Abstract
Pax6 is a pivotal regulator of eye development throughout Metazoa, but the direct upstream regulators of vertebrate Pax6 expression are unknown. In vertebrates, Pax6 is required for formation of the lens placode, an ectodermal thickening that precedes lens development. Here we show that the Meis1 and Meis2 homeoproteins are direct regulators of Pax6 expression in prospective lens ectoderm. In mice, Meis1 and Meis2 are developmentally expressed in a pattern remarkably similar to Pax6 and their expression is Pax6-independent. Biochemical and transgenic experiments reveal that Meis1 and Meis2 bind a specific sequence in the Pax6 lens placode enhancer that is required for its activity. Furthermore, Pax6 and Meis2 exhibit a strong genetic interaction in lens development, and Pax6 expression is elevated in lenses of Meis2-overexpressing transgenic mice. When expressed in embryonic lens ectoderm, dominant-negative forms of Meis down-regulate endogenous Pax6. These results contrast with those in Drosophila, where the single Meis homolog, Homothorax, has been shown to negatively regulate eye formation. Therefore, despite the striking evolutionary conservation of Pax6 function, Pax6 expression in the vertebrate lens is uniquely regulated.
Keywords: Pax6, Meis1, Meis2, lens
The central, evolutionarily conserved role of Pax6 in eye development is illustrated by its mutational analysis in numerous organisms. Humans carrying PAX6 loss-of-function alleles exhibit a range of ocular phenotypes including aniridia, anophthalmia, and Peters' anomaly (Ton et al. 1991; Glaser et al. 1992; Hanson et al. 1994). In Drosophila, loss-of-function mutations in one of two Pax6 genes, eyeless (ey), result in the complete absence of photoreceptor formation (Quiring et al. 1994), while Toy, the protein encoded by the second fly Pax6 gene, twin of eyeless (toy), directly regulates ey by binding to the ey eye-specific enhancer (Czerny et al. 1999). Strikingly, when misexpressed in their respective organisms, toy, ey, and Xenopus Pax6 can each initiate the entire molecular cascade culminating in ectopic eye formation (Halder et al. 1995; Chow et al. 1999; Czerny et al. 1999). Although Toy directly regulates ey, the gene duplication that created two Drosophila Pax6 genes appears specific to insect evolution (Czerny et al. 1999). Hence, this regulatory model does not extrapolate to vertebrates, and the direct molecular regulators of vertebrate Pax6 expression remain unknown.
In vertebrates, the function of Pax6 in lens development has been revealed by the mouse Small eye (Sey) Pax6 null mutation. In Sey/Sey homozygotes, eye development arrests after formation of the optic vesicle but before lens placode induction (Hogan et al. 1988; Hill et al. 1991). In addition, tissue-specific gene targeting and embryologic experiments reveal that Pax6 functions autonomously in the prospective lens ectoderm for lens placode formation (Fujiwara et al. 1994; Ashery-Padan et al. 2000; van Raamsdonk and Tilghman 2000). Collectively, these studies indicate that, during vertebrate oculogenesis, the transition from pre-placodal ectoderm to lens placode is one of the earliest developmental steps for which Pax6 function is required.
Although the direct upstream molecular regulators of Pax6 expression are unknown, several genes have been shown to act earlier than Pax6 in regulating vertebrate eye development. Abrogation of Otx2, Rx, or Xtll in each case results in a failure in optic vesicle development (Acampora et al. 1995; Mathers et al. 1997; Hollemann et al. 1998). In Lhx2 mutant mice, the optic vesicle fails to contact the surface ectoderm, and Pax6 is not expressed in Lhx mutant presumptive lens ectoderm (Porter et al. 1997). However, these genes are either expressed prior to Pax6 in head ectoderm (Otx2), or are expressed only in the optic vesicle (Lhx, Rx, Xtll), and therefore are likely to affect Pax6 only indirectly.
The inhibition of several signaling cascades is also known to disrupt vertebrate eye development. For example, Pax6 expression is initiated in the prospective lens placode ectoderm of Bmp7−/− mouse embryos, but expression is not maintained (Wawersik et al. 1999). This is reminiscent of the function of the Drosophila Bmp family member, decapentaplegic (dpp), which is required for early patterning events in eye imaginal discs in the fly (Chen et al. 1999). In addition, disruption of fibroblast growth factor (FGF) signaling activity reduces Pax6 lens placodal expression in the mouse, and genetic evidence suggests that the FGF and BMP7 pathways work synergistically to regulate Pax6 during lens development (Faber et al. 2001). Other signaling pathways are also implicated. An antisense study has shown that depletion of retinoic acid by inhibition of retinal binding protein results in a failure of lens placode formation, similar to the Sey phenotype (Bavik et al. 1996). In addition, Wingless (wg), a Wnt family protein, is involved in the correct initiation of the morphogenetic furrow in Drosophila eye development (Treisman and Rubin 1995), and misexpression of a vertebrate Wnt receptor, Xfz3, leads to ectopic Pax6 expression and eye formation in Xenopus (Rasmussen et al. 2001). Lastly, injection of insulin-like growth factor (IGF) RNA into Xenopus oocytes promotes anterior neural fate and also induces ectopic eyes (Pera et al. 2001).
In medaka fish, Sox3 overexpression induces Pax6 expression and leads to a transformation of ectodermal tissue into lens (Köster et al. 2000), and ectopic lens and retina form when medaka fish and Xenopus blastomeres are injected with Six3 RNA or Six6 RNA, respectively (Oliver et al. 1996; Bernier et al. 2000). However, while Pax6 expression is induced in these ectopic ocular structures, Six3 and Six6 are unlikely to directly activate Pax6 expression, because thus far both have been shown to act mainly as transcription repressors in eye development (Zuber et al. 1999; Kobayashi et al. 2001; Zhu et al. 2002).
Therefore, in contrast to information about factors that reside genetically upstream of Pax6, our knowledge about the specific transcription factors that directly regulate Pax6 expression in the developing eye remains limited. One approach to this problem is to focus on the molecular analysis of tissue-specific Pax6 enhancers that have been identified by studies in transgenic mice (Xu et al. 1999). Recent work has uncovered a number of such enhancers, including a specific 107-bp minimal element residing 3.7 kb upstream of the Pax6 P0 promoter, which directs a dynamic expression in the prospective mouse lens (Williams et al. 1998; Kammandel et al. 1999). This enhancer becomes active as the lens placode develops from the head ectoderm, and its activity persists into adulthood in lens and corneal epithelium.
In vertebrates, including human, mouse, quail, and fugu fish, this 107-bp element is remarkably conserved not only in sequence but also in function (Williams et al. 1998; Kammandel et al. 1999; Plaza et al. 1999). For example, a 12-kb fugu fish sequence containing this element exhibits lens enhancer activity in transgenic mice (Kammandel et al. 1999). In addition, the functional significance of this enhancer element is further demonstrated by genetic deletion experiments, which result in diminished Pax6 expression as well as lens developmental defects (Dimanlig et al. 2001). We have sought to dissect the genetic hierarchy of eye specification by focusing on the direct molecular regulators of Pax6 expression. Our results demonstrate that the TALE (three amino acid loop extension) homeoproteins, Meis1 and Meis2, directly bind to the Pax6 lens placode enhancer and control Pax6 expression during early vertebrate lens induction.
