Abstract
Chromatin-remodeling complexes couple ATP hydrolysis to alterations in histone–DNA interactions and nucleosome mobility, allowing transcription factors access to chromatin. Here, we use triple-helix strand-displacement assays, DNA length-dependent ATPase assays, and DNA-minicircle ATPase assays to establish that RSC, as well as its isolated ATPase subunit Sth1, are DNA translocases. RSC/Sth1 ATPase activity is stimulated by single-stranded DNA, suggesting that Sth1 tracks along one strand of the DNA duplex. Each RSC complex appears to contain a single molecule of Sth1, and isolated Sth1 is capable of nucleosome remodeling. We propose that the remodeling enzyme remains in a fixed position on the octamer and translocates a segment of DNA (with accompanying DNA twist), which breaks histone–DNA contacts and propagates as a wave of DNA around the octamer. The demonstration of DNA translocation presented here provides a mechanistic basis for this DNA wave. To test the relative contribution of twist to remodeling, we use nucleosomes containing nicks in precise locations to uncouple twist and translocation. Nucleosomes bearing nicks are remodeled less efficiently than intact nucleosomes. These results suggest that RSC and Sth1 are DNA translocases that use both DNA translocation and twist to remodel nucleosomes efficiently.
Keywords: RSC, Sth1, chromatin, nucleosome, translocation, SWI/SNF
Chromatin is dynamic, and is remodeled to affect changes in gene expression. Chromatin remodeling involves altering the structure, positioning, and modification state of nucleosomes, the primary repeating unit of chromatin structure. Nucleosomes can repress transcription by blocking important cis regulatory sequences such as enhancer-binding sites, the TATA box, or the transcription initiation site (Owen-Hughes and Workman 1994). Nucleosomes are often repositioned during the activation process, enabling transcription factors access to their sites of action (Almer et al. 1986; Côté et al. 1994). In addition, many covalent modifications occur on nucleosomes—such as acetylation, methylation, and phosphorylation (Strahl and Allis 2000; Jenuwein and Allis 2001)—that may mediate the binding of factors that further affect chromatin structure and the transcriptional state. Although these modifications play a crucial role in specifying factor recruitment, they have little effect on the intrinsic stability or mobility of nucleosomes.
Structural alteration and repositioning of nucleosomes is performed by large multiprotein complexes, termed remodelers, which consist of four classes: SWI/SNF, ISWI, CHD, and Mi-2/NURD (Vignali et al. 2000). Our studies focus on the biochemical properties of RSC, an essential and abundant SWI/SNF-family remodeler from Saccharyomyces cerevisiae(Cairns et al. 1996). RSC is composed of 15 proteins, five of which are highly similar to subunits of the yeast SWI/SNF complex, the founding member of the family (Laurent et al. 1992; Tsuchiya et al. 1992; Yoshinaga et al. 1992; Cairns et al. 1996; Cao et al. 1997; Treich and Carlson 1997). Both yeast complexes are highly similar to human SWI/SNFa and SWI/SNFb complexes in composition and activities (Imbalzano et al. 1994; Kwon et al. 1994; Wang et al. 1996; Xue et al. 2000). All remodeling complexes contain a DNA-dependent ATPase subunit, and the primary sequence of the ATPase domain itself is highly similar among these classes, suggesting that they all use a similar mechanism (Boyer et al. 2000). However, the domains outside the ATPase domain, as well as associated subunits, are quite different and help specify each class. ATP hydrolysis is required for remodeling activity, as mutations that abolish ATP hydrolysis likewise abolish remodeling (Côté et al. 1994; Tsukiyama and Wu 1995). For the SWI/SNF, ISWI, and CHD classes, the ATPase subunit has been isolated, and in each case displays a modest level of nucleosome remodeling in vitro, showing that the ATPase is the catalytic engine for remodeling (Corona et al. 1999; Phelan et al. 1999, 2000; Travers 1999; Tran et al. 2000). RSC contains the DNA-dependent ATPase Sth1 (Laurent et al. 1992; Tsuchiya et al. 1992; Cairns et al. 1996), but the remodeling properties of this protein in isolation have not been reported.
A central feature of remodelers is their ability to mobilize nucleosomes. Repositioning of histone octamers can occur along the same DNA segment by sliding (Hamiche et al. 1999; Längst et al. 1999; Whitehouse et al. 1999; Längst and Becker 2001), or to an unlinked DNA segment by octamer transfer (Lorch et al. 1999, 2001; Phelan et al. 2000). In addition, octamer eviction has been observed when the underlying sequence contains multiple binding sites for a transcriptional activator and both the remodeler and the activator are present at high levels (Owen-Hughes and Workman 1996).
The stability of the nucleosome is attributed to the sum of its 14 histone–DNA contacts (Luger et al. 1997). A central question for remodelers is how ATP hydrolysis is converted into a mechanical force that can break these contacts. All remodeler ATPases are members of the SF2 superfamily of DNA-dependent ATPases, and many of the other members are proven DNA helicases (Eisen et al. 1995). However, helicase activity (DNA duplex separation) has not been shown for any remodeler, nor are single-stranded regions of DNA generated during remodeling (Côté et al. 1998). However, remodelers in the SWI/SNF and ISWI classes do display DNA twisting activity, suggesting that the twisting of DNA on the histone octamer surface may provide the mechanical force for breaking histone–DNA contacts to create a small segment of DNA that is lifted from the octamer surface (Varga-Weisz and Becker 1998; Havas et al. 2000; Flaus and Owen-Hughes 2001; Gavin et al. 2001). In addition, remodelers have been proposed to impose structural alterations in the octamer that alter histone–DNA interactions and transiently expose DNA segments, allowing factors to bind or the DNA to reposition on the octamer surface (Kingston and Narlikar 1999; Narlikar et al. 2001). Finally, a role for DNA tracking/translocation by remodelers has been proposed, with the remodeler either moving along the DNA around the nucleosome, or remaining fixed at the nucleosome entry/exit site (Kadonaga 1998; Lorch et al. 1998; Travers 1999), but experiments testing for translocation by remodelers have not been reported. The data presented here establish DNA translocation as a property of RSC and Sth1, and suggests that the twisting of DNA that accompanies translocation augments remodeling efficiency. This work supports a model for remodeling that accommodates DNA translocation, DNA twist, and conformational change.
Results
Isolation and characterization of Sth1 and RSC
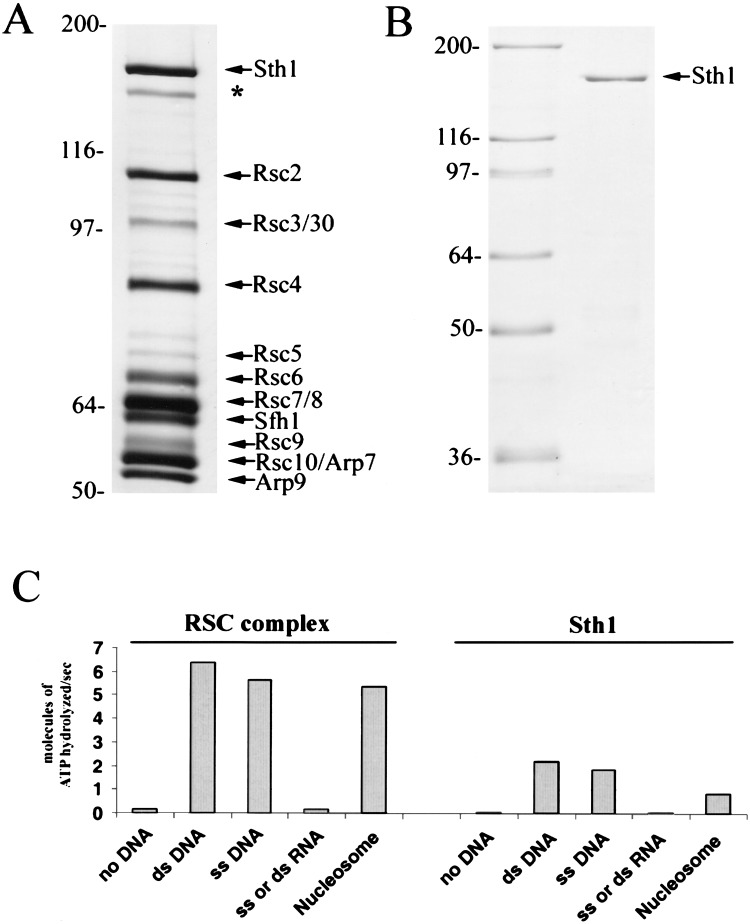
RSC was purified by tandem-affinity purification (TAP; Puig et al. 2001) using a yeast strain expressing an Rsc2 derivative that contains the calmodulin-binding peptide and the IgG-binding portion of protein A in tandem at the C terminus. TAP purification produced a RSC preparation of high yield, purity, and activity (Fig. 1A). RSC ATPase activity was stimulated almost equally by single-stranded DNA, double-stranded DNA, or mononucleosomes (Fig. 1C). To isolate Sth1, the ATPase from the RSC complex, it was overproduced as a derivative in yeast with two Flag epitopes and a 7×-histidine tag (Fig. 1B). Western analysis verified that purified Sth1 lacks other RSC components (data not shown). STH1 is an essential gene, and genetic experiments showed that this tagged Sth1 derivative fully complements an sth1Δ null mutation, indicating that this modification does not significantly alter activity (data not shown). The ATPase activity of purified Sth1 with single- or double-stranded DNA was 2.5-fold below that displayed by intact RSC (normalized for Sth1 molecules; Fig. 1C), similar to the reduction in ATPase activity observed for the isolated Brg1 subunit, the ATPase of human SWI/SNF (Phelan et al. 1999, 2000). This reduced activity may be caused by a reduction in percent active molecules, the lack of other RSC components (lowering specific activity), or a combination of these two factors. Neither RSC nor Sth1 can use 100- to 200-base (or base pair) single- or double-stranded homopolymeric RNA (Fig. 1C). In addition, mononucleosomes are not as effective as naked DNA in stimulating the ATPase activity of Sth1, showing a five- to sixfold reduction relative to RSC (Fig. 1C), which suggests that other components of RSC assist Sth1 in accessing the DNA on nucleosomes. As presented below, isolated Sth1 is capable of nucleosome remodeling, although at reduced efficiency.
