Abstract
The metazoan transcription elongation factor P-TEFb (CDK-9/cyclin T) is essential for HIV transcription, and is recruited by some cellular activators. P-TEFb promotes elongation in vitro by overcoming pausing that requires the SPT-4/SPT-5 complex, but considerable evidence indicates that SPT-4/SPT-5 facilitates elongation in vivo. Here we used RNA interference to investigate P-TEFb functions in vivo, in the Caenorhabditis elegans embryo. We found that P-TEFb is broadly essential for expression of early embryonic genes. P-TEFb is required for phosphorylation of Ser 2 of the RNA Polymerase II C-terminal domain (CTD) repeat, but not for most CTD Ser 5 phosphorylation, supporting the model that P-TEFb phosphorylates CTD Ser 2 during elongation. Remarkably, although heat shock genes are cdk-9-dependent, they can be activated when spt-4 and spt-5 expression is inhibited along with cdk-9. This observation suggests that SPT-4/SPT-5 has an inhibitory function in vivo, and that mutually opposing influences of P-TEFb and SPT-4/SPT-5 may combine to facilitate elongation, or insure fidelity of mRNA production. Other genes are not expressed when cdk-9, spt-4, and spt-5 are inhibited simultaneously, suggesting that these genes require P-TEFb in an additional mechanism, and that they and heat shock genes are regulated through different P-TEFb-dependent elongation pathways.
Keywords: Transcription, C. elegans, P-TEFb, CDK9, DSIF, SPT5
The largest subunit of RNA polymerase II (Pol II) is distinguished by a unique C-terminal domain (CTD) which undergoes a cycle of phosphorylation and dephosphorylation during transcription. The CTD consists of a consensus (YSPTSPS) that is repeated 52 times in human and 42 times in Caenorhabditis elegans (Corden 1990). Pol II is recruited to promoters in a hypophosphorylated form, and then shortly after transcription begins the CTD becomes phosphorylated (Dahmus 1996). CTD phosphorylation is thought to mark a critical mechanistic transition, during which RNA polymerase clears the promoter and shifts into an elongation mode in which the polymerase is resistant to pausing (Payne et al. 1989; Weeks et al. 1993; O'Brien et al. 1994; Akhtar et al. 1996). The CTD also integrates transcription and processing of the nascent mRNA, by recruiting mRNA capping, splicing, cleavage, and polyadenylation complexes directly to the transcribing polymerase (Hirose and Manley 2000; Fong and Bentley 2001). As transcription progresses the CTD is dephosphorylated, so that Pol II is eventually able to repeat the transcription cycle (Dahmus 1996; Komarnitsky et al. 2000; Schroeder et al. 2000; Cho et al. 2001).
During the transcription cycle, the CTD repeat is phosphorylated on both Ser 2 and Ser 5. Initially, Ser 5 is phosphorylated by the cyclin-dependent kinase (CDK) component of the general factor TFIIH: Kin28 (Saccharomyces cerevisiae) or CDK-7 (metazoans) (Hengartner et al. 1998; Komarnitsky et al. 2000). Ser 5 phosphorylation predominates near the promoter, and allows the CTD to recruit activities which place the 5′ cap on the pre-mRNA (Komarnitsky et al. 2000; Rodriguez et al. 2000; Schroeder et al. 2000). Less is understood about the role of CTD Ser 2 phosphorylation, which may generally predominate on elongating Pol II (Komarnitsky et al. 2000). In metazoan embryos, CTD Ser 2 phosphorylation levels correlate closely with transcription activity (Seydoux and Dunn 1997; Tenenhaus et al. 1998; Palancade et al. 2001; Walker et al. 2001). In S. cerevisiae however, this CTD Ser 2 phosphorylation is mediated primarily by the CDK Ctk1, which promotes elongation and is required for some cellular processes, but not for viability (Sterner et al. 1995; Lee and Greenleaf 1997; Patturajan et al. 1999; Cho et al. 2001).
The metazoan kinase CDK-9 and its cyclin partners (T1, T2 or K) form the transcription factor P-TEFb, which prevents transcription from stalling in vitro (Marshall and Price 1995; Price 2000). CDK-9 is similar to and may be functionally related to yeast Ctk1. Stimulation of elongation by P-TEFb is required for human immunodeficiency virus (HIV) transcription, during which P-TEFb is recruited directly to the nascent mRNA by the transactivator Tat (Mancebo et al. 1997; Yang et al. 1997; Zhu et al. 1997; Wei et al. 1998; Price 2000). P-TEFb preferentially phosphorylates CTD Ser 2 in vitro, but in association with Tat P-TEFb also phosphorylates Ser 5, and may thereby bypass the requirement for CDK-7 (Chen and Zhou 1999; Zhou et al. 2000). P-TEFb is recruited by cellular activators that include NF-κB and c-Myc (Kanazawa et al. 2000; Barboric et al. 2001; Eberhardy and Farnham 2001; Lee et al. 2001), and it is present at sites of active transcription in Drosophila (Lis et al. 2000). Treatment of mammalian cells with a pharmacological inhibitor of CDK-9 appears to block most Pol II transcription (Chao and Price 2001), but P-TEFb functions have not been investigated in a genetic system. It is an important issue for HIV therapeutics to understand how broadly P-TEFb is required for transcription of cellular genes, because CDK-9 inhibition is a preeminent strategy for interfering with Tat (Mancebo et al. 1997; Flores et al. 1999; Chao et al. 2000).
Transcription proceeds independently of P-TEFb in vitro in the absence of the factor DSIF, which cooperates with the negative factor NELF to inhibit elongation (Wada et al. 1998a,b; Yamaguchi et al. 1999, 2002; Renner et al. 2001). While this argues that P-TEFb is required simply to overcome DSIF/NELF-dependent pausing, DSIF increases elongation in vitro under conditions that enhance pausing, suggesting that it has a dual function (Wada et al. 1998a). The bulk of in vivo evidence also suggests that DSIF facilitates elongation, possibly in concert with P-TEFb. DSIF consists of the proteins SPT-4 and SPT-5 (Wada et al. 1998a), which yeast and zebrafish genetic experiments indicate are positive elongation factors (Hartzog et al. 1998; Kaplan et al. 2000; Lindstrom and Hartzog 2001; Keegan et al. 2002). SPT-5 is related to Escherichia coli NusG, an antiterminator that increases elongation (Wada et al. 1998a). Accordingly, SPT-5 is associated with elongating RNA Pol II in vivo (Andrulis et al. 2000; Kaplan et al. 2000) and is required for HIV Tat to stimulate elongation (Wu-Baer et al. 1998; Kim et al. 1999; Parada and Roeder 1999; Ivanov et al. 2000; Bourgeois et al. 2002). To understand how P-TEFb promotes transcription elongation, it is critical to determine whether an inhibitory effect of SPT-4/SPT-5 can be identified in vivo, and to elucidate how P-TEFb interacts functionally with SPT-4/SPT-5 in vivo.
We have performed the first genetics-based study of P-TEFb functions, in the C. elegans embryo. Using RNA interference (RNAi), we investigated the requirements for P-TEFb for CTD Ser 2 and Ser 5 phosphorylation, and for early embryonic transcription. We also tested whether depletion of SPT-4/SPT-5 relieves any requirements for P-TEFb. Our results indicate that P-TEFb is required for phosphorylation of CTD Ser 2 but not for most Ser 5 phosphorylation. In apparent contrast to yeast Ctk1, P-TEFb is also broadly essential for transcription in the early embryo. Simultaneous inhibition of spt-4 and spt-5 restored heat shock gene transcription in cdk-9(RNAi) embryos, suggesting that P-TEFb counteracts an inhibitory function of SPT-4/SPT-5 in vivo. Significantly, other genes still required P-TEFb when spt-4 and spt-5 were inhibited, suggesting that they and heat shock genes maintain transcription elongation through different P-TEFb-dependent pathways.