Results
Identification of the Meis homeoproteins that bind to the Pax6 lens enhancer
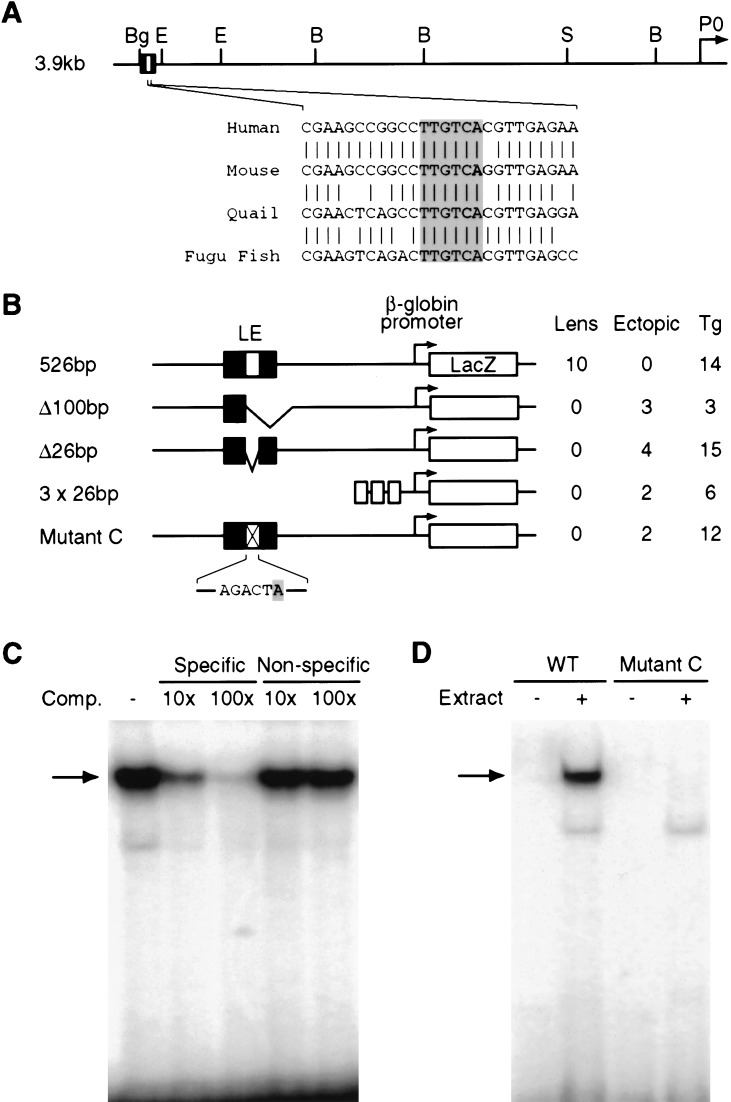
Using transient transgenic analyses in embryonic day E12.5 mouse embryos, we identified a 26-bp sequence within the 107-bp Pax6 lens enhancer element that is necessary but not sufficient for embryonic Pax6 lens and corneal expression (Fig. 1A,B). This same 26-bp sequence was also capable of binding factors present in E12.5 mouse embryonic head nuclear extracts in an electrophoretic mobility shift assay (EMSA; Fig. 1C). In addition, mutational analysis of the 26-bp fragment revealed that five evolutionarily conserved nucleotides were necessary for both gel shift complex formation and for lacZ transgene expression in the developing lens and cornea (Fig. 1B, D, mutant C). This result suggested that the nuclear factor binding these 5 bp in vitro and the factor required for Pax6 lens placode enhancer activity in vivo were potentially the same.
Figure 1.
Identification of a 26-bp sequence essential for Pax6 lens ectoderm expression. (A, top) The mouse Pax6 locus extending 3.9 kb upstream of the Pax6 P0 promoter, along with the previously determined 107-bp minimal lens ectoderm enhancer located between −3803 and −3696 bp upstream of the mouse Pax6 P0 promoter (Williams et al. 1998; Kammandel et al. 1999). (Bottom) Conservation of the 26-bp sequence (−3759 to −3734 bp) within the 107-bp minimal element, including six invariant base pairs (shaded) comprising the Meis homeodomain binding site. (B) Transient transgenic mouse analysis of the cis-regulatory requirements for Pax6 lens ectoderm expression. Constructs and the number of independent transgenic lines (Tg) exhibiting lens ectoderm expression (Lens) and/or ectopic expression (Ectopic) are shown. The 107-bp minimal lens ectoderm enhancer resides in the center of the 526-bp transgene sequence (−3969 to −3443 bp). These experiments identify 26 bp that are necessary (e.g., construct Δ26 bp) but not sufficient (e.g., construct 3 × 26 bp) for 526-bp fragment lens enhancer activity in vivo. Five of the six base pairs in the invariant Meis binding site were altered in mutant C, which also fails to support lens ectoderm expression in vivo. (C) Specific DNA complex formation on the 26-bp sequence with nuclear extracts prepared from E12.5 mouse embryonic heads. Complex formation (arrow) is competed by specific but not by non-specific oligonucleotides at 10× and 100× molar ratios. (D) Concordant with the transgenic experiments in B, nuclear extracts bind the wild-type 26-bp probe (WT) but not the mutant C probe.
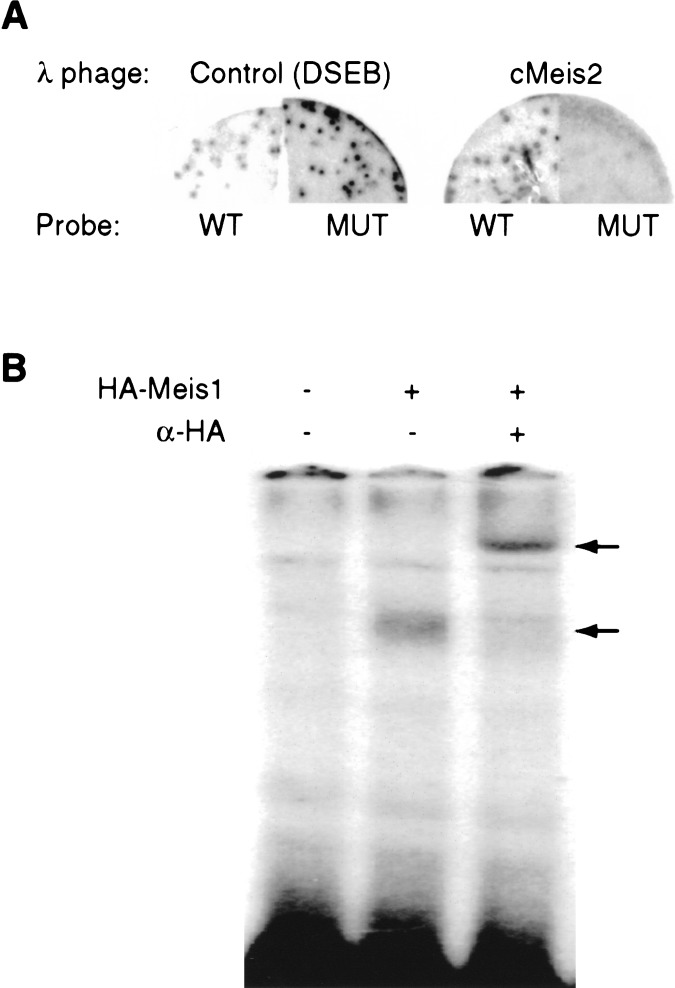
To identify the responsible factor, the 26-bp element was used to screen a λgt11 chicken embryonic lens cDNA expression library, resulting in the isolation of a cDNA clone encoding the chicken TALE homeoprotein Meis2 (Moskow et al. 1995; Nakamura et al. 1996). Whereas phage clones expressing DSEB (a nonspecific DNA binding protein) bound both wild-type and the 5-bp substitution mutant C probes equally well, Meis2-expressing clones only bound the wild-type sequence (Fig. 2A). Because the critical 5-bp are part of the consensus DNA-binding sequence for Meis1, 5′-TGACA(G/A)-3′ (Chang et al. 1997), we examined whether Meis1 could also bind the 26-bp lens placode enhancer. When in vitro translated Meis1 bearing an HA epitope tag was tested for binding to the 26-bp DNA probe, specific binding was observed. In addition, when an anti-HA antibody was added to the gel shift reactions, a more abundant supershift complex formed that ran with the expected lesser mobility (Fig. 2B). Therefore, both Meis1 and Meis2 can bind the 26-bp Pax6 lens placode enhancer in vitro.
Figure 2.
Meis1 and Meis2 binding to the 26-bp Pax6 lens element sequence. (A) After blotting λgt11 clones to nitrocellulose and probing with wild-type or mutant radioactive ds-oligonucleotides, phage clones expressing DSEB (a non-specific DNA binding protein; Stuempfle and Floros 1997) bind both wild-type and mutant C probes (left), whereas chicken Meis2-expressing clones specifically bind the wild-type 26-bp probe but not the mutant C probe (right). (B) Mouse Meis1 protein binds the 26-bp element. HA-tagged Meis1 forms a gel-shift complex with the 26-bp probe (lower arrow), that is supershifted (upper arrow) when anti-HA antibody is added.
