Abstract
The pseudomalignant nature of the placenta prompted us to search for tumor suppressor gene hypermethylation, a phenomenon widely reported in cancer, in the human placenta. Nine tumor suppressor genes were studied. Hypermethylation of the Ras association domain family 1 A (RASSF1A) gene was found in human placentas from all three trimesters of pregnancy but was absent in other fetal tissues. Hypermethylation of Rassf1 was similarly observed in placentas from the rhesus monkey but not the mouse. An inverse relationship between RASSF1A promoter methylation and gene expression was demonstrated by bisulfite sequencing of microdissected placental cells and immunohistochemical staining of placental tissue sections using an anti-RASSF1A antibody. Treatment of choriocarcinoma cell lines, JAR and JEG3, by 5-aza-2′-deoxycytidine and trichostatin A led to reduction in RASSF1A methylation but increased expression. These observations extend the analogy between the primate placenta and malignant tumors to the epigenetic level.
Successful placentation is critical for human prenatal development. Placentation is a complex process involving a series of orchestrated events including cytotrophoblast differentiation, uterine invasion, and remodeling of the uterine vasculature.1 Placental development and trophoblast differentiation share many similarities with the process of tumorigenesis. Analogies have been drawn between placental tissues and malignancies in terms of their biological behavior, such as rapid proliferation and invasiveness2 and gene expression profiles,3 for example, the expression of angiogenetic factors4 and certain proto-oncogenes.5 The placenta has thus been described as being pseudomalignant in nature.6 We hypothesized that the analogy might extend to an epigenetic level.
Although the epigenetic phenomena of genomic imprinting7 and X chromosome inactivation8,9 have been well studied, few studies have systematically investigated the methylation status of tumor suppressor genes (TSGs) in the human placenta. TSG silencing by gene promoter hypermethylation is a well-recognized mechanism associated with the pathogenesis of malignancies.10 We aimed to investigate whether the promoters of some of the TSGs might similarly be methylated in the placenta and started by studying the methylation status of nine TSGs in human placental tissues.
Materials and Methods
Sample Collection
The study and the collection of human clinical samples were approved by the respective institutional review boards. Informed consent was sought from each subject. First trimester placental tissues were collected immediately after elective pregnancy terminations. Third trimester placental tissues were collected after elective cesarean delivery of uncomplicated pregnancies. Maternal peripheral blood samples (12 ml of ethylenediaminetetraacetic acid) were collected just before the performance of obstetrics procedures. Fetal tissue biopsies were obtained from two second trimester fetuses aborted spontaneously and confirmed to be karyotypically normal. For animal studies, pregnant mice (ICR) and rhesus monkeys were sacrificed near term. Murine placental tissues (E18.5) were obtained from pregnant mice at the Laboratory Animal Services Center of The Chinese University of Hong Kong with institutional animal ethics approval. Among the seven placentas collected, two were bisulfite sequenced individually and the remaining five were pooled before bisulfite sequencing. Rhesus tissues were obtained from the Guangdong-Zhaoqing Laboratory Animal Research Center in China using procedures stipulated by the Convention on International Trade in Endangered Species of Wild Fauna and Flora and the animal ethics committee at The Chinese University of Hong Kong. Peripheral blood samples were collected from the two pregnant rhesus monkeys. Placenta, liver, and heart tissues were collected from the fetuses of each pregnancy.
Sample Processing and DNA Extraction
Blood samples were centrifuged at 1600 × g for 10 minutes at 4°C. After the removal of the supernatant, the peripheral blood cell portion was recentrifuged at 2500 × g. Any residual plasma was further removed. DNA was extracted from peripheral blood cells using the Nucleon Blood DNA extraction kit (GE Health Care-Biosciences, Little Chalfont, UK) and from placental tissues using the QIAamp tissue kit (Qiagen, Hilden, Germany), each according to the manufacturer’s instructions.
Laser-Captured Microdissection
Formalin-fixed, paraffin-embedded tissue blocks from first-trimester placentas were retrieved from the block bank of the Department of Anatomical and Cellular Pathology, The Chinese University of Hong Kong. Tissue sections of 10 μm each were mounted onto membrane slides. After deparaffinization with xylene and ethanol, the sections were lightly stained with hematoxylin. Microdissection was performed using the PALM MicroLaser Systems (P.A.L.M. Microlaser Technologies AG, Bernreid, Germany) according to the manufacturer’s instructions. DNA was extracted from the microdissected samples as previously described.11
Isolation of Cytotrophoblast Progenitors
The cytotrophoblast progenitors were isolated by a previously described method.12,13 These cells are the population of cytotrophoblasts that have initiated differentiation along the invasive pathway. Thus, they are unlike the villous cytotrophoblasts, which are structurally part of the floating villi and are noninvasive in nature. Briefly, six to eight first trimester placentas were pooled and processed collectively. After removal of the syncytium, the cells were released by sequential enzyme digestion using collagenase followed by trypsin digestion. The cells were enriched by Percoll centrifugation with further purification through the removal of leukocytes using Dynabeads CD45 (Dynal Biotech LLC, Brown Deer, WI). DNA was extracted from the isolated cells using the Promega Wizard Genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer’s instructions.
Cell Lines
Trophoblast-derived choriocarcinoma cell lines JAR14 and JEG315 were purchased from the American Type Culture Collection (Manassas, VA). JAR was maintained in RPMI 1640 medium with 2 mmol/L l-glutamine adjusted to contain 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mmol/L HEPES, and 1.0 mmol/L sodium pyruvate (90%) and supplemented with fetal bovine serum (10%). JEG3 was maintained in Eagle’s minimum essential medium with 2 mmol/L l-glutamine and Earle’s balanced salt solution adjusted to contain 1.5 g/L sodium bicarbonate, 0.1 mmol/L nonessential amino acids, and 1.0 mmol/L sodium pyruvate (90%) and supplemented with fetal bovine serum (10%). The cells were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Bisulfite Conversion
Bisulfite converts unmethylated cytosine into uracil while leaving methylated cytosine unchanged.16 Extracted DNA samples were bisulfite converted using the CpGenome Universal DNA modification kit (Chemicon, Temecula, CA) according to the manufacturer’s instructions. For each conversion reaction, 1 μg of DNA was incubated at 50°C for 16 hours after the addition of reagent I.