Results
CDK-9 and cyclin T required for early embryonic development
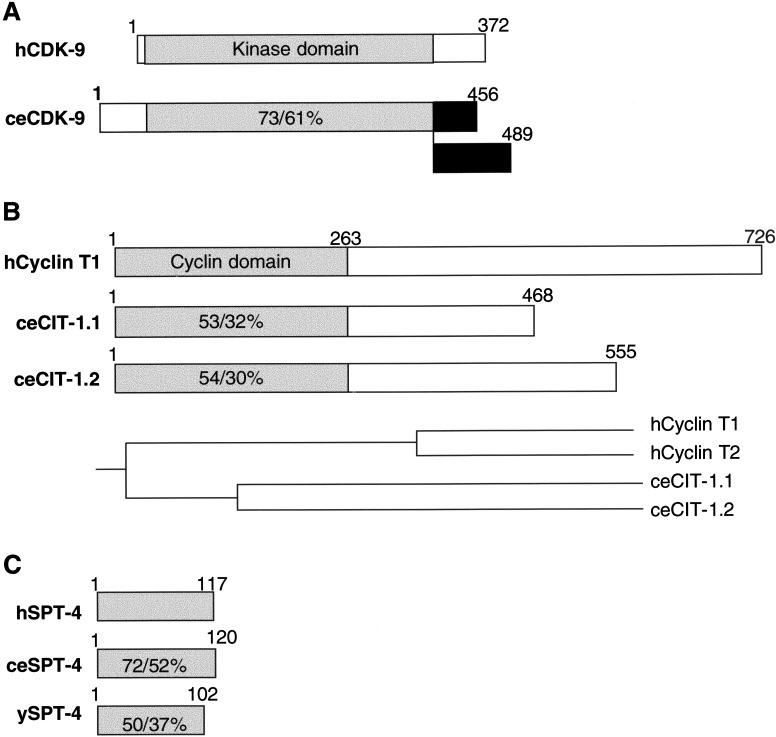
Well conserved unambiguous orthologs of each P-TEFb and DSIF component gene are present in the C. elegans genome. cdk-9 encodes a predicted kinase that is highly related to human CDK-9 (Fig. 1A), and includes the signature PITALRE motif (Grana et al. 1994; Price 2000). Two closely related C. elegans cyclin T genes, cit-1.1 and cit-1.2 (Fig. 1B), encode predicted proteins that are each slightly more similar to human cyclin T1 than T2, but neither contains the motif through which HIV Tat binds human cyclin T1 (Price 2000). The respective human and C. elegans cyclin T gene pairs each appear to have arisen independently from an ancestral gene (Fig. 1B). C. elegans also encodes a single conserved ortholog each of spt-4 (Fig. 1C) and spt-5 (Hartzog et al. 1998; Wada et al. 1998a; Guo et al. 2000). Each of these C. elegans genes re-identifies its human or yeast counterparts as its closest relative in GenBank databases.
Figure 1.
Predicted C. elegans (ce) CDK-9, cyclin T, and SPT-4 proteins. (A) ceCDK-9, with similarity/identity to human (h) CDK-9 within the kinase domain indicated. Expressed cDNA sequences predict existence of two CDK-9 isoforms derived from alternative splicing of C-terminal exons (in black). (B) ceCIT-1.1 and ceCIT-1.2, compared with hcyclin T1, with similarity/identity to hcyclin T1 indicated. A phylogenic tree based on sequence comparison is shown at the bottom of the panel. (C) ceSPT-4 and ySPT-4 are compared with hSPT-4 as in (A).
The early C. elegans embryo provides an advantageous system for studying transcription in vivo. C. elegans embryonic development is initiated by maternally derived proteins and mRNAs, and then new mRNA transcription begins around the four-cell stage (Seydoux and Dunn 1997; Newman-Smith and Rothman 1998). Because RNAi inhibits both maternal and zygotic expression of early embryonic genes (Fire et al. 1998), it is possible to deplete expression of a transcription factor of interest starting at the beginning of embryogenesis. When embryonic mRNA transcription is inhibited, maternal gene products maintain viability until approximately the 100-cell stage, making it feasible to manipulate expression of even essential transcription factors in these living cells (Powell-Coffman et al. 1996; Dantonel et al. 2000; Kaltenbach et al. 2000; Walker et al. 2001). To determine whether the CDK-9 protein is present in the early embryo, we examined its expression by antibody staining. Using conditions optimized for early embryos, CDK-9 was readily apparent in interphase nuclei from the two-cell stage until morphogenesis (Fig. 2A; data not shown). CDK-9 was also detectable in adult germline and oocyte nuclei (data not shown), suggesting that it is maternally expressed.
Figure 2.
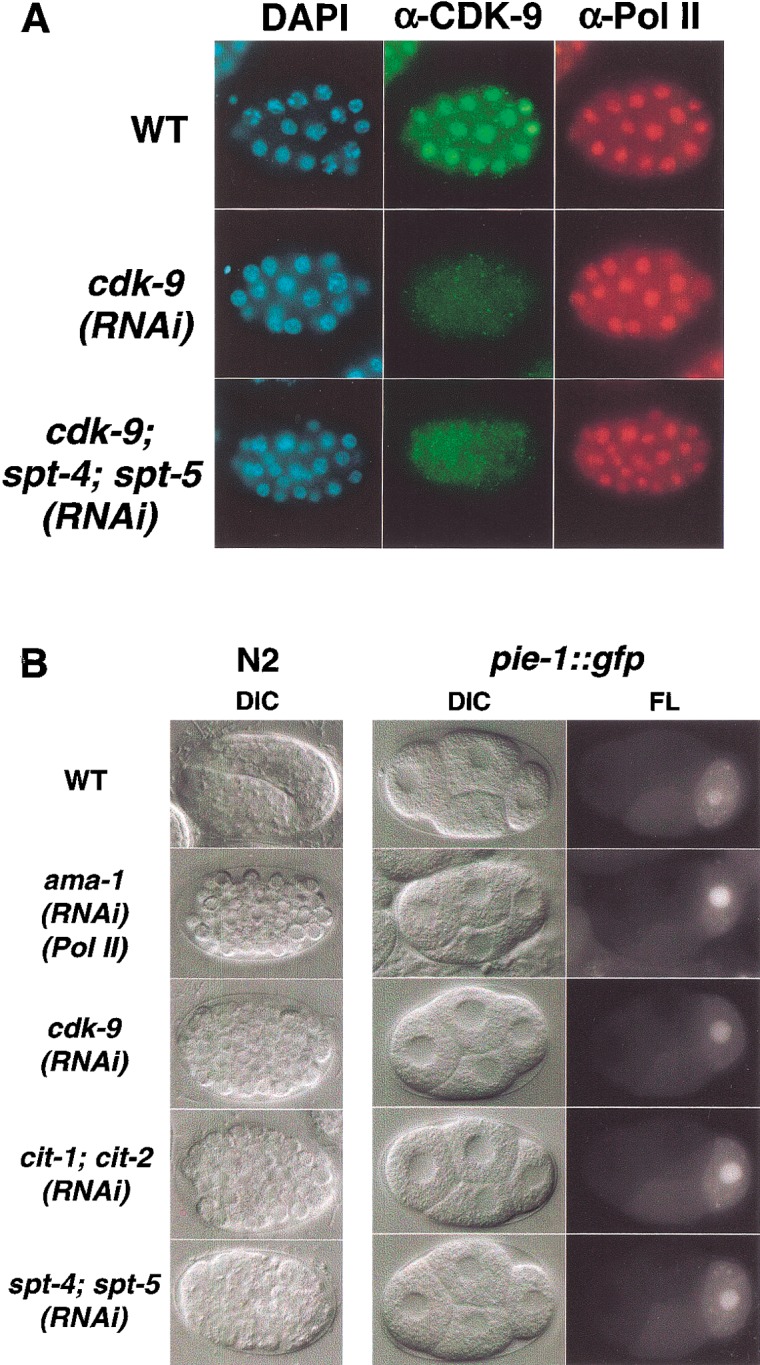
Phenotypic analysis of RNAi embryos. (A) Expression of CDK-9 and RNA Pol II. Representative wild-type and RNAi embryos (indicated in rows) were stained with DAPI to visualize DNA, or with the indicated antibodies. The CDK-9 protein was not detected in any cdk-9(RNAi) or cdk-9; spt-4; spt-5(RNAi) embryos. The Pol II large subunit AMA-1 was detected with an antibody against the unphosphorylated CTD (8WG16). (B) Terminal and early cell division phenotypes of RNAi embryos. Embryos produced by N2 (wild-type) or pie-1∷gfp mothers were examined by differential interference (DIC) or fluorescence (FL) microscopy. Typical wild-type or RNAi embryos are shown in rows, as indicated to the left. In the leftcolumn, terminally arrested RNAi embryos are compared with a wild-type embryo that is continuing to develop. The right two columns show four-cell WT and RNAi pie-1∷gfp embryos. These RNAi embryos were indistinguishable from wild-type with respect to each aspect of PIE-1∷GFP germline and subcellular localization, including the presence of PIE-1 in germline RNA-protein P granules. Anterior is to the left.