Figure 1.
Purification and ATPase activities of RSC and Sth1. (A) Purified RSC. RSC was purified from strain BCY211, which expresses TAP-tagged Rsc2 from the chromosomal RSC2 locus. Harvesting and purification were performed as described in Puig et al. (2001) and Materials and Methods. Small RSC subunits Rsc12–15, which stain weakly with silver, are present but not shown. (*) Degradation product of Sth1. (B) Purified Sth1. Sth1 was purified from strain BCY243, which bears a chromosomally integrated Gal4 overproduction system (B. Cairns, unpubl.) and a high-copy plasmid directing the synthesis of Flag-tagged Sth1 from the GAL1 promoter. Soluble Sth1 was purified by ion exchange (SP), Ni-NTA, and M2-Flag affinity columns. Western analysis verified the absence of RSC subunits in the purified material (data not shown). (C) ATPase properties of RSC and Sth1. Turnover numbers (Vmax) for RSC and Sth1 with various nucleic acid substrates: (ds DNA) double-stranded plasmid (BSCR, 3 kb); (ss DNA) single-stranded phagemid (BSCR, 3 kb); (ss or ds RNA) polyI · polyC double-stranded RNA, polyG single-stranded RNA, or polyU single-stranded RNA; (Nucleosomes) intact mononucleosomes (167 bp) as described in Fig. 6. Values are the average of at least three experiments, and all are within a standard error of ±5%.
Sth1 is monomeric in soluble RSC
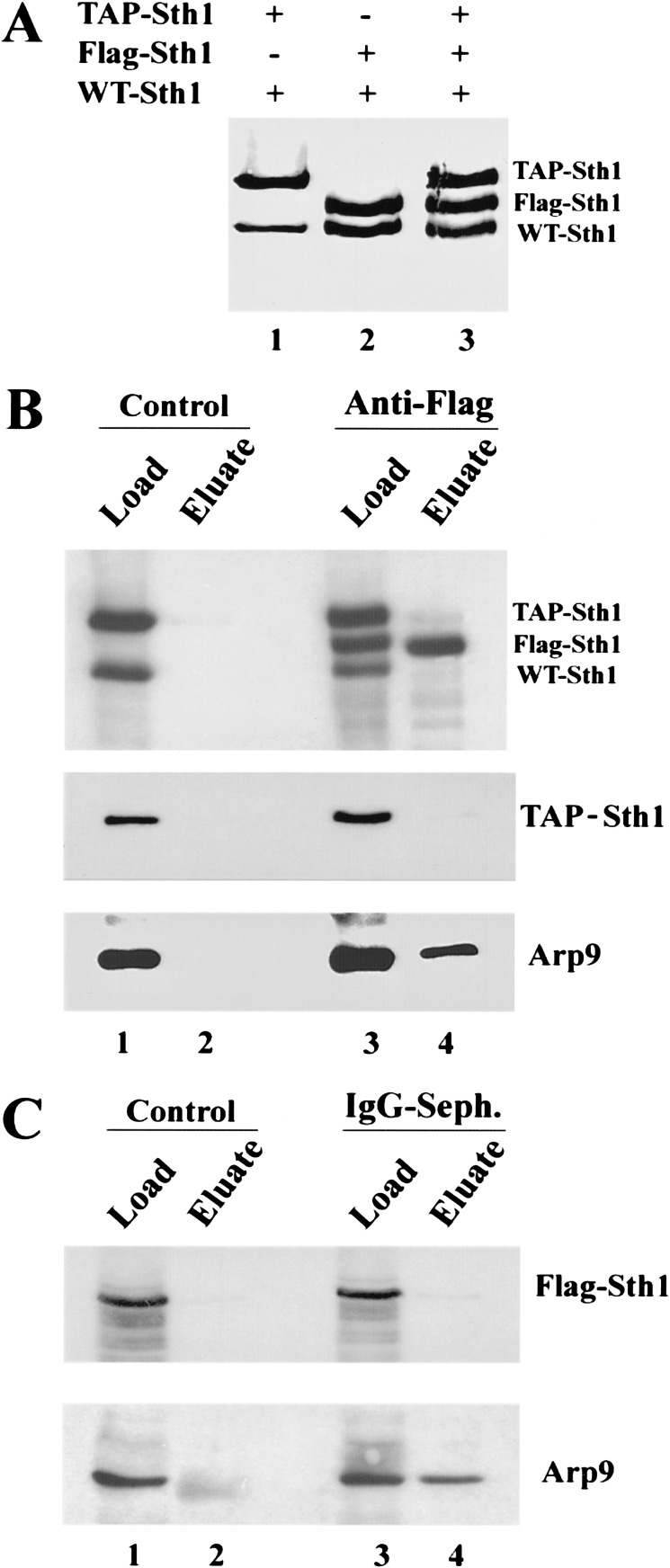
DNA helicases are commonly believed to function as dimers or hexamers (Lohman and Bjornson 1996), but recent experiments suggest that a subset function as monomers (Mechanic et al. 1999; Velankar et al. 1999). Knowing the oligomeric state of a translocase is important for understanding its mechanism of action on DNA. To determine the oligomeric state of Sth1 in the RSC complex, we tested for coimmunoprecipitation of STH1 derivatives bearing different epitope tags. First, we isolated a diploid strain bearing three different alleles of Sth1: wild-type (untagged), Protein A-tagged, and Flag-tagged. The former two alleles were present at their endogenous chromosomal location and thus were expressed from their own promoters, whereas the gene encoding the Flag-tagged allele was expressed from a plasmid containing the methionine-repressible MET15 promoter. These tagged alleles fully complement an sth1Δ null strain (data not shown).
A diploid strain bearing all three alleles was grown under conditions that maintained expression of Flag-tagged Sth1 at levels within twofold of the chromosomal wild-type and TAP-tagged STH1 alleles, as revealed by Western analysis using anti-Sth1 polyclonal antibodies (Fig. 2A, lane 3). Strains bearing only one tagged allele verified the identity and migration of each Sth1 derivative (Fig. 2A). Immunoprecipitation of Flag–Sth1 efficiently precipitates Flag–Sth1 and the RSC component Arp9 (and Rsc3, not shown), but does not coprecipitate either wild-type or TAP-tagged Sth1 (Fig. 2B). Likewise, immunoprecipitation of protein A coprecipitates Arp9 (and Rsc3, not shown), but not Flag-tagged Sth1 (Fig. 2C; data not shown). Here, the protein A epitope of TAP–Sth1 is cleaved off with TEV protease, allowing the elution of associated proteins. Thus, the presence of TAP-tagged Sth1 cannot be established; only the absence of Flag-tagged Sth1 can be verified. These results strongly suggest that Sth1 is present as a monomer in the RSC complex. However, this does not rule out the association of more than one RSC complex on the same nucleosome during remodeling.
Figure 2.
