Abstract
Maintenance of progenitor cell properties in development is required for proper organogenesis of most organs, including those derived from the endoderm. FGF10 has been shown to play a role in both lung and pancreatic development. Here we find that FGF10 signaling controls stomach progenitor maintenance, morphogenesis and cellular differentiation. Through a characterization of the initiation of terminal differentiation of the three major gastric regions in the mouse, forestomach, corpus and antrum, we first describe the existence of a “secondary transition” event occurring in mouse stomach between E15.5-E16.5. This includes the formation of terminally differentiated squamous cells, parietal, chief a nd gastric endocrine cells from a pre-patterned gastric progenitor epithelium. Expression analysis of both FGF and Notch signaling components suggested a role of these networks in such progenitors, which was tested through ectopically expressing FGF10 in the developing posterior stomach. These data provide evidence that gastric gland specification and progenitor cell maintenance is controlled by FGF10. The glandular proliferative niche was disrupted in pPDX-FGF10FLAG mice leading to aberrant gland formation, and endocrine and parietal cell differentiation was attenuated. These effects were paralleled by changes in Hes1, Shh, and Wnt6 expression, suggesting that FGF10 acts in concert with multiple morphogenetic signaling systems during gastric development.
Keywords: endoderm, stomach, FGF10, FGFR2, Notch, Hes1, differentiation, Sox2, chief cells, parietal cells, SPEM
Introduction
Development of the mammalian gut is a highly orchestrated process involving a multitude of morphogenetic processes, tissue interactions, and cell differentiation events. Of the tissues derived from the gut endoderm, development of the budding organs such as the lung, liver and pancreas is best characterized, and several gene products and signaling systems have been described securing regional identity and morphogenetic development of these organs (Bellusci et al., 1997; Costa et al., 2001; Jensen, 2004; Lemaigre and Zaret, 2004). In contrast, the morphogenesis of the main gut tract, composed of esophagus, stomach and intestinal system is less well characterized, and most studies related to cell differentiation within these regions have focused on the behavior of the gut endoderm in the normal adult, or transformed state, through investigations of stomach and intestinal cancer.
Gastric mesenchyme is essential for survival of the epithelium in vitro and tissue-grafting experiments have shown that a diffusible factor from the mesenchyme is critical for stomach epithelial development in the chicken (Koike and Yasugi, 1999). Recombination with forestomach mesenchyme did not change the fate of glandular epithelium and vice versa after E11.5 in the mouse (Fukamachi et al., 1979), suggesting that the primary specification of the gastric epithelium occurs before E11.5. Factors that influence this primary specification include the mesenchymal-expressed Hoxa5 (Aubin et al., 2002) and Barx1, a homeobox gene that act upstream of Wnt-antagonists (Kim et al., 2005). In addition to this primary specification of the stomach, a secondary cell specification and glandular formation is required to form the mature stomach. Differentiation of specific cell lineages and invaginations of epithelium to form glands begins already in the embryo, but mature glands are not formed until after birth. Sonic hedgehog expressed in the epithelium has been shown to regulate gland development in the adult mouse stomach (van den Brink et al., 2001), and BMP (Narita et al., 2000) and Notch signaling (Matsuda et al., 2005) is critical for gland formation in the chicken proventricular stomach. Yet, we do not know the identity of any mesenchymal expressed morphogen(s) being responsible for regulating epithelial cell differentiation in the embryonic stomach.
Several studies have revealed significant sharing of gene function along the gut. We and others previously showed that the Notch signaling system, via Hes1, acts as a unified regulatory mechanism controlling the development of endodermal endocrine cells in the lung (Ito et al., 2000), stomach (Jensen et al., 2000), pancreas (Jensen et al., 2000), small intestine (Jensen et al., 2000; Milano et al., 2004; van Es et al., 2005) and colon (Jensen et al., 2000). Furthermore, this signaling system might be critical for proper differentiation from several, if not all, of the endodermal stem-cell populations in the gut throughout life (van Es et al., 2005; Milano et al., 2004; Zecchini et al., 2005; Collins et al., 2004; Apelqvist et al., 1999). Also, Ngn3 has been shown to be necessary for endocrine development in both the pancreas (Gradwohl et al., 2000), intestine (Jenny et al., 2002), and stomach (Jenny et al., 2002; Lee et al., 2002).