We used RNAi to inhibit CDK-9 and cyclin T expression in the early embryo. In cdk-9(RNAi) embryos, the cdk-9 mRNA was substantially depleted (described below) and CDK-9 antibody staining was eliminated (Fig. 2A), suggesting a severe reduction in CDK-9 expression. As a benchmark for a general transcription defect, we inhibited expression of the Pol II large subunit gene ama-1 (Powell-Coffman et al. 1996; Walker et al. 2001). To assess accumulation of maternal mRNA, we monitored early cell division patterns and performed parallel experiments in a transgenic strain that expresses a fusion of the maternally derived germline protein PIE-1 to green fluorescent protein (GFP). PIE-1∷GFP recapitulates the endogenous PIE-1 localization pattern, which depends upon at least 20 maternal genes (Tenenhaus et al. 1998; Reese et al. 2000). Like ama-1(RNAi) embryos, cdk-9(RNAi) embryos arrested development at ∼100 cells and lacked signs of differentiation, but their maternal PIE-1∷GFP expression and localization patterns appeared normal at every stage (Fig. 2B; data not shown). Early cell division timing and cleavage planes were also generally normal in cdk-9(RNAi) embryos, except for the cell cycle period of the two E cell daughters (E2 cells), which give rise to the endoderm. When mRNA transcription is broadly inhibited, the E2 cell cycle length is shortened from 45 min to ∼22 min, resulting in a failure of gastrulation (Powell-Coffman et al. 1996; Walker et al. 2001). The E2 cells similarly divided after 22 min in cdk-9(RNAi) embryos (data not shown). Lack of CDK-9 in the embryo thus does not detectably affect maternal mRNA stores, but may significantly impair new mRNA transcription.
Inhibition of cit-1.1 and cit-1.2 individually by RNAi did not impair development to adulthood or fertility (data not shown). In contrast, simultaneous RNAi inhibition of both cyclin T genes resulted in an early undifferentiated embryonic arrest similarly to inhibition of cdk-9 or ama-1 (Fig. 2B). This suggests that the two C. elegans cyclin T genes function redundantly in the early embryo, and is consistent with broad involvement of P-TEFb in embryonic transcription.
Dependence of CTD serine 2 phosphorylation on P-TEFb
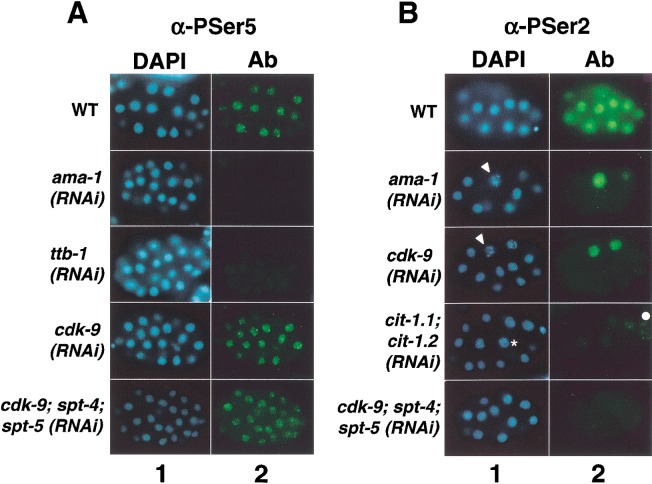
The patterns and intensity of Pol II CTD phosphorylation appear to parallel overall transcription activity in the C. elegans embryo (Seydoux and Dunn 1997; Tenenhaus et al. 1998; Walker et al. 2001). We assayed CTD Ser 5 phosphorylation in wild-type and RNAi embryos by staining with the P-CTD antiserum (Schroeder et al. 2000), which recognizes a pattern indistinguishable from that obtained with the phospho-Ser 5 antibody H14 (Seydoux and Dunn 1997; Walker et al. 2001). We detected phospho-Ser 2 with the H5 antibody (Bregman et al. 1995; Seydoux and Dunn 1997; Patturajan et al. 1998; Cho et al. 2001). For clarity, we refer to P-CTD and H5 as α-PSer5 and α-PSer2, respectively. Phosphorylation of CTD serines 5 and 2 is first detected in embryonic nuclei at the three-to-four-cell stage, concurrently with the onset of transcription in somatic cells (Seydoux and Dunn 1997). α-PSer5 staining then appears in a generalized punctate nucleoplasmic pattern in somatic cells (Fig. 3A), and as two discrete nuclear foci in the transcriptionally silent germline precursor (data not shown; Seydoux and Dunn 1997; Walker et al. 2001). Nuclear α-PSer2 staining is uniformly robust in wild-type somatic cells, but is undetectable in the early germline (Fig. 3B; data not shown; Seydoux and Dunn 1997; Walker et al. 2001).
Figure 3.
CDK-9 and cyclin T are required for phosphorylation of Ser 2 of the Pol II CTD repeat, but not for most Ser 5 phosphorylation. In A and B, representative wild-type or RNAi embryos that are actively undergoing cell division are shown in rows, as indicated. Embryos were stained with DAPI to visualize DNA (column 1), and with α-P-Ser5 (A) or α-P-Ser2 (B) antibodies (Ab; column 2). (A) CTD Ser5 phosphorylation levels are not detectably reduced when CDK-9 is depleted by RNAi. α-P-Ser5 staining was comparable to wild-type at each stage in 100% of cdk-9(RNAi) embryos, but was dramatically reduced when transcription initiation was inhibited by depletion of the essential factor TFIIB [ttb-1(RNAi) embryos]. (B) CTD Ser2 phosphorylation is not detectable in embryos lacking P-TEFb. Throughout their development until terminal arrest, in 100% of cdk-9(RNAi), cit-1.1; cit-1.2(RNAi), and cdk-9; spt-4; spt-5(RNAi) embryos, α-P-Ser2 staining in interphase nuclei was not detectable above the background seen in ama-1(RNAi) embryos. Within each embryo set, in mitotic cells α-P-Ser2 also detected a cross-reactive epitope that does not derive from Pol II (see text). Examples of nuclei in early and late stages of mitotic chromosome condensation are indicated by an asterisk and arrowheads, respectively. No nuclear α-P-Ser2 staining is detectable in the wild-type germline precursor, which is not visible in the focal plane shown. Some α-P-Ser2-stained germline cells have weak perinuclear staining deriving from cross-reactivity of the secondary antibody with P granules (Walker et al. 2001). A white dot denotes a germline cell within the focal plane.