Methylation-Specific Polymerase Chain Reaction (MSP)
The methylation status of nine human TSGs, namely, RASSF1A, CDKN2A (p14), CDKN2A (p16), CDKN2B, DAPK1, GSTP1, MGMT, CDH1, and PTEN was investigated by MSP.16 More specifically, we targeted the 5′-CpG island of RASSF1, which is the CpG island previously shown to be responsible for the transcriptional control of the A isoform of RASSF1, RASSF1A.17 For each MSP, 80 ng of bisulfite-converted DNA were amplified by the polymerase chain reaction (PCR) primers18,19,20,21,22,23,24 specific to either the methylated or the unmethylated DNA sequences, using a GeneAmp PCR core reagent kit (Applied Biosystems, Foster City, CA). The primer sequences are listed in Table 1. All assays were run in a reaction volume of 25 μl consisting of 1× buffer II, 200 μmol/L of each dNTPs, 200 nmol/L of each primer, and 1 U of AmpliTaq Gold. The MgCl2 concentrations used for each assay are listed in Supplemental Table 1A (available online at http://ajp.amjpathol.org). Genomic DNA universally methylated by SssI methylase and no template controls were also amplified with each set of reactions. The thermal profiles used for the methylation-specific PCR assays are listed in Supplemental Table 1B (available online at http://ajp.amjpathol.org).
Table 1.
Sequences of PCR Primers
| Oligo | Forward | Reverse | Product length (bp) | Reference (if any) |
|---|---|---|---|---|
| Methylation-specific PCR | ||||
| RASSF1A-M | 5′-GTGTTAACGCGTTGCGTATC-3′ | 5′-AACCCCGCGAACTAAAAACGA-3′ | 82 | 18 |
| RASSF1A-U | 5′-TTTGGTTGGAGTGTGTTAATGTG-3′ | 5′-CAAACCCCACAAACTAAAAACAA-3′ | 94 | 18 |
| p14-M | 5′-GTGTTAAAGGGCGGCGTAGC-3′ | 5′-AAAACCCTCACTCGCGACGA-3′ | 122 | 19 |
| p14-U | 5′-TTTTTGGTGTTAAAGGGTGGTGTAGT-3′ | 5′-CACAAAAACCCTCACTCACAACAA-3′ | 132 | 19 |
| p16-M | 5′-TTATTAGAGGGTGGGGCGGATCGC-3′ | 5′-GACCCCGAACCGCGACCGTAA-3′ | 150 | 20 |
| p16-U | 5′-TTATTAGAGGGTGGGGTGGATTGT-3′ | 5′-CAACCCCAAACCACAACCATAA-3′ | 151 | 20 |
| CDKN2B-M | 5′-GCGTTCGTATTTTGCGGTT-3′ | 5′-CGTACAATAACCGAACGACCGA-3′ | 148 | 20 |
| CDKN2B-U | 5′-TGTGATGTGTTTGTATTTTGTGGTT-3′ | 5′-CCATACATAACCAAACAACCAA-3′ | 154 | 20 |
| DAPK1-M | 5′-GGATAGTCGGATCGAGTTAACGTC-3′ | 5′-CCCTCCCAAACGCCGA-3′ | 98 | 21 |
| DAPK1-U | 5′-GGAGGATAGTTGGATTGAGTTAATGTT-3′ | 5′-CAAATCCCTCCCAAACACCAA-3′ | 106 | 21 |
| GSTP1-M | 5′-AGTTGCGCGGCGATTTC-3′ | 5′-GCCCCAATACTAAATCACGACG-3′ | 140 | 22 |
| GSTP1-U | 5′-TTTTTGGTTAGTTGTGTGGTGATTTT-3′ | 5′-CACTCCACCCCAATACTAAATC- ACA-3′ | 152 | |
| MGMT-M | 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ | 5′-GCACTCTTCCGAAAACGAAACG-3′ | 81 | 23 |
| MGMT-U | 5′-TTTGTGTTTTGATGTTTGTAGGTT- TTTGT-3′ | 5′-AACTCCACACTCTTCCAAAAAC- AAAAC-3′ | 93 | 23 |
| CDH1-M | 5′-TTAGGTTAGAGGGTTATCGCGT-3′ | 5′-TAACTAAAAATTCACCTACCGAC-3′ | 116 | 20 |
| CDH1-U | 5′-TAATTTTAGGTTAGAGGGTTATTGT-3′ | 5′-CACAACCAATCAACAACACA-3′ | 97 | 20 |
| PTEN-M | 5′-GGTTTCGGAGGTCGTCGGC-3′ | 5′-CCGCAACCGAATAATAACTAC- TACGACG-3′ | 158 | Modified from 24 |
| TEN-U | 5′-GTTTGGGTTTTGGAGGTTGTTGGT-3′ | 5′-AAACCACAACCAAATAATAAC- TACTACAACA-3′ | 166 | Modified from 24 |
| Real-time quantitative methylation-specific PCR | ||||
| RASSF1A-qM | 5′-CGTGTTAACGCGTTGCGTATC-3′ | 5′-TACGTAACAAACCCCACGAAC- TAAAAACG-3′ | 104 | |
| RASSF1A-qU | 5′-GGTTGGAGTGTGTTAATGTGTTGT- GTATT-3′ | 5′-TACATAACAAACCCCACAAAC- TAAAAACA-3′ | 112 | |
| Bisulfite sequencing (human) | ||||
| RASSF1A | ||||
| HsPromoter | 5′-GGGTTTTATAGTTTTTGTATTT- AGGTTTTT-3′ | 5′-CAACTCAATAAACTCAAACTCC- CCC-3′ | 201 | |
| HsExon1 | 5′-GGGGAGTTTGAGTTTATTGAGTTG-3′ | 5′-CTACCCCTTAACTACCCCTTCC-3′ | 297 | |
| PTEN | ||||
| PTENexon1 | 5′-GGYGGYGGTTGGTATATTT-3′ | 5′-ACAAAAACTACTAATAACRAAAC- TTCTTCTA-3′ | 334 | |
| Bisulfite sequencing (mouse) | ||||
| Rassf1 | ||||
| MmPromoter | 5′-AGGGGTTTTTTGAAAGGGTTTA- TTTT-3′ | 5′-CCAACTCRCRTAATTCAATAAAT- TCTAACTCC-3′ | 253 | |
| MmExon1 | 5′-GTTATGTYGGYGGAGTTAGAA- TTTATTGAATTA-3′ | 5′-CCCRCAACTATAATACTACCTCC- CTTTCC-3′ | 315 | |
| Bisulfite sequencing (rhesus) | ||||
| Rassf1 | ||||
| RhPromoter | 5′-AGGAAGGGTAAGGTTGGGGTTT-3′ | 5′-CCRCAACTCAATAAACTCAAAC- TCC-3′ | 250 | |
| RhExon1 | 5′-TYGGYGGAGTTTGAGTTTATTGAGTTG-3′ | 5′-CTACCCCTTAACTACCCCTTCC-3′ | 301 | |
| Real-time RT-PCR for RASSF1A mRNA | ||||
| RASSF1A-mRNA | 5′-GTCGTGCGCAAAGGCCT-3′ | 5′-GCGCGGCAGCGGTAGT-3′ | 70 |
Abbreviations: M, methylated target; U, unmethylated target.
