Abstract
We report a cross-species expression profiling analysis of the human, mouse, and rat male meiotic transcriptional program, using enriched germ cell populations, whole gonads, and high-density oligonucleotide microarrays (GeneChips). Among 35% of the protein-coding genes present in rodent and human genomes that were found to be differentially expressed between germ cells and somatic controls, a key group of 357 conserved core loci was identified that displays highly similar meiotic and postmeiotic patterns of transcriptional induction across all three species. Genes known to be important for sexual reproduction are significantly enriched among differentially expressed core loci and a smaller group of conserved genes not detected in 17 nontesticular somatic tissues, correlating transcriptional activation and essential function in the male germ line. Some genes implicated in the etiology of cancer are found to be strongly transcribed in testis, suggesting that these genes may play unexpected roles in sexual reproduction. Expression profiling data further identified numerous conserved genes of biological and clinical interest previously unassociated with the mammalian male germ line.
Keywords: bioinformatics, spermatogenesis, transcriptome
Meiosis and gametogenesis are critical processes in the transmission of genetic material to subsequent generations during sexual reproduction. A large number of genes involved in this process have been characterized in model organisms, such as budding and fission yeast (1–4) and nematodes (5), but comparatively few essential and specifically expressed loci are known in mammals, particularly humans (6, 7). In males, mitotically growing spermatogonia develop into meiotic spermatocytes that give rise to haploid spermatids which differentiate into mature sperm. This pathway is controlled in part by somatic Sertoli cells that physically interact with germ cells and communicate with them by hormonal cues (for review, see refs. 8–11). Many of the loci required for meiotic landmark events such as recombination and gamete formation are known to be expressed only in cells capable of undergoing the process, but the regulatory network that confers germ-line-specific transcription in higher eukaryotes is poorly understood, especially at the mitotic and meiotic stages (12).
Recent studies have identified a large number of genes active during gametogenesis using testicular expressed sequence tag libraries, serial analysis of gene expression (SAGE) and microarray profiling of enriched germ cell and testis samples from rodents (13, 14). These array studies covered approximately one third of the genes currently known to be encoded by the mouse and rat genomes and showed that differentiating germ cells up-regulate hundreds of transcripts (15–17), including many that encode germ-line-specific products (18). Although numerous mRNAs have also been detected in mature human sperm, regulation of gene expression underlying germ cell development and the roles these transcripts may play during embryogenesis remain unclear (19). Statistical analysis, using data from the Gene Ontology Consortium (20), shows that genes known or predicted to be important for meiosis and reproduction are significantly enriched among loci expressed in testicular cell types (13). Thus, profiling experiments likely help identify factors important for the meiotic developmental pathway in both yeast (3, 4) and mammals (14).
In this study, enriched mitotic, meiotic, and postmeiotic germ cells were compared with somatic Sertoli cells to select differentially expressed mouse and rat loci. Among them, a group of conserved core genes were identified whose expression profiles in meiotic and postmeiotic germ cells are highly similar between rodents and human. Numerous potentially testis-specific loci were found by comparing the transcriptional activity of differentially expressed genes in testis to 17 somatic control tissues. Using whole-genome arrays covering all currently annotated genes in rodent and human genomes together with a comprehensive cross-species experimental approach revealed several hundred novel genes likely to be involved in the mammalian meiotic process. A graphical display of the expression data is available from the GermOnline database (www.germonline.org) that can be searched for individual genes as well as groups of loci (21). Additional information, including analysis software and raw data, is available from the authors upon request [see supporting information (SI) Results].
Results and Discussion
Defining the Mammalian Testicular Expression Program.
mRNA from highly enriched populations of rodent somatic Sertoli cells and mitotic spermatogonia, meiotic spermatocytes and postmeiotic round spermatids as well as isolated seminiferous tubules and total testis samples were analyzed by using high density oligonucleotide microarrays [Fig. 1; see also SI Table 1]. Total and cRNA quality was found to be very homogenous (SI Fig. 5 a and b) and the overall reproducibility of the data was excellent as shown by a distance matrix (SI Fig. 5c). The transcripts corresponding to GeneChip probe set IDs were annotated by using information provided by the GeneChip manufacturer and additional data retrieved by links to external databases (SI Fig. 6 a and b).