Depletion of the CDK-9 kinase resulted in dramatically different effects on the levels of embryonic CTD Ser 2 and CTD Ser 5 phosphorylation. When the Pol II large subunit AMA-1 is depleted by RNAi, embryonic staining with multiple Pol II antisera is reduced to background (Seydoux and Dunn 1997; Walker et al. 2001), as is α-PSer5 staining (Fig. 3A). As reported previously (Seydoux and Dunn 1997), in ama-1(RNAi) embryos a cross-reactive mitotic epitope is detected where chromosome condensation is apparent, but specific α-PSer2 staining in interphase nuclei is abolished (Fig. 3B). When genes involved in transcription initiation are inhibited by RNAi, α-PSer5 and α-PSer2 staining levels are generally decreased in parallel (Walker et al. 2001). For example, α-PSer5 and α-PSer2 staining is reduced to background when the essential initiation factor TFIIB (ttb-1) is depleted (Fig. 3A; Walker et al. 2001). In cdk-9(RNAi) embryos, AMA-1 (Pol II) protein levels appeared normal (Fig. 2A), but the α-PSer2 signal was reduced to background in all interphase cells at all stages, as in ama-1(RNAi) embryos (Fig. 3B). Specific α-PSer2 staining was also eliminated in cit-1.1; cit-1.2(RNAi) embryos, but was normal when cit-1.1 and cit-1.2 were inhibited individually (Fig. 3B; data not shown). In striking contrast, in cdk-9(RNAi) embryos, α-PSer5 staining was not distinguishably different from wild-type (Fig. 3A). We conclude that P-TEFb is required for phosphorylation of CTD Ser 2, but not for the bulk of embryonic CTD Ser 5 phosphorylation.
P-TEFb is required broadly for gene expression
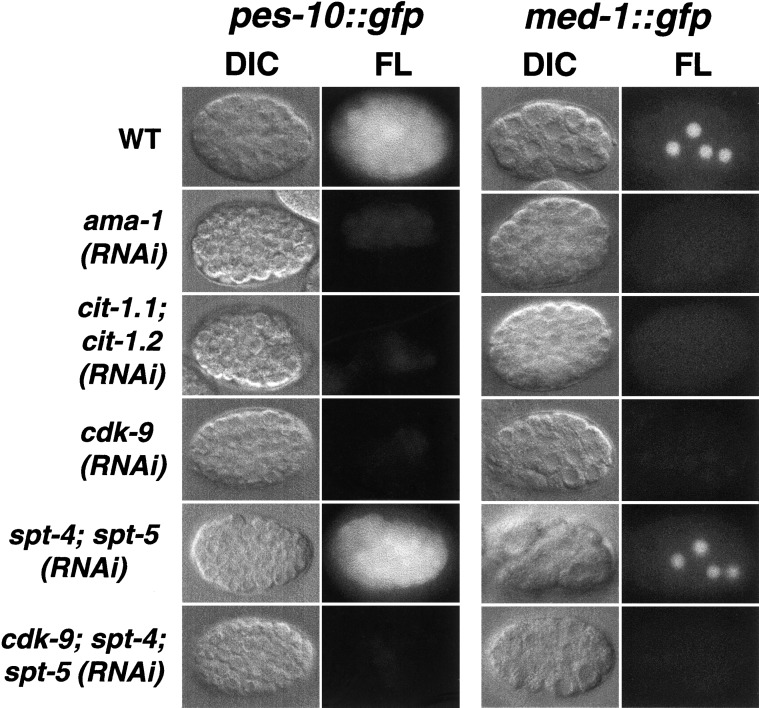
To investigate the importance of P-TEFb for transcription of individual genes, we performed RNAi experiments in C. elegans strains that carry reporter transgenes in which regulatory regions are intact, and coding regions are fused to GFP. These reporters are expressed in the early embryo in parallel to the corresponding endogenous genes. We first examined expression of med-1, which specifies mesendodermal lineages, and pes-10; each of these genes is expressed as transcription initiates (Seydoux and Fire 1994; Maduro et al. 2001). In cdk-9(RNAi) embryos, PES-10∷GFP and MED-1∷GFP expression was reduced to very low levels that were characteristic of ama-1(RNAi) embryos (Fig. 4). cdk-9 was similarly required for induction of the heat shock gene hsp-16.2 (Fig. 5A), a finding which is notable because S. cerevisiae heat shock gene transcription does not require ctk1 (Patturajan et al. 1999). cdk-9 was also required for expression of let-858 and rps-5, which are common to yeast and metazoans, and was essential at multiple genes which are specific to metazoans (Table 1). The latter group includes the broadly expressed genes cki-2 and sur-5 (Gu et al. 1998; Hong et al. 1998), and the developmental specification genes end-1 (Zhu et al. 1998), pha-4 (Horner et al. 1998; Kalb et al. 1998), and elt-5 (Koh and Rothman 2001). Simultaneous inhibition of cit-1.1 and cit-1.2 similarly prevented reporter gene expression (Figs. 4, 5A; Table 1), suggesting that both CDK-9 and cyclin T are broadly required for embryonic transcription.
Figure 4.
P-TEFb is required for expression of early embryonic genes. Wild-type (WT) and RNAi embryos (designated in rows) that were derived from transgenic GFP reporter strains were analyzed by DIC and fluorescence (FL) microscopy (as indicated above columns). These embryos are representative of the entire population analyzed in each of multiple independent experiments, in which >40 embryos were scored.
Figure 5.
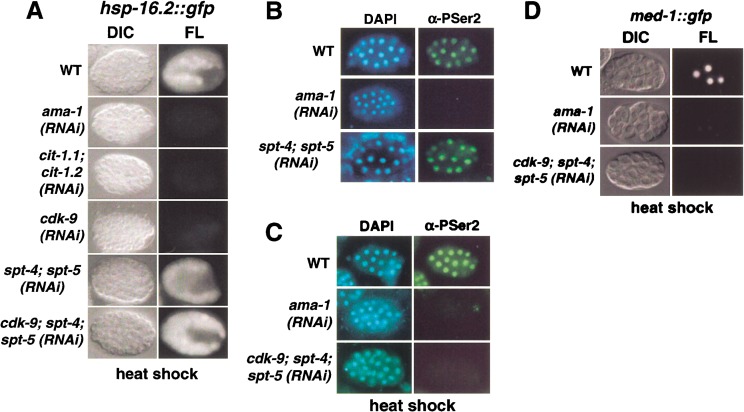
Requirement for P-TEFb is relieved at a heat shock gene by inhibition of spt-4 and spt-5. (A) Wild-type (WT) and RNAi hsp-16.2∷gfp embryos (designated in rows) were analyzed as in Fig. 4. Levels of HSP-16.2∷GFP expression varied within sets of wild-type, spt-4; spt-5(RNAi), and cdk-9; spt-4; spt-5(RNAi) embryos, but those shown correspond to representative differences between WT and RNAi embryos. (B) α-P-Ser2 staining in spt-4; spt-5(RNAi) embryos, analyzed as in Fig. 3. No mitotic cells are in focus in the ama-1(RNAi) (Pol II large subunit) embryo, which is shown to indicate background levels. (C) α-P-Ser2 staining is undetectable in heat shocked cdk-9; spt-4; spt-5(RNAi) embryos, which were analyzed as in (B). The α-P-Ser2 staining levels in heat shocked WT embryos were not detectably different from controls that were not heat shocked (data not shown). Some cross-reactive germline P granule staining is apparent in the ama-1(RNAi) embryo. (D) Heat shock did not induce MED-1∷GFP expression in cdk-9; spt-4; spt-5(RNAi) embryos, which were analyzed in parallel to the controls shown as in Fig. 4.
Table 1.
CDK-9/Cyclin T is broadly required for early embryonic gene expression
| Experiment
|
END-1::GFP (endoderm)
|
ELT-5::GFP (ectoderm)
|
PHA-4::GFP (pharynx)
|
CKI-2::GFP (cell
cycle)
|
SUR-5::GFP (MAP
kinase pathway)
|
LET-858::GFP (Conserved eukaryotic)
|
RPS-5::GFP (Conserved eukaryotic)
|
|---|---|---|---|---|---|---|---|
| WT | + | + | + | + | + | + | + |
| ama-1(RNAi) | − | − | − | − | − | − | − |
| cdk-9(RNAi) | − | − | − | − | − | − | − |
| cit-1.1; cit-1.2(RNAi) | − | ND | ND | ND | − | − | ND |
Reporter strains were scored as + when GFP was expressed at wild-type levels. They were scored as − when GFP was uniformly undetectable, or present at comparable trace levels in ama-1(RNAi), cdk-9(RNAi), and cit-1.1; cit-1.2(RNAi) embryos. In each experiment, more than 40 embryos were analyzed from at least six different injected P0 animals. Genes that are common to yeast and metazoans are referred to as “conserved eukaryotic.” ND, not done.
