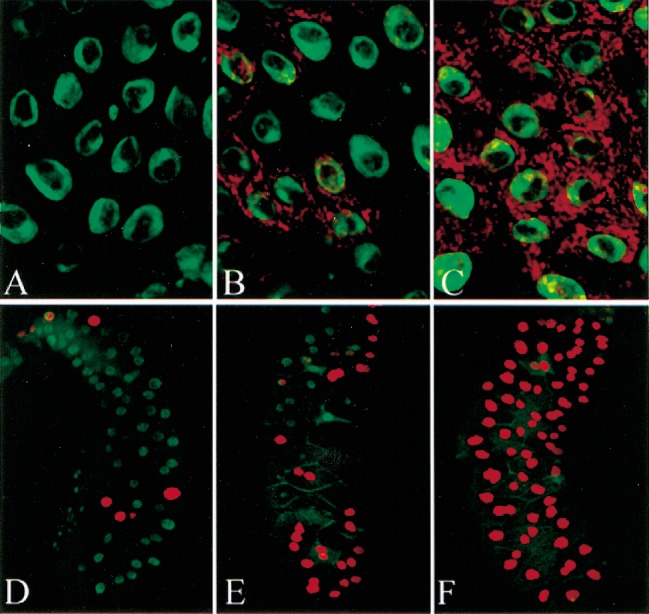
ParpCH1 and Sir205327 have opposite dominant effects on the variegated expression of GAL4/UAS constructs. The variegated expression of an Arm-Gal4-driven UAS-Tim17B -DsRed construct (A,B) or a UAS-Sir2-DsRed construct (D–E) is modified by background genotype. In a ParpCH1/+ background (A,D), expression is strongly reduced compared to expression in a wild-type background (B,E). Similar variegated expression of the same constructs driven by 69B-GAL4 is almost completely suppressed in a Sir205327/+ background (C,F). Green: DNA.
Discussion
PARP regulates chromatin structure during development
PARP is a conserved protein known to play critical roles that help restore and maintain genomic integrity (for review, see de Murica and Shall 2000). By identifying lethal Parp mutations, we showed that Drosophila Parp also plays an essential role during the lifecycle in the absence of external stresses. Many genes have been identified that act in both DNA repair and during development (Baker et al. 1976; Gatti and Baker 1989). However, the phenotype of Parp mutants differs from those of other genes in this class, which typically produce third instar larvae deficient in diploid tissue as a result of defects in the mitotic cell cycle.
Our experiments suggest that Drosophila Parp plays a special and fundamental role in organizing chromatin on a global scale. Parp mutant cells lack nucleoli and contain unusually nuclease-accessible repetitive sequences. Both the heavy expression of Parp-e and Parp-I in oocytes and early embryos and the early onset of these defects suggest that a major role for PARP occurs as development begins. At fertilization, the zygote genome is quiescent and unregionalized, but during the final cleavage divisions heterochromatin becomes distinguishable from euchromatin, nucleoli form, and specific gene transcription begins. Zygotic PARP activity may be needed to carry out these changes, which are reminiscent of the amphibian “mid-blastula transition.” The strong enrichment of epitope-tagged PARP in nucleoli and on heterochromatin is consistent with such a role. When Parp function is limited by a declining maternal pool, chromatin may not regionalize normally, stunting further development.
Our observations argue that the role of Parp is not limited to the initial stages of development, however. Programmed changes in chromatin organization continue after blastoderm formation in concert with cell differentiation (for review, see Hagstrom and Schedl 1997). The effects of reducing Parp expression later in embryonic development using RNAi, and the influence of Parp dosage on GAL4/UAS variegation, indicate that it also participates in organizing chromatin domains during later embryonic and larval growth. PARP plays a positive role in expressing euchromatic UAS constructs, because reduced Parp dosage enhances the variegation of these transgenes. Furthermore, Parp function is likely to be specially required for larval metamorphosis, because up to 50% of mutant larvae were arrested at precisely this stage. Thus, PARP influences both the expression and silencing of particular euchromatic and heterochromatic sequences at diverse times during Drosophila development.