Meis1 and Meis2 are expressed in the lens placode independent of Pax6
To ascertain whether Meis1 and Meis2 were expressed at the correct time and place to be endogenous regulators of Pax6, we performed whole-mount RNA in situ hybridizations and section immunohistochemistry in E9.5–E10.5 mouse embryos. Meis1 was strongly expressed in the developing lens placode and surrounding ectoderm, whereas Meis2 was expressed at much lower levels (Fig. 3A). No signals were detected in the eye for three other TALE homeodomain genes, Meis3, Prep1, and TGIF, despite strong expression elsewhere (data not shown). Antibodies specific to Meis1 or to Meis2 detected similar levels of both proteins in lens pre-placodal and placodal ectoderm, and the combined expression patterns of both Meis proteins matches that of Pax6 in the lens and cornea from E9.5 to E12.5 (Fig. 3B; data not shown). The specificity of the Meis1 and Meis2 antibodies was demonstrated by their distinct immunohistochemical patterns in hindbrain and nasal placode, respectively (Fig. 3C). Moreover, in contrast to genes such as Six3, Msx2, and Dach1 whose expression in pre-lens placodal ectoderm is Pax6-dependent (Ashery-Padan et al. 2000; Purcell 2001), Meis1 and Meis2 pre-placodal ectodermal expression is Pax6-independent, as the ectodermal expression of both proteins was maintained in Sey1Neu/Sey1Neu embryos null for Pax6 function (Fig. 3B). This molecular epistasis places Meis expression either upstream of or parallel to Pax6 in the lens-forming regulatory hierarchy.
Figure 3.
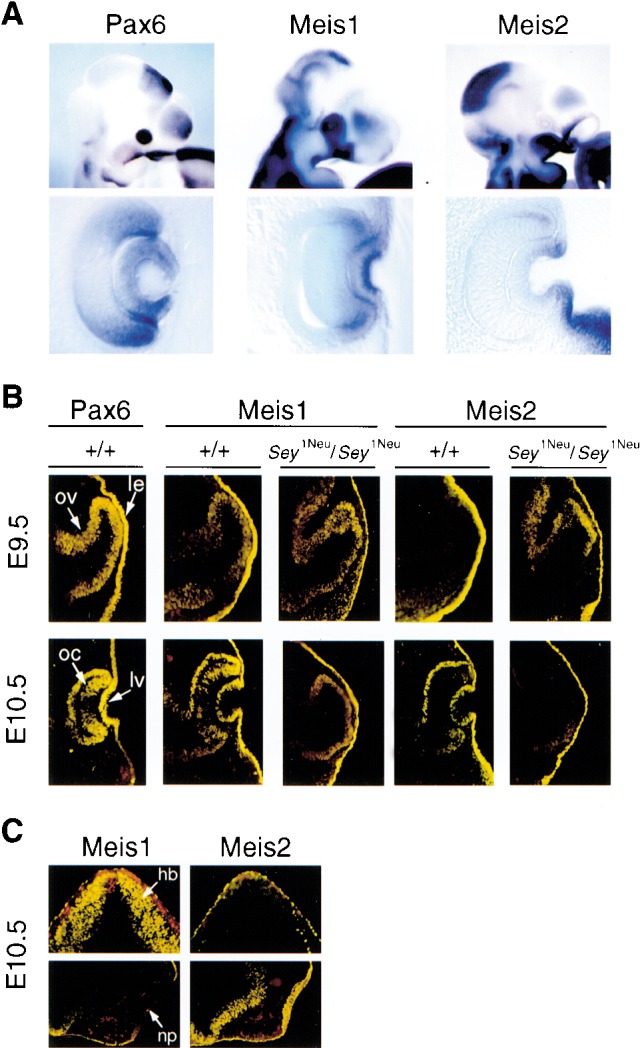
Meis1 and Meis2 are expressed in prospective lens placode ectoderm. (A) Whole-mount mRNA in situ hybridization shows strong Meis1 expression in the eye in E10.5 mouse embryos. Note that the Meis1 ocular expression domain extends beyond the Pax6 expression domain, while Meis2 is expressed at much lower levels in the eye at this developmental stage. (B) Meis1 and Meis2 proteins are expressed in the lens ectoderm of wild-type and Sey1Neu/Sey1Neu embryos at both E9.5 and E10.5 (yellow immunofluorescence). Optic vesicle and optic cup expression of Meis1 and Meis2 is also retained in Sey1Neu/Sey1Neu embryos. (oc) Optic cup; (ov) optic vesicle; (le) prospective lens ectoderm; (lv) lens vesicle. (C) Specificity of the Meis1 and Meis2 antibodies. Antibodies directed against Meis1 and Meis2 were tested on adjacent E10.5 mouse cryosections by immunohistochemistry. Meis1 but not Meis2 is strongly expressed in hindbrain, whereas Meis2 but not Meis1 is strongly expressed in nasal placode. These and other results (Toresson et al. 2000) support the specificity of these antibodies. (hb) Hindbrain; (np) nasal placode.
Meis binding is required for the Pax6 lens enhancer activity
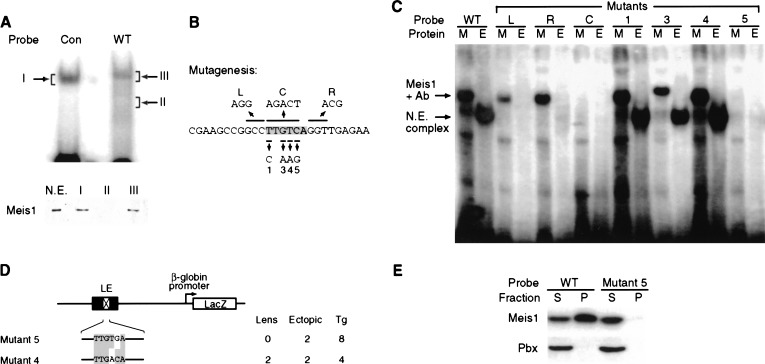
Next, we sought to establish whether Meis was a component of the EMSA complex identified when E12.5 mouse embryonic head nuclear extracts bound the 26-bp lens ectoderm regulatory element. Following gel shift analysis and complex detection by autoradiography, regions of the gel corresponding to either the shifted complex or areas with no complex were excised (Fig. 4A, top), eluted, and analyzed by SDS-PAGE followed by Western blotting using the anti-Meis1 polyclonal antibody (Fig. 4A, bottom). This experiment indicated that Meis1 was indeed present in the EMSA complex that forms when nuclear extracts are incubated with the 26-bp sequence.
Figure 4.
Meis1 binds the 26-bp element in a higher order nuclear complex to regulate Pax6 lens ectoderm expression. (A) Meis1 is part of a nuclear complex that forms on the Pax6 26-bp probe. (Top) EMSA was performed after nuclear extracts were incubated with either a 26-bp probe containing a Pbx/Meis consensus site (Con; Chang et al. 1997) or with the 26-bp wild-type Pax6 probe (WT). (Bottom) Western blot showing Meis1 is present in eluates from gel slices containing complexes (I and III), but not from a control gel slice (II). Nuclear extract (N.E.) was included as a positive control. (B) Scheme showing design of point mutations in the 26-bp sequence. The Meis binding site is shaded. Mutations were designed based on Chang et al. (1997) and behaved as expected, with the exception of mutant 4 which behaved permissively in our experiments. (C) Mutational analysis of the Meis binding site and its flanking sequences. Probes corresponding to wild-type (WT) and mutant 26-bp sequences were tested in EMSA for complex formation with in vitro translated HA tagged Meis1 plus anti-HA antibody (M) or with nuclear extract (E). [Anti-HA antibody was added because the Meis–DNA complex appears stronger when bound by antibody (Fig. 2B). This explains the slower mobility of Meis1 complexes relative to those involving nuclear extracts, which include Meis1 plus other components.] Mutants L and R abrogate nuclear extract complex formation but not Meis binding, whereas mutant C abolishes both. Point mutations in the Meis binding site (mutants 1, 3, and 4) have little or no effect on binding except for mutant 5, which (similar to mutant C) abolishes both Meis1 binding and nuclear extract complex formation. (D) Transgenic analysis of mutants 4 and 5 confirms that Meis binding is required for activity of the Pax6 lens ectoderm enhancer in vivo. The number of independent transgenic lines (Tg) exhibiting lens ectoderm expression (Lens) and/or ectopic expression (Ectopic) is shown. (E) Pbx1–3 are not present in the nuclear extract complex that binds the 26-bp element. When nuclear extract complexes were precipitated with the 26-bp probe (WT), only Meis1 is detected in the pellet (P), with Pbx1–3 immunoreactivity remaining in the supernatant (S). Mutant 5 precipitates neither Meis nor Pbx1–3.