P-TEFb overcomes inhibition by SPT-4 and SPT-5 in vivo
To investigate the functional relationship between P-TEFb and SPT-4/SPT-5, we first analyzed requirements for spt-4 and spt-5 during early embryogenesis. Like cdk-9(RNAi) embryos, spt-4; spt-5(RNAi) embryos failed to gastrulate (data not shown). They arrested development with >300 cells and showed many signs of differentiation, including presence of birefringent granules (gut), twitching (muscle), and some morphogenesis (Fig. 2B; data not shown), suggesting that transcription was not completely blocked. Accordingly, in spt-4; spt-5(RNAi) embryos, α-PSer2 staining levels were approximately normal (Fig. 5B). spt-5(RNAi) embryos had very similar phenotypes, but most spt-4(RNAi) embryos survived to hatching (data not shown). The developmental arrest phenotypes of spt-4; spt-5(RNAi) embryos were highly consistent, and accompanied by depletion of the target mRNAs as detected by RT-PCR (data not shown), suggesting that spt-4 and spt-5 expression was significantly reduced in these RNAi embryos.
P-TEFb is required in vitro to overcome transcription pausing that depends upon SPT-4/SPT-5 (Wada et al. 1998b; Renner et al. 2001). If the importance of P-TEFb in vivo derives largely from this effect, it would be predicted that a reduction in spt-4 and spt-5 expression would relieve requirements for P-TEFb during early embryonic transcription. To test this model, we inhibited spt-4 and spt-5 along with cdk-9 by RNAi, using procedures in which individual and total input RNA concentrations were carefully controlled (Materials and Methods). PES-10∷GFP and MED-1∷GFP were expressed robustly in spt-4; spt-5(RNA) embryos, but were inhibited in all cdk-9; spt-4; spt-5(RNAi) embryos (Fig. 4). In addition, cdk-9; spt-4; spt-5(RNAi) embryos were indistinguishable from cdk-9(RNAi) embryos not only in their overall levels of CTD Ser 2 and Ser 5 phosphorylation, but also in their appearance upon terminal arrest (Fig. 3), suggesting that they were subject to a severe transcription block. Together, these findings suggest that depletion of SPT-4 and SPT-5 did not bypass requirements for cdk-9 at many embryonic genes.
Significantly, however, our analysis of heat shock gene transcription uncovered an inhibitory effect of SPT-4/SPT-5 in cdk-9(RNAi) embryos. In spt-4; spt-5(RNAi) embryos, the HSP-16.2∷GFP reporter was induced to wild-type levels by heat shock (Fig. 5A). Although HSP-16.2∷GFP was not induced above background in cdk-9(RNAi) embryos, in response to heat shock it was expressed robustly in 50%–70% of cdk-9; spt-4; spt-5(RNAi) embryos (Fig. 5A; data not shown). HSP-16.2∷GFP induction was also restored in ∼50% of cdk-9; spt-5(RNAi) embryos, but to moderately lower levels (data not shown). The cdk-9 mRNA and CDK-9 protein were comparably depleted in cdk-9(RNAi) and cdk-9; spt-4; spt-5(RNAi) embryos (Figs. 2A, 6A, lanes 2,4,6; Materials and Methods), indicating that this restoration of HSP-16.2∷GFP expression did not arise from a failure of cdk-9 RNAi. In contrast, in heat-shocked cdk-9; spt-4; spt-5(RNAi) embryos, MED-1∷GFP was not expressed above the low levels apparent in ama-1(RNAi) embryos, and CTD Ser 2 phosphorylation was undetectable (Fig. 5C,D). These last findings suggest that the HSP-16.2∷GFP expression observed in cdk-9; spt-4; spt-5(RNAi) embryos did not arise from the heat shock response stimulating an alternative pathway of CTD Ser 2 phosphorylation, or a more general recovery of transcription.
Figure 6.
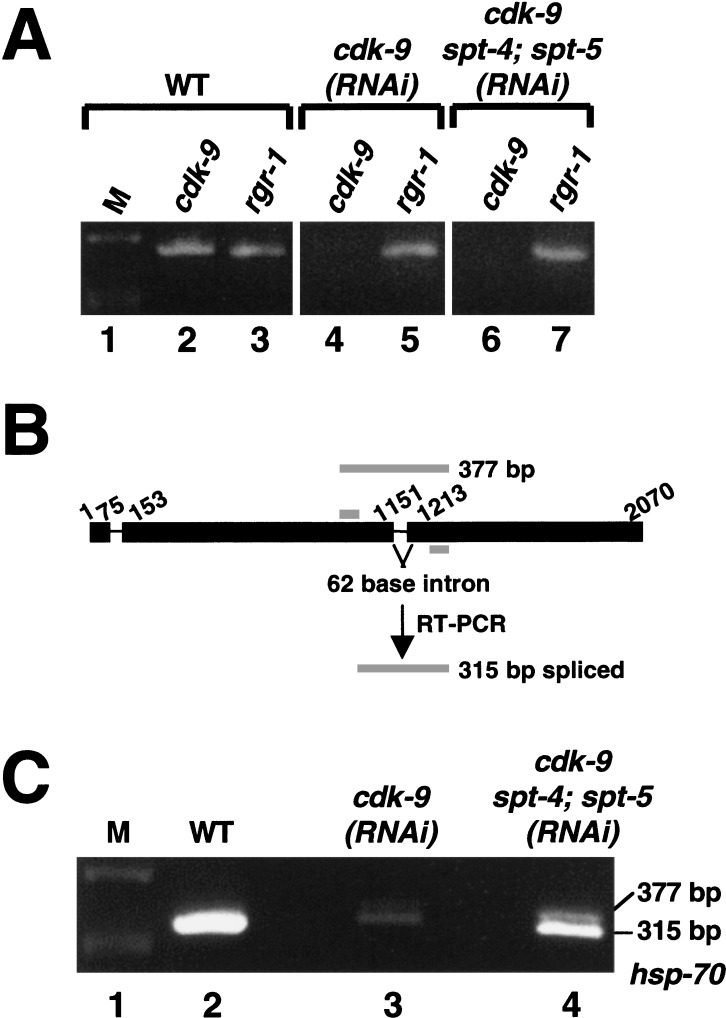
Restoration of endogenous hsp-70 transcription in cdk-9(RNAi) embryos by depletion of SPT-4/SPT-5. (A) Specific depletion of cdk-9 mRNA in RNAi embryos. Total RNA from the indicated embryo sets was assayed by RT-PCR to detect expression of cdk-9 (lanes 2,4,6), and a control mRNA (rgr-1; lanes 3,5,7). Each PCR primer set spanned an intron (data not shown). Products were analyzed on an agarose gel stained with ethidium bromide. DNA size markers are designated as M. (B) The hsp-70 gene, with exons indicated by a thick line. Primers used for RT-PCR (gray bars) flank intron 2. Sizes of predicted spliced and unspliced products are shown. (C) Endogenous hsp-70 expression. The samples analyzed in B were assayed by RT-PCR for hsp-70 RNA, using the primers shown in A. The hsp-70 mRNA (315-bp fragment) was present at markedly reduced levels in cdk-9(RNAi) embryos, but was significantly restored in cdk-9; spt-4; spt-5(RNAi) embryos. The 377-bp species in lane 4 corresponds to the unspliced hsp-70 sequence. It could not be established whether this product derived from incompletely processed RNA, because some samples contained trace amounts of genomic DNA (data not shown).
To investigate expression of a second heat shock gene in these sets of RNAi embryos, we assayed production of the endogenous hsp-70 mRNA by RT-PCR. We used primers that were located well within the gene and spanned an intron, so that they would detect the presence of long, processed transcripts (Fig. 6B). Compared to wild-type, in cdk-9(RNAi) embryos levels of the hsp-70 RT-PCR product were dramatically reduced (Fig. 6C, lanes 2,3). In contrast, a considerable level of endogenous hsp-70 mRNA was produced in cdk-9; spt-4; spt-5(RNAi) embryos (Fig. 6C, lane 4). We conclude that when SPT-4 and SPT-5 levels are depleted, these heat shock genes can be transcribed independently of P-TEFb.