We and others recently described the effects of over-expressing the FGFR2- IIIb ligand FGF10 in pancreatic epithelium (Norgaard et al., 2003; Hart et al., 2003). FGF10 is normally expressed by the distal-most pancreatic mesenchyme, and this mesenchymal/epithelial signaling is critical for pancreatic epithelial growth (Bhushan et al., 2001). However, FGF10 also plays a role in coordinating epithelial-derived cell differentiation, as increased FGF10 signaling leads to an almost complete abrogation of all differentiated pancreatic cell types: endocrine, exocrine and ductal (Norgaard et al., 2003; Hart et al., 2003). As this effect is comparable to that observed during increased Notch-signaling (Hald et al., 2003; Murtaugh et al., 2003) and given that pancreatic expression of notch signaling components was increased in the pPDX-FGF10 mouse (Norgaard et al., 2003), it is plausible that FGF10 may exert control of differentiation via the Notch pathway. The requirement of Notch signaling in FGF10-mediated suppression of pancreatic differentiation was recently demonstrated by Miralles et al. using an explant culture model (Miralles et al., 2006).
In the present study we questioned if such a genetic link would be conserved in stomach development. We first define the temporal onset of the gastric secondary transition and demonstrate the spatial expression of FGF10/FGFR2 and Notch components at this time. These results suggest a role in mesenchymal-to-epithelial signaling of the FGF10/FGFR2 functional pair and a role of downstream Notch signaling in epithelial progenitor maintenance. Taking advantage of the expansion of the Pdx1-promoter domain into the stomach and duodenal regions, we have here analyzed the specific effects on stomach development in the presence of increased levels of FGF10 signaling as provided by Pdx1-FGF10 transgenic mice. We find that the stomach endodermal regions behave with an overall similar mechanism to the pancreatic epithelium in that cell differentiation is suppressed, although with selective effects on specific cell lineages. Consequently, stomach morphological development is severely disrupted. We also conclude that similar to the pancreas, FGF10 induces expression of the Notch target gene Hes1, indicating a conserved regulatory network.
Materials and Methods
Transgenic mouse derivation and embryo isolation
FVB mice with transient ectopic expression of FGF10 controlled by the Pdx1-promoter were generated as described in Norgaard et al (2003) by oocyte injection of a pPdx1- FGF10FLAG fragment (Norgaard et al., 2003). Date of transfer was set at E0.5 and the embryonic gut was harvested in ice-cold PBS under stereomicroscope at day E12.5, E14.5 or E18.5 for tissue or RNA. Genotyping was performed by PCR using primers specific for the transgenic construct (Supp. table 1). A total of n=3 E12.5 transgenic (TG) embryos, n=3 E14.5 TG embryos, and n=6 E18.5 TG embryos were analyzed in this study. Non-transgenic littermates (WT) were used as controls throughout.
Histology
Tissues were fixed in 4% PFA o.n, transferred to 30% glucose for 2 hours and 15% sucrose in 50% OCT for 1 hour followed by 1 hour of 100% OCT before being quick frozen in OCT. Immunohistochemistry was performed on 6 μm frozen slides. Slides were dried at 37C, microwaved for 2x 5 min. in citrate buffer pH 6 and washed in PBS. Slides were then treated with 3% hydrogen peroxide for 5 min, washed in PBS and blocked with TSA blocking buffer (Perkin Elmer) for 1 hour. Primary antibodies were applied o.n. The next morning, slides were washed in PBS and incubated with either appropriate secondary antibody conjugated to Texas red/Cy2/AMCA (Jackson ImmunoResearch) for 1 hour or biotinylated secondary antibody from Zymed (Histostain) for 30 min. After washes in PBS, slides incubated with biotinylated secondary antibody were incubated with streptavidin peroxidase conjugate (Zymed) for 15 min, washed in PBS and inc ubated with 1:50 FITC-conjugated tyramide signal amplification (TSA) reagent (Perkin Elmer) for 10 min. All slides were mounted in 30% glycerol in PBS.