Cloning and Bisulfite Genomic Sequencing
Each bisulfite-converted DNA sample was subjected to PCR by primers that did not discriminate between methylated and unmethylated sequences, using a GeneAmp PCR core reagent kit (Applied Biosystems). The primer sequences are listed in Table 1, and reaction conditions are listed in Supplemental Tables 1C and 1D (available online at http://ajp.amjpathol.org). Each subsequent PCR product was TA-cloned into pGEM-Teasy vector25 (Promega) for transformation into Escherichia coli strain JM109, according to the manufacturer’s instructions. Clones were picked randomly and colony PCR was then performed using vector primers T7 and SP6 to amplify the cloned inserts. Cycle sequencing was performed using BigDye version 1.1 (Applied Biosystems) and an automated capillary DNA sequencer Genetic Analyzer 3100 (Applied Biosystems). The sequences obtained were aligned and compared using SeqScape software (Applied Biosystems). The completeness of bisulfite conversion was first confirmed before scoring. The CpG sites sequenced as cytosine or thymine residues were scored as methylated or unmethylated, respectively. The methylated site frequency was calculated for each sample by dividing the total number of methylated sites over all cloned CpG sites.
Real-Time Quantitative MSP
The relative proportion of RASSF1A sequences that were methylated and unmethylated among the tissue biopsies obtained from the second trimester abortuses was quantified by real-time quantitative MSP,26 using PCR primers and TaqMan probes specific for the methylated and unmethylated sequences, respectively. The PCR was performed using a TaqMan PCR Core Reagent kit (Applied Biosystems) on an ABI 7900 HT sequence detection system (Applied Biosystems). The primer, probe, and standard calibration sequences are listed in Tables 1 and 2, and reaction conditions are listed in Supplemental Table 1E (available online at http://ajp.amjpathol.org). Target quantities were determined by a calibration curve prepared by serial dilutions of known concentrations of single-stranded synthetic DNA oligonucleotides (Proligo, Singapore) specific to the methylated and unmethylated amplicons. A methylation index for each sample was determined by dividing the copies of methylated molecules by the total copies of methylated and unmethylated molecules. Genomic DNA universally methylated by SssI methylase and no template controls were also amplified with each set of reactions.
Table 2.
Sequences of Fluorescent Probes and Calibration Standards
| Name | Sequence |
|---|---|
| Real-time quantitative methylation-specific PCR | |
| RASSF1A-MP | (FAM)−5′-TACGGTAGTTGGTTTTTGGTCGTGGTTATCGTTTTTAGTT-3′-(TAMRA) |
| RASSF1A-MS | 5′-GTTGGAGCGTGTTAACGCGTTGCGTATCGCGCGGGGTATCGCGTGTAATTTTATACGGTA- GTTGGTTTTTGGTCGTGGTTATCGTTTTTAGTTCGCGGGGTTCGTTACGT-3′ |
| RASSF1A-UP | (FAM)−5′-TATGGTAGTTGGTTTTTGGTTGTGGTTATTGTTTTTAGTT-3′-(TAMRA) |
| RASSF1A-US | 5′-GTTGGAGTGTGTTAATGTGTTGTGTATTGTGTGGGGTATTGTGTGTAATTTTATATGGTA- GTTGGTTTTTGGTTGTGGTTATTGTTTTTAGTTTGTGGGGTTTGTTATGT-3′ |
| Real-time RT-PCR for RASSF1A mRNA | |
| RASSF1A-mRNA-P | (FAM)- 5′-CAGTGCGCGCATTGCAAGTTCACC-3′-(TAMRA) |
| RASSF1A-mRNA-S | 5′-ATCAAGTCGTGCGCAAAGGCCTGCAGTGCGCGCATTGCAAGTTCACCTGCCACTACCGCT- GCCGCGCGCT−3′ |
Abbreviations: M, methylated target; U, unmethylated target; P, fluorescent probe; S, calibration standard; FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Immunohistochemical Staining of RASSF1A
Mouse monoclonal [3F3] anti-RASSF1A antibody (ab23950; Abcam, Cambridge, UK) was used. Serial sections (4 μm thick) of first trimester placental tissues were prepared, and antigen retrieval was performed using Bond Epitope Retrieval Solution 2 on the Bond-max automated immunostainer (Vision BioSystems, Mount Waverley, Australia) at 100°C for 25 minutes. Staining was performed according to a standard protocol in the immunostainer. Anti-RASSF1A antibody was used at a dilution of 1:150. A polymer detection system was selected to avoid the problem of nonspecific endogenous biotin staining. A normal liver tissue was used as a positive control, and negative controls were performed by replacing the antibody with Tris buffered saline. The stained slides were evaluated in five fields (×400) under light microscope by two independent observers. The proportion of the examined cells with positive staining was expressed as a percentage ranging from 0 to 100%. The rates of positive staining determined by each observer were averaged.