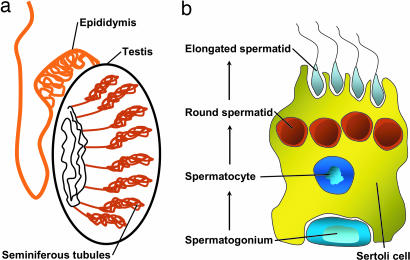
Fig. 1.
Schematic representation of the mammalian testis. (a) Drawing of the testis containing seminiferous tubules and the epididymis. (b) Sertoli nurse cell associated with a mitotic spermatogonium, a meiotic pachytene spermatocyte, and round and elongated postmeiotic spermatids.
Genes whose expression signals varied significantly between Sertoli cells, germ cells, tubules, and whole-organ controls were designated as “differentially expressed in testis” (DET) (this abbreviation is also used in the BioMart query form of GermOnline). This approach identified 9,280 and 5,895 probe sets that map to 7,066 and 5,140 unique protein-coding mouse and rat genes, respectively, representing ≈36% of the transcripts detected by the rodent GeneChips (22, 23). The differentially expressed probe sets were further classified by using a hierarchical clustering algorithm (SI Fig. 7 a and b). Somatic Sertoli cells and mitotic spermatogonia show similar profiles, distinguishing them from meiotic spermatocytes and postmeiotic spermatids clustered together with tubules and total testis samples (top dendrogram). This finding is in agreement with earlier observations using similar rat samples (15). Subsequently, human chondrocytes and smooth vascular muscle cells were compared with enriched pachytene spermatocytes and postmeiotic round spermatids, yielding 6,499 probe sets that correspond to 5,119 unique genes as differentially expressed between somatic controls and germ cells (SI Fig. 7c). Hierarchical clustering identified somatic, meiotic and postmeiotic expression classes (SI Fig. 7c, left dendrogram). As expected, the expression signals of differentially expressed genes distinguished the human somatic from the germ cell samples (SI Fig. 7c, upper dendrogram).
Next, Mus musculus was selected as the reference species to determine the number of clusters within the testicular expression program because extensive annotation data are available, its sample set includes four testicular cell types, and it yields the largest number of differentially expressed genes. A clustering algorithm (partitioning around medoids [PAM]) was used to separate mouse genes into four empirically determined groups that best fit the cell populations analyzed. Seven thousand sixty-six genes were thus classified into broad somatic (1,684), mitotic (2,581), meiotic (1,809), and postmeiotic (1,435) expression clusters showing transcriptional peaks in somatic Sertoli cells, mitotic spermatogonia, meiotic spermatocytes and postmeiotic round spermatids, respectively (SI Fig. 7d). Note that this classification is based on the strongest level of induction and does not necessarily reflect cell-type specific expression.
Identification of Conserved and Differentially Expressed Meiotic Genes.
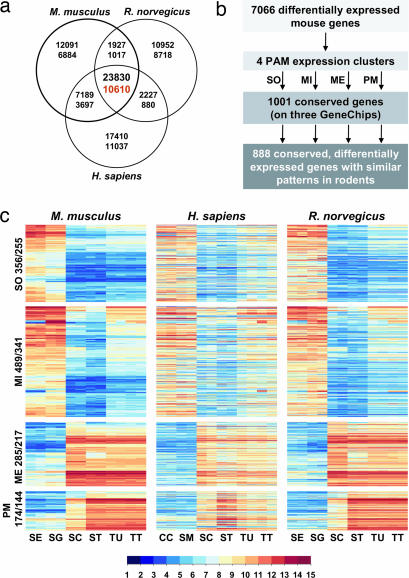
To classify genes as “conserved and differentially expressed in testis” (CDET) (this abbreviation is also used in the BioMart query form of GermOnline), we first assembled a list of 10,610 mouse, rat and human homologs for which expression data were available (Fig. 2a) and identified 1,001 loci as being conserved at the peptide level and differentially expressed in all three species (Fig. 2b). Finally, a group of 888 conserved genes showing very similar patterns in rodents (correlation coefficient of >0.8) was identified and distributed among the four clusters previously found to be differentially expressed in testis (DET). We made two striking observations. First, the majority of rodent genes showing strong expression in enriched somatic Sertoli cells and mitotic spermatogonia were not, or only barely, detected in adult tubules or total testis samples (Fig. 2c). This underlines the importance of using highly purified populations when studying gene expression in cells that constitute only a small fraction of the total testicular cell mass. Second, most of the human orthologs of 255 somatic and 341 mitotic mouse and rat genes showed clear expression in nontesticular somatic controls. This result implies that the vast majority of the genes expressed in Sertoli cells and spermatogonia are also active in nonreproductive tissues such as chondrocytes and smooth vascular muscle. Our experiment also identified 217 meiotic and 144 postmeiotic genes, using enriched rodent spermatocytes and spermatids. These genes often appear to be specific for meiotic and postmeiotic cells, because in most cases they are not expressed in Sertoli cells and spermatogonia. Moreover, their human orthologs display strong germ cell expression, whereas they show no or only very weak signals in the somatic controls. As expected, the data obtained with purified meiotic and postmeiotic germ cell populations were confirmed by using isolated seminiferous tubules and total testis samples in all three species, because their testicular cell mass predominantly consists of meiotic and postmeiotic germ cells.