PARP may act by modifying chromosomal proteins
Enzymes that add or remove phosphoryl, acetyl, or methyl groups on specific residues of histones are known to control key aspects of chromatin structure during differentiation and development (Wolffe and Hayes 1999; Strahl and Allis 2000). In particular, heterochromatin assembly on both repeated and unique DNA is defined at least in part by a code of particular histone modifications (Nakayama et al. 2001). In mammals, histones and other chromosomal proteins are subject to modification by PARP (Tanuma et al. 1983; Krupitza and Cerrutti 1988; for review, see Althaus et al. 1995). We observed a strong reduction in the levels of protein ADP-ribosylation in PARP mutants. Many of the modified proteins detectable with antibodies that recognize protein-ADP(ribosyl) groups are located along chromosomes, and are particularly enriched in nucleoli and in the heterochromatic chromocenter, regions strongly affected by Parp mutations. These observations support the idea that PARP acts on Drosophila chromatin by ADP-ribosylating chromatin proteins.
PARP-e autoregulates the activity of a complex Parp gene located within heterochromatin
Our structural characterization of the Parp gene reveals that both the gene itself and its surrounding chromosomal region are complex. The Parp locus is localized in 3R heterochromatin near band h55, where it spans at least 150 kb. At least two promoters are utilized, and the upstream promoter, Pm2, produces a transcript encoding a novel protein isoform, PARP-e, primarily during oogenesis and early larval development. Four other genes reside nearby and are transcribed in the same direction. In contrast, most of the DNA located outside gene exons consists of diverse transposable elements that are oriented opposite to the genes, perhaps as a result of selection to minimize the disruptive effects of transposon-encoded transcription and splicing signals. Much remains to be learned about the number, structure, regulation, and evolution of heterochromatic genes (Cook and Karpen 1994; for review, see Weiler and Wakimoto 1995). The Parp region may now serve as a valuable model for detailed studies of these issues.
Our experiments suggest that Parp itself is subject to novel regulatory mechanisms. ParpCH1 likely disrupts Parp-e transcription from Pm2, but homozygotes also have greatly reduced levels of Parp-I mRNA and of PARP activity, despite the fact that Pm1 is located at least 75 kb downstream from the ParpCH1 insertion site. Thus, Parp-e production appears to be essential for transcription of Parp-I from Pm1. It is difficult to rule out the existence of additional promoters or splice forms of PARP transcripts. However, the fact that expression of a cDNA encoding PARP-e rescues lethality, Parp-I mRNA production, and ADP ribosyl-transferase activity argues strongly that PARP-e autoregulates Parp transcription. Indeed, Parp-e expression may be rate-limiting for Parp-I transcription, because overproduced Parp-e from the rescue construct was associated with elevated levels of Parp-I mRNA (Fig. 6C).
There are two basic ways in which Parp-e might control Parp-I transcription. PARP-e may simply function as a factor that activates transcription from Pm1. Alternatively, it may function by a novel mechanism related to its action on heterochromatin. The Pm1 promoter and surrounding sequences may need to acquire a compact, heterochromatic chromatin state for activity. Zygotic PARP-e produced near the onset of development would facilitate heterochromatin formation, thereby activating Pm1 and Parp-I production. Simultaneously, this chromatin transition might shut off or limit Parp-e production from Pm2. Such a feedback switch would link PARP production to the chromatin state and might represent a mechanism utilized by other heterochromatic genes. Two other such genes were recently shown to require the heterochromatin-specific HP-1 protein to be efficiently expressed (Lu et al. 2000).
PARP may remodel and maintain chromatin domains
Previous studies of the role played by PARP during DNA repair have led to a model of how it acts on chromatin (for review, see Zeigler and Oei 2001). Following DNA damage, inactive PARP-I protein located near the damaged region binds to DNA breaks, activating the catalytic site, and begins to transfer ADP-ribose groups to the chromatin proteins located in the immediate vicinity and to the PARP automodification domain. The modified proteins are released from the DNA, allowing repair enzymes to access the damaged region. When repair is complete, the ADP-ribosyl groups are removed by a specific glycosylase, and the disrupted chromatin reassembles. During this time, automodified PARP may serve as a local storage site for the dissociated chromatin proteins, preventing them from diffusing away and mixing with general pools (Althaus 1992). The local nature of the disruption may help to ensure that repair does not inadvertently lead to alterations in the preexisting state of chromatin programming.