Discussion
P-TEFb, a broadly required elongation factor
In this first genetics-based study of P-TEFb, we determined that C. elegans P-TEFb is broadly required for embryonic transcription. In cdk-9(RNAi) embryos, terminal arrest phenotypes were indistinguishable from those caused by lack of the essential transcription factors ama-1 or ttb-1 (Fig. 2B; Walker et al. 2001), and expression of each gene that we analyzed was reduced to background levels (Figs. 4, 5A, 6C; Table 1). Simultaneous inhibition of the cyclin T genes cit-1.1 and cit-1.2 had similar effects (Figs. 2B, 4, 5A; Table 1), suggesting that in the embryo these cyclin T proteins function redundantly. This broad requirement for P-TEFb suggests that a significant number of transcription activators or coactivators may recruit P-TEFb directly. P-TEFb-dependent elongation pathways might be especially important in rapidly dividing early embryonic cells, but we consider it more likely that P-TEFb is also generally essential for mRNA transcription in other metazoan cell types, a model that is consistent with previous pharmacologic inhibitor studies (Chao and Price 2001). Although Tat-dependent transcription is particularly sensitive to CDK-9 inhibition (Chao et al. 2000), treatment with highly specific CDK-9 inhibitors may significantly impair transcription of important host cell genes.
C. elegans P-TEFb also appears to be essential for most embryonic CTD Ser 2 phosphorylation. RNAi embryos in which either CDK-9 or cyclin T was depleted lacked detectable specific α-PSer 2 staining, comparably to ama-1(RNAi) embryos (Fig. 3B). Depletion of CDK-9 did not appear to reduce the overall level of CTD Ser 5 phosphorylation however (Fig. 3A), although we cannot exclude the possibility that some embryonic Ser 5 phosphorylation might be P-TEFb-dependent. Our results suggest that P-TEFb is the major embryonic CTD Ser 2 kinase, and that CTD Ser 2 phosphorylation by P-TEFb may be generally important for metazoan transcription. An analysis of two genes in S. cerevisiae suggests that during transcription, the CDK-9-related kinase Ctk1 phosphorylates Ser 2 after Ser 5 phosphorylation has occurred (Komarnitsky et al. 2000; Cho et al. 2001). In the C. elegans embryo, appearance of both the α-PSer 5 and α-Ser 2 CTD phosphoepitopes depends on the CTD Ser 5 kinase CDK-7 (Wallenfang and Seydoux 2002), and on essential initiation factors such as ttb-1 (Fig. 3A; Walker et al. 2001). Our finding that P-TEFb may not be required for most CTD Ser 5 phosphorylation (Fig. 3A) suggests that a sequential ordering of CTD Ser 5 and Ser 2 phosphorylation may be a general characteristic of Pol II transcription (Fig. 7).
Figure 7.
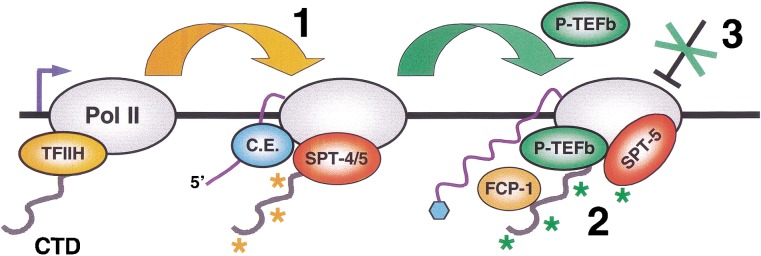
P-TEFb-dependent elongation mechanisms. Details and references are given in the text. (1) The TFIIH kinase (CDK-7; Kin28) phosphorylates CTD Ser 5 (orange asterisks) upon promoter clearance, and independently of P-TEFb. Pol II is then susceptible to DSIF (SPT-4/5)-dependent pausing. This phosphorylation also recruits capping enzyme (C.E.) to the CTD. (2) P-TEFb phosphorylates CTD Ser 2 and SPT-5 (green asterisks), and relieves inhibition by SPT-4/5. A speculative possibility is that SPT-4/5 may thereby be converted into a positively acting form. This mechanism may enhance the efficiency of elongation or mRNA processing by monitoring the presence or activity of factors required for these events. P-TEFb, the phosphatase FCP-1, and SPT-5 may remain associated with active elongating polymerase complexes, either continuously or intermittently, suggesting that SPT-5 may be regulated by opposing effects of P-TEFb and FCP-1. It is not known whether SPT-4 is then present. Heat shock genes can be expressed independently of cdk-9 when SPT-4 and SPT-5 are depleted by RNAi. (3) At most other genes, P-TEFb appears to be required in an additional mechanism. This mechanism may involve the release of a distinct, possibly chromatin-associated barrier to elongation, or an enhancement of cotranscriptional mRNA processing.
In S. cerevisiae, the CTD Ser 2 kinase Ctk1 is not essential for viability (Sterner et al. 1995; Patturajan et al. 1999), in apparent contrast to the importance of C. elegans P-TEFb for embryonic transcription. Yeast genes on average are shorter than metazoan genes, but some genes analyzed here are less than 2 kb in length (med-1; hsp-16.2), arguing against a significant difference in intrinsic elongation requirements. Alternatively, differences in biological contexts may be involved, or in S. cerevisiae a different kinase might compensate for Ctk1 at some genes or substrates. A candidate is the related kinase Bur1, which promotes elongation but preferentially phosphorylates CTD Ser 5 (Murray et al. 2001). Finally, given that mRNA processing is coupled to elongation (Hirose and Manley 2000), a broader requirement for a CTD Ser 2 kinase in metazoans might be related to the greater relative importance of mRNA splicing.
Functional antagonism between P-TEFb and SPT-4/SPT-5 in vivo
In light of the considerable evidence that SPT-4/SPT-5 promotes transcription elongation (as discussed at the beginning of this paper), it seems paradoxical that in vitro SPT-4/SPT-5 induces polymerase pausing that must be overcome by P-TEFb (Wada et al. 1998b; Renner et al. 2001). Here we obtained in vivo evidence to support the latter model. In cdk-9(RNAi) embryos, transcription of two different heat shock genes was significantly restored when spt-4 and spt-5 were also inhibited by RNAi (Figs. 5A, 6C). This recovery of heat shock gene expression in spt-4; spt-5; cdk-9(RNAi) embryos was not associated with a global activation of an alternative CTD Ser 2 phosphorylation pathway by heat shock (Fig. 5C), strongly suggesting that removal of an inhibitory effect of SPT-4/SPT-5 was involved. HSP-16.2∷GFP expression was restored to a lesser extent in spt-5; cdk-9(RNAi) embryos, indicating that both spt-4 and spt-5 contribute to this inhibition (data not shown). Although these effects could derive from unanticipated functions of P-TEFb and SPT-4/SPT-5, a simpler explanation is that these factors have opposing influences on transcription. Biochemical evidence indicates that P-TEFb counteracts SPT-4/SPT-5 by phosphorylating the CTD (Wada et al. 1998b; Yamaguchi et al. 1999; Renner et al. 2001), but its phosphorylation of SPT-5 might also be important (Ivanov et al. 2000). By demonstrating an antagonistic relationship between cdk-9 and spt-4/spt-5 directly in a genetic system, our data strongly support the model that overcoming SPT-4/SPT-5-dependent inhibition is a critical function of P-TEFb. At the same time, the model that SPT-4/SPT-5 has both negative and positive effects may explain why we did not observe elevated HSP-16.2∷GFP expression in spt-4; spt-5(RNAi) embryos (Fig. 5A).