Primary antibodies and dilutions used in this study includes: Goat α-FGF10, 1:200, Abcam, USA; Mouse HRP-conjugated α-FLAG, M2, 1:1000 (TSA), Sigma, MO, USA; Rabbit α-IDX1 (PDX1), Dr. Joel Habener, Boston, MA, USA, 1:2000; rat α-E-cadherin, ECCD2, 1:200, Zymed, CA, USA; rabbit α-β-catenin Ab-1, 1:100, Neomarkers/Labvision, CA, USA; mouse α-human smooth muscle actin, clone1A4, 1:500, DAKO, Denmark; rabbit α-chromogranin A, 18-0094, 1:200, Zymed, CA, USA; Sheep α-human Pepsinogen II, K90132S, 1:200, Biodesign, ME, USA; rabbit α-H,K-ATPase α-unit (C terminal), AB1674, 1:100, Chemicon, USA; rabbit α-secretin, H-067-04, 1:300, Phoenix, CA, USA; mouse-α-glucagon, K79BB10, 1:200 Sigma, St. Louis, MO; rabbit-α-somatostatin, 1:100, DAKO, Denmark; goat-α-ghrelin, sc-10368, 1:1000, Santa Cruz Biotech, CA, USA; rabbit-α-CCK, 1:200, Immunostar, USA; rabbit-α-gastrin 1:200, Neomarkers-Labvision, CA, USA; rabbit α-p-Histone-H3, 06-570, 1:100, Upstate, VA, USA; rabbit α-PCNA, FL-261, 1:150, Santa Cruz Biotech., CA, USA; rabbit α-Bek/FGFR2 (C17, recognizes both splice forms), 1:800 (TSA), Santa Cruz Biotech., CA, USA; rabbit α-Hes1 (Lee et al., 2005), 1:500 (TSA); rabbit-α-Sox2, 1:800 (TSA), Abcam, USA; Nkx-6.1, 1:500 (Jensen et al., 1996), Denmark; Rhodamine-Dolichos Biflorus Agglutin (DBA), 1:200, Vector Laboratories, CA, USA ; Fluorescein-Ulex Euopaeus Agglutin I (UEA), 1:200, Vector Laboratories, CA, USA. Nuclei were stained for 5 min with 1:2000 Hoechst where needed for morphometric studies.
In-situ hybridization was performed on 6μm frozen slides using digoxygenin-labeled RNA probes. The RNA probes were generated by in-vitro transcription from linearized template DNA as described in (Norgaard et al., 2003). DNA templates were derived from the following clones: FGF10 in pCR4 (own); Hes1 in pBS-SKII (Dr. R. Kageyama, Uni. Kyoto, Japan); Notch1 in pBK-CMV (Dr. J. Hald, Denmark); Notch2 in pBK-CMV (Dr. J. Hald, Denmark ); Jagged1 in pCR4 (own); Jagged2 in pSKII (Dr. G. Weinmaster, UCLA); CDX2 in pT7T3D-pac (IMAGE clone 437757, Invitrogen); IFABP in bluntII-topo (Dr. L. Sussel, UCHSC, USA); TFF2 in pSPORT1 (IMAGE clone 6431145, Open Biosystems); Wnt6 in pCMV-SPORT6 (IMAGE clone 6511061, Open Biosystems); Shh in Bluescript II-SK (Dr. A.P. McMahon, Harvard, USA).
Hybridized probe was visualized using anti-Dig-AP-conjugated antibodies (Roche) as previously described (Norgaard et al., 2003) followed by NBT-BCIP substrate color reaction (Roche).
Slides were analyzed using an Olympus BX51 microscope station equipped with Nomarski interference and images where obtained through a Pixera CL600 camera. At least 3 E12.5, 3 E14.5 and 3 E18.5 TG embryos from different litters were characterized by histology. 3 WT littermates were included per time point for pair wise comparisons. The transgenic phenotypes of the three replicates were similar although not identical.
Morphometry
Morphometric analysis was done using ImagePro v. 4.5 (Media Cybernetics) software. Digital images were saved from 5–10 non-overlapping FOVs at 10x or 20x for each slide, and at least 3 slides distributed across the stomach were counted for each animal. Epithelial area was quantified by area integration based on E cadherin staining. For mean epithelial area per slide, the following number of slides distributed across the stomach were measured: E18.5: 4 slides/80 total sto mach slides, E14.5: 3 slides/35 total, E12.5: 3 slides/15 total. Cell numbers were counted automatically based on Hoechst staining, where a limited watershed algorithm was applied to separate overlapping nuclei. Specialized cell types were counted manually.