Oligomeric state of Sth1 in RSC. Coimmunoprecipitation analysis was performed using whole-cell extracts and either anti-Flag agarose or IgG-Sepharose (for TAP tag). (A) Western analysis using anti-Sth1 antisera of extracts from three diploid strains bearing different tagged STH1 alleles. All three strains contain a wild-type STH1 allele. BCY241 (lane 1) also bears an integrated TAP-tagged STH1 allele, BCY240 (lane 2) also contains a Flag-tagged allele present on a centromeric plasmid (p1170.STH1), and BCY242 (lane 3) contains both the integrated TAP-tagged allele and p1170.STH1. (B) Precipitation of Flag-tagged Sth1. (Control) Anti-Flag beads with BCY241 extract. Anti-Flag precipitation was performed with BCY242 extract. (C) Precipitation of BCY242 extract with IgG. (Control) IgG with BCY240 extract.
Remodelers as ATP-dependent translocases
Sth1 belongs to the DEAD/H-box family of the SF2 superfamily of DNA helicases (Laurent et al. 1992; Eisen et al. 1995; Lohman and Bjornson 1996). However, purified SWI/SNF and RSC do not show classical DNA helicase activity. That is, they do not catalyze DNA duplex strand separation, as detected by the displacement of a DNA oligonucleotide hybridized to a complementary single-stranded DNA template (Côté et al. 1994; data not shown). However, DNA helicases have two properties: ATP-dependent DNA translocation and DNA duplex strand separation. In the case of the monomeric DNA helicase PcrA, these properties can be uncoupled by mutation, as certain PcrA mutants lack duplex unwinding but maintain DNA translocation (Soultanas et al. 2000). In addition, PcrA can translocate along a single strand of DNA (Dillingham et al. 2000, 2002), and the crystal structure of PcrA shows the binding of its translocation domain to one strand of the DNA (Velankar et al. 1999). Importantly, ATP hydrolysis by PcrA is apparently coupled to DNA translocation (Dillingham et al. 2002). Using PcrA as a model, we reasoned that remodeler ATPases might possess DNA translocation activity but not separate the strands of the duplex, as remodeling does not involve the creation of single-stranded regions (Côté et al. 1998). Indeed, DNA duplex separation is only required for processes in which one strand is used as a template, such as DNA/RNA synthesis or DNA repair. Three assays were applied to test for DNA translocation: DNA length-dependent ATPase activity, single-stranded DNA-minicircle analysis, and triple-helix strand displacement.
Sth1 and RSC display DNA length-dependent ATPase activity
If RSC hydrolyzes ATP to translocate along DNA, then its ATPase activity may be proportional to DNA length, in contrast to ATPases that are stimulated by DNA binding alone. In previous work, a set of short oligonucleotides was used to determine the minimum length for appreciable RSC ATPase activity (∼25 bases), but the basis for this length dependence was not addressed (Cairns et al. 1996). To more clearly determine whether DNA length dependence was related to remodeler translocation, ATPase activity was correlated to DNA length using a large range of single- and double-stranded DNA molecules. These experiments were modeled on previous work showing that the ATPase activity of certain helicases is proportional to DNA length over a limited range (Young et al. 1994). This observation can be explained as follows: when the rate of DNA binding (or more likely, the transition to translocation) is slow relative to the rate of translocation, and ATPase activity is coupled to translocation, then the observed ATPase activity is proportional to DNA length. Here, short fragments of DNA should display a lower apparent ATPase activity than long fragments, owing to premature release of the DNA following translocation through the end of the fragment (termination-release). Termination-release would be followed by a period of inactivity that lasts until the enzyme binds and begins translocation on another DNA molecule. Thus, the actual curve of ATPase activity versus DNA length is predicted to have three phases: (1) a region representing the minimum DNA length for initial DNA binding, where ATPase activity is very low; (2) a region where ATPase activity is proportional to DNA length; and (3) a region where increasing the DNA length does not increase ATPase activity. Here, the length at which the maximal ATPase velocity (Vmax) is reached signifies the processivity limit for the translocase.
To ensure that our experiments were performed at substrate concentrations that elicited maximal velocity, we determined the Km (in micromolar nucleotide) values for RSC and Sth1 for single- and double-stranded DNA molecules. The Km values for RSC are 0.36 ± 0.02 μM and 0.34 ± 0.02 μM for single- or double-stranded DNA, respectively, whereas values for Sth1 are 0.62 ± 0.04 μM and 0.46 ± 0.02 μM, respectively, using single- or double-stranded DNA (phagemid Bluescript, 3 kb). Each DNA was then tested at a uniform nucleotide concentration of 30 μM (not oligonucleotide concentration). Thus, reactions containing longer oligonucleotides contain proportionately fewer DNA molecules than reactions containing shorter oligonucleotides, but all contain identical nucleotide concentrations. Control experiments verified that 30 μM nucleotide is at least 30-fold above the concentration that elicits Vmax for oligonucleotides that are of sufficient length to bind RSC/Sth1 (see below).
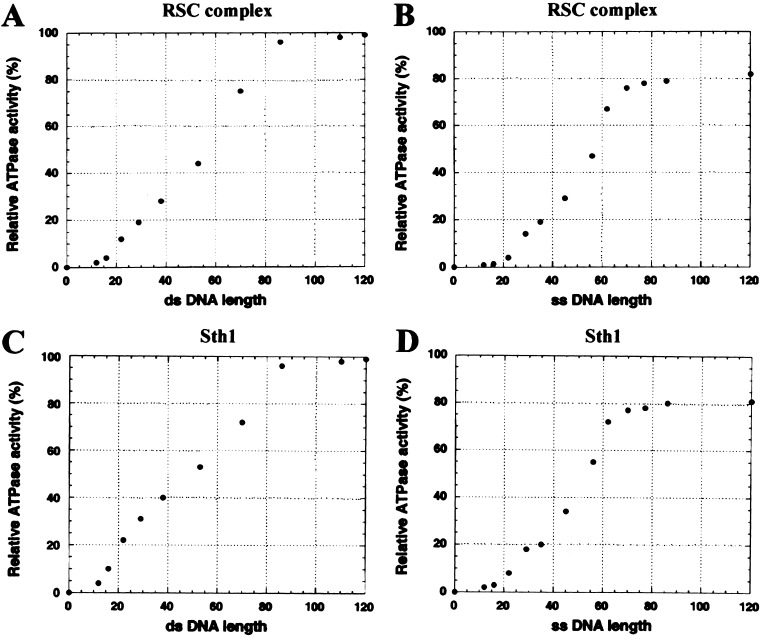
The ATPase activities of RSC and Sth1 display the three-phase response described above: double-stranded DNA molecules shorter than 15 bp are relatively inert, those in the range of 15 to 85 bp show ATPase activity proportional to length, and those longer than 85 bases provide no additional stimulation (Fig. 3A,C). Similar results are observed with single-stranded DNA, with the lower and upper length limits for ATPase activity at ∼20 and ∼75 bases, respectively (Fig. 3B,D). These results suggest that RSC/Sth1 binds to DNA of 15–20 bases, translocates, and then terminates translocation after ∼80 bases unless an end is reached. Consistent with this interpretation, we do not observe binding of RSC to DNA molecules shorter than 18 bases, the lower limit for eliciting ATPase activity (data not shown). Notably, the length of DNA bound to the PcrA translocase in the cocrystal structure is ∼17 bases (Velankar et al. 1999), consistent with our observations. Double-stranded DNA templates are only slightly more effective in stimulating ATPase activity than are single-stranded templates of equal length, consistent with RSC/Sth1 tracking along one strand of the duplex, but showing a slight preference for its presumed natural substrate, double-stranded DNA.
Figure 3.
Dependence of RSC and Sth1 ATPase activity on DNA length. The maximal velocity of ATPase activity was determined with RSC or Sth1 as described in Materials and Methods. Values are the average of at least three experiments and are reported relative to the Vmax observed with double-stranded plasmid DNA (BSCR, 3 kb). The standard error with double-stranded DNAs is ±5%; with single-stranded DNAs, ±3%. (A) Length dependence of RSC with double-stranded DNA. (B) Length dependence of RSC with single-stranded DNA. (C) Length dependence of Sth1 with double-stranded DNA. (D) Length dependence of Sth1 with single-stranded DNA.