We propose that the role played by PARP in DNA repair, as described above, represents just one instance of a general function PARP carries out to reprogram chromatin at multiple points during the lifecycle. Inactive PARP molecules located in many chromosome regions may be subject to activation by particular developmental and environmental stimuli in addition to DNA damage. Following such stimulation, activated PARP would catalyze the dissociation of chromosomal proteins in the affected domain. Introducing new or differentially modified chromosomal proteins to the affected site in conjunction with PARP activation would cause the local chromatin state to be specifically altered when ADP-ribosyl residues are subsequently cleaved and the dissociated proteins reassembled. Such a mechanism would allow chromatin remodeling to be precisely limited to particular chromosome regions by spatially controlling the sites of PARP activation and protein delivery. It might also explain many previous observations concerning the transcriptional role of PARP and its interaction with transcription factors.
Our results suggest that PARP acts to maintain certain chromatin domains as well as to remodel them. For example, copia sequences in animals that had already formed heterochromatin became nuclease-sensitive when PARP levels were gradually reduced in developing embryos with the use of RNAi. Even when enzymatically inactive, PARP molecules remain associated with many chromosome regions and may play essential structural roles. Disruption of these roles may be responsible for some of the effects caused by loss of the enzymatically inactive PARP-e isoform, and for some of the deleterious effects of PARP-I overexpression. Our findings emphasize the importance of learning more about the properties of PARP molecules within specific chromosome regions and how they change during chromatin reprogramming. Finally, our results suggest ways in which manipulating PARP molecules might allow chromatin reprogramming to be experimentally controlled.
Materials and methods
Drosophila strains and genetics
Genetic markers are described in Flybase (1999), and stocks were obtained from the Bloomington Stock Center except as indicated. The CH(3)1 and CH(3)4 strains were generated in a single P-element mutagenesis screen (Zhang and Spradling 1994). CH(3)1 was found to be viable in combination with Df(3R)10-65; Southern blotting indicated that this deletion does not remove PARP coding sequences (data not shown). y w67c23(2) was used as the host for transformation. The driver strain for the rescue experiments was arm-GAL44 (Sanson et al. 1996). The 69B-GAL4 driver is described by Brand and Perrimon (1993). Sir205237 was constructed by Karpen and Spradling (1992) and is described by Rosenberg and Parkhurst (2002). Balancer chromosomes carrying Kr-GFP were used to differentiate homozygous mutant embryos and larvae: TM3, Sb, P{w+, Kr-GFP}, and CyO, P{w+, Kr-GFP} (Casso et al. 2000). Imprecise excision of CH(3)1 was carried out as described (Zhang and Spradling 1994).
Developmental timing measurements
Embryos were collected on grape juice/agar plates for 2 h at 25°C, aged 10–12 h, and subsets were subsequently analyzed each 1–2 h during daytime for 4–8 d. Larval stages were identified by mouth hook and/or posterior appendage morphology.
Construction of transgenic Drosophila
For the rescue experiments, pP{w+, UAST-PARP-I} was constructed by cloning the NotI/KpnI fragment encoding PARP-I from cDNA LD02455 into the pUAST vector. pP{w+, UAST-PARP-e} was constructed by fusing a NotI/KpnI fragment encoding PARP-e from cDNA GM10715 into pUAST. To detect protein localization in vivo, PARP-I cDNA was fused to DsRed (Clontech Laboratories) in pP{w+, UAST-PARP-I-DsRed}, and PARP-e was fused to EGFP (Clontech Laboratories) in pP{w+, UAST-PARP-e-EGFP}. To study the variegation of UAS constructs, we built pP{w+, UAST-Tim17b-DsRed}, which contains a Tim17b cDNA fused inframe to DsRed (Clontech Laboratories) in pUAST. Transformation was as described (Spradling and Rubin 1982), with modifications (Prokhorova et al. 1994).