Treatment of JAR and JEG3 with 5-Aza-2′-Deoxycytidine (5-Aza-CdR) and Trichostatin A (TSA)
A methyltransferase inhibitor, 5-aza-CdR, was used to demethylate RASSF1A. After the seeding of JAR and JEG3, when the cells were observed to be at 20 to 30% confluence, 1 μmol/L or 3 μmol/L 5-aza-CdR (Sigma-Aldrich, St. Louis, MO) was added to the medium. The medium and 5-aza-CdR were replaced every 24 hours. JAR was kept for 96 hours and JEG3 for 72 hours. To extend the effect of 5-aza-CdR, an inhibitor of histone deacetylases, TSA (Sigma-Aldrich) was also added in some cases. For JAR incubated in 1 μmol/L 5-aza-CdR, either 0, 100, or 500 ng/ml TSA was added 24 hours before the cells were harvested. For JEG3 incubated in 1 μmol/L 5-aza-CdR, either 0, 50, or 100 ng/ml TSA was added 7 hours before the cells were harvested. DNA and RNA were isolated from the harvested cells using the QIAamp tissue kit (Qiagen) and Trizol reagents (Invitrogen, Carlsbad, CA), respectively.
Real-Time Reverse Transcriptase (RT)-PCR
Expression levels of RASSF1A in the choriocarcinoma cell lines, JAR and JEG3, were determined before and after demethylation. Total RNA was extracted with Trizol reagent (Invitrogen) followed by DNase I treatment (Invitrogen) according to the manufacturer’s instructions. RASSF1A mRNA was quantified using one-step real-time RT-PCR27 and normalized to the corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA concentration. The primer, probe, and standard calibration sequences are listed in Tables 1 and 2, and reaction conditions for the RASSF1A RT-PCR assay are listed in Supplemental Table 1F (available online at http://ajp.amjpathol.org). The RT-PCR assays were set up according to the manufacturer’s instructions (EZ rTth RNA PCR reagent set; Applied Biosystems) and performed in a ABI Prism 7300HT (Applied Biosystems).
Results
RASSF1A Hypermethylation in Human Placental Tissues
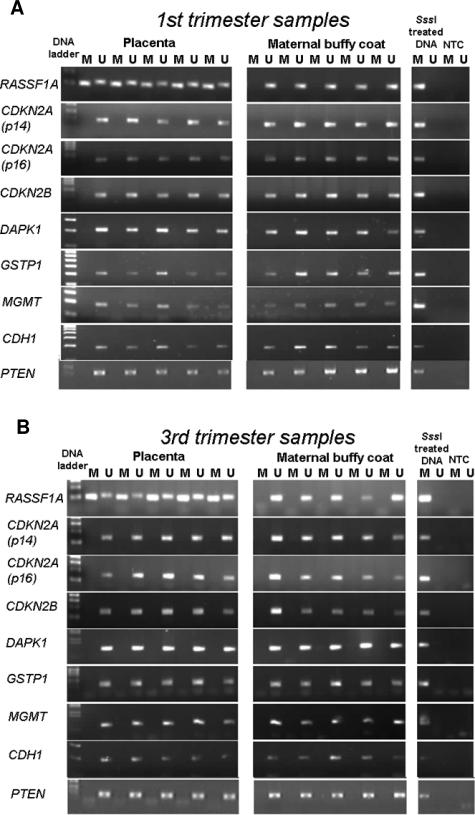
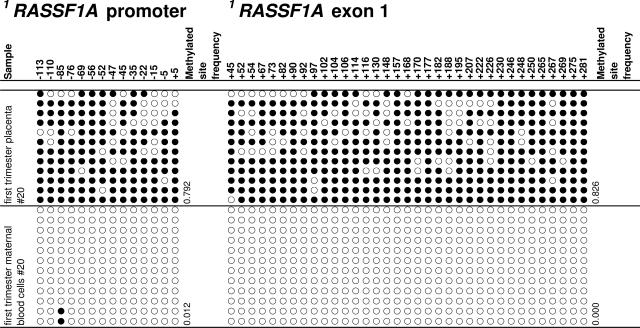
The methylation status of nine TSGs in human placental tissues was studied by MSP. Methylation of RASSF1A was observed in all first (n = 5) and third trimester (n = 5) placentas examined but not in the blood samples collected from pregnant women (Figure 1). To confirm these data, cloning and bisulfite sequencing were performed. RASSF1A hypermethylation was observed in the placental tissues while maternal blood cells were unmethylated. The data of one representative first trimester case are shown in Figure 2, whereas the data from all cases are shown in Supplemental Tables 2 and 3 (available online at http://ajp.amjpathol.org). For the first trimester placental samples, the methylated site frequency (number of methylated sites/all cloned CpG sites) ranged from 0.423 to 0.851 for the RASSF1A promoter and 0.826 to 0.938 for exon 1 of RASSF1A. For the third trimester placental samples, the methylated site frequency ranged from 0.619 to 0.929 for the RASSF1A promoter and 0.705 to 0.926 for exon 1 of RASSF1A. The methylated site frequencies for all of the studied maternal blood cell samples were less than 0.002. In all cases, the methylated site frequency of the placental tissue was higher than that of the corresponding blood cell sample.
Figure 1.
MSP analysis of nine TSGs in first (A) and third (B) trimester human placental tissues. Maternal blood cells collected from the corresponding pregnancies were used as controls for the unmethylated sequence, whereas SssI methylase-treated DNA was used as positive control for the methylated sequence. Genes are identified by the official symbol according to the HUGO gene nomenclature. M, methylated; U, unmethylated; NTC, no template control.
Figure 2.
Cloning and bisulfite sequencing of the RASSF1A CpG island in first trimester placental tissues and maternal blood cells. Results for one placental tissue sample and its corresponding blood cell sample are shown. Five cases each from the first and third trimesters of pregnancy were studied, and results are shown in Supplemental Tables 2 and 3, respectively (available online at http://ajp.amjpathol.org). Circles along one column represent one CpG site (filled circles, methylated; open circles, unmethylated). Each row represents one clone. The analyzed CpG sites are named according to the RASSF1 (Homo sapiens) GenBank accession NM_007182 with the start codon of its protein coding sequence as position +1. The first 1RASSF1 CpG site (−113) corresponds to chr3:50353354 (reverse strand) of the human genome in the UCSC Genome Browser (May 2004 assembly, hg17).