A model that accommodates both positive and negative roles is that SPT-4/SPT-5 monitors recruitment of elongation or mRNA processing factors at the Pol II complex (Andrulis et al. 2000; Orphanides and Reinberg 2002). By enforcing this transcription “checkpoint,” SPT-4/SPT-5 may enhance the efficiency of elongation, or the fidelity of cotranscriptional mRNA processing (Fig. 7). This scheme is consistent with evidence that SPT-5 directly promotes mRNA capping (Wen and Shatkin 2000). SPT-5 may also promote elongation by acting as an antiterminator (Bourgeois et al. 2002), presumably after SPT-4/SPT-5-dependent pausing has been relieved by P-TEFb. It has been proposed that SPT-4/SPT-5 and NELF together induce 5′ pausing of Pol II prior to elongation or capping (Andrulis et al. 2000; Orphanides and Reinberg 2002), but the evidence that SPT-4/SPT-5 is opposed by P-TEFb suggests that SPT-4/SPT-5-dependent pausing might also occur more distally within a gene (Fig. 7). In yeast, CTD Ser 2 phosphorylation predominates >500 bp from the promoter, where Ctk1 and the CTD phosphatase Fcp1 engage in an ongoing process of CTD Ser 2 phosphorylation and dephosphorylation (Komarnitsky et al. 2000; Cho et al. 2001). SPT-5 similarly appears to be present along actively transcribed metazoan genes (Andrulis et al. 2000; Kaplan et al. 2000). If SPT-5-dependent pausing is regulated by a dynamic modulation of CTD Ser 2 phosphorylation (Fig. 7), this process may monitor recruitment of elongation or mRNA processing factors until late stages of the transcription cycle.
P-TEFb-dependent pathways of elongation control
cdk-9; spt-4; spt-5(RNAi) embryos expressed two heat shock genes, but not the early genes med-1 or pes-10 (Figs. 4, 5A,D, 6C). cdk-9; spt-4; spt-5(RNAi) embryos also were not distinguishably different in their terminal arrest phenotype from cdk-9(RNAi) or ama-1(RNAi) embryos, suggesting that they failed to transcribe many or possibly most other genes. In yeast, heat shock genes differ from most cellular genes in that they can be transcribed independently of the CTD, and do not require activation mechanisms that depend upon kin28 or certain Mediator components (Lee and Lis 1998; McNeil et al. 1998). Our findings indicate that heat shock genes are also distinct in their requirements for P-TEFb, suggesting that metazoans are able to sustain transcription elongation through two different P-TEFb-dependent pathways.
What distinguishes these P-TEFb-dependent postinitiation pathways from each other? It is possible that heat shock genes are highly resistant to SPT-4/SPT-5-dependent pausing, and that sufficient residual SPT-4/SPT-5 remained in cdk-9; spt-4; spt-5(RNAi) embryos to prevent expression of med-1, pes-10, and most other embryonic genes. This seems unlikely however, given that spt-4; spt-5(RNAi) embryos were characterized by consistent and severe developmental abnormalities (Fig. 2B). Alternatively, at most cellular genes P-TEFb may be required in an SPT-4/SPT-5-independent mechanism that was not predicted by in vitro experiments that were performed using naked DNA. For example, P-TEFb may be needed to overcome an SPT-4/SPT-5-independent elongation block that is specific to a chromatin environment. Consistent with this model, the FACT complex is required along with P-TEFb for elongation to occur on chromatin templates in vitro (Orphanides et al. 1998; Wada et al. 2000). P-TEFb might also activate other positive elongation factors. Alternatively, evidence that the phosphorylated CTD stimulates certain mRNA processing steps (Hirose and Manley 2000) suggests that these steps may be facilitated by P-TEFb, an effect that could increase elongation rates (Fong and Zhou 2001). Elucidation of this P-TEFb-dependent mechanism, and of why it is regulated differently at heat shock genes, should provide important and perhaps unexpected insights into postinitiation levels of transcription regulation.
Materials and methods
C. elegans and bioinformatics
C. elegans strains were maintained as described (Brenner 1974). The wild-type strain was N2. GFP reporter strains were generously provided by many laboratories, as cited previously (Walker et al. 2001). C. elegans orthologs of genes that encode CDK-9, T-type cyclins, and SPT-4 were identified by searching WORMpep or genomic databases (Sanger Centre) with human or S. cerevisiae protein sequences. C. elegans SPT-5 was identified previously (Hartzog et al. 1998; Wada et al. 1998a; Guo et al. 2000). Alignments were produced by Megalign (DNAStar). Both cdk-9 open reading frames (ORFs) H25P06.2A and H25P06.2B are represented in EST databases. Previously reported observations obtained in large-scale RNAi screens concur with our findings that RNAi inhibition of either cit-1.1 or cit-1.2 individually has no effect on development, and that spt-5(RNAi) leads to 100% embryonic lethality (Gonczy et al. 2000; Maeda et al. 2001). The cit-1.1, cit-1.2, spt-4, spt-5, and hsp-70 ORFs are F44B9.4, F44B9.3, F54C4.2, K08E4.1, and C12C8.1, respectively.
Immunostaining and fluorescence analysis
The P-CTD (α-PSer5) (Schroeder et al. 2000), H5 (α-PSer2) (Babco), 8WG16 (α-unP CTD) (Babco), and POL 3/3 (Bellier et al. 1997) antibodies were used to detect forms of the Pol II large subunit. α-PSer5 and α-PSer-2 staining were performed as in Seydoux and Dunn (1997), and POL 3/3 and 8WG16 staining as in Walker et al. (2001). It was reported that at extremely high antigen concentrations, α-PSer2 shows some cross-reactivity in vitro with the CTD phosphorylated at Ser 5 (Cho et al. 2001). α-PSer2 completely fails to detect the characteristic nuclear phospho-Ser 5 foci that are present in C. elegans and Drosophila embryonic germline precursors, however (Seydoux and Dunn 1997; Walker et al. 2001), indicating that this weak cross-reactivity does not affect our findings. Two rabbit antisera were raised against the CDK-9 peptide RPPKRPNTEHAQEPPKR, with a carboxyl terminal Cys added. For CDK-9 staining, freeze-cracked embryos were fixed in 3% paraformaldehyde and then methanol, and washed with PBT (1× phosphate buffered saline, 1% Triton X-100, 1% BSA) prior to staining with affinity-purified CDK-9 antiserum. CDK-9 antibody staining was competed by the immunogenic but not heterologous peptides (data not shown). For GFP analysis, embryos were transferred to 2% agarose pads. Images were captured with a Zeiss AxioSKOP2 microscope and AxioCam digital camera, and GFP or antibody staining intensities were compared over a range of exposure times. Pixel intensities were standardized using Adobe Photoshop 5.0.
RNAi analysis
cdk-9 (yk443b10), cit-1.2 (yk426d10), spt-4 (yk254b1), and spt-5 (yk72d11) cDNAs were obtained from Yuji Kohara (National Institute of Genetics, Japan). A cit-1.1 cDNA fragment was amplified by PCR from a C. elegans cDNA library [gift of Marc Vidal (Dana-Farber Cancer Center, Boston, MA)]. Sense and antisense RNA strands were synthesized using the Ribomax kit (Promega), then annealed and injected at 0.33–1.0 μg/μL into young adults (two to eight embryos). Embryos were collected after at least 24 h because uniform populations of terminally arrested spt-4; spt-5(RNAi), cdk-9(RNAi), and cit-1.1; cit-1.2(RNAi) embryos appeared at 18, 22, and 24 h after injection, respectively. In heat shock experiments, hsp-16.2∷gfp RNAi and control embryos were collected at least 24 h after injection, then incubated at 37°C for 1 min in 10 μL of M9. Fluorescence was examined 1 h later. Embryos that were not heat shocked were monitored in parallel to confirm their developmental phenotypes. In comparison to other heat shock protocols, this procedure minimized effects of heat shock on embryonic development, but yielded maximal expression of HSP-16.2∷GFP. To analyze expression of endogenous hsp-70, N2 and RNAi worms were heat shocked as above, then allowed to recover at room temperature for 10 min prior to RNA preparation.