To test whether Meis bound DNA in the EMSA complex, we conducted a detailed mutational analysis of the Meis-binding site. In addition to the 5-bp substitution of the Meis DNA-binding site (mutant C), we also engineered individual substitutions in four of the six bases comprising the Meis site (mutants 1, 3, 4, and 5; Fig. 4B). When tested by EMSA, an exact concordance was observed between mutations that interfere with Meis1 binding (mutants C and 5) and those that abolish complex formation (Fig. 4C). Conversely, individual nucleotide substitutions in the Meis site that failed to affect Meis1 binding (mutants 1, 3, and 4) also failed to affect complex formation. We therefore conclude that formation of the nuclear extract complex detected by EMSA involves direct sequence specific DNA recognition by Meis.
Next, we investigated whether specific Meis binding is also necessary for Pax6 lens enhancer activity in vivo. Based on our in vitro results, we chose to test mutants 4 and 5, two adjacent point mutations with opposite effects on Meis binding. By site directed mutagenesis, we made the same changes (T → A for mutant 4, C → G for mutant 5) in a 526-bp Pax6 fragment containing the lens ectoderm enhancer. As predicted, none of the eight transient transgenic mice carrying the disruptive mutant 5 showed reporter expression in the lens, whereas two transient transgenic lines harboring the permissive mutant 4 completely preserved lens enhancer activity (Fig. 4D). These experiments provide compelling evidence that Meis, acting by direct DNA binding, is an endogenous regulator of the Pax6 lens ectoderm regulatory element.
Although Meis is essential for formation of a nuclear extract complex on the 26-bp element, other factors also appear to be present in the EMSA complex. When residues immediately flanking the Meis binding site were changed (Fig. 4B, mutants L and R), both mutant DNA probes supported binding by Meis1 but not by the nuclear extract (Fig. 4C). This result indicates that formation of the complex detected by EMSA likely involves additional components binding each side of the Meis site. Members of the Pbx and Hox families are currently the only known binding partners for Meis, but the 26-bp sequence does not contain DNA sequence motifs previously shown to function as Hox and Pbx binding sites (Chang et al. 1997; Mann and Affolter 1998). To further test whether Pbx1, Pbx2, or Pbx3 was part of the Meis-containing complex, we precipitated the complex from E12.5 mouse embryonic head nuclear extracts using the 26-bp probe coupled to agarose beads (Fig. 4E). Meis1 was present in the pellet brought down by the wild-type but not the mutant 5 probe, whereas for both probes Pbx1–3 immunoreactivity remained in the supernatant (Fig. 4E). Therefore, the Meis complex on this sequence element appears to involve novel cofactors other than Pbx.
Modulation of Meis activity alters Pax6 lens expression in vivo
To test the functional significance of the Meis-Pax6 regulatory interaction in vivo, we undertook two additional experimental strategies. First, we prepared permanent lines of transgenic mice expressing Meis1 or Meis2 under the control of the 526-bp Pax6 fragment containing the lens element (Fig. 5A). Significantly, 2 out of 2 lines of Meis2-overexpressing transgenic mice exhibited cataracts (Fig. 5B), a phenotype also observed in Pax6-overexpressing mice (Schedl et al. 1996). In contrast, 3 out of 3 lines of Meis1 transgenic mice did not exhibit an ocular phenotype, despite comparable levels of transgene expression. Indeed, when pools of E13.5 lens RNA from wild-type and Meis2-overexpressing transgenic mice were analyzed by quantitative real-time RT–PCR, the lens RNA isolated from the Meis2 transgenic mice contained 2.3 ± 0.3-fold (mean ± S.D.) more Pax6 transcripts than that from wild-type mice (Fig. 5C).
Figure 5.
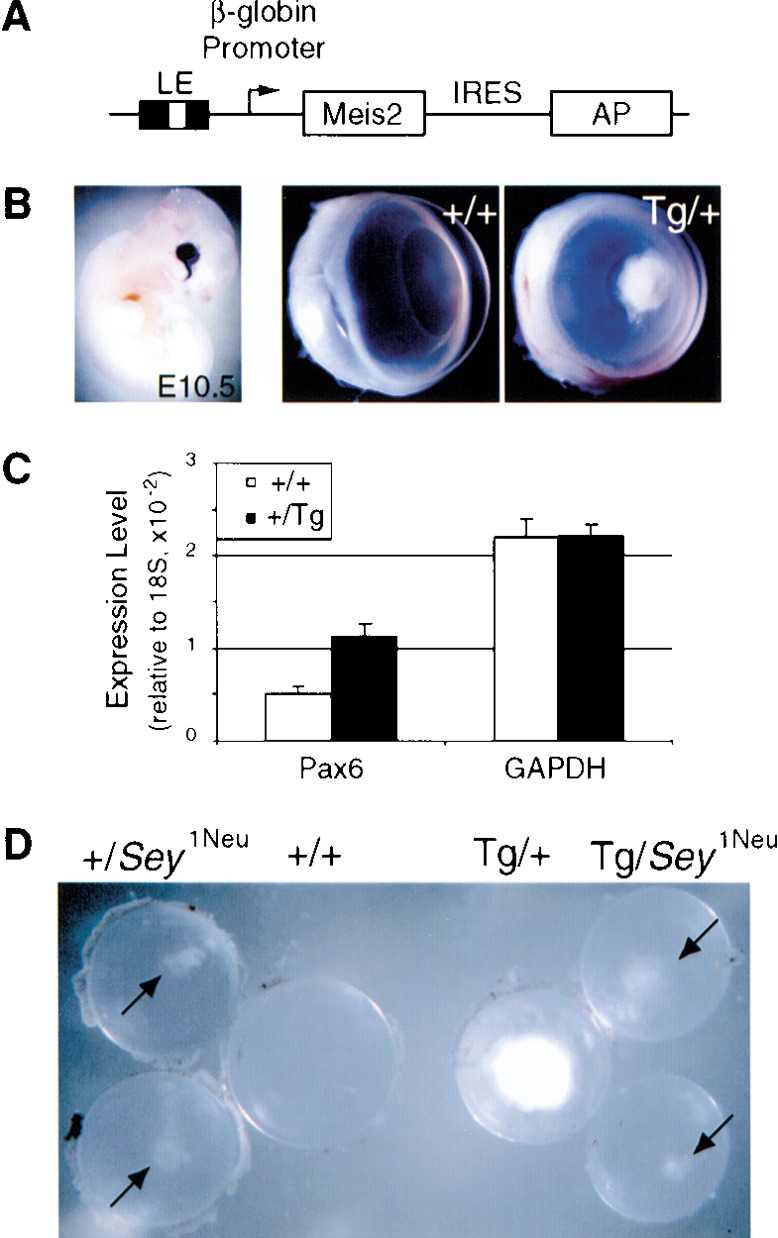
Pax6–Meis2 genetic interaction. (A) Construct for Meis2 overexpression in embryonic lens ectoderm. (LE) 526-bp Pax6 fragment containing the lens ectoderm enhancer; (β-globin) human β-globin promoter; (IRES) internal ribosome entry site; (AP) alkaline phosphatase reporter gene. (B) Transgene activity was scored at E10.5 by AP activity (left). Wild-type and Meis2 overexpression phenotypes are shown (middle and right, respectively). Cataracts were clearly visible in 1-month-old FVB/N × C3H/HeN Meis2 transgenic mice. Cataracts are also present in the FVB/N strain in which the transgene was generated (data not shown). (C) RNA quantitation by real time RT–PCR shows that Pax6 expression is elevated by 2.3 ± 0.3-fold (mean ± S.D.) in E13.5 lenses collected from Meis2-transgenic embryos relative to wild-type lenses, while GAPDH expression remains constant. Expression levels are shown relative to 18S RNA expression. (D) Lenses from 1-month-old F1 littermates from aSey1Neu/+ × Tg-Meis2/+ intercross, showing suppression of Tg-Meis2 cataracts in a Sey1Neu/+ background. Genotypes are indicated above the lenses.