ATPase properties of Sth1 and RSC with DNA minicircles
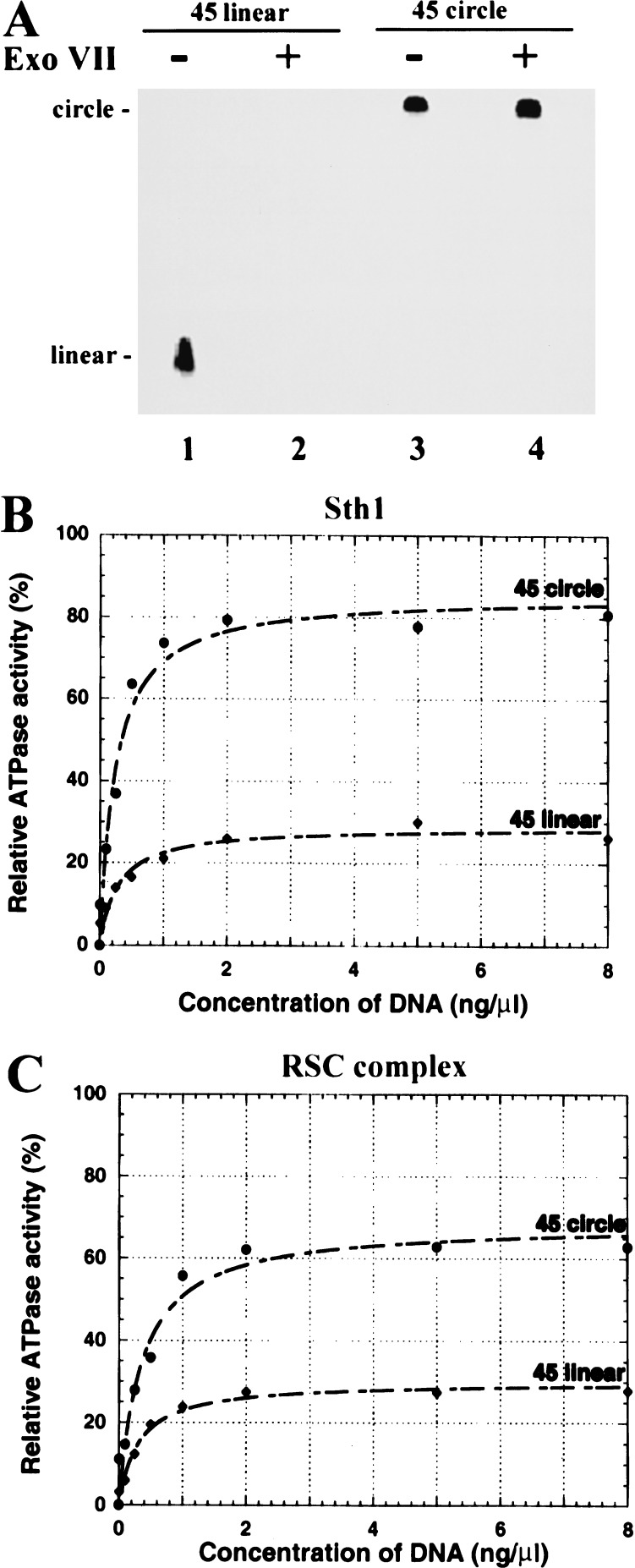
We reasoned that if premature termination-release underlies the lower ATPase activity observed with short DNA molecules, then a relatively higher ATPase activity should be observed with short circular DNA molecules compared with identical linear molecules. Here, circular DNA may mimic a DNA of infinite length and simply revolve through the translocase. To test this, we developed an ATPase assay involving DNA minicircles. Because of the rigidity of double-stranded DNA, circles of <120 bp cannot be formed (without DNA-bending agents), whereas single-stranded DNA has no conformational restraints or persistence length (Hagerman 1988). Therefore, we prepared single-stranded DNA minicircles of 45 or 30 bases, verified their integrity by resistance to exonuclease digestion (Fig. 4A; data not shown), and tested their activity in ATPase assays.
Figure 4.
ATPase activity with single-stranded DNA minicircles. (A) Integrity of the DNA minicircle. Integrity was verified by resistance to ExoVII. Linear or circular 45-mer DNA (300 ng) was treated with ExoVII, separated on a 7.5% polyacrylamide gel, and visualized by ethidium bromide staining. A negative of the image is provided. Identical susceptibility and resistance were obtained with the linear and circular 30-mer, respectively (data not shown). (B,C) ATPase activity of Sth1 (B) or RSC complex (C) with linear (closed diamonds) or circular (closed circles) 45-mers. Values are the average of at least three experiments and are reported relative to the Vmax elicited by double-stranded plasmid DNA (BSCR, 3 kb). Standard errors are reported in the text.
Consistent with translocation, circular 45-mers are significantly more effective at stimulating ATPase activity than the identical linear molecule; activity is increased 2.5-fold with Sth1 and 2.2-fold with RSC (Fig. 4B,C). Importantly, the Vmax levels approach those observed with long single-stranded DNA (note that all values are reported relative to Vmax with double-stranded plasmid DNA). The differences are not caused by substrate preference because the observed Km values for linear-versus-circular templates are nearly identical: the Km values for RSC with linear and circular 45-mers are 1.04 ± 0.13 μM and 1.01 ± 0.11 μM, respectively, and the Km values for Sth1 with linear and circular 45-mers are 0.82 ± 0.05 μM and 0.76 ± 0.06 μM, respectively. In contrast, both linear and circular 30-mers were poor stimulators of ATPase activity: the Vmax values for RSC were 14% and 20%, respectively, and for Sth1 were 18% and 25%, respectively. We suggest that the difference in the 45-mer as compared with the 30-mer reflects the minimum length required to both bind and commit to processive translocation.
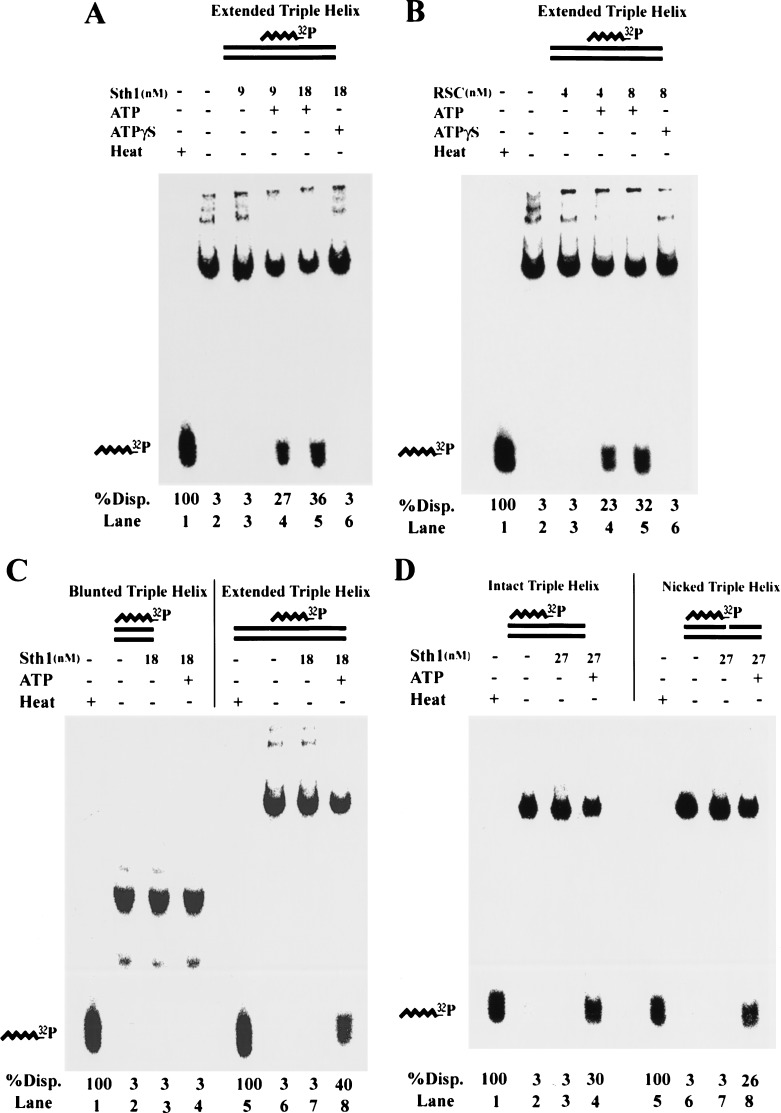
Sth1 and RSC displace a triple helix
Triple-helix strand displacement has been used to show translocation by the endonuclease EcoAI or by SV40 T-antigen (Kopel et al. 1996; Firman and Szczelkun 2000). The assay relies on the propensity for a homopurine–homopyrimidine repeat to form a three-strand triple helix (H-DNA) with a complementary homopyrimidine oligonucleotide at moderately low pH (pH 5.5; Htun and Dahlberg 1989). When shifted to near neutral pH (pH 7–8), the triple helix remains stably bound. However, if the third strand is displaced, it will not reform a triple helix at neutral pH. In the triple helix, the homopyrimidine oligonucleotide occupies the major groove of the mirror repeat, with each base of the oligonucleotide forming a Hoogsteen base pair with the Watson–Crick base pairs (van Dongen et al. 1999). The basis for the translocation assay is the displacement of the homopyrimidine oligonucleotide (bound to the duplex by weaker Hoogsteen base pairs) as the translocase proceeds through the triple helix.