Lack of RASSF1A Hypermethylation in Nonplacental Fetal Tissues
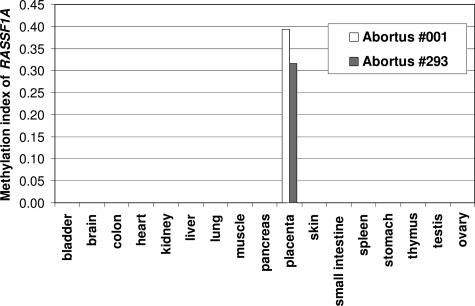
To explore whether methylation of RASSF1A was also observable in tissues of the fetus proper, real-time quantitative MSP28 analyses were applied to a panel of tissues obtained from two second trimester (gestational ages, 20.7 and 23 weeks) abortuses. Remarkably, hypermethylation of RASSF1A was only detected in the placental tissues (Figure 3).
Figure 3.
Real-time quantitative MSP analysis of RASSF1A in a panel of fetal and placental tissues from two abortuses. The methylation index was calculated by dividing the concentration of the methylated sequence by the sum of methylated and unmethylated sequences.
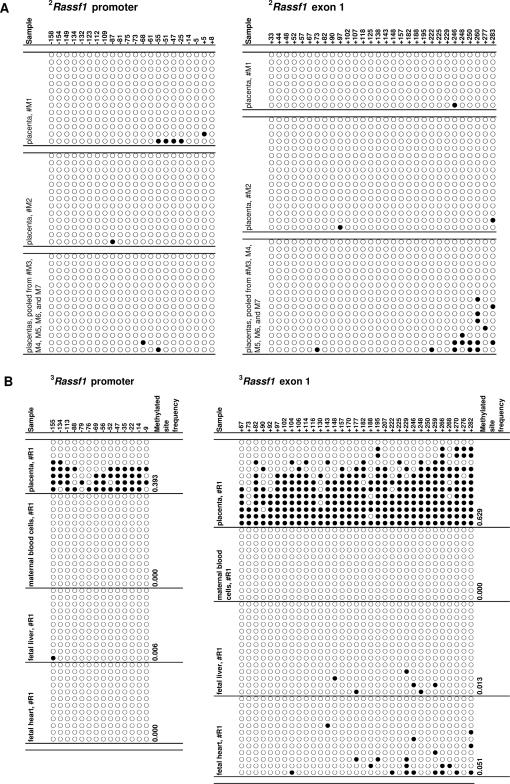
Hypermethylation of Rassf1 in Rhesus Monkey but Not Murine Placental Tissues
Homozygous Rassf1 knockout mice have been found to be fertile and free from reproductive consequence, which suggests that this gene is not essential for murine placental development.29 We studied whether Rassf1 hypermethylation might be observable in the murine placenta. With institutional animal ethics approval, DNA from seven E18.5 mouse (Mus musculus) placentas were subjected to cloning and bisulfite sequencing and revealed the lack of Rassf1 hypermethylation (Figure 4A). To explore whether Rassf1 hypermethylation might be observed in other primates, cloning and bisulfite sequencing was performed on term placental tissues collected from two rhesus monkeys (Macaca mulatta). Remarkably similar to the human placenta, Rassf1 hypermethylation was observed in the rhesus placental tissues but not in the blood cells of the pregnant monkeys or the liver and heart tissues of the fetus (Figure 4B and Supplemental Table 4, see http://ajp.amjpathol.org).
Figure 4.
Cloning and bisulfite sequencing of the Rassf1 CpG island in murine and rhesus placentas (filled circles, methylated; open circles, unmethylated). A: Seven E18.5 mouse placentas from one litter were studied. Two placentas were analyzed independently, and the remaining five were pooled. 2The murine Rassf1 genomic sequence is derived based on aligning the RASSF1A protein sequence for Homo sapiens (GenBank accession: NP_009113) with the genome of Mus musculus (UCSC genome browser, August 2005 assembly, mm7). Thus, the CpG sites are named with the start codon of the RASSF1A protein sequence (Homo sapiens) as position +1. The first CpG site (−158) corresponds to Chr9:107280886 (forward strand) of mm7. B: Tissues from the placenta, liver, and heart of two rhesus monkey fetuses and the corresponding maternal blood cells were analyzed. Data from the pregnancy of one rhesus monkey are shown here and that for the remaining case are shown in Supplemental Table 4 (available online at http://ajp.amjpathol.org). 3The rhesus Rassf1 genomic sequence is derived based on aligning the RASSF1A protein sequence for Homo sapiens (GenBank accession: NP_009113) with the genome of Macaca mulatta (UCSC genome browser, January 2005 assembly, Mmul_0.1). Thus, the CpG sites are named with the start codon of the RASSF1A protein sequence (Homo sapiens) as position +1. The first CpG site (−155) corresponds to SCAFFOLD10178:43405 (forward strand) of Mmul_0.1.
RASSF1A Hypermethylation in Microdissected Trophoblast Cells
We investigated the relationship between placental RASSF1A methylation and gene expression. To localize the placental cell types exhibiting RASSF1A hypermethylation, cloning and bisulfite sequencing were performed on pools of first trimester villous cytotrophoblasts, syncytiotrophoblasts, and stroma isolated by microdissection. To study the trophoblast population not associated with floating villi, cytotrophoblast progenitors that have initiated differentiation along the invasive pathway were isolated by sequential enzyme digestion.13 The methylated site frequencies were calculated for each of the cell populations and were found to be 0.755 in the villous cytotrophoblasts, 0.688 in the cytotrophoblast progenitors, 0.570 in the syncytiotrophoblasts, and 0.284 in the stromal cells (Supplemental Table 5, see http://ajp.amjpathol.org).
RASSF1A Expression in Human Placental Cells
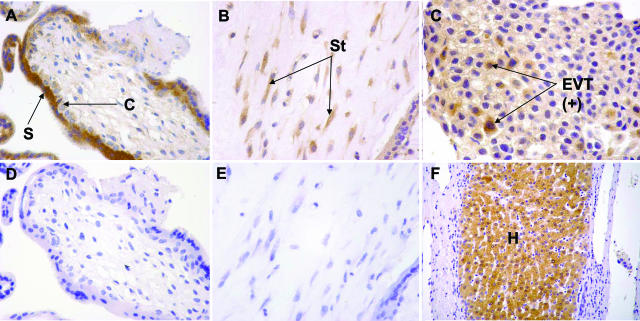
To investigate RASSF1A expression in the placental cell types studied above, we subjected sections of placental tissues (n = 13) from the first trimester of pregnancy to immunohistochemical staining by an anti-RASSF1A antibody (Abcam). Positive staining was observed in 10% of villous cytotrophoblasts, 14% of extravillous trophoblasts, 47% of syncytiotrophoblasts, and 66% of stromal cells (Figure 5, A–F). In essence, RASSF1A expression was highest in the stromal cells and lowest in the cytotrophoblasts, which were the cell populations with the least and the highest degree of RASSF1A methylation, respectively.