Next, to test for a genetic interaction between Meis2 and Pax6, we intercrossed the FVB/N inbred Tg-Meis2 line 4979 with C3H/HeN inbred Sey1Neu/+ mice haploinsufficient for Pax6. The FVB/N × C3H/HeN F1 offspring should be heterozygous at all loci and hence genetically uniform with respect to background. One hundred percent of F1 mice carrying the Tg-Meis2 allele alone exhibited dense cataracts (n = 30), whereas the phenotypic severity in all Tg-Meis2, Sey1Neu/+ mice (n = 7) was dramatically reduced (Fig. 5D). The attenuation of the Meis2 overexpression phenotype by reduced Pax6 gene dosage further supports a model whereby Meis2 overexpression causes cataracts by elevating Pax6 expression.
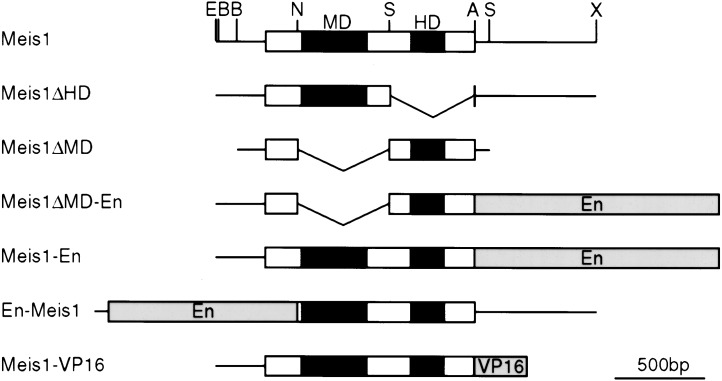
In a second approach, murine Meis1a cDNA constructs, analogous to known dominant-negative Drosophila and Xenopus Meis isoforms (Dibner et al. 2001; Inbal et al. 2001), were introduced into the chicken embryonic eye-forming region together with a GFP expression plasmid by in ovo electroporation (Fig. 6), and cells that had taken up the Meis/GFP expression plasmids (GFP+) were analyzed for Pax6 protein expression by confocal microscopy. The electroporation process itself does not interfere with Pax6 expression, as shown by the abundant Pax6+/GFP+ cells in the lens and corneal ectoderm when only the control GFP expression plasmid was introduced (Fig. 7A; Table 1). Similarly, neither Meis1, Meis2 nor a Meis1 homeodomain linked to the Engrailed repressor domain (Meis1ΔMD-En) altered Pax6 expression (Table 1; Fig. 7B). In contrast, when full or nearly full length Meis1-Engrailed repressor constructs (Meis1-En or En-Meis1, respectively) were introduced, Pax6 lens placode expression was strongly and consistently down-regulated (Fig. 7A,B; Table 1). Such repression was specific, as Pax6− cells still expressed AP2α and Pbx (Fig. 7C). Interestingly, while GFP+ cells with reduced Pax6 expression were initially present in both the lens placode and the surrounding ectoderm, at later time points only the corneal ectoderm contained GFP+/Pax6− cells (Fig. 7A,B). This result agrees with findings in (Pax6Sey/Sey-Neu ↔ Pax6+/+) mouse chimeras, where Pax6−/− cells were excluded from the developing lens in a cell-autonomous manner (Collinson et al. 2000, 2001). Of note, misexpression of Meis1 or Meis1 fused with the activation domain of VP16 (Meis1-VP16) in ectoderm outside the eye-forming region did not produce ectopic Pax6 expression (data not shown), presumably because other regulatory factors required for Pax6 expression are limiting. Nonetheless, these in vivo experiments establish that the Pax6 lens placode enhancer element is a natural target for Meis1 and Meis2, which functionally regulate Pax6 expression in vivo.
Figure 6.
Meis constructs used in electroporation are based on the cDNA for Meis1a (kindly provided by Dr. M. Cleary; Jacobs et al. 1999). The En or VP16 domains confer repressor or activator activity, respectively upon the fusion proteins. (MD) Meis domain; (En) Drosophila Engrailed repressor domain; (HD) Meis Homeodomain; (VP16) activation domain of virion protein 16 of herpes simplex virus. (For detailed information on the constructs or their preparation, contact R. Maas.)
Figure 7.
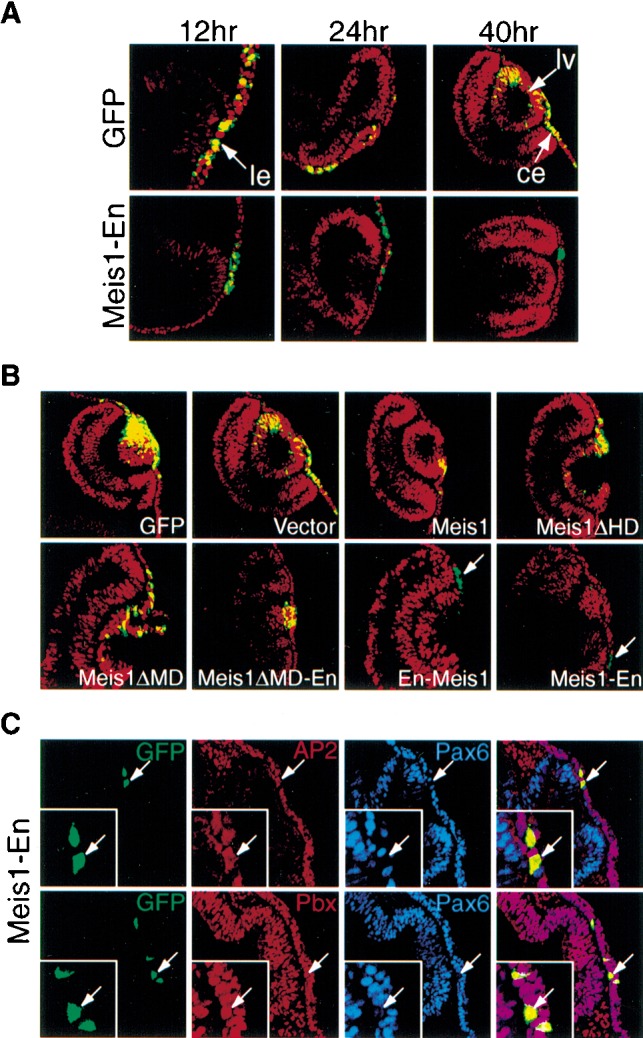
Meis1-En represses Pax6 lens ectoderm expression in vivo. (A, top) In chick embryos, endogenous Pax6 expression (red fluorescence) is not altered by electroporation of a Control GFP-expressing vector (green fluorescence), as evident by extensive overlapping expression (yellow). (Bottom) In contrast, following co-electroporation of Meis1-En (10-fold molar excess of a construct expressing Meis1 fused to the Drosophila Engrailed repressor domain) and GFP, endogenous Pax6 nuclear expression in the prospective lens ectoderm is reduced by 12 h and 24 h, and Pax6−/GFP+ cells are excluded from the lens vesicle by 40 h. (ce) Corneal ectoderm; (le) lens ectoderm; (lv) lens vesicle. (B) Requirement of the full-length Meis1-Engrailed repressor domain protein to repress Pax6 expression. When chicken embryos were electroporated with GFP plus various Meis contructs (the latter present in 10-fold molar excess) and analyzed after 24 h, only En-Meis1 and Meis1-En repressed Pax6 expression. Pax6+ cells are red, Meis construct expressing cells are green (due to GFP), and Pax6+/GFP+ cells are yellow. (C) Meis1-Engrailed proteins do not repress AP2α or Pbx1–3 expression. After electroporation, Meis1-Engrailed positive cells (GFP+, green) still express AP2α (red) and Pbx (red), but not Pax6 (blue).
Table 1.