Genomic mapping and sequencing
A physical map of the Parp region was constructed using cDNA libraries (Rubin et al. 2000) and a P1 library (Kimmerly et al. 1996) from the Berkeley Drosophila Genome Project (BDGP). Clone DS09016 was subcloned into pTZ19R using XbaI or EcoRI digestion and sequenced. The following cDNAs were sequenced and used to express PARP isoforms: LD02455 (Parp-I) and GM10715 (Parp-e). In addition, 13 other Parp cDNAs were fully sequenced. To determine the location of transcribed exons in the Parp region (Fig. 2), we also fully sequenced the following cDNAs: SD15682 (TK), RE01394 (Tim23), CK01513 (Tim17b), and LP13698 (no ORF).
Fluorescent in situ hybridization (FISH)
Mithotic chromosomes were prepared as described by Laverty (2001). Probe preparation by nick translation, pretreatment, hybridization, and signal detection were performed as described (Dej and Spradling 1999). cDNA LD02455 was used to detect Parp coding sequences, and the PZ element without rosy gene sequences (Karpen and Spradling 1992) was used to detect the PZ insertion in CH(3)1.
Double-stranded RNA interference (dsRNAi)
RNAi was prepared as described by Kennerdell and Carthew (1998). The following regions were targeted: 269–864 for GM10715, 1–604 for LD02455, and 485–891 for copia. DsRNAi was injected into the posterior region of precellular blastoderm embryos at a concentration 0.5 μg/μL, and the embryos were allowed to develop for an appropriate period under oil in a humid chamber prior to analysis.
RT-PCR and Northern blot
Total RNA was isolated using Trizol reagent (Gibco BRL), precipitated twice with 3M LiCl, and treated with amplification-grade Dnase I (Gibco BRL) and poly(A)-containing RNA purified using a MessageMarker kit (Gibco BRL). The SuperScript Preamplification System (Gibco BRL) was then used to synthesize cDNA and for RT-PCR. The following primers were used to distinguish PARP-I and PARP-e transcripts: P-I (5′-AAATAAT AAATGTCTTGAAATTG-3′) for PARP-I, P-e (5′-GTCTTGAT TTTGTGTATACCG-3′) for PARP-e, and R4 (5′-TTTTATGA AACCAATTCG-3′) for both. Total Parp transcripts were detected using D1 (5′-GTGTCGTGGATGTGAAC-3′) and R2 (5′-TTGGAATTCTGGATTTTG-3′), which target a common coding region within the DNA binding domain. Copia-specific transcripts were detected using 5′-copia (5′-CCGTTTGATGG CGAGAAGTACGCGATTTGG-3)‘ and 3′-copia (5′-CCATC GTAACACGAAGGCAATGTGATC-3′), which target part of ORF1. For the Northern blot analysis, at least of 2.5 μg of poly(A) RNA from second instar larvae was used for each lane. The PARP probe was from the DNA binding domain, and an rp49 probe was used as a control.
Nuclease sensitivity assays
Embryos were collected on grape juice/agar plates for 2 h at 25°C and aged for 10–12 h, after which subsets were analyzed at frequent intervals during the next 4–8 d. The micrococcal nuclease sensitivity of purified nuclei was determined as described by Wu (1989) and Quivy and Becker (1997) with minor modifications (available on request). Controls showing the absence of endogenous nuclease activity were carried out, and the levels of micrococcal nuclease used were calibrated for each stock.
Immunohistochemistry and fluorescence microscopy
Tissues were fixed and stained with primary and secondary antibodies as described (Grieder et al. 2000) and examined by confocal microscopy using a Leica TCS-NT microscope. Primary antibodies were: mouse monoclonal (mAb) Aj1 (1:100) and anti-fibrillarin (1:200) from J. Gall (Department of Embryology, Carnegie Institution of Washington, Baltimore, MD); and mouse mAb 10H (1:20–50) from Dr. Manfred Frey (Steinbeis-Transferzentrum fur Angewandte Biologische Chemie). 10H specifically recognizes ADP-ribose polymers (Kawamitsu et al. 1984). Nuclear staining by 10H such as that shown in Figure 1F was abolished in Parp mutant larvae, further confirming the specificity of this reagent (data not shown). Mouse Alexa-568 (Molecular Probes) (1:400) was used as a secondary antibody.
pADPr assay
Embryos were collected and nuclei purified as for the nuclease sensitivity assay. Nuclei were incubated in nuclear buffer (Quivy and Becker 1997) containing 0.1 mCi/mL [32P]-NAD (Amersham) for 15 min at room temperature. Then nuclei were washed twice in nuclear buffer, collected by centrifugation, and preheated for 3 min. The protein gel was then processed, dried, and subjected to autoradiography.