Fig. 2.
The core testicular transcriptome in mammals (CDET). (a) Venn diagram of conserved probesets (top number) and corresponding genes (bottom number) represented on each GeneChip. (b) Summarizes the filtration and clustering procedure. (c) Heatmaps of expression patterns in four clusters for three species as indicated. Numbers of probe sets and corresponding genes in each expression cluster are shown at the left. Sample names are Sertoli cells (SE), spermatogonia (SG), spermatocytes (SC), spermatids (ST), tubules (TU), and total testis (TT). Human samples include chondrocytes (CC) and vascular smooth muscle (SM) cells. To produce a multispecies heatmap where each line contains the mouse, human, and rat homologs, only the mouse genes were clustered, whereas the data for corresponding human and rat orthologs are displayed on the same line. Log2-transformed signals are shown according to the scale bar.
Tissue Profiling Analysis Reveals a Complement of Potentially Specific Loci.
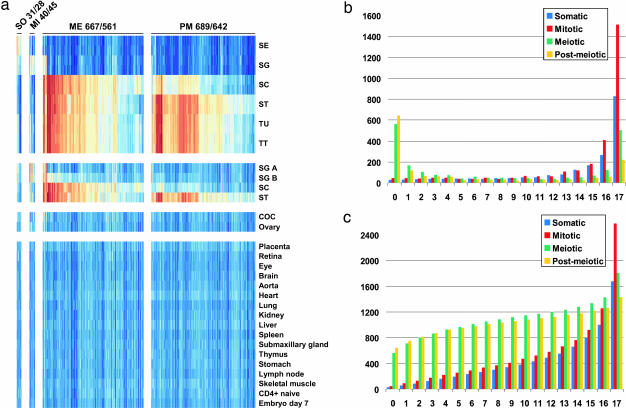
The comparison of rodent with human samples suggested that most genes active in somatic Sertoli cells and mitotic spermatogonia are also expressed in a wide range of other cell-types (such as chondrocytes and smooth vascular muscle), whereas loci transcribed in meiotic spermatocytes and postmeiotic spermatids tend to be specific for the germ line. To identify putative testis-specific genes we therefore assembled a representative set of data from 24 normal mouse tissues available from the National Center for Biotechnology Information GeneOmnibus (www.ncbi.nlm.nih.gov/geo) (SI Fig. 8 and SI Table 2). This set includes five mouse testicular samples (A- and B-type spermatogonia, pachytene spermatocytes, postmeiotic round spermatids and total testis), one female reproductive cell type (cumulus-oocyte complex), one female tissue type (ovary) and 17 somatic controls. Among 1,269 transcripts not detected in any somatic controls only small groups were classified as somatic (28) and mitotic (45), whereas the vast majority fell into meiotic (561) and postmeiotic (642) clusters (Fig. 3a and SI Fig. 8). Analysis of the numbers of genes detected in the germ line and up to 17 somatic tissues shows that the majority of genes falling into the somatic and mitotic clusters are widely expressed, whereas most genes classified as meiotic or postmeiotic are not detected in any somatic controls (Fig. 3b). Accordingly, the total number of genes identified increases massively in the case of somatic and mitotic genes but only moderately in the case of meiotic and postmeiotic genes, when expression in one or several somatic controls is permitted (Fig. 3c).
Fig. 3.