Multiplex RT-PCR
RNA was isolated from the stomachs of each of two transgenic E18.5 embryos from different litters and two littermate WT controls. A time series of RNA from E14.5–E18.5 stomachs was isolated from CD1 embryos by careful dissection of the stomach into glandular stomach and forestomach. Stomachs from 4 embryos were pooled for E14.5 samples, 3 embryos were pooled for E15.5 samples, and 2 embryos were pooled for each of the samples at E16.5, E17.5 and E18.5. Tissue was homogenized immediately in TriZol reagent (Invitrogen), and RNA was isolated according to manufacturers’ protocol. cDNA was synthesized from 2 μg RNA in a 50μl reaction with 7μg random primer (Invitrogen), 40 μM dNTPs, 1x Promega opti buffer, 0.1M DTT, 40 units RNAsin (Fisher) and 200 units M-MLV RT enzyme (Fisher) at 37C for 60 min.
Semi-quantitative multiplex RT-PCR analysis was done essentially as described in (Jensen et al., 1996). PCR reactions with app. 0.05 μg cDNA in a 22 μl reaction with 10x buffer incl. MgCl (Promega), 8mM dATP, dGTP and dTTP, 4mM dCTP, 20μM of each primer (up to 6 primers in each reaction), 0.5U Taq polymerase (Promega) and 4 mM α-32P-dCTP were run for 19 or 24 cycles depending on target abundance. The reactions were analyzed by polyacrylamide gel electrophoresis, and the intensity of the individual bands was quantified using ImageQuant software version 6.0 (Molecular Dynamics). All values are expressed as mean +/− SEM relative to the internal standard. Primers were designed according to Genebank reports and the sizes of the amplicons were 150–300 bp. Primer sequences are included in supplementary table 1.
Results
A secondary transition of the murine stomach occurs at E15.5–E16.5
The murine stomach develops from a homogenous pseudo-stratified epithelium, but differentiates into several different regions. At E17.5 the stomach consists of a forestomach region with squamous epithelium and a glandular region with stratified epithelium. These two regions can be easily distinguished from the exterior morphology of the stomach (Fig. 1A). The glandular stomach can be further divided into the corpus and the antrum based on differentiated cell lineages. In order to determine the exact onset of terminal cell differentiation in the developing glandular stomach, we performed multiplex RT-PCR for markers of differentiated stomach cell lineages. Analyzing for expression of maturing endocrine cells (Chromogranin A), zymogenic chief cells (pepsinogen C), parietal cells (H+/K+-ATPase) and pre-pit cells (trefoil factor 1 and 2) we found complete absence of expression at embryonic day 14.5, whereas all are highly expressed at E16.5 (Fig. 1C). The pan-endocrine marker chromogranin A is present at low levels at E15.5 but expression doub les on E16.5 and remains constant for the remainder of embryonic development. The parietal cell marker H+/K+-ATPase is likewise undetectable until E16.5, peaking at E17.5. Pepsinogen expressed by the zymogenic chief cell is weakly detectable at E15.5 but increases dramatically on E16.5. Finally, trefoil factor (TFF) 1 is strongly expressed from E16.5 onwards, while TFF2 is weakly expressed at E15.5, increases dramatically on E16.5 and remains more or less constant during embryonic development. These results were confirmed by immunohistochemistry for Pepsinogen C, H+/K+-ATPase and Chromogranin A (Fig. 1B and D). All three markers were completely absent in E15.5 stomach, while abundant positive cells were found in E16.5 stomach. The A-P patterning into forestomach, corpus and antrum is already evident at the histological level at this age as a region-specific distribution of the three cell types was apparent: Chromogranin A positive endocrine cells were distributed throughout the stomach glandular epithelium, Pepsinogen C positive zymogenic cells were only present in the antrum, while H+/K+-ATPase positive parietal cells were only found in the corpus (Fig. 1B and 1D). Only one immature gland noticed in this study in the corpus-antrum transition region contained both pepsinogen and H+/K+-ATPase positive cells (Fig. 1D inset).
Figure 1. The secondary transition of the stomach.