We formed a triple helix of 40 bp, located centrally on a 190-bp double-stranded DNA, using a 40-base homopyrimidine oligonucleotide. This triple helix is highly stable when shifted to pH 7–8, although it is released efficiently by heating briefly at 90°C (Fig. 5, lane 1). Pure RSC or Sth1 shows comparable ATP-dependent triple-helix displacement when normalized for ATPase activity (Fig. 5A,B). ATP hydrolysis is required, because no displacement is observed with ATPγS (Fig. 5A,B, lane 6). For both Sth1 and RSC, displacement is proportional to the amount of enzyme added and the reaction time, consistent with the substrate representing a single species (data not shown). However, this displacement could be caused by direct binding and disruption of the triple helical structure itself rather than translocation through it. To address this, we isolated and tested the 40-bp triple helix used above. No displacement of the third strand was observed with Sth1 (Fig. 5C) or RSC (data not shown), strongly suggesting that RSC or Sth1 binds to the flanking duplex DNA, and invades the triple helix by translocation.
Figure 5.
RSC and Sth1 displace a triple helix. Triple-helix substrates consisting of a 40-base triple helical region were prepared as described in Materials and Methods. Triple helices were center-positioned on a 190-bp duplex DNA, end-positioned on a 114-bp duplex DNA, or prepared without duplex extension. Substrates were treated as indicated at 30°C for 30 min (or heated briefly at 90°C, labeled Heat) and separated in a 15% polyacrylamide gel. Displacement of the 32P-labeled third strand as a percentage of total displacement (%Disp.) was quantified by PhosphorImager analysis. (A) Displacement of a center-positioned triple helix by Sth1. (B) Displacement of a center-positioned triple helix by RSC. (C) Sth1 cannot displace an isolated triple helix. Identical results were obtained with 8 nM RSC (data not shown). (D) Displacement of an end-positioned triple helix by Sth1 is not affected by the presence of a nick near the duplex/triplex junction. Intact substrate or an identical substrate bearing a nick 4 bp from the duplex/triplex junction was used. Here, reactions were performed at 30°C for 1 h to compensate for lower efficiency compared with center-positioned substrates. Identical results were obtained with 8 nM RSC (data not shown).
However, triple-helix displacement might also result from an ATP-dependent conformational change in the extended duplex region (such as DNA twisting) that propagates to the triplex region. We note that these substrates lack a constrained end, and therefore are free to rotate, which should dissipate twist and prevent propagation to the triplex region. Nevertheless, we tested this by using substrates that either have or lack a nick in the phosphodiester backbone of one of the duplex DNA strands near the duplex/triplex junction, which should prevent twist or conformational propagation as it imparts rotational and conformational flexibility to the DNA. We formed a triple helix of 40 bp at one end of a 114-bp duplex DNA (identical in sequence to those described above) that either lacks or contains a nick within 4 bases of the duplex/triplex junction (Fig. 5D). Interestingly, nearly identical levels of third-strand displacement were observed with Sth1 (Fig. 5D) or RSC (data not shown), a result inconsistent with twist/conformation propagation but consistent with translocation through the nick and/or along the intact strand. We note that displacement for end-positioned triplex substrates is two- to threefold less efficient (when normalized for enzyme concentration and time), consistent with our interpretation that the duplex extension is used for invasion, as it is present on only one side of the end-positioned substrates.
Sth1 and RSC remodel recombinant yeast nucleosomes
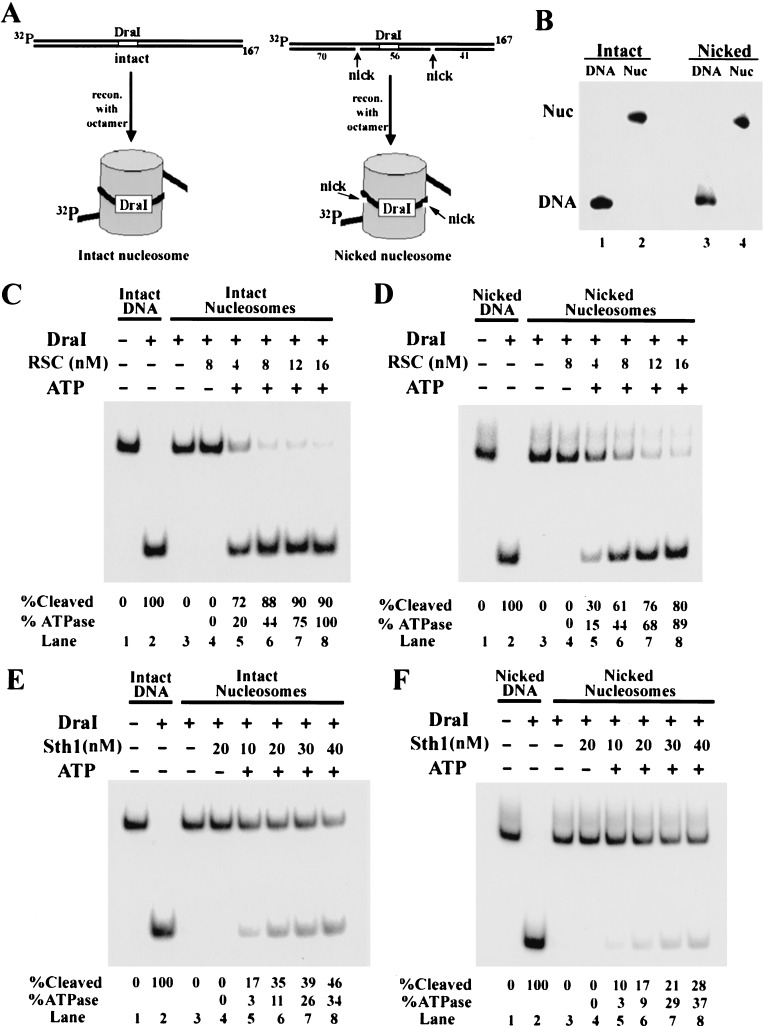
We have shown previously that yeast RSC remodels rat liver nucleosomes (Cairns et al. 1996; Lorch et al. 1998), but its action on yeast nucleosomes has not been tested. To prepare yeast nucleosomes, recombinant versions of each of the four yeast histones were expressed in Escherichia coli and purified to homogeneity. The histones were then assembled into octamers and reconstituted into mononucleosomes using radiolabeled 5S DNA (167 bp), a strong nucleosome positioning sequence that contains a recognition site for the endonuclease DraI near the dyad of the nucleosome (Fig. 6A, left panel). When assembled into a nucleosome, the DraI site is protected from DraI cleavage by the underlying histone octamer.
Figure 6.
Effect of DNA nicks on nucleosome remodeling efficiency. Recombinant yeast octamers were assembled into radiolabeled intact or nicked mononucleosomes (167 bp) and purified as described in Materials and Methods. The 5S positioning sequence leaves 17 bp outside the nucleosome on the labeled end, and 4 bp on the unlabeled end. The nicks are positioned 23 and 33 bases upstream and downstream of the center of the DraI site (6 bp). (A) Reconstitution strategy and location of the DraI cleavage site relative to the DNA nicks. (B) Purified nucleosome substrates. Purified nucleosomes or DNA alone were separated on a 4% polyacrylamide gel and visualized by autoradiography. (C–F) Remodeling efficiency of intact and nicked nucleosomes. Where indicated, reactions contain DraI (20 units), ATP (1 mM), and nucleosomes (30 ng; 12 nM). All ATPase values provided are relative to activity with the RSC complex and intact nucleosomes (panel C, lane 8) so that direct comparisons may be made. Shown is a representative experiment, but the ATPase and cleavage values are the average of three experiments.
To assess the chromatin-remodeling activity of RSC and Sth1 on yeast nucleosomes, the ability of RSC and Sth1 to permit cleavage by DraI was monitored. We find that RSC and Sth1 render the nucleosome susceptible to DraI cleavage in an ATP-dependent manner (Fig. 6C,E, lanes 4–8), whereas reactions containing ATPγS display no cleavage (data not shown). However, Sth1 is five- to sixfold less active than RSC, consistent with our ATPase results with nucleosomes (Fig. 1C) and previous results with Brg1 (Phelan et al. 1999). Therefore, Sth1 possesses the ATPase and remodeling properties of RSC, although at reduced efficiency.