In performing RNAi on cdk-9, spt-4, and spt-5 simultaneously, care was taken to control for injected RNA concentrations, because differences in input RNA levels can influence the efficiency of RNAi. These dsRNAs were mixed to 1:1:1 to a total concentration of 1.0 μg/μL. In parallel experiments, the phenotypic effects of administering these individual dsRNAs on their own were comparable when they were injected at either 0.33 or 1.0 μg/μL, or at 0.33 μg/μL but brought to a total RNA concentration of 1.0 μg/μL with an unrelated dsRNA (glp-1) (as judged by terminal phenotype and PES-10∷GFP, MED-1∷GFP, and HSP-16.2∷GFP expression; data not shown). In multiple experiments, RT-PCR indicated that in cdk-9(RNAi) and cdk-9; spt-4; spt-5(RNAi) embryos, the cdk-9 mRNA was reproducibly depleted in parallel to a similar extent (>10–100 fold reduction).
RT-PCR
Each RT-PCR reaction included total embryonic RNA prepared from 10 gravid hermaphrodites using Tri-reagent (Sigma). RT-PCR was performed essentially as described (Longman et al. 2000), using the Superscript One Step system (GIBCO BRL) and 40 cycles of amplification, followed by analysis on an ethidium bromide-stained agarose gel. RT-PCR primers used were 5′-951-hsp-70: GGTTGAAAAGGCACTTCGTGATGCAAAAAC; 3′-1327-hsp-70: CTTGAATAGAGACACCAGGCTGATTGTCTGC; 5′-518-cdk-9: GGATCGTGCAACTTTCTACTTGGTAATGGC; 3′-1050-cdk-9: GGTTCAGAAGACATTGCCGACCATAATGGC; 5′-1075-rgr-1: CCTTGAAGCTGCTGAACCAAGATTAGAAGTG; 3′-1585-rgr-1: CCTCCAACTGAATGAACTGAGCTCGGTCTCC.
Acknowledgments
We thank Grace Gill, Steve Buratowski, Craig Kaplan, and Blackwell lab members for helpful discussions of, or critically reading this manuscript. We are also grateful to David Bentley for the P-CTD antiserum, and to Olivier Bensaude for alerting us when cdk-9 appeared in C. elegans sequence databases. This work was supported by grants from the March of Dimes to T.K.B., from the NIH to T.K.B. (GM62891) and Y.S. (GM58012), and from the Lalor Foundation to E.Y.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL blackwell@cbr.med.harvard.edu; FAX (617) 278-3153.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.999002.
References
- Akhtar AX, Faye G, Bentley DL. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Bellier S, Dubois MF, Nishida E, Almouzni G, Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol Cell Biol. 1997;17:1434–1440. doi: 10.1128/mcb.17.3.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 tat by preventing premature RNA release at terminator sequences. Mol Cell Biol. 2002;22:1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DBX, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, Peterlin BM, Price DH. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhou Q. Tat activates human immunodeficiency virus type 1 transcriptional elongation independent of TFIIH kinase. Mol Cell Biol. 1999;19:2863–2871. doi: 10.1128/mcb.19.4.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Dantonel JC, Quintin S, Lakatos L, Labouesse M, Tora L. TBP-like factor is required for embryonic RNA polymerase II transcription in C. elegans. Mol Cell. 2000;6:715–722. doi: 10.1016/s1097-2765(00)00069-1. [DOI] [PubMed] [Google Scholar]
- Eberhardy SR, Farnham PJ. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J Biol Chem. 2001;276:48562–48571. doi: 10.1074/jbc.M109014200. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Flores O, Lee G, Kessler J, Miller M, Schlief W, Tomassini J, Hazuda D. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc Natl Acad Sci. 1999;96:7208–7213. doi: 10.1073/pnas.96.13.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bentley DL. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: Different functions for different segments of the CTD. Genes & Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Echeverri G, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Grana X, De Luca A, Sang N, Fu Y, Claudio PP, Rosenblatt J, Morgan DO, Giordano A. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc Natl Acad Sci. 1994;91:3834–3838. doi: 10.1073/pnas.91.9.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Orita S, Han M. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LET-60 Ras activity during vulval induction. Mol Cell Biol. 1998;18:4556–4564. doi: 10.1128/mcb.18.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Yamaguchi Y, Schilbach S, Wada T, Lee J, Goddard A, French D, Handa H, Rosenthal A. A regulator of transcriptional elongation controls vertebrate neuronal development. Nature. 2000;408:366–369. doi: 10.1038/35042590. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao S-M, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in caenorhabditis elegans. Genes & Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-3α, β, γ homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Kaltenbach L, Horner MA, Rothman JH, Mango SE. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell. 2000;6:705–713. doi: 10.1016/s1097-2765(00)00068-x. [DOI] [PubMed] [Google Scholar]
- Kanazawa S, Okamoto T, Peterlin BM. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Morris JR, Wu C, Winston F. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes & Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Lee DH, Koos DS, Ho RK, Stainier DY, Yelon D. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebrafish embryo. Development. 2002;129:1623–1632. doi: 10.1242/dev.129.7.1623. [DOI] [PubMed] [Google Scholar]
- Kim JB, Yamaguchi Y, Wada T, Handa H, Sharp PA. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol Cell Biol. 1999;19:5960–5968. doi: 10.1128/mcb.19.9.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- Lee DK, Duan HO, Chang C. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J Biol Chem. 2001;276:9978–9984. doi: 10.1074/jbc.M002285200. [DOI] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J Biol Chem. 1997;272:10990–10993. doi: 10.1074/jbc.272.17.10990. [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Hartzog GA. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics. 2001;159:487–497. doi: 10.1093/genetics/159.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Longman D, Johnstone IL, Caceres JF. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 2000;19:1625–1637. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3β homolog is mediated by MED-1 and -2 in C. elegans. Mol Cell. 2001;7:475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high- throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Mancebo HSY, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV tat transcriptional activation in vivo and in vitro. Genes & Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- McNeil JB, Agah H, Bentley D. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes & Dev. 1998;12:2510–2521. doi: 10.1101/gad.12.16.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S, Udupa R, Yao S, Hartzog G, Prelich G. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol Cell Biol. 2001;21:4089–4096. doi: 10.1128/MCB.21.13.4089-4096.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman-Smith ED, Rothman JH. The maternal-to-zygotic transition in embryonic patterning of Caenorhabditis elegans. Curr Opin Genet Dev. 1998;8:472–480. doi: 10.1016/s0959-437x(98)80120-2. [DOI] [PubMed] [Google Scholar]
- O'Brien TX, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Palancade B, Bellier S, Almouzni G, Bensaude O. Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. J Cell Sci. 2001;114:2483–2489. doi: 10.1242/jcs.114.13.2483. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Conrad NK, Bregman DB, Corden JL. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J Biol Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- Payne JM, Laybourn PJ, Dahmus ME. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- Powell-Coffman JA, Knight J, Wood WB. Onset of C. elegans gastrulation is blocked by inhibition of embryonic transcription with an RNA polymerase antisense RNA. Dev Biol. 1996;178:472–483. doi: 10.1006/dbio.1996.0232. [DOI] [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes & Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 1994;120:2823–2834. doi: 10.1242/dev.120.10.2823. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Lee JM, Hardin SE, Greenleaf AL. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for inhibition of gene expression and PIE-1 localization in the embryonic germ lineage of caenorhabditis elegans. Dev Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998a;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998b;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Orphanides G, Hasegawa J, Kim DK, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, et al. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- Walker AK, Rothman JH, Shi Y, Blackwell TK. Distinct requirements for C.elegans TAF(II)s in early embryonic transcription. EMBO J. 2001;20:5269–5279. doi: 10.1093/emboj/20.18.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallenfang MR, Seydoux G. cdk-7 is required for mRNA transcription and cell cycle progression in C. elegans embryos. Proc Natl Acad Sci. 2002;99:5527–5532. doi: 10.1073/pnas.082618399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang S-M, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity loop-specific binding to Tar RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Cap methyltransferase selective binding and methylation of GpppG-RNA are stimulated by importin-alpha. Genes & Dev. 2000;14:2944–2949. doi: 10.1101/gad.848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV- 1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Gold MO, Tang DN, Lewis DE, Aguilar-Cordova E, Rice AP, Herrmann CH. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes & Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes & Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]