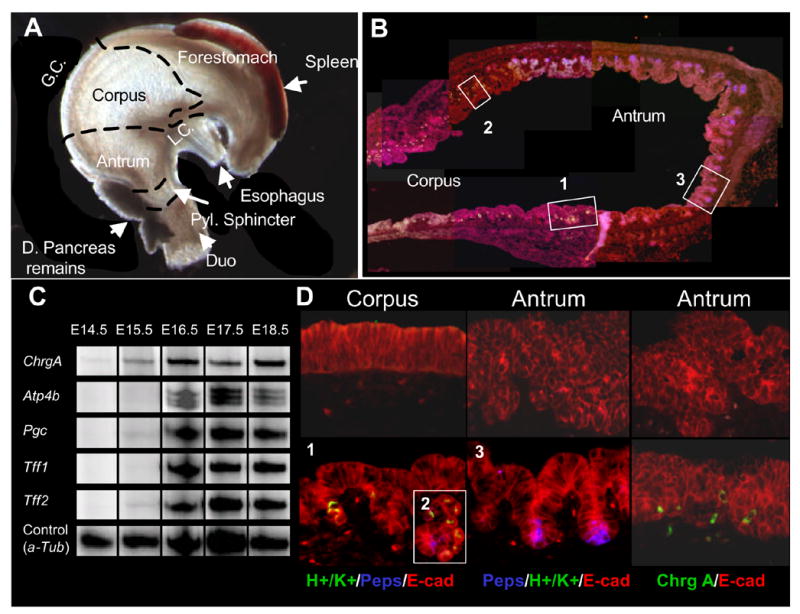
A: Regions of the stomach at E17.5. G.C: greater curvature, L.C. Lesser curvature. B: Co-staining of H+/K+-ATPase (green), Pepsinogen C (blue) and E-cadherin in E16.5 stomach. The numbers refer to pictures in D). C: Multiplex RT-PCR of expression of Chromogranin A (Chrg A), H+/K+ ATPase (Atp4b), pepsinogen C (Pgc) and Trefoil Factor 1 and 2 (Tff1 and Tff2). a-tubulin (a-tub) was used as an internal control. The shown bands are representative examples from two experiments. Ages analyzed included E14.5, E15.5, E16.5 E17.5 and E18.5. (n=2/gestational time point). D: Immunohistochemistry for H+/K+ ATPase and Pepsinogen C or Chromogranin A co-stained with E-cadherin at E15.5 (top row) and E16.5 (bottom row). Numbers refer to positions in B). Insert 2 shows one forming gland with both pepsinogen and H+/K+ ATPase expressing cells. This was the only one found (n=3).
We note that for both antrum and corpus, morphogenetic development of the gastric pit-gland unit is only rudimentary at E15.5–E16.5, with shallow epithelial invaginations (Fig 1B and 1D). Consequently, gastric cytodifferentiation is initially independent of a complete pit-gland unit morphogenesis. In contrast to the glandular regions, the mouse forestomach appears to generate only a single differentiated cell type, easily distinguishable through the presence of a stratified squamous epithelium, not found elsewhere in the stomach. Inspecting the developmental profile of the mouse forestomach between E14.5–E18.5 we conclude that squamous cell development occurs concomitantly with onset of terminal differentiation in the glandular regions, between E15.5–E16.5, as the forestomach epithelium changes from pseudostratified to squamous during this time (data not shown). Thus, the stomach epithelium goes through a secondary transition in a temporally and spatially coordinated fashion in the mouse.