Tissue profiling identifies potentially testis-specific genes. (a) Heatmap showing expression in mouse germ cells and somatic controls of genes expressed specifically in Sertoli cells or mitotic, meiotic, and postmeiotic germ cells. The loci are organized into four expression clusters across species as shown. The corresponding probe set and gene numbers per cluster are indicated. Sample names are abbreviated as in Fig. 2 except for A-type spermatogonia (SG A), B-type spermatogonia (SG B), and cumulus-oocyte complex (COC). (b) Shown is the specific number of probe sets (y axis) falling into four expression clusters for which expression was detected in none of the somatic controls (0) or in 1–17 tissues as indicated (x axis). (c) Shows the total sum of probe-sets identified in testis (0) or in testis and 1–17 somatic controls. Expression clusters are shown in blue (SO, somatic), red (MI, mitotic), green (ME, meiotic), and yellow (PM, postmeiotic) as indicated in the legend.
To identify genes that are “conserved, differentially expressed, and specific to testis” (CDEST) (this abbreviation is also used in the BioMart query form of GermOnline) we asked which members of CDET were not detected in any of 17 somatic nontesticular control tissues. Using extremely stringent selection criteria, 80 genes were detected in the somatic (2), mitotic (3), meiotic (42), or postmeiotic (33) expression clusters (SI Fig. 9 a and b). When using profiling data for the identification of potentially important genes, one needs to bear in mind that their expression as detected by microarrays may not be absolutely testis-specific. For example, the conserved recombination enzyme Spo11, which is highly expressed in spermatocytes and essential for normal mouse spermatogenesis (24, 25), is absent from the CDEST group because it is also detected in thymus and CD4+ cells (see GermOnline). However, the biological significance for this finding is unclear, because Spo11 does not appear to play a role in immunological processes (26).
The somatic and mitotic core testis-specific clusters shown in SI Fig. 9a include genes involved in Sertoli cell gene expression and lipid transport (Gata4, Abca1) and a transcription factor essential for mammalian testis differentiation (Dmrt1). The meiotic cluster contains genes known or predicted to be involved in meiotic cell cycle progression (Aurkc, Ccna1, Spdy1), spermatogenesis (Acrbp, Adam2, Adam18, Pla2g6, Ribc2, Tcfl5), and sperm motility (Ppp3r2, Smcp, Spag6). The postmeiotic cluster shown in SI Fig. 9b contains genes involved in cell growth regulation (Socs7), regulation of transcription (Ankrd5), spermatogenesis (Fscn3, Spag4l), spermiogenesis (Odf1, Odf3, Tnp2), sperm motility (Akap3) and fertility (Mmel1/Nl1, Spaca1, Spaca3, Zpbp). The results confirm and extend expressed sequence tag data obtained with pooled mouse spermatocytes (4933401K09Rik, 4933417C16Rik, 9630025C22) or testicular tissue (4930524B15Rik, 4930550C14Rik, 4933417A18Rik). Thus they suggest roles in male meiosis and gametogenesis for the genes corresponding to these expressed sequence tags.
Validating Expression Profiles by Chromosomal Localization.
It was observed that the mammalian X chromosome is enriched for loci involved in brain development and genes expressed in mitotic spermatogonia, whereas it is devoid of loci showing peak expression in meiotic spermatocytes (for review, see ref. 27). The latter phenomenon is thought to be due to meiotic sex chromosome inactivation, a process causing wide-spread transcriptional silencing of X and Y from the meiotic prophase onward until late stages of spermiogenesis (28, 29). Our results based on arrays that cover all currently known rodent genes confirm and extend earlier reports, because loci in the rodent mitotic and somatic expression clusters are clearly enriched on the X chromosome, whereas, remarkably, not one of 1,809 mouse and 1,413 rat genes showing peak expression in meiotic spermatocytes was X-linked (SI Fig. 10). Moreover, among 1,263 human homologs of mouse genes in the meiotic expression class, none (apart from 10 probe sets with erroneous annotation) were on the X chromosome. These results extend a similar chromosomal localization analysis carried out by others to the genome-wide level (30). Importantly, they validate the meiotic expression cluster containing genes that show peak transcriptional induction in spermatocytes.
Correlating Germ-Line Expression and Reproductive Function.