Remodeling is less efficient on nicked nucleosomes
Models for nucleosome remodeling involving DNA twisting have been proposed, as they provide an attractive mechanism for disrupting histone–DNA contacts (Havas et al. 2000; Gavin et al. 2001). Here, remodelers may remain in a fixed position on the nucleosome and rotate the DNA duplex. However, DNA helix rotation may also occur as a consequence of DNA translocation (Janscak and Bickle 2000). If the remodeler translocates by tracking along one strand and remains in a fixed position on the histone octamer, the DNA must undergo one full rotation for every 10.6 bp of translocation. The DNA exiting the translocase will be underwound, and the DNA approaching the translocase will be overwound. For a mononucleosome, the DNA in the linker region would be free to rotate (dissipating twist); however, the DNA passing through the ATPase would be constrained by the remodeler and the contacts of the DNA on the octamer surface (see Fig. 7 and Discussion). Therefore, the disruption of histone–DNA contacts by translocation may involve both a translational and a twist component.
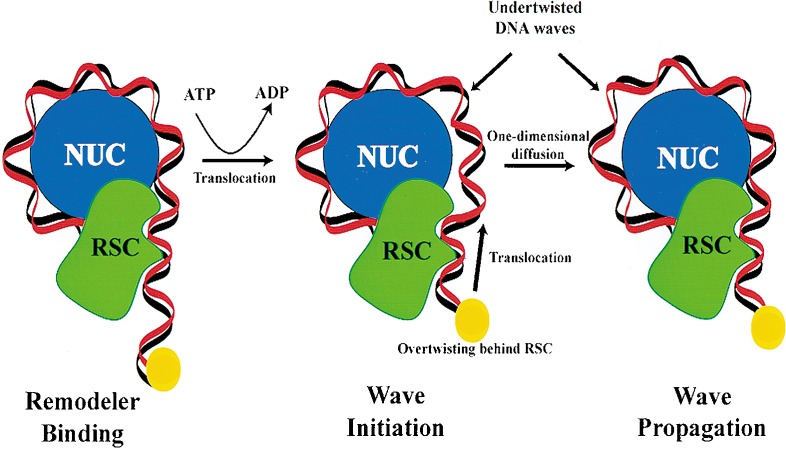
Figure 7.
A model for DNA translocation in nucleosome remodeling and mobility. RSC engages the nucleosome (for clarity, only half is shown) at or near the entry/exit site and uses the energy of ATP hydrolysis to translocate a segment of DNA, depicted by the movement of a fixed point on the DNA (yellow sphere). This segment bears both a translational and a twist component that breaks histone–DNA contacts, and may propagate through the nucleosome by one-dimensional diffusion. DNA-binding factors may have access to and bind the DNA wave during propagation. Resolution of the wave at the other entry/exit site results in a new translational position, causing octamer sliding (not shown).
To address the relative contributions of twist and translocation to the remodeling process, an assay was used that uncouples twist from translocation during the remodeling process by using nicked nucleosomes as remodeling substrates. Mononucleosomes were formed on a 167-bp 5S DNA template that contains nicks in two defined locations, positioned 23 bases upstream and 33 bases downstream of the center of the DraI site (Fig. 6A,B). These nicks are predicted to prevent the propagation of twist to the DraI site by allowing free rotation of the DNA about the phosphodiester backbone of the intact strand. The efficiency of remodeling intact or nicked substrates was quantified based on the extent of DraI cleavage in the presence of increasing amounts of RSC or Sth1. We find that at low enzyme concentrations, the presence of nicks reduces remodeling efficiency two- to threefold for both RSC and Sth1 (Fig. 6C–F, cf. lane 5 of each). However, remodeling proceeds to near completion in the presence of excess RSC (Fig. 6C,D, lane 8). Importantly, intact and nicked substrates were comparable in their ability to elicit ATPase activity over this concentration range, suggesting that although ATP hydrolysis is occurring on nicked nucleosomes, remodeling is not as efficient. Furthermore, gel shift analysis shows that RSC binds indistinguishably to intact and nicked nucleosomes (Kd ≈ 5 nM in the presence of ATP; data not shown). These results strongly suggest that DNA twist does not have to be propagated through the entire nucleosome for remodeling to occur, but does promote remodeling efficiency.
Discussion
Here, we provide three lines of evidence that RSC and its isolated ATPase Sth1 are DNA translocases: triple-helix displacement, length-dependent ATPase activity, and enhanced ATPase activity on DNA minicircles. The ability of RSC and Sth1 to displace a triple helix provides strong evidence for DNA translocation. Importantly, displacement is only observed when the triple-stranded region bears significant flanking duplex DNA, and is not observed with the isolated triple-helix region. In addition, triple-helix displacement occurs on substrates bearing a nick in one strand of the DNA near the duplex/triplex junction, arguing against a conformational change such as twist being propagated to the triplex from the duplex region. These results suggest that the enzyme initiates translocation on the duplex DNA and then displaces the third strand by invasion. The modest efficiency of this reaction is likely owing to the length of the triple-helical region (40 bp), and a possible reduction in RSC or Sth1 processivity caused by the third strand, which may impede translocation.
Also consistent with translocation, Sth1 and RSC hydrolyze ATP in proportion to DNA length. Although formally possible, we do not favor the interpretation that RSC/Sth1 displays these properties because it binds to multiple sites over a range of 80 bases, with each site adding in a uniform and additive way to ATPase velocity; multiple binding sites should result in cooperative interaction and result in a lower Km for longer substrates. However, we find that all DNA substrates regardless of length show nearly identical Km values, suggesting a single binding site. We suggest that the lack of increased ATPase activity above 85 bases reflects a processivity limit of ∼70 bases (subtracting the binding site of ∼15 bases), an interpretation supported by our experiments with DNA minicircles, described below. This length is considerably longer than the average length of DNA between nucleosomes in S. cerevisiae (∼10 bp) or metazoans (30–50 bp), raising the possibility that RSC can slide an octamer these distances without disengagement. In addition, the other subunits of the RSC complex may serve to increase binding to, or processivity on, nucleosomes. However, isolated Sth1 lacks these associated proteins and may show reduced processivity or remodeling efficiency as a consequence, which may explain the five- to sixfold reduction in remodeling efficiency of Sth1 compared to RSC.
The effectiveness of single-stranded DNA in stimulating ATPase activity strongly suggests that translocation occurs along one strand of the duplex. Still, single-stranded DNA is slightly less potent than duplex DNA, eliciting a lower Vmax (84%) and displaying an upper limit of length dependence at 75 bases. However, single-stranded DNA is much more flexible than duplex DNA, and may adopt conformations that limit the processivity of the enzyme and reduce the observed maximal ATPase velocity.
To test our DNA translocation processivity interpretation, we developed an ATPase assay using single-stranded DNA minicircles. This model predicts that with short DNA molecules, RSC/Sth1 reaches the end of the template before its normal processivity limit, terminates ATP hydrolysis, and then releases from the substrate. We reasoned that a short circular template might mimic a template of infinite length (or at least one longer than the processivity limit) as the template may simply revolve through the enzyme until it reaches its intrinsic processivity limit. The results with the circular 45-mer were clear, as the Vmax was increased two- to threefold, to levels approaching the Vmax obtained with much longer DNA templates. However, the circular 30-mer stimulated the ATPase activity of RSC/Sth1 minimally. We suggest that circular 45-mers are handled differently than 30-mers by RSC; the 30-mer may not be able to accommodate possible shape changes in RSC/Sth1 (including the DNA step size) necessary for processive translocation, as the end-to-end length is equivalent to only 15 bases.
Over the past few years, several models for nucleosome mobility and remodeling have been proposed (see above), and the three most prevalent are DNA twisting, DNA translocation, and structural alteration of the octamer. We emphasize that these models are not mutually exclusive, and that important elements from all three models may be combined to arrive at the true mechanism. One model suggests that remodelers engage the nucleosome at or near the point at which DNA enters the nucleosome, and generate a wave of DNA derived from the linker that invades the nucleosome (Lorch et al. 1998; Travers 1999; Flaus and Owen-Hughes 2001; Längst and Becker 2001). The evidence for DNA translocation presented here provides a mechanistic basis for this DNA wave. We envision RSC/Sth1 to remain in a fixed position, bound to both the histone octamer and the DNA at the entry site (Fig. 7). Indeed, previous experiments with remodelers suggest their presence at or near the entry/exit site in the absence of ATP (Lorch et al. 1998; Längst and Becker 2001). ATP hydrolysis may then initiate translocation, which causes translational movement of the DNA accompanied by DNA twist; the DNA emitting from the remodeler (between the remodeler and the first histone–DNA contact) becomes undertwisted (>10.6 bp/turn) owing to threading of the DNA through the remodeler (Fig. 7). In this region, the DNA length increases, whereas the number of turns remains constant. This combination of translation and twist provides the energy necessary for breaking the initial histone–DNA contact. The presence of RSC/Sth1 at a fixed position on the nucleosome confines the twist to the DNA between the remodeler and the histone–DNA contact, and therefore cannot be dissipated through simple rotation. This action initiates a DNA wave; a segment of DNA that cannot lie in its preferred position on the nucleosome because of either its length or twist properties.