FGF10 and FGFR2 are expressed complementarily in the gastric mesenchyme and epithelium during embryonic development
The temporal and spatial specificity of cell differentiation in the stomach suggests that cell differentiation is tightly regulated here. Given the role of FGF10 in pancreatic progenitor maintenance, we speculated that FGF signaling might play a role in regulating the secondary transition of the stomach as well. First, we identified members of the FGF-family expressed during the late embryonic development of the stomach (E14.5–E18.5) using a multiplex RT-PCR screen of all known FGF-family genes in the mouse (FGF1 to FGF23). Here we find that FGF1-3, FGF7, FGF9 and FGF10 were expressed at or above 20% of our control gene, TATA box binding protein (TBP), at one data point (Fig. 2A), but FGF10 was the only FGF expressed at high levels before the secondary transition. In-situ hybridization for FGF10 confirmed the multiplex analysis and showed the confinement of FGF10 expression to the gastric mesoderm with strongest expression posterior (Fig. 3A, and Supp. Figure 1A and 2). FGF10 is an FGFR2-IIIb ligand, and was previously found to be expressed in the pre-differentiated mouse stomach at E11.5 well before cytodifferentiation (Bhushan et al., 2001). Fgfr2-IIIb mRNA was expressed at a constant level throughout the stages analyzed, except for E14.5 expression that was higher in glandular regions (where the Fgfr2-IIIb/TBP volume ratio of 2.5 compared to ratios of 1–1.7, Fig. 2B). Using an antibody that detects both splice forms we found expression of FGFR2 protein in the pre-forestomach and pre-antrum epithelium at E15.5 (Fig. 4A). Only negligible staining could be discerned in the mesenchyme or in the epithelium of the pre-corpus region. Expression data based on in-situ hybridization at E14.5 showed epithelial localization of Fgfr2IIIb mRNA, as well as Fgfr3 and Fgfr4 (Supp. Fig. 1B), confirming the immunohistochemical analysis and establishing that the FGFR2IIb isoform is expressed here. At E18.5 FGFR2 protein expression in the forestomach epithelium was confined to the basal stem cell layer. Expression of FGFR2 had moved into the corpus region where it is localized to the forming glands, and expression was persisting in the antrum, with strongest expression in the forming glands (Fig. 4B). The region-specific expression of FGFR2 makes it likely that FGF signaling is involved in spatial patterning as well as temporal regulation of stomach development. In order to validate the gastric patterning of FGFR2, we used the gastric HMG-box containing transcription factor Sox2, as this is only expressed within the anterior-most endoderm with a posterior boundary at the pyloric sphincter (Tsukamoto et al., 2004). Immunohistochemistry for Sox2 on a consecutive slide confirmed the gastric identity of the FGFR2 expressing epithelium. Also, Sox2 expression was reduced in the corpus mirroring FGFR2 expression (Fig. 4A). We conclude that similar to the pancreas and lung, the FGF10/FGFR2 pair is expressed in the stomach in a manner consistent with mesenchymal-to-epithelial signaling.
Figure 2. Cell signaling during normal stomach development.
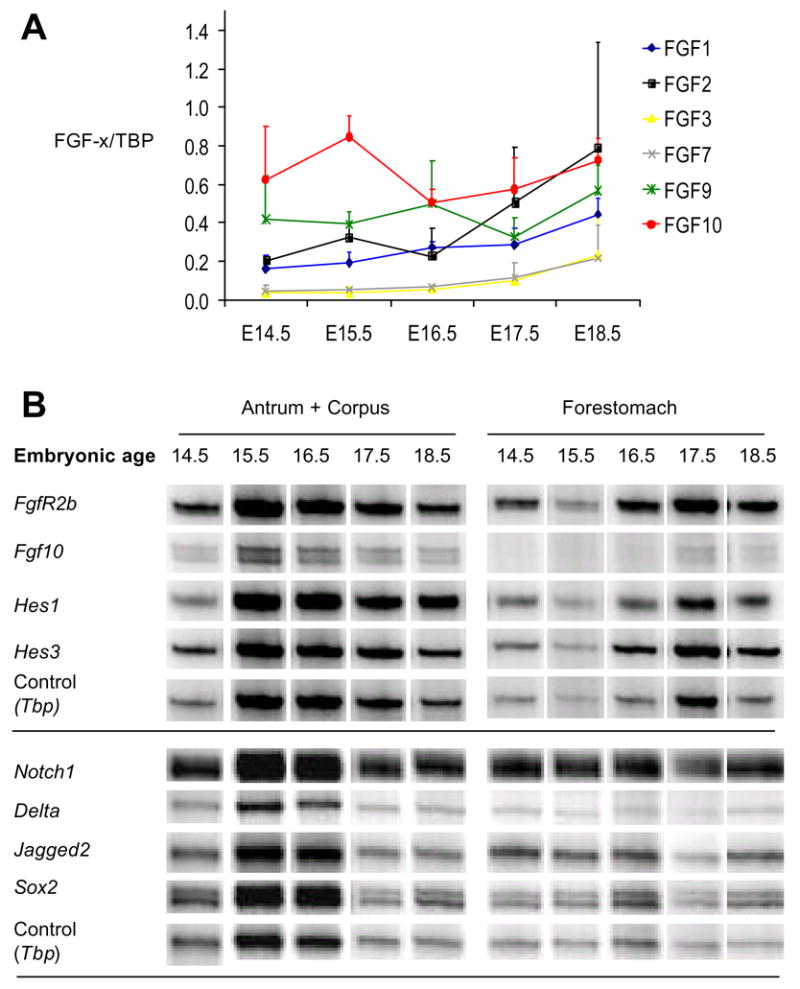
A: Semi quantitative multiplex RT-PCR of the expression of FGFs in the glandular stomach. Y-axis shows quantification of intensity of FGF-x bands relative to TBP +/− SEM. (n=3). B: Semi-quantitative multiplex RT-PCR of the expression of Fgf10, the FGF10 receptor FgfR2b, the Notch signaling components Hes1, Hes3, Notch1, Delta and Jagged2 as well as the gastric transcription factor Sox2. The shown bands are representative examples from n=2 experiments with exception of Fgf10 and FgfR2b RT-PCR that was performed four times. Tbp was used as internal control.