Figure 5.
Immunohistochemical staining for RASSF1A. Sections of first trimester placental tissues showing a chorionic villus (A), stroma (B), and a cell island composed primarily of extravillous trophoblasts (C). S, syncytiotrophoblast; C, cytotrophoblast; St, stromal cell; and EVT(+), extravillous trophoblast with positive staining. The corresponding negative controls for A and B are shown as D and E, respectively. Positive staining for RASSF1A in hepatocytes is shown in F and used as a positive control. H, hepatocytes. Brown color, immunohistochemistry staining; blue color, hematoxylin counterstain. Original magnifications: ×400 (A–C); ×200 (F).
Demethylation of RASSF1A in Choriocarcinoma Cell Lines
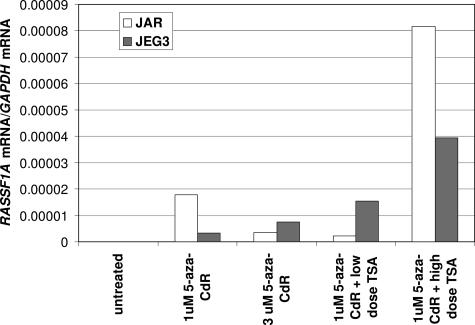
The difficulty in culturing primary human trophoblast cells prompted us to further investigate the relationship between RASSF1A methylation and gene expression in two choriocarcinoma cell lines, JAR14 and JEG3.15 Bisulfite sequencing indicated that the RASSF1A CpG island was heavily methylated for both cell lines (Supplemental Table 6, see http://ajp.amjpathol.org). RASSF1A mRNA was undetectable in both cell lines by real-time reverse transcriptase (RT)-PCR (Figure 6). Treatment of these cell lines with 5-aza-2′-deoxycytidine, with and without TSA resulted in re-expression of RASSF1A mRNA (Figure 6) with 28.2 to 77.1% and 36.89 to 50.64% reduction in the methylated site frequencies for JAR and JEG3, respectively (Supplemental Table 6, see http://ajp.amjpathol.org). This experiment thus demonstrated the reciprocal relationship between promoter methylation and gene expression at the RASSF1A locus in malignant cells of the trophoblastic lineage.
Figure 6.
Expression of RASSF1A mRNA in JAR and JEG3 after treatment with 5-Aza-CdR and TSA. RASSF1A and GAPDH mRNA levels were measured by real-time RT-PCR. Low-dose TSA: 100 ng/ml, 24 hours for JAR; 50 ng/ml, 7 hours for JEG3; high-dose TSA: 500 ng/ml, 24 hours for JAR; 100 ng/ml, 7 hours for JEG3.
Discussion
In summary, we have demonstrated that hypermethylation of RASSF1A could be observed in the human placenta. RASSF1 contains the most frequently methylated TSG promoter in human cancers.29 Up to 37 tumor types have been reported to harbor RASSF1A hypermethylation but rarely in the nontumorous tissue types studied to date.30 Recently, gene promoter methylation in a tissue-specific manner has been observed in genes with tissue-specific expression patterns.31,32,33 However, differential methylation in a tissue-specific manner has not been reported for RASSF1A. We have studied a panel of 17 fetal tissues and maternal blood cells, but hypermethylation of RASSF1A was found only in the placenta. RASSF1A hypermethylation was observed in every one of the studied human placental tissues.
Previous studies investigating the role of TSG methylation in choriocarcinomas and hydatidiform moles have used normal placentas for baseline comparison.34,35 Xue and colleagues34 reported the lack of hypermethylation in hypermethylated in cancer 1 (HIC1), TIMP metallopeptidase inhibitor 3 (TIMP3), cadherin 1, type 1, E-cadherin (CDH1), glutathione S-transferase pi (GSTP1), death-associated protein kinase 1 (DAPK1), and cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) (p16) in normal placentas. We have also studied the methylation status of the latter four genes and the results were concordant. Chen and colleagues,35 on the other hand, reported the presence of PTEN hypermethylation in 3 of 21 normal placentas examined. These data are in contrast to ours (Figure 1). We further performed bisulfite sequencing and failed to find evidence of PTEN hypermethylation in the five first and third trimester placentas studied (Supplemental Table 7, see http://ajp.amjpathol.org). Although the difference in sample size may explain the discrepancy in data, it is noteworthy that the earlier study35 was based on the use of methylation-sensitive restriction enzyme digestion followed by nested PCR. Although a positive control for the methylated sequence and a blank control were included in the analysis, the use of a control for the restriction digestion of the unmethylated sequence was not reported.35
Genomic imprinting is another possible reason for certain genes to be partially methylated in the placenta. For genomic imprinting, monoallelic methylation contributing to ∼50% of the sequenced clones to be methylated would be expected.36 In general, the methylated site frequencies in the placental tissues were much greater than 0.500 for RASSF1A. Thus, RASSF1A hypermethylation in placental tissues is unlikely to be attributable to imprinting. Formal exclusion of imprinting control would entail the demonstration of biallelic expression of RASSF1A.
We believe that the study findings may be of relevance to investigators studying the biology of RASSF1A and postulate that RASSF1A hypermethylation in the human placenta may play a significant biological role based on the following lines of evidence. RASSF1A hypermethylation was consistently observed in all studied placental tissues from all three trimesters of pregnancy. We have demonstrated a relationship between its expression and promoter methylation. The potential biological significance of this phenomenon in primate placentation could be inferred by its conservation in the placenta of the rhesus macaque but not in the murine placenta. Recently, a study on nasopharyngeal carcinoma reported that RASSF1A expression modulates the expression of inhibitor of DNA binding 2 (ID2).37 Incidentally, ID2 has been reported to be an important helix-loop-helix protein that regulates cytotrophoblast differentiation and function.38 Thus, there exists a possibility that RASSF1A may indeed play a significant biological role in cytotrophoblast development through its effects on ID2. Further studies should therefore be directed to address if ID2 expression in placental tissues is similarly modulated by RASSF1A, as in the case for nasopharyngeal carcinoma cells.