Our experiments with nicked nucleosomes suggest that the force of translocation may be the most important component of the DNA wave. As double-stranded DNA can be considered a semirigid rod over short distances, translocation itself may provide sufficient mechanical force to break histone–DNA contacts. This creates an intermediate with a potential energy higher than the initial remodeler–nucleosome complex, with the additional energy stored in DNA topology and the broken histone–DNA contact(s). Now, the DNA that was formerly present in the linker DNA has the opportunity to bind to the recently exposed octamer contacts. This may be the basis for DNA wave propagation; histone–DNA contacts are broken at the leading edge of the wave and reformed at the lagging edge (Fig. 7). These alternative wave positions may represent conformers of roughly equal potential energy, allowing the wave to propagate by one-dimensional diffusion. Although a wave present on the nucleosome can, in principle, move in either direction, the energy in the wave can only be dissipated following exit from the nucleosome. Directionality may be imposed by the presence of RSC/Sth1 continually pumping DNA waves from one side of the nucleosome. Furthermore, the DNA wave may transiently uncover the binding site for a transcription factor, and the energy of factor binding may compensate for the energy lost through the disruption of histone–DNA contacts. Thus, the DNA wave may be trapped by the transcription factor, providing a mechanistic basis for factors to load onto the surface of a nucleosome.
Our data suggest a role for DNA twist in remodeling efficiency. However, recent experiments question the role of DNA twist in remodeling (Längst and Becker 2001). This study analyzed a different remodeler (ISWI) in a unidirectional octamer sliding assay. ISWI catalyzed efficient sliding of randomly nicked nucleosomes, and showed enhanced sliding with nucleosomes containing a single nick placed just outside of the entry/exit site (Längst and Becker 2001). Several explanations may reconcile these observations with our work and the work of others on the role of DNA twist (Havas et al. 2000; Gavin et al. 2001). For example, these assays measure different aspects of remodeling (octamer sliding versus DNA accessibility), and DNA twist may play a different role in these assays. In addition, it cannot be ruled out that nucleosomes bearing nicks are structurally identical to intact nucleosomes, and may behave differently in certain assays. Furthermore, ISWI and RSC may differ in their translocation properties and therefore generate translocated DNA segments with different length or twist properties. Indeed, ISWI and RSC behave differently in many biochemical assays (Tsukiyama and Wu 1995; Ito et al. 1997; Cairns 1998; Varga-Weisz and Becker 1998). Finally, a role for translocation and twist does not rule out an additional role for conformational changes in the octamer during the remodeling process. For example, binding of remodelers and the strain imposed on the octamer by DNA twist and translocation may simultaneously impart a conformational change to the octamer that may alter histone–DNA interactions and/or affect wave initiation or propagation. Further mechanistic work is required to understand precisely how ATP hydrolysis mobilizes nucleosomes and contributes to the important and diverse roles remodelers play in chromatin transitions and transcriptional regulation.
Materials and methods
Purification of RSC
RSC was purified from BCY211, which expresses TAP-tagged Rsc2 from the chromosomal RSC2 locus. Cells were grown in 2× YPD, and the soluble extract was used to purify RSC, essentially as described in Puig et al. (2001), with an added nucleic acid precipitation step performed with the whole-cell extract using 0.1% (v/v) polyethylenimine and 400 mM KOAc.
Purification of Sth1
An Sth1 derivative bearing a 2× Flag N-terminal tag and a 7× Histidine C-terminal tag was prepared by PCR tagging. It was fully sequenced, and then cloned into the high-copy plasmid p904.STH1, containing the galactose-controlled GAL1-10 promoter. Strain BCY243 bears GAL4 under the control of the GAL1-10 promoter integrated at the LYS2 locus, inducing high levels of the Gal4 activator in the presence of galactose (B. Cairns, unpubl.). Cultures were grown in raffinose to an OD600 of 0.8, galactose (2% w/v final) was added, and growth was continued at room temperature for 12 h. Cell harvesting and extract preparation, and nucleic acid removal were performed as described previously for the RSC complex (Cairns et al. 1996). Sth1 was purified using SP Fast-flow (Pharmacia), Ni-NTA (QIAGEN), and M2-FLAG (Sigma) columns (for details, refer to Supplementary Material at http://www.genesdev.org).
DNA-dependent ATPase assays
ATP hydrolysis was quantified by a colorimetric assay by monitoring the production of a phosphomolybdate complex in the presence of malachite green as described previously (Cairns et al. 1994). All reactions used substrates at a nucleotide concentration of 30 μM, except when the Km was determined. The double-stranded form of the phagemid BSCR (Strategene) was used as the standard substrate. For oligonucleotide sequences, refer to Supplementary Material at http://www.genesdev.org.
Immunoprecipitations
Extracts were prepared as described previously (Cairns et al. 1999). Each immunoprecipitation was performed with 300 μg of whole cell extract and 25 μL of beads for 2 h in a buffer containing 50 mM Tris-Cl (pH 7.5), 10% (v/v) glycerol, 2 mM EDTA, 100 mM NaCl, and 0.05% NP-40. Precipitates were recovered and were washed in IP buffer containing 250 mM NaCl. Proteins bound to Flag beads were recovered by incubating with Flag peptides, whereas proteins bound to IgG sepharose were recovered by cleavage with TEV protease. SDS-PAGE gels were immunoblotted to PVDF membranes and developed. Anti-Sth1 antisera (Covance Inc.) was generated in rabbits using purified full-length Sth1 protein.
DNA minicircle preparation
Single-stranded oligonucleotides of 45 (or 30) bases were end-to-end ligated using a 16-base (or 12-base) oligonucleotide complementary to the eight (or six) terminal bases with T4 DNA ligase. (For oligonucleotide, refer to Supplementary Material at http://www.genesdev.org.) The closed circular single-stranded DNA forms were separated from other products of ligation using a 7.5% denaturing polyacrylamide gel and purified by gel extraction using standard methods. The integrity of the DNA minicircles was verified by resistance to ExoVII (GIBCO-BRL). Linear or purified circular DNA (300 ng) was exposed to 1 U of ExoVII at 37°C overnight, separated on an 7.5% polyacrylamide gel, and visualized by ethidium bromide staining.
Triple-helix displacement assay
All double-stranded DNAs for triple-helix construction contain a 40-bp d(GA) · d(TC) tract. The intact and nicked end-positioned substrates (114 bp) and the isolated triple helix (40 bp) were prepared by hybridizing complimentary oligonucleotides. Nicked end-positioned substrate used a nonphosphorylated oligonucleotide that was not ligated, leaving a nick 4 bases from what will become the duplex/triplex junction. The intact substrate used a phosphorylated oligonucleotide and was ligated using T4 ligase. The substrates were analyzed using a denaturing polyacrylamide gel to ensure integrity, and were purified following electrophoresis in 15% native polyacrylamide gel. Double-stranded DNA for center-positioned triplexes (190 bp) was prepared by PCR using a BSCR plasmid containing the a centrally located 40-bp d(GA) · d(TC) tract. The third strand, consisting of a 40-nt single-stranded homopyrimidine repeat (dTC)20, was end-labeled with [γ-32P]ATP at the 5′ end. Substrates were formed by incubating the labeled (dTC)20 with a twofold molar excess of double-stranded DNA, in a buffer containing 33 mM Tris-acetate (pH 5.5), 66 mM potassium acetate, 100 mM NaCl, 10 mM MgCl2, and 0.4 mM spermine (Kopel et al. 1996). Triple-helix displacement assays were carried out at 30°C in a reaction buffer containing 36 mM Tris-acetate (pH 7.3), 20 mM KOAc, 8 mM MgCl2, 5% (v/v) glycerol, 0.5 mM DTT, and (when indicated) 1 mM ATP. Reactions (25 μL) contained 30 ng of triple-helix DNA and 80 ng (18 nM) of Sth1 or 200 ng (8 nM) of RSC. After 30 min, reactions were quenched with 0.5 % SDS, 15 mM EDTA, 3% (v/v) glycerol, 2 mM Tris-HCl (pH 8.0), and 25 ng of unlabeled (dTC)20. Products were resolved on a 15% native polyacrylamide gel and analyzed using Image Quant (Molecular Dynamics Inc.). For oligonucleotide sequences, refer to Supplementary Material at http://www.genesdev.org.