Acknowledgments
We thank the Berkeley Drosophila Genome Project and Dr. J. Gall for providing materials. Doug Koshland, Daniella Drummond-Barbosa, Jim Wilhelm, Yurii Chinenov, and Horacio Frydman contributed valuable comments on the manuscript. Research expenses were defrayed in part by N.I.H. grant GM27875 to A.C.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL spradling@ciwemb.edu; FAX (410) 243-6311.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1003902.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Althaus FR. Poly ADP-ribosylation: a histone shuttle mechanism in DNA excision repair. J Cell Sci. 1992;102:663–670. doi: 10.1242/jcs.102.4.663. [DOI] [PubMed] [Google Scholar]
- Althaus FR, Bachmann S, Hofferer L, Kleczkowska HE, Malanga M, Panzeter PL, Realini C, Zweifel B. Interactions of poly(ADPribose) with nuclear proteins. Biochimie. 1995;77:423–432. doi: 10.1016/0300-9084(96)88155-7. [DOI] [PubMed] [Google Scholar]
- Amé JC, Rolli V, Scureiber V, Niedergang C, Apiou F, Decker P, Muller S, Hoger T, Ménissier-de Murica J, de Murica G. PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Baker BS, Boyd JB, Carpenter AT, Green MM, Nguyen TD, Ripoll P, Smith PD. Genetic controls of meiotic recombination and somatic DNA metabolism in Drosophila melanogaster. Proc Natl Acad Sci. 1976;73:4140–4144. doi: 10.1073/pnas.73.11.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow AL, van Drunen CM, Johnson CA, Tweedie S, Bird A, Turner BM. dSIR2 and dHDAC6: Two novel, inhibitor-resistant deacetylases in Drosophila melanogaster. Exp Cell Res. 2001;265:90–103. doi: 10.1006/excr.2001.5162. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling machines: Similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- Casso D, Ramirez-Weber F, Kornberg TB. GFP-tagged balancer chromosomes for Drosophila melanogaster. Mech Dev. 2000;91:451–454. doi: 10.1016/s0925-4773(00)00248-3. [DOI] [PubMed] [Google Scholar]
- Cook KR, Karpen GH. A rosy future for heterochromatin. Proc Natl Acad Sci. 1994;91:5219–5221. doi: 10.1073/pnas.91.12.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Menissier-de Murcia J. Functional association of poly(ADP-ribose) polymerase with DNA polymerase alpha-primase complex: A link between DNA strand break detection and DNA replication. Nucleic Acids Res. 1998;26:1891–1898. doi: 10.1093/nar/26.8.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej KJ, Spradling AC. The endocycle controls nurse cell polytene chromosome structure during Drosophilaoogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- de Murcia G, Shall S. From DNA Damage and Stress Signalling to Cell Death: poly (ADP-Ribosylation) reactions. New York, NY: Oxford University Press; 2000. [Google Scholar]
- Farkas G, Leibovitch BA, Elgin SC. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophilagenome projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. http://flybase.bio.indiana.edu/ . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V, Alberts B. Studies of nuclear and cytoplasmic behavior during the five mitotic cycles that precede gastrulation in Drosophilaembryogenesis. J Cell Sci. 1983;61:51–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes & Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Gerasimova T, Corces V. Chromatin insulators and boundaries: Effects on transcription and nuclear organization. Annu Rev Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- Grieder NC, de Cuevas M, Spradling AC. The fusome organizes the microtubule network during oocyte differentiation in Drosophila. Development. 2000;127:4253–4264. doi: 10.1242/dev.127.19.4253. [DOI] [PubMed] [Google Scholar]
- Hagstrom K, Schedl P. Remembrance of things past: Maintaining gene expression patterns with altered chromatin. Curr Opin Genet Dev. 1997;7:814–821. doi: 10.1016/s0959-437x(97)80045-7. [DOI] [PubMed] [Google Scholar]