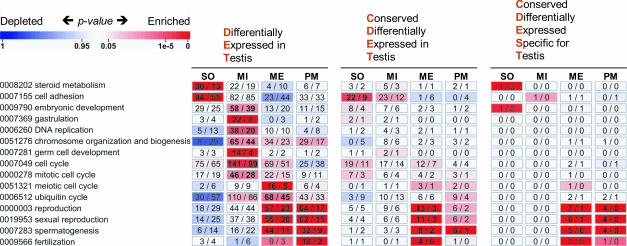
To study the correlation between DET and germ-line function, we used controlled vocabulary from the Gene Ontology Consortium (Fig. 4). Such analysis demonstrates that genes showing peak expression in somatic Sertoli cells are involved in biological processes, such as steroid metabolism (P = 5 × 10−6; 30 genes annotated) and cell adhesion (2 × 10−7; 94), whereas genes strongly induced in mitotic spermatogonia were associated with the cell cycle (P = 4 × 10−6; 141 genes annotated), germ cell development (2 × 10−5; 14), chromosome organization and biogenesis (6 × 10−4; 65), and DNA replication (6 × 10−5; 38). Interestingly, mitotic spermatogonia express many genes involved in embryonic development (P = 7 × 10−4; 58 genes annotated) and more specifically gastrulation (10−9; 22), the process where ecto-, meso-, and endoderm are established during early embryogenesis. The meiotic class covering spermatocytes contains genes required for reproduction (P = 8 × 10−11; 57 genes annotated), spermatogenesis (10−15; 44), the meiotic cell cycle (10−5; 16), and protein degradation by the ubiquitin cycle (3 × 10−4; 68). Genes induced in postmeiotic spermatids are involved in reproduction (P = 5 × 10−6; 64 genes annotated), spermatogenesis (9 × 10−7; 32), and fertilization (3 × 10−5; 10). Complementary results were obtained by searching the mouse data for gene ontology (GO) terms from the ontologies “cellular component” and “molecular function.” Importantly, similar patterns of enrichment were observed in the rat dataset (the human data were not further explored because the sample set lacked Sertoli cells and spermatogonia). The clusters defined by the CDET expression class were enriched for nearly the same GO terms as the initial complement of loci identified on the basis of differential testicular expression (Fig. 4). Only relevant GO terms such as sexual reproduction (6 × 10−6; 6) and spermatogenesis (7 × 10−6; 5) were significantly enriched in the meiotic cluster as defined by the CDEST class (Fig. 4).
Fig. 4.
Biological Process GO term enrichment in four expression clusters containing genes differentially expressed in testis (DET), conserved and differentially expressed in testis (CDET), or conserved, differentially expressed, and specific to testis (CDEST). These abbreviations are also used in the BioMart query form of GermOnline. Probe set and corresponding gene numbers are given above each map. Each cluster is matched with enriched GO terms from the ontology “biological process” that are ordered according to peak expression in somatic (SO), mitotic (MI), meiotic (ME), and postmeiotic (PM) clusters. Numbers of genes associated with a specific GO term and enriched in each cluster are given within rectangles in bold as observed and as expected. A color code indicates overrepresentation (red) and underrepresentation (blue) as indicated in the scale bar.
Taken together, the results clearly demonstrate that the expression clusters correlate well with known processes, functions and chromosomal localization patterns relevant for sexual reproduction. It is noteworthy that the core class of conserved meiotic and postmeiotic mouse loci contains 165 loci where no or extremely little information is available about their biological functions. Our findings enable us to infer roles for known genes not previously detected in male gonads and to provide clues for potential reproductive functions for many as yet poorly characterized rodent and human loci (see also GermOnline). For example, among the mitotic expression cluster showing peak expression in spermatogonia we identified loci involved in processes important for embryonic development, such as gastrulation (Hmgb1, Ppap2b, Smad4), somitogenesis (Pcdh8), and limb morphogenesis (Fbn2, Ptch1, Ski, Twist1). Furthermore, a combination of expression data and domain analysis suggests possible roles for D930005D10Rik in testicular signaling (RA; see the European Bioinformatics Institute (EBI) InterPro database at www.ebi.ac.uk/interpro for definitions of IPR000159; PDZ, IPR001478; and SMAD-FHA, IPR008984) and transcriptional regulation (forkhead-associated, IPR000253). Similarly, it predicts 4933401K09Rik to be involved in spindle pole body formation (homology with inner plaque spindle pole body component NUF1 from S. cerevisiae) and it implies a role for 4931407G18Rik in fertilization [similar to Zonadhesin (NM_011741.1), which is required for binding of sperm to the zona pelludica (31)]. Likewise, 4632434I11Rik may encode a protein that binds nucleic acids (OB fold, IPR008994), whereas 4933417K04Rik, 4930579A10Rik, 4921513E08Rik, and 1700095F04Rik are predicted to be part of oligomeric complexes formed in spermatocytes and/or spermatids (coiled-coil domain). Efforts are currently under way to experimentally test some of these predictions.