Repression of Pax6 by Meis 1-En constructs in chick embryonic lens ectoderm.
| Construct
|
Pax6+
cells/GFP+ cells
(%)
|
No.experiments
|
|---|---|---|
| GFP alone | 612/666 (92%) | 3 |
| Meis1 + GFP | 154/177 (88%) | 2 |
| Meis1ΔMD-En + GFP | 36/40 (90%) | 1 |
| En-Meis1 + GFP | 106/540 (20%) | 5 |
| Meis1-En + GFP, 12 h | 35/162 (22%) | 6 |
| Meis1-En + GFP, 24 h | 13/77 (17%) | 5 |
| Meis1-En + GFP, 40 h | 4/33 (12%) | 2 |
Constructs were electroporated into E2 chick embryos (H&H stages 9–10) at 10-fold molar excess relative to co-electroporated GFP expression plasmid, increasing the likelihood that GFP+ cells also took up the test construct. Embryos were harvested and stained for endogenous Pax6 expression at 24 h unless otherwise noted. (GFP) Green fluorescent protein; (En) Drosophila Engrailed repressor domain.
Discussion
Meis homeoproteins directly regulate Pax6 lens placode expression
We have presented several lines of evidence demonstrating that the Meis1 and Meis2 homeobox transcription factors are direct upstream regulators of Pax6 expression in the developing lens ectoderm. These include: (1) Meis1 and Meis2 proteins are coexpressed with Pax6 in the developing lens placode; (2) Meis binds the Pax6 lens enhancer as part of a nuclear protein complex in vitro; (3) Meis binding is required for Pax6 lens enhancer activity in transgenic mice; (4) Meis2 interacts genetically with Pax6 and leads to increased Pax6 expression in developing lens; (5) dominant-negative Meis1 represses Pax6 expression in vivo.
Although Meis2 was the Meis family member initially isolated from the chick embryonic lens expression library, subsequent experiments suggested that Meis1 transcripts are more abundant in developing lens ectoderm than Meis2 transcripts. However, both Meis proteins were readily detected by immunohistochemistry using specific antibodies, and Meis2 transgenics exhibited cataracts associated with up-regulated Pax6 expression, whereas Meis1 transgenics did not. On the other hand, the in ovo experiments suggest that Meis1 can also modulate Pax6 expression in vivo. Therefore, although our data do not exclude qualitatively different molecular functions for Meis1 and Meis2, on the basis of the Meis2 transgenic mouse experiment, at least Meis2 must function in vivo in a positive regulatory capacity.
Recent experiments have suggested that the 107-bp enhancer is not the only cis-acting element controlling Pax6 expression in the lens. This is highlighted by the genetic deletion of this element in the mouse, which results in a delay in lens development and produces moderate lens abnormalities (Dimanlig et al. 2001). In contrast, a conditional knockout of the Pax6 gene specific to the lens placode completely abolishes lens formation (Ashery-Padan et al. 2000). This phenotypic discrepancy has been attributed to the residual Pax6 placodal expression that is observed in the enhancer null mice, a result that suggests the existence of additional lens enhancer(s) elsewhere in the Pax6 locus (Dimanlig et al. 2001). Consistent with this idea, the Sey Pax6 null mutation can be rescued by a 420-kb yeast artificial chromosome (YAC) transgene containing the human PAX6 locus, but not by a shorter 310-kb YAC (Kleinjan et al. 2001). The genomic fragment absent from the 310-kb YAC contains a number of tissue-specific enhancers, including a 2.5-kb element (denoted the “SIMO” element) that is capable of directing reporter gene expression in the lens. Interestingly, we have found five Meis consensus sites within the SIMO element, four of which are clustered in a 200-bp sequence with 99% conservation between human and mouse (X. Zhang and R. Maas, unpubl.). This raises the possibility that Meis homeoproteins bind both the 107-bp and the SIMO lens elements, thereby controlling the overall level of Pax6 expression in the lens. This model is supported by the study of Meis2+/− and compound Meis1+/−, Meis2+/− mice, both of which exhibit significant ocular phenotypes involving the lens (R. Richel, T. Hisa, N. Jenkins, and N. Copeland, pers. comm.).
Nature of the Meis complex
Other roles for Meis have been identified in vertebrate hindbrain development and limb morphogenesis (Capdevila et al. 1999; Mercader et al. 1999; Dibner et al. 2001; Waskiewicz et al. 2001; Choe et al. 2002; Maeda et al. 2002). In both processes, Meis acts as a cofactor for Pbx protein by modulating the latter's transcription activity. This is accomplished through a direct Meis–Pbx protein–protein interaction, which is required for both nuclear translocation and for synergistic binding of the Meis–Pbx complex to its target DNA sequence (Chang et al. 1997; Rieckhof et al. 1997). In this study of Meis function in eye development, we have also uncovered a potential role for Meis interacting partner(s) in regulating Pax6 gene expression. However, several lines of evidence suggest that, in this particular case, Pbx proteins are unlikely to be the relevant Meis interactors. First, Pbx protein, as assessed by a Pbx1–3 pan-reactive antibody, is absent from the Meis containing complex formed on the Pax6 lens enhancer. Second, Pbx does not bind to the sequence adjacent to the Meis site, even though this sequence is also required for the Pax6 lens enhancer activity (X. Zhang and R. Maas, unpubl.). Finally, Pbx1 knockout mice exhibit no obvious eye phenotype (Selleri et al. 2001), and mutation in the predominant Pbx gene expressed during early zebrafish development, Pbx4, also failed to cause any eye abnormality (Popperl et al. 2000). Together, these results raise the interesting possibility that an unknown factor assembles with Meis homeoproteins on the Pax6 lens placode element. In fact, study of Meis activity in zebrafish hindbrain development has suggested that the N-terminal domain of Meis3 may bind to an auxiliary factor in addition to Pbx (Choe et al. 2002). Identification of these unknown potential Meis interactors should further illuminate Meis function during vertebrate eye development.
Comparison to the ey hierarchy in Drosophila
Despite the striking evolutionary conservation of the Pax6 pathway (Pichaud et al. 2001), it is interesting to contrast our result with Drosophila, where the sole Meis homolog, Homothorax (Hth), suppresses eye formation in the ventral half of the eye and has been proposed to delimit the eye field (Pai et al. 1998). Meis/Homothorax function in controlling anterior–posterior embryonic patterning and proximo–distal limb development is conserved between Drosophila and vertebrates (Rieckhof et al. 1997; Kurant et al. 1998; Pai et al. 1998; Capdevila et al. 1999; Mercader et al. 1999; Dibner et al. 2001; Waskiewicz et al. 2001; Choe et al. 2002; Maeda et al. 2002). Nevertheless, the fact that Meis1 and Meis2 are expressed throughout the developing lens, retina, and cornea further indicates that Meis1 and Meis2 cannot play exclusively repressive roles in vertebrate eye development. In addition, although the Meis binding site in the Pax6 lens enhancer is conserved from fish to human, it does not appear to be present in the eye imaginal disc enhancer of the Drosophila Pax6 homolog eyeless (Hauck et al. 1999; Xu et al. 1999). In fact, when the eyeless enhancer was introduced into transgenic mice, it reproduced endogenous Pax6 expression in retina and spinal cord, but not in lens (Xu et al. 1999). Therefore, the mechanism of Pax6 regulation in the vertebrate lens placode differs in at least one respect from that in the Drosophila eye imaginal disc. Although Pax6 function is evolutionarily conserved, potentially indicating a monophyletic origin for the eye (Gehring and Ikeo 1999), it is attractive to suggest that divergent mechanisms regulating Pax6 expression also exist. These enhancer-specific differences may underlie the unique aspects of oculogenesis in vertebrates and invertebrates.
Materials and Methods
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as described (Dignam et al. 1983). EMSA reactions were carried out as described in a buffer containing 10 mM Na-HEPES (pH 7.4), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 25 ng/μL poly(dIdC) (Epstein et al. 1994). HA-Meis1 plasmid (kindly provided by Dr. P.Knoepfler, University of California at San Diego, La Jolla, CA) was transcribed and translated in vitro using a kit (Promega). The HA antibody (12 CA5) was from Boehringer Mannheim.
Southwestern screen
The chicken lens cDNA λgt11 library, a generous gift of Professor K. Yasuda (Nara Institute of Science and Technology, Japan), was previously used to isolate L-Maf, and screening was performed as described (Ogino and Yasuda 1998).