On the other hand, if RASSF1A plays a significant role in placental development, its methylation status may be altered in certain placental pathologies and is certainly another direction of investigation worth pursuing. We have conducted a preliminary study investigating RASSF1A hypermethylation in placental tissues collected from preeclamptic pregnancies. RASSF1A hypermethylation was similarly observed in such cases (unpublished data). Further investigation is being conducted to evaluate whether the degree of methylation and RASSF1A expression was different between normal and preeclamptic placentas.
In relation to the cross-species comparison, both the mouse and human placenta are classified as hemochorial. The murine placenta has commonly been studied as a model system for human placentation.39 Despite their similarities, there are notable differences between the murine and human placentas.40 In fact, the former is more specifically classified as hemotrichorial whereas the latter is hemomonochorial in nature.41 Thus, it may not be surprising to see the epigenetic differences between the placentas of the two species as found in this study. Interestingly, a recent study also reported the lack of evolutionary conservation of imprinted genes between the extraembryonic tissues of mouse and human.42 Although functional differences between human and nonhuman primate placentas have also been reported,43 the conservation of Rassf1 hypermethylation suggests that the rhesus placenta may be a better model for studying the epigenetic mechanisms in the human placenta than that of the mouse.43 Our study and that by Monk and colleagues42 highlight that epigenetics may add another dimension to the study of species evolution.
Last, the systematic difference in the methylation profile of RASSF1A in human placentas and maternal blood cells offer the opportunity for the development of a universal fetal DNA marker for maternal plasma detection. In recent years, research has shown that during pregnancy, nucleic acids of fetal origin can be detected in the plasma of pregnant women.44 Fetal DNA contributes some 5% of the total DNA detectable in maternal plasma with the rest contributed by maternal sources.45 Investigators have reported the aberrant increase in fetal DNA concentrations in the plasma of women whose pregnancy was complicated by preeclampsia, fetal aneuploidies, preterm labor, and so on.46 However, those studies were based on the detection of Y-chromosomal sequences that could readily be distinguished from the maternal background DNA as fetal-derived, but limiting those potential applications to pregnancies involving male fetuses only. The bulk of the maternal DNA background has been hypothesized to be derived from the maternal blood cells whereas the placenta was the predominant source of fetal nucleic acids in maternal plasma.47,48 Hence, the differences in the methylation profile of RASSF1A in the placenta and maternal blood cells could be exploited for the development a fetal DNA marker detectable in the plasma of all pregnancies through the detection of hypermethylated RASSF1A sequences.49
In conclusion, the present work has opened up numerous lines of future research including 1) the extension of the search of placental hypermethylation to all known TSGs, 2) the role of this phenomenon in placental biology and development, 3) the possibility of aberrant epigenetic alterations in pregnancy-associated disorders, and 4) the utility of a universal circulating fetal DNA marker for noninvasive prenatal assessments. The elucidation of these issues will be expected greatly to advance our understanding of the epigenetics and biology of the placenta.
Supplementary Material
Footnotes
Address reprint requests to Y.M. Dennis Lo, Department of Chemical Pathology, The Chinese University of Hong Kong, Room 38023, 1/F Clinical Sciences Building, Prince of Wales Hospital, 30-32 Ngan Shing St., Shatin, New Territories, Hong Kong Special Administrative Region, China. E-mail: loym@cuhk.edu.hk.
Supported by the Research Grants Council of the Hong Kong Special Administrative Region (China) (Earmarked Research Grant CUHK 4437/05M) and the Li Ka Shing Foundation (Chair Professorship Scheme to Y.M.D.L.).
R.W.K.C. and S.S.C.C. contributed equally to this work.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Y.M.D.L. wishes to declare that a patent application describing the prenatal diagnostic applications of the methylation changes described in this article has been filed. Y.M.D.L. also wishes to declare that he is a shareholder of Plasmagene Biosciences Limited.
References
- Janatpour MJ, Utset MF, Cross JC, Rossant J, Dong J, Israel MA, Fisher SJ. A repertoire of differentially expressed transcription factors that offers insight into mechanisms of human cytotrophoblast differentiation. Dev Genet. 1999;25:146–157. doi: 10.1002/(SICI)1520-6408(1999)25:2<146::AID-DVG9>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Bischof P, Meisser A, Campana A. Biochemistry and molecular biology of trophoblast invasion. Ann NY Acad Sci. 2001;943:157–162. doi: 10.1111/j.1749-6632.2001.tb03799.x. [DOI] [PubMed] [Google Scholar]
- Janneau JL, Maldonado-Estrada J, Tachdjian G, Miran I, Motte N, Saulnier P, Sabourin JC, Cote JF, Simon B, Frydman R, Chaouat G, Bellet D. Transcriptional expression of genes involved in cell invasion and migration by normal and tumoral trophoblast cells. J Clin Endocrinol Metab. 2002;87:5336–5339. doi: 10.1210/jc.2002-021093. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Genbacev O, Fisher SJ. The human placenta remodels the uterus by using a combination of molecules that govern vasculogenesis or leukocyte extravasation. Ann NY Acad Sci. 2003;995:73–83. doi: 10.1111/j.1749-6632.2003.tb03211.x. [DOI] [PubMed] [Google Scholar]
- Ohlsson R. Growth factors, protooncogenes and human placental development. Cell Differ Dev. 1989;28:1–15. doi: 10.1016/0922-3371(89)90018-x. [DOI] [PubMed] [Google Scholar]