Male Germ-Line Development and Somatic Cancer.
De-repression of germ-line genes in nontesticular malignancies [cancer/testis (CT) genes] has been associated with hallmarks of cancer, such as genetic instability, immortality, and invasiveness (31). For many CT genes broadly classified as being expressed in the testis, the dataset reported here reveals peak expression in Sertoli or germ cells. This includes loci strongly expressed in Sertoli cells, spermatogonia, and somatic controls (Lip1, Maged1, Maged2) and others that were found to be induced in spermatocytes (Adam2, Tdrd1) or spermatids (Cage1, Mageb5, Ssx) but that failed to be expressed to detectable levels in 17 somatic controls (see SI Fig. 9 a and b). Peak expression in postmeiotic germ cells of human Cage1 (33) and its rodent orthologs (mouse Cage1: MGI:1918463; rat Ctag3 RGD:1306102) is consistent with the protein being a component of the acrosome, an organelle required for sperm–egg fusion (34) (SI Fig. 11). To identify novel CT gene candidates a detailed analysis of our germ cell data together with results from the Expression Project for Oncology is currently under way (F.C. and M.P., unpublished data).
Expression profiling of testicular cells further reveals links between male germ cells and various human somatic malignancies in unexpected ways. For example, the core meiotic expression cluster contains human Nmes1, a putative tumor suppressor gene strongly expressed in esophageal mucosa and down-regulated in corresponding cancer tissues (35). Nmes1 and its mouse (MGI:3034182) and rat (RGD:1359583) orthologs are highly and reproducibly expressed in meiotic spermatocytes and in postmeiotic round spermatids. This marks out Nmes1 as a factor potentially involved in male germ cell proliferation and differentiation. Very strong expression of Mum1 [melanoma associated antigen (mutated) 1] in germ cells was observed across human and rodent orthologs (MGI:1915364; Mum1_predicted, RGD:1308340), suggesting a novel role in spermatogenesis for this gene known to be involved in the progression of B-cell lymphoma/leukemia (36) (SI Fig. 11 and GermOnline). These cancer-related loci are interesting examples for which our data predict roles in male meiosis and gametogenesis.
Summary and Conclusions.
In this article we report a cross-species whole-genome expression profiling analysis of the male germ line in mammals. Using high-density oligonucleotide microarrays and purified testicular cells together with whole-gonad samples, we determined the rodent testicular differential expression program and the core male meiotic transcriptome in mouse, rat, and human. A tissue profiling experiment, using selected somatic controls, enabled us to identify genes that appear to be specifically expressed in testicular germ cell populations. This approach thus yielded hundreds of conserved genes for which highly reliable and reproducible expression data were obtained in all three species during the critical meiotic and postmeiotic stages of sexual reproduction. The outcome of the work reported here provides the most comprehensive map of the mammalian testicular expression program at a cell-type specific level available to date. Moreover, it reveals a complement of conserved core loci predicted to be involved in meiosis and gametogenesis in the male. The dataset is thus an asset to the scientific and medical communities for ultimately gaining insight into the molecular events leading to human reproductive pathologies and de-repression of testicular genes in cancer.
Materials and Methods
Detailed descriptions of materials and methods are provided in SI Materials and Methods.
Feature-level data files (CEL) and RMA-normalized files corresponding to the Sertoli cells (SE), spermatogonia (SG), spermatocytes (SC), spermatids (ST), tubules (TU), and total testis (TT) samples are available at the European Bioinformatics Institute ArrayExpress public data repository (accession no. E-TABM-130). CEL feature level data files of all samples including human chondrocytes (CC) and smooth vascular muscle (SM) controls are available from the authors upon request.
Supplementary Material
Acknowledgments
We thank R. E. Esposito for critical reading, H. Leroy (Institut de Recherche en Informatique et Systèmes Aléatoires) for GermOnline administration and R. Schlapbach (Functional Genomics Center Zurich) for human chondrocytes and smooth vascular muscle data. The work was supported by the Swiss Institute of Bioinformatics (C.N.-W. and A.G.), a fellowship from Institut National de la Santé et de la Recherche Médicale (A.D.R.), Région Bretagne (A.D.R.), and National Institutes of Health Grant R01 HD34915 (to D.J.W.).