Figure 3. Notch components in stomach development.
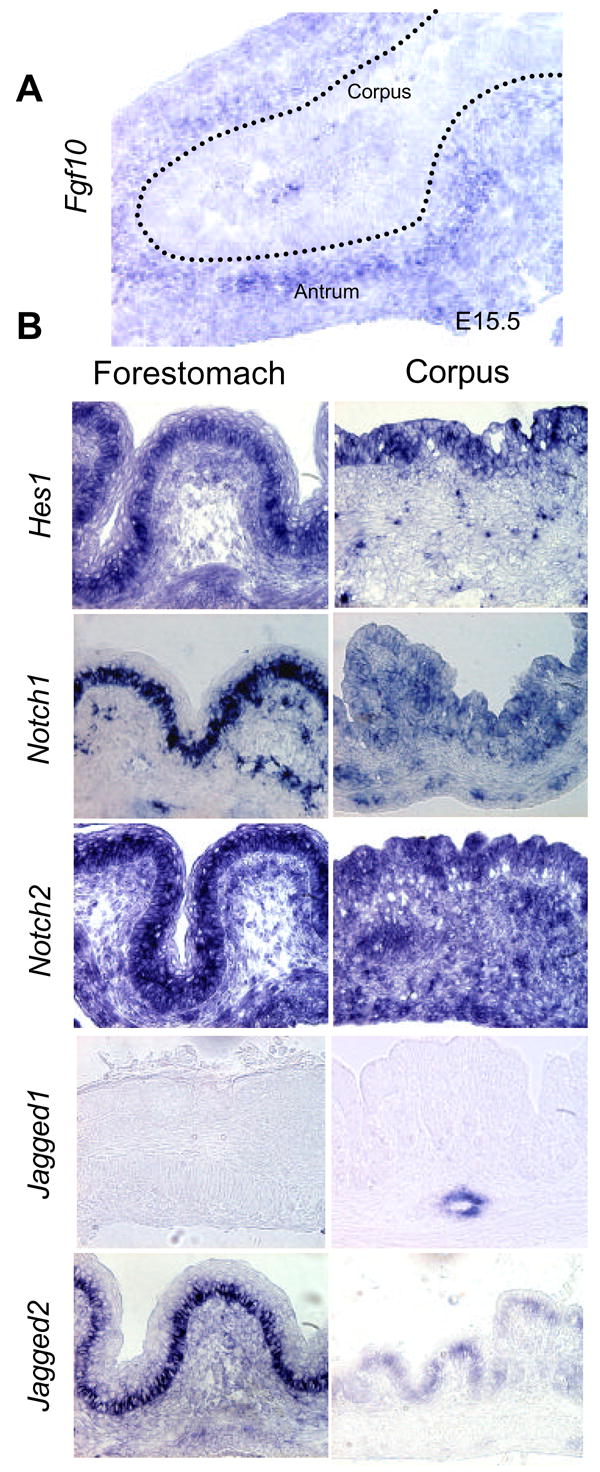
A: In-situ hybridization for FGF10 sho ws mesenchymal expression in an E15.5 stomach. B: In situ hybridization for Hes1, Notch1, Notch2, Jagged1 and Jagged2 in forestomach and fundus of E18.5 stomach
Figure 4. FGFR2 distribution in the stomach.