- Strickland S, Richards WG. Invasion of the trophoblasts. Cell. 1992;71:355–357. doi: 10.1016/0092-8674(92)90503-5. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Moore T, Detmar J, Lewis A, Hemberger M, Jammes H, Kelsey G, Roberts CT, Jones H, Constancia M. Epigenetics and imprinting of the trophoblast—a workshop report. Placenta. 2006;27:S122–S126. doi: 10.1016/j.placenta.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Axelman J, Jeppesen P. Differential X reactivation in human placental cells: implications for reversal of X inactivation. Am J Hum Genet. 2005;77:355–364. doi: 10.1086/432815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Ferguson-Smith AC. Developmental biology: the X-inactivation yo-yo. Nature. 2005;438:297–298. doi: 10.1038/438297a. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Chan AS, To KF, Lo KW, Mak KF, Pak W, Chiu B, Tse GM, Ding M, Li X, Lee JC, Huang DP. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from southern Chinese. Cancer Res. 2000;60:5365–5370. [PubMed] [Google Scholar]
- Librach CL, Werb Z, Fitzgerald ML, Chiu K, Corwin NM, Esteves RA, Grobelny D, Galardy R, Damsky CH, Fisher SJ. 92-kd type IV collagenase mediates invasion of human cytotrophoblasts. J Cell Biol. 1991;113:437–449. doi: 10.1083/jcb.113.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier JF, Weier HU, Jung CJ, Gormley M, Zhou Y, Chu LW, Genbacev O, Wright AA, Fisher SJ. Human cytotrophoblasts acquire aneuploidies as they differentiate to an invasive phenotype. Dev Biol. 2005;279:420–432. doi: 10.1016/j.ydbio.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Pattillo RA, Hussa RO, Huang WY, Delfs E, Mattingly RF. Estrogen production by trophoblastic tumors in tissue culture. J Clin Endocrinol. 1972;34:59–61. [PubMed] [Google Scholar]
- Kohler PO, Bridson WE. Isolation of hormone-producing clonal lines of human choriocarcinoma. J Clin Endocrinol Metab. 1971;32:683–687. doi: 10.1210/jcem-32-5-683. [DOI] [PubMed] [Google Scholar]
- Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56:722–727. [PubMed] [Google Scholar]
- Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- Lo KW, Kwong J, Hui AB, Chan SY, To KF, Chan AS, Chow LS, Teo PM, Johnson PJ, Huang DP. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61:3877–3881. [PubMed] [Google Scholar]
- Esteller M, Tortola S, Toyota M, Capella G, Peinado MA, Baylin SB, Herman JG. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 2000;60:129–133. [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, Jen J, Herman JG, Sidransky D. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000;60:892–895. [PubMed] [Google Scholar]
- Jerónimo C, Usadel H, Henrique R, Oliveira J, Lopes C, Nelson WG, Sidransky D. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst. 2001;93:1747–1752. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]
- Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- Zysman MA, Chapman WB, Bapat B. Considerations when analyzing the methylation status of PTEN tumor suppressor gene. Am J Pathol. 2002;160:795–800. doi: 10.1016/S0002-9440(10)64902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res. 1999;59:3899–3903. [PubMed] [Google Scholar]
- Ng EKO, Tsui NBY, Lam NY, Chiu RWK, Yu SC, Wong SC, Lo ES, Rainer TH, Johnson PJ, Lo YMD. Presence of filterable and nonfilterable mRNA in the plasma of cancer patients and healthy individuals. Clin Chem. 2002;48:1212–1217. [PubMed] [Google Scholar]
- Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 2005;65:92–98. [PubMed] [Google Scholar]
- Pfeifer GP, Dammann R. Methylation of the tumor suppressor gene RASSF1A in human tumors. Biochemistry (Mosc) 2005;70:576–583. doi: 10.1007/s10541-005-0151-y. [DOI] [PubMed] [Google Scholar]
- Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–3508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- Strathdee G, Sim A, Brown R. Control of gene expression by CpG island methylation in normal cells. Biochem Soc Trans. 2004;32:913–915. doi: 10.1042/BST0320913. [DOI] [PubMed] [Google Scholar]
- Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci USA. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue WC, Chan KY, Feng HC, Chiu PM, Ngan HY, Tsao SW, Cheung AN. Promoter hypermethylation of multiple genes in hydatidiform mole and choriocarcinoma. J Mol Diagn. 2004;6:326–334. doi: 10.1016/S1525-1578(10)60528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ye D, Xie X, Lu W, Zhu C, Chen X. PTEN promoter methylation and protein expression in normal early placentas and hydatidiform moles. J Soc Gynecol Invest. 2005;12:214–217. doi: 10.1016/j.jsgi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Epigenetic status of human embryonic stem cells. Nat Genet. 2005;37:585–587. doi: 10.1038/ng1556. [DOI] [PubMed] [Google Scholar]
- Chow LS, Lam CW, Chan SY, Tsao SW, To KF, Tong SF, Hung WK, Dammann R, Huang DP, Lo KW. Identification of RASSF1A modulated genes in nasopharyngeal carcinoma. Oncogene. 2006;25:310–316. doi: 10.1038/sj.onc.1209001. [DOI] [PubMed] [Google Scholar]
- Janatpour MJ, McMaster MT, Genbacev O, Zhou Y, Dong J, Cross JC, Israel MA, Fisher SJ. Id-2 regulates critical aspects of human cytotrophoblast differentiation, invasion and migration. Development. 2000;127:549–558. doi: 10.1242/dev.127.3.549. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Malassiné A, Frendo JL, Evain-Brion D. A comparison of placental development and endocrine functions between the human and mouse model. Hum Reprod Update. 2003;9:531–539. doi: 10.1093/humupd/dmg043. [DOI] [PubMed] [Google Scholar]
- Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod Biol Endocrinol. 2004;2:46. doi: 10.1186/1477-7827-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci USA. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golos TG. Pregnancy initiation in the rhesus macaque: towards functional manipulation of the maternal-fetal interface. Reprod Biol Endocrinol. 2004;2:35. doi: 10.1186/1477-7827-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- Lo YMD, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu RWK, Lo YMD. The biology and diagnostic applications of fetal DNA and RNA in maternal plasma. Curr Top Dev Biol. 2004;61:81–111. doi: 10.1016/S0070-2153(04)61004-0. [DOI] [PubMed] [Google Scholar]
- Lui YYN, Chik KW, Chiu RWK, Ho CY, Lam CW, Lo YMD. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin Chem. 2002;48:421–427. [PubMed] [Google Scholar]
- Ng EKO, Tsui NBY, Lau TK, Leung TN, Chiu RWK, Panesar NS, Lit LCW, Chan KW, Lo YMD. mRNA of placental origin is readily detectable in maternal plasma. Proc Natl Acad Sci USA. 2003;100:4748–4753. doi: 10.1073/pnas.0637450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KCA, Ding C, Gerovassili A, Yeung SW, Chiu RWK, Leung TN, Lau TK, Chim SSC, Chung GTY, Nicolaides KH, Lo YMD. Hypermethylated RASSF1A in maternal plasma: a universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin Chem. 2006;52:2211–2218. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.