- Hanai M, Uchida M, Kobayashi S, Miwa M, Uchida K. Genomic organization of Drosophilapoly(ADP-ribose) polymerase and distribution of its mRNA during development. J Biol Chem. 1998;273:11881–11886. doi: 10.1074/jbc.273.19.11881. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M. Cellular memory of transcriptional states by Polycomb-group proteins. Semin Cell Dev Biol. 1999;10:227–235. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- Karpen G, Spradling AC. Analysis of subtelomeric heterochromatin in the Drosophilaminichromosome Dp1187 by single P element insertional mutagenesis. ITLGenetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamitsu H, Hoshino H-O, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Hanai S, Yokota T, Hayashi T, Poltronieri P, Miwa M, Uchida K. An alternative form of poly(ADP-ribose) polymerase in Drosophila melanogasterand its ectopic expression in rat-1 cells. Biochem Biophys Res Commun. 1998;251:35–40. doi: 10.1006/bbrc.1998.9403. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W, Stultz K, Lewis S, Lewis K, Lustre V, Romero R, Benke J, Sun D, Shirley G, Martin C, et al. A P1-based physical map of the Drosophilaeuchromatic genome. Genome Res. 1996;6:414–430. doi: 10.1101/gr.6.5.414. [DOI] [PubMed] [Google Scholar]
- Krupitza G, Cerruti P. Poly(ADP-ribosyl)ation of histones in intact human keratinocytes. Biochemistry. 1988;28:4054–5060. doi: 10.1021/bi00435a063. [DOI] [PubMed] [Google Scholar]
- Laverty, Todd. 2001. In situ hybridization using Digoxigenin labeled probes. http://www.fruitfly.org/about/methods/cytogenetics.html.
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menissier-de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) polymerase binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Poirier GG, de Murcia G, Jongstra-Bilen J, Niedergang C, Mandel P. Poly(ADP-ribosyl)ation of polynucleosomes cause relaxation of chromatin structure. Proc Natl Acad Sci. 1982;79:3423–3427. doi: 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova AV, Voloshina MA, Shostak NG, Barskii VE, Golubovskii MD. Preparation and primary genetic analysis of Drosophila melanogastertransformants line w‘lz(b)/XXywf, containing mini-white genes, integrated in the genome during P-element-dependent transformation. Genetika (Moscow) 1994;30:874–878. [PubMed] [Google Scholar]
- Quivy JP, Becker PB. Genomic footprinting of Drosophilaembryo nuclei by linker tag selection LM-PCR. Methods. 1997;11:171–179. doi: 10.1006/meth.1996.0403. [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Parkhurst SM. DrosophilaSir2 is required for heterochromatic silencing and by euchromatic Hairy/E(Spl) bHLH repressors in segmentation and sex determination. Cell. 2002;109:447–458. doi: 10.1016/s0092-8674(02)00732-8. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Hong L, Brokstein P, Evans-Holm M, Frise E, Stapleton M, Harvey DA. A Drosophilacomplementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- Sanson B, White P, Vincent JP. Uncoupling cadherin-based adhesion from wingless signaling in Drosophila. Nature. 1996;383:627–630. doi: 10.1038/383627a0. [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 2001;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophilagerm line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tanuma S, Johnson JD, Johnson GS. ADP-ribosylation of chromosomal proteins and mouse mammary tumor virus gene expression. J Biol Chem. 1983;258:15371–15377. [PubMed] [Google Scholar]
- Uchida K, Hanai S, Ishikawa K, Ozawa Y, Uchida M, Sugimura T, Miwa M. Cloning of cDNA encoding Drosophilapoly(ADP-ribose) polymerase: Leucine zipper in the auto-modification domain. Proc Natl Acad Sci. 1993;90:3481–3485. doi: 10.1073/pnas.90.8.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes & Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Analysis of hypersensitive sites in chromatin. Methods Enzymol. 1989;170:269–289. doi: 10.1016/0076-6879(89)70052-5. [DOI] [PubMed] [Google Scholar]
- Zhang P, Spradling AC. Insertional mutagenesis of Drosophilaheterochromatin with single P elements. Proc Nat Acad Sci. 1994;91:3539–3543. doi: 10.1073/pnas.91.9.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler M, Oei SL. A cellular survival switch: Poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays. 2001;23:543–548. doi: 10.1002/bies.1074. [DOI] [PubMed] [Google Scholar]