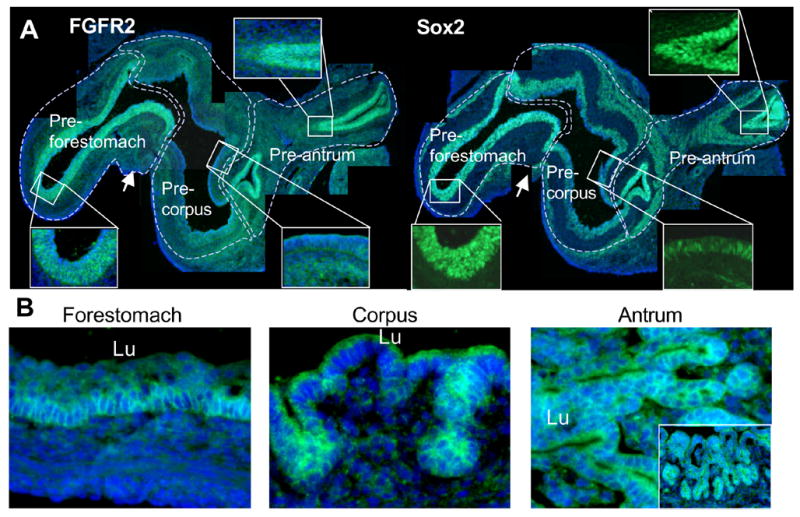
A: Immunostaining for FGFR2 and Sox2 in E15.5 stomach FGFR2 is expressed in the epithelium in the pre-forestomach and pre-antrum regions at E15.5. Arrow points to the esophagus. The Sox2 antibody displays a specific nuclear staining in the gastric epithelium and a nonspecific cytoplasmic staining in mesenchymal cells. B: Immunostaining for FGFR2 in E18.5 stomach. FGFR2 is expressed in the basal layer of the forestomach epithelium, the forming glands in the corpus and in all epithelial cells in the antrum at E18.5. Picture showing antrum is rotated 90 degrees left and has an insert showing a lower magnification of the same region (not rotated). Nuclei are stained with Hoechst
Notch signaling components are expressed in embryonic stomach
Notch signaling is downstream of FGF-signaling in the embryonic pancreas (Norgaard et al., 2003) and has been shown to control gastric enteroendocrine development, through activation of the bHLH factor Hes1. Expression of Notch signaling components have been addressed during chicken stomach development, where both cNotch1 and cNotch2, cHairy 1 (orthologous to Hes1) and cHairy2 (orthologous to Hes2) are predominantly confined to the epithelial compartment (Matsuda et al., 2005). Mouse embryonic stomach has not been similarly analyzed, although RT-PCR analysis of Hes1, Hes3, Notch1, Jagged1 and Jagged2 has demonstrated their expression in the embryonic mouse stomach at E18.5 (Jensen et al., 2000). We here demonstrate expression of Hes1 mRNA in the embryonic stomach throughout development (Fig. 2B) and confinement of Hes1 mRNA to the embryonic gastric epithelium at E18.5 (Fig. 3B). We also find that abundant expression of the transcriptional regulator Hes3 occur throughout stomach development (Fig. 2B) and that Notch1 is expressed in the embryonic stomach epithelial progenitors in the mouse (Fig. 2B and 3B). Given that Norgaard et al. suggested a role for Jagged-type ligands during FGF10 stimulated notch signaling in the pancreas, we were particularly interested in the spatial pattern of Jagged1 and Jagged2 expression. Jagged1 was not expressed by stomach epithelium, but instead confined to vessels (Fig. 3B). In contrast, Jagged2 was expressed in a completely overlapping manner with Notch1/Hes1 in the gastric epithelium in antrum and corpus, and basal population of the forestomach epithelium (Fig. 2B and 3B).
Generation of pPdx1-FGF10FLAG transgenic mice
To address the functional role of FGF10 signaling in gastric development we generated transient transgenic embryos through injection of the full length, C-terminally FLAG-tagged FGF10 coding region under control of 4.5 kb upstream Pdx1 proximal promoter (Norgaard et al., 2003; Stoffers et al., 1999; Apelqvist et al., 1997; Li and Edlund, 2001) (