In situ hybridization and immunostaining
Whole-mount mRNA in situ hybridization and immunohistochemistry were performed as described (Wawersik et al. 1999). Antisense probes were generated from cDNAs for Meis1–3 (kindly provided by Dr. N. Copeland, NCI, Frederick), TGIF, Prep1, and Pax6. Meis1 and Meis2 antibodies were generous gifts from Dr. A. Buchburg (Jefferson Medical College, Philadelphia, PA). These antibodies were made to 16-residue N-terminal peptides of Meis1 and Meis2, which differ at a single position (Swift et al. 1998). Nevertheless, they recognize Meis1 or Meis2 with a high degree of specificity in DNA mobility shift assays, in Western blot assays, in immunostained transfected cells, and in mouse tissue sections (Fig. 3C; data not shown). The specificity of these antibodies has also been noted by others (Toresson et al. 2000). Affinity purified polyclonal rabbit Pax6 antibody (PRB-278P) was obtained from Covance (Richmond, CA).
Protein extraction after EMSA
The gel slice from EMSA was boiled with 50 μL of loading buffer (12.5 mM Tris at pH 6.8, 2% SDS, 20% glycerol, 0.002% bromphenol blue, 10% 2-mercaptoethanol) for 5 min and incubated at 4°C overnight. Eluted proteins were resolved by SDS-PAGE, transferred to nitrocellulose, incubated with antibody, and detected by chemiluminescence (Pierce). This strategy was used because the Meis1 antibody does not function in a supershift protocol.
DNA pull down assay
Five microliter protein preparations were incubated with 200 ng of biotinylated double-stranded DNA in 20 μL of EMSA binding buffer for 30 min on ice. The reactions were then mixed with 5 μL of Ultralink streptavidin-agarose (Pierce) for 10 min, washed 3 times, and analyzed by Western blotting. The biotin labels were on the 5′ end of oligonucleotides 5′-AATTGTAGATC GAAGCCGGCCTTGTCAGGTTGAGAA-3′ (wild type) and 5′-AATTGTAGATCGAAGCCGGCCTTGTGAGGTTGAGA A-3′ (mutant). The rabbit polyclonal Pbx antibody (sc-888x, lot B150, Santa Cruz Biotechnology) recognizes a C-terminal epitope identical between human PBX1, PBX2, and PBX3. An additional Pbx family member with a more divergent C terminus, Pbx4, has been reported recently, but its RNA expression is not detected in mouse embryos between E7.0–E10.5 (Wagner et al. 2001).
Transgenic overexpression
The transgenic expression vector was constructed by placing the 526-bp Pax6 fragment (−3969 to −3442 bp) containing the lens enhancer, and the human β-globin promoter, 5′ to the Meis2b cDNA (kindly provided by Dr. P. Chambon, Institut de Biologique, Strasbourg, France), followed by an IRES-alkaline phosphatase (AP) cassette (kindly provided by Dr. T. Lufkin, Mt. Sinai School of Medicine, New York, NY). Transgenic mice were generated by pronuclear injection on a FVB/N background, and lens-specific transgene expression was verified by AP activity and RT–PCR. Meis1 transgenics (three independent permanent lines) did not exhibit a lens phenotype, whereas analogous Meis2 transgenics (two independent permanent lines) did, even though transgene expression levels appear similar as judged by alkaline phosphatase reporter activity. This is possibly because when tested for ability to stimulate Hox and Pbx mediated transactivation of a synthetic reporter (a gift of Dr. M. Cleary; Jacobs et al. 1999), Meis1 was reproducibly weaker than Meis2 (data not shown). For the genetic interaction experiment, Sey1Neu/+ mice were maintained in a C3H/HeN background. Although the Pax6 gene is not deleted in Sey1Neu/Sey1Neu mice, the PST domain encoded by the Sey1Neu allele is devoid of transactivating function in transient transfection assays (Glaser et al. 1992). In addition, the Sey1Neu/+ ocular phenotype closely resembles that of SeyH/+ embryos, which carry a deletion of the Pax6 locus (Hill et al. 1991).
In ovo electroporation
In ovo electroporation was performed as described (Momose et al. 1999). The Meis constructs were cloned in pIRES2-EGFP (Clontech), a CMV-based expression vector with an internal GFP reporter, and protein expression was verified in cell culture. However, because the IRES2-EGFP signal was too weak to be easily detected after cryosection, we co-electroporated an excess of Meis construct (3 μg/μL) with pCS2-GFP (0.3 μg/μL; kindly provided by Dr. C. Cepko, Department of Genetics, Harvard Medical School, Boston, MA). Sections were processed for immunohistochemistry with antibodies against Pax6 (Developmental Hybridoma Bank), AP2α, and Pbx1–3 (both from Santa Cruz).
Quantitative real time RT–PCR
E13.5 lenses were collected from 18 litters of Tg-Meis2/+ × +/+ matings. RNA was extracted from lenses pooled by genotype from 85 +/+ and 74 +/Tg embryos for quantitative RT–PCR analysis (Bio-Rad iCycler IQ; Invitrogen, no. 10928-042). Pax6 cDNA was detected with primers 5′-CTACCAGCCAATCCCACAGC-3′, 5′-TTCGGCCCAACATGGAAC-3′ (Invitrogen), and probe 5′-(6-FAM)CACCACACCT-GTCTCCTCCTT CACATCA(BHQ-1)-3′ (Biosearch) and normalized against 18S expression. Experiments were repeated two times, each in triplicate.
Computational analysis
Composite binding sites for Pbx1-Meis1, Pbx-Prep1 and Meis-Hoxa-9 have also been described (Chang et al. 1997; for review, see Mann and Affolter 1998). None of these conform to sequences on the Pax6 26-bp element. That Homothorax is the sole Drosophila melanogaster Meis family member was verified by a BlastP homology search of the fly genome for sequences homologous to the Meis-Homothorax (MH) domain. The 212-bp D. melanogaster ey eye enhancer sequence (Hauck et al. 1999; GenBank accession no. AJ131630) was searched for the Meis1 DNA recognition consensus sequence, 5′-TGACA(A/G)-3′ (Chang et al. 1997). No 5- or 6-bp matches were identified.
Acknowledgments
We thank Drs. Connie Cepko and Shahla Vahabi (Harvard Medical School) for use of the in ovo electroporation apparatus, and Drs. Neal Copeland (NCI, Frederick) and Henry Sun (Academia Sinica, Taipei) for communicating unpublished results. A.F. was an HHMI summer student during the course of these studies. This work was supported by NIH RO1 EY10123 (R.M.), and by Postdoctoral Training Grant in the Molecular Bases of Eye Diseases NIH T32 EY07145 (X.Z.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL maas@rascal.med.harvard.edu; FAX (617) 732-5123.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1007602.