Histone expression and octamer reconstitution
The yeast histone ORFs HHT1, HHF1, HTA1, and HTB1 were PCR-amplified (for primer sequences, refer to Supplementary Material at http://www.genesdev.org) and cloned into pET11a (Stratagene). Clones were verified by DNA sequencing. Proteins were overexpressed in BL21-Codonplus RIL cells (Stratagene) with 0.2 mM IPTG at an OD600 of 0.4 for several hours. Histones were purified from inclusion bodies and reconstituted into octamers as described (Luger et al. 1999), except the size-exclusion chromatography step was eliminated from the histone purification.
Nucleosome assembly
The 167-bp EcoRI–ScaI fragment from pIC5x207(EX) (Meersseman et al. 1991) was subcloned into pBSCR.KS (Stratagene), forming p872.5S, which was used as a template to PCR-amplify (for primer sequences, refer to Supplementary Material at http://www.genesdev.org) the Xenopus 5S RNA gene, with one primer radiolabeled with [γ-32P]ATP. For nicked nucleosomes, nicks were placed at defined positions by hybridization of three oligonucleotides (70 bases, 56 bases, and 41 bases) to an intact complementary radiolabeled 167-base single strand of DNA (derived by single-stranded rescue and cleavage of BSCR). The nicks were positioned 23 and 33 bases upstream and downstream of the center of the DraI site. Histidine-tagged yeast Nap1 (yNAP1) was overexpressed from pET28 in BL21-Codonplus RIL cells (Stratagene) as described for the histones, and purified chromatographically using Ni2+-NTA agarose (QIAGEN) and Q-Sepharose (Pharmacia). Nucleosomes were reconstituted by mixing equimolar amounts (determined by A280, A260, and gel analysis) of yNAP1, histone octamer, and DNA in 10 mM HEPES (pH 7.6), 50 mM KCl, 5 mM MgCl2, 10% (w/v) glycerol, and 0.5 mg/mL BSA with rotation at 30°C for 4 h. Supernatants were loaded onto a 12-mL sucrose gradient (10%–30%) and centrifuged in a SW41 rotor at 41,000 rpm for 24 h. Fractions were analyzed on a 4% native gel (15:1 acryl:bis) containing 2 mM MgCl2, 5% (w/v) glycerol in 0.5× TBE; nucleosome-containing fractions were pooled and concentrated using a Centricon-10 (Amicon).
Remodeling assays
Remodeling reactions containing 30 ng of nucleosomal DNA in a buffer of 20 mM Tris-acetate (pH 7.9), 50 mM KOAc, 10 mM MgOAc, 1 mM DTT, and 0.1 mg/mL BSA, with or without 1 mM ATP and 20 U of DraI, were allowed to proceed at 30°C for 1 h. The reactions were then quenched with 10 mM EDTA, followed by DNA extraction and electrophoresis on an 8% polyacrylamide gel. DraI cutting was quantified using PhosphoImager analysis.
Yeast strains
The strain for RSC isolation was BCY211: ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 pep4Δ∷HIS3 prb1Δ∷LEU2 RSC2-TAP∷TRP1 prcΔ∷HISG cir0. The strain for Sth1 isolation was BCY243: ade2-1 can1-100 his3-11,15 leu2-3,112 lys2Δ trp1-1 ura3-1 cir+ pep4Δ∷HIS3 prb1Δ∷LEU2 bar1Δ∷HISG lys2∷Gal1/10-GAL4 with plasmid [p904.STH1]. The strains for determining the oligomeric state of Sth1 were BCY242: ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 pep4Δ∷HIS3/pep4Δ∷HIS3 prb1Δ∷LEU2/prb1Δ∷LEU2 STH1-TAP∷TRP1/STH1 prc1Δ∷HISG/PRC lys2∷GAL1-10-GAL4/LYS2 with plasmid [p1170.STH1]; BCY240: ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 pep4Δ∷HIS3 prb1Δ∷LEU2 lys2∷GAL1/10-GAL4 containing plasmid [p1170.STH1]; and BCY241: ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 pep4Δ∷HIS3/pep4Δ∷HIS3 prb1Δ∷LEU2/prb1Δ∷LEU2 STH1-TAP∷TRP1/STH1 prc1Δ∷HISG/PRC1 lys2∷GAL1-10-GAL4/LYS2.
Acknowledgments
We thank Bob Schackmann for oligonucleotide syntheses, Roger Kornberg for the TAP-tagged Rsc2 strain, Janet Lindsley and James Stray for the GAL1-regulated expression plasmids, Voot Yin for the cloning of Flag-tagged Sth1, Toshio Tsukiyama for the yNAP1 expression plasmid and octamer preparation protocol, and Tim Formosa for comments on the manuscript. B.R.C. is an Assistant Investigator with the Howard Hughes Medical Institute and an Investigator at the Huntsman Cancer Institute. This work was funded by the National Institutes of Health GM60415 (A.S.), the Huntsman Cancer Institute (J.W. and CA24014 for the support of core facilities), and the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL brad.cairns@hci.utah.edu; FAX (801) 585-6410.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.995002.
References
- Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J Biol Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling machines: Similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- Cao Y, Cairns BR, Kornberg RD, Laurent BC. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Längst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- Côté J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- Côté J, Peterson CL, Workman JL. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: Measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- ————— Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry. 2002;41:643–651. doi: 10.1021/bi011137k. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- Gavin I, Horn PJ, Peterson CL. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol Cell. 2001;7:97–104. doi: 10.1016/s1097-2765(01)00158-7. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Janscak P, Bickle TA. DNA supercoiling during ATP-dependent DNA translocation by the type I restriction enzyme EcoAI. J Mol Biol. 2000;295:1089–1099. doi: 10.1006/jmbi.1999.3414. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kingston RE, Narlikar GJ. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- Kopel V, Pozner A, Baran N, Manor H. Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res. 1996;24:330–335. doi: 10.1093/nar/24.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- Längst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- Längst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Laurent BC, Yang X, Carlson M. An essential Saccharomyces cerevisiaegene homologous to SNF2 encodes a helicase-related protein in a new family. Mol Cell Biol. 1992;12:1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC–nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- ————— RSC unravels the nucleosome. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- Mechanic LE, Hall MC, Matson SW. Escherichia coliDNA helicase II is active as a monomer. J Biol Chem. 1999;274:12488–12498. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- Meersseman G, Pennings S, Bradbury EM. Chromatosome positioning on assembled long chromatin. Linker histones affect nucleosome placement on 5S rDNA. J Mol Biol. 1991;220:89–100. doi: 10.1016/0022-2836(91)90383-h. [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Workman JL. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:403–441. [PubMed] [Google Scholar]
- ————— Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 1996;15:4702–4712. [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol Cell Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Soultanas P, Dillingham MS, Wiley P, Webb MR, Wigley DB. Uncoupling DNA translocation and helicase activity in PcrA: Direct evidence for an active mechanism. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. Chromatin modification by DNA tracking. Proc Natl Acad Sci. 1999;96:13634–13637. doi: 10.1073/pnas.96.24.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I, Carlson M. Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:1768–1775. doi: 10.1128/mcb.17.4.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Uno M, Kiguchi A, Masuoka K, Kanemori Y, Okabe S, Mikayawa T. The Saccharomyces cerevisiae NPS1 gene, a novel CDC gene which encodes a 160 kDa nuclear protein involved in G2phase control. EMBO J. 1992;11:4017–4026. doi: 10.1002/j.1460-2075.1992.tb05495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- van Dongen MJ, Doreleijers JF, van der Marel GA, van Boom JH, Hilbers CW, Wijmenga SS. Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix. Nat Struct Biol. 1999;6:854–859. doi: 10.1038/12313. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB. Chromatin-remodeling factors: Machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes & Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- Xue Y, Canman JC, Lee CS, Nie Z, Yang D, Moreno GT, Young MK, Salmon ED, Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga SK, Peterson CL, Herskowitz I, Yamamoto KR. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- Young MC, Schultz DE, Ring D, von Hippel PH. Kinetic parameters of the translocation of bacteriophage T4 gene 41 protein helicase on single-stranded DNA. J Mol Biol. 1994;235:1447–1458. doi: 10.1006/jmbi.1994.1100. [DOI] [PubMed] [Google Scholar]