Abbreviation
- GO
gene ontology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the European Bioinformatics Institute (EBI) ArrayExpress database, www.ebi.ac.uk/arrayexpress (accession no. E-TABM-130).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701883104/DC1.
References
- 1.Deutschbauer AM, Williams RM, Chu AM, Davis RW. Proc Natl Acad Sci USA. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enyenihi AH, Saunders WS. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K. Curr Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 4.Rabitsch KP, Toth A, Galova M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, et al. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 5.Colaiacovo MP, Stanfield GM, Reddy KC, Reinke V, Kim SK, Villeneuve AM. Genetics. 2002;162:113–128. doi: 10.1093/genetics/162.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy A, Matzuk MM. Reproduction. 2006;131:207–219. doi: 10.1530/rep.1.00530. [DOI] [PubMed] [Google Scholar]
- 7.Matzuk MM, Lamb DJ. Nat Cell Biol. 2002;4(Suppl):s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 8.Griswold MD. Semin Cell Dev Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 9.Jegou B. Int Rev Cytol. 1993;147:25–96. [PubMed] [Google Scholar]
- 10.Wolgemuth DJ, Lele KM, Jobanputra V, Salazar G. Int J Androl. 2004;27:192–199. doi: 10.1111/j.1365-2605.2004.00480.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhao GQ, Garbers DL. Dev Cell. 2002;2:537–547. doi: 10.1016/s1534-5807(02)00173-9. [DOI] [PubMed] [Google Scholar]
- 12.Maclean JA, II, Wilkinson MF. Curr Top Dev Biol. 2005;71:131–197. doi: 10.1016/S0070-2153(05)71005-X. [DOI] [PubMed] [Google Scholar]
- 13.Wrobel G, Primig M. Reproduction. 2005;129:1–7. doi: 10.1530/rep.1.00408. [DOI] [PubMed] [Google Scholar]
- 14.Lin YN, Matzuk MM. Semin Reprod Med. 2005;23:201–212. doi: 10.1055/s-2005-872448. [DOI] [PubMed] [Google Scholar]
- 15.Schlecht U, Demougin P, Koch R, Hermida L, Wiederkehr C, Descombes P, Pineau C, Jegou B, Primig M. Mol Biol Cell. 2004;15:1031–1043. doi: 10.1091/mbc.E03-10-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz N, Hamra FK, Garbers DL. Proc Natl Acad Sci USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shima JE, McLean DJ, McCarrey JR, Griswold MD. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 18.Eddy EM. Recent Prog Horm Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- 19.Miller D, Ostermeier GC, Krawetz SA. Trends Mol Med. 2005;11:156–163. doi: 10.1016/j.molmed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gattiker A, Niederhauser-Wiederkehr C, Moore J, Hermida L, Primig M. Nucleic Acids Res. 2007;35:D457–D462. doi: 10.1093/nar/gkl957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 23.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 24.Romanienko PJ, Camerini-Otero RD. Mol Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 25.Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Mol Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 26.Klein U, Esposito G, Baudat F, Keeney S, Jasin M. Eur J Immunol. 2002;32:316–321. doi: 10.1002/1521-4141(200202)32:2<316::AID-IMMU316>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 27.Graves JA. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Dev Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 30.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. Nat Genet. 2004;36(6):642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 31.Hardy DM, Garbers DL. J Biol Chem. 1995;270:26025–26028. doi: 10.1074/jbc.270.44.26025. [DOI] [PubMed] [Google Scholar]
- 32.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 33.Park S, Lim Y, Lee D, Cho B, Bang YJ, Sung S, Kim HY, Kim DK, Lee YS, Song Y, Jeoung DI. Biochim Biophys Acta. 2003;1625:173–182. doi: 10.1016/s0167-4781(02)00620-6. [DOI] [PubMed] [Google Scholar]
- 34.Alsheimer M, Drewes T, Schutz W, Benavente R. Eur J Cell Biol. 2005;84:445–452. doi: 10.1016/j.ejcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhou J, Wang H, Lu A, Hu G, Luo A, Ding F, Zhang J, Wang X, Wu M, Liu Z. Int J Cancer. 2002;101:311–316. doi: 10.1002/ijc.10600. [DOI] [PubMed] [Google Scholar]
- 36.Uranishi M, Iida S, Sanda T, Ishida T, Tajima E, Ito M, Komatsu H, Inagaki H, Ueda R. Leukemia. 2005;19:1471–1478. doi: 10.1038/sj.leu.2403833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.