References
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2−/−mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121:3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes & Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavik C, Ward SJ, Chambon P. Developmental abnormalities in cultured mouse embryos deprived of retinoic acid by inhibition of yolk-sac retinol binding protein synthesis. Proc Natl Acad Sci. 1996;93:3110–3114. doi: 10.1073/pnas.93.7.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier G, Panitz F, Zhou X, Hollemann T, Gruss P, Pieler T. Expanded retina territory by midbrain transformation upon overexpression of Six6 (Optx2) in Xenopusembryos. Mech Dev. 2000;93:59–69. doi: 10.1016/s0925-4773(00)00271-9. [DOI] [PubMed] [Google Scholar]
- Capdevila J, Tsukui T, Rodriquez Esteban C, Zappavigna V, Izpisua Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol Cell. 1999;4:839–849. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- Chang CP, Jacobs Y, Nakamura T, Jenkins NA, Copeland NG, Cleary ML. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol Cell Biol. 1997;17:5679–5687. doi: 10.1128/mcb.17.10.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Halder G, Zhang Z, Mardon G. Signalling by the TGFβ homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999;126:935–943. doi: 10.1242/dev.126.5.935. [DOI] [PubMed] [Google Scholar]
- Choe SK, Vlachakis N, Sagerstrom CG. Meis family proteins are required for hindbrain development in the zebrafish. Development. 2002;129:585–595. doi: 10.1242/dev.129.3.585. [DOI] [PubMed] [Google Scholar]
- Chow R, Altmann C, Lang R, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Collinson J, Hill R, West J. Different roles for Pax6 in the optic vesicle and facial epithelium mediate early morphogenesis of the murine eye. Development. 2000;127:945–956. doi: 10.1242/dev.127.5.945. [DOI] [PubMed] [Google Scholar]
- Collinson JM, Quinn JC, Buchanan MA, Kaufman MH, Wedden SE, West JD, Hill RE. Primary defects in the lens underlie complex anterior segment abnormalities of the Pax6heterozygous eye. Proc Natl Acad Sci. 2001;98:9688–9693. doi: 10.1073/pnas.161144098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Dibner C, Elias S, Frank D. XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevisembryos. Development. 2001;128:3415–3426. doi: 10.1242/dev.128.18.3415. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimanlig PV, Faber SC, Auerbach W, Makarenkova HP, Lang RA. The upstream ectoderm enhancer in Pax6has an important role in lens induction. Development. 2001;128:4415–4424. doi: 10.1242/dev.128.22.4415. [DOI] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Uchida T, Osumi-Yamashita N, Eto K. Uchida rat (rSey): A new mutant rat with craniofacial abnormalities resembling those of the mouse Seymutant. Differentiation. 1994;57:31–38. doi: 10.1046/j.1432-0436.1994.5710031.x. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton D, Maas R. Genomic structure, evolutionary conservation and aniridia mutations in the human PAX6gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring W. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hanson IM, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutations at the PAX6locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- Hauck B, Gehring WJ, Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc Natl Acad Sci. 1999;96:564–569. doi: 10.1073/pnas.96.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BLM, Ton CT, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Hirst EM, Horsburgh G, Hetherington CM. Small eye (Sey): a mouse model for the genetic analysis of craniofacial abnormalities. Development. 1988;103 Suppl:115–119. doi: 10.1242/dev.103.Supplement.115. [DOI] [PubMed] [Google Scholar]
- Hollemann T, Bellefroid E, Pieler T. The Xenopus homologue of the Drosophila gene taillesshas a function in early eye development. Development. 1998;125:2425–2432. doi: 10.1242/dev.125.13.2425. [DOI] [PubMed] [Google Scholar]
- Inbal A, Halamachi N, Dibner C, Frank D, Salzberg A. Genetic evidence for the transcriptional-activating function of Homothorax during adult fly development. Development. 2001;128:3405–3413. doi: 10.1242/dev.128.18.3405. [DOI] [PubMed] [Google Scholar]
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Distinct cis-essential modules direct the time-space pattern of the Pax6gene activity. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Schedl A, Quinlan RA, Danes S, van Heyningen V. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Nishikawa K, Suzuki T, Yamamoto M. The homeobox protein Six3 interacts with the Groucho corepressor and acts as a transcriptional repressor in eye and forebrain formation. Dev Biol. 2001;232:315–326. doi: 10.1006/dbio.2001.0185. [DOI] [PubMed] [Google Scholar]
- Köster R, Kühnlein R, Wittbrodt J. Ectopic Sox3 activity elicits sensory placode formation. Mech Dev. 2000;95:175–187. doi: 10.1016/s0925-4773(00)00356-7. [DOI] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, Salzberg A. Dorsotonals/homothorax, the Drosophilahomologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Maeda R, Ishimura A, Mood K, Park E, Buchberg A, Daar I. Xpbx1b and Xmeis1b play a collaborative role in hindbrain and neural crest gene expresssion in Xenopusembryos. Proc Natl Acad Sci. 2002;99:5448–5453. doi: 10.1073/pnas.082654899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Hox proteins meet more partners. Curr Opin Genet Dev. 1998;8:423–429. doi: 10.1016/s0959-437x(98)80113-5. [DOI] [PubMed] [Google Scholar]
- Mathers P, Grinberg A, Mahon K, Jamrich M. The Rxhomeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Azpiazu N, Serrano A, Morata G, Martinez C, Torres M. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402:425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- Momose T, Tonegawa A, Takeuchi J, Ogawa H, Umesono K, Yasuda K. Efficient targeting of gene expression in chick embryos by microelectroporation. Dev Growth Differ. 1999;41:335–344. doi: 10.1046/j.1440-169x.1999.413437.x. [DOI] [PubMed] [Google Scholar]
- Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Jenkins NA, Copeland NG. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13:2235–2242. [PubMed] [Google Scholar]
- Ogino H, Yasuda K. Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science. 1998;280:115–118. doi: 10.1126/science.280.5360.115. [DOI] [PubMed] [Google Scholar]
- Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- Pai C, Kuo T, Jaw T, Kurant E, Chen C, Bessarab D, Salzberg A, Sun Y. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes & Dev. 1998;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, Wessely O, Li SY, De Robertis EM. Neural and head induction by insulin-like growth factor signals. Dev Cell. 2001;1:655–665. doi: 10.1016/s1534-5807(01)00069-7. [DOI] [PubMed] [Google Scholar]
- Pichaud F, Treisman J, Desplan C. Reinventing a common strategy for patterning the eye. Cell. 2001;105:9–12. doi: 10.1016/s0092-8674(01)00292-6. [DOI] [PubMed] [Google Scholar]
- Plaza S, Saule S, Dozier C. High conservation of cis-regulatory elements between quail and human for the Pax-6gene. Dev Genes Evol. 1999;209:165–173. doi: 10.1007/s004270050240. [DOI] [PubMed] [Google Scholar]
- Popperl H, Rikhof H, Chang H, Haffter P, Kimmel CB, Moens CB. lazarus is a novel pbx gene that globally mediates hox gene function in zebrafish. Mol Cell. 2000;6:255–267. doi: 10.1016/s1097-2765(00)00027-7. [DOI] [PubMed] [Google Scholar]
- Porter F, Drago J, Xu Y, Cheema S, Wassif C, Huang S, Lee E, Grinberg A, Massalas J, Bodine D, et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Purcell P. Genes involved in early development of the mouse cranial sensory placodes. Ph.D. thesis. Boston, MA: Harvard University; 2001. [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring W. Homology of the eyeless gene of Drosophila to the Small eyegene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Rasmussen J, Deardorff M, Tan C, Rao M, Klein P, Vetter M. Regulation of eye development by frizzled signaling in Xenopus. Proc Natl Acad Sci. 2001;98:3861–3866. doi: 10.1073/pnas.071586298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie ND. Influence of PAX6gene dosage on development: overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- Selleri L, Depew MJ, Jacobs Y, Chanda SK, Tsang KY, Cheah KS, Rubenstein JL, O'Gorman S, Cleary ML. Requirement for Pbx1in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- Stuempfle KJ, Floros J. Caution is advised when cDNA expression libraries are screened by southwestern methodologies. Biotechniques. 1997;22:260–264. doi: 10.2144/97222bm13. [DOI] [PubMed] [Google Scholar]
- Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2) Mol Cell Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton C, Hirvonen H, Miwa H, Weil M, Monaghan P, Jordan T, van Heyningen V, Hastie N, Meijers-Heijboer H, Drechsler M. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridiaregion. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Toresson H, Parmar M, Campbell K. Expression of Meis and Pbxgenes and their protein products in the developing telencephalon: Implications for regional differentiation. Mech Dev. 2000;94:183–187. doi: 10.1016/s0925-4773(00)00324-5. [DOI] [PubMed] [Google Scholar]
- Treisman J, Rubin G. Wingless inhibits morphogenetic furrow movement in the Drosophilaeye disc. Development. 1995;121:3519–3527. doi: 10.1242/dev.121.11.3519. [DOI] [PubMed] [Google Scholar]
- van Raamsdonk C, Tilghman S. Dosage requirement and allelic expression of PAX6during lens placode formation. Development. 2000;127:5439–5448. doi: 10.1242/dev.127.24.5439. [DOI] [PubMed] [Google Scholar]
- Wagner K, Mincheva A, Korn B, Lichter P, Popperl H. Pbx4, a new Pbxfamily member on mouse chromosome 8, is expressed during spermatogenesis. Mech Dev. 2001;103:127–131. doi: 10.1016/s0925-4773(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Waskiewicz AJ, Rikhof HA, Hernandez RE, Moens CB. ZebrafishMeis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development. 2001;128:4139–4151. doi: 10.1242/dev.128.21.4139. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Purcell P, Rauchman M, Dudley A, Robertson E, Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–188. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- Williams S, Altmann C, Chow R, Hemmati-Brivanlou A, Lang R. A highly conserved lens transcriptional control element from the Pax-6gene. Mech Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- Xu P, Zhang X, Heaney S, Yoon A, Michelson A, Maas R. Regulation of Pax6expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- Zhu CC, Dyer MA, Uchikawa M, Kondoh H, Lagutin OV, Oliver G. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- Zuber M, Perron M, Philpott A, Bang A, Harris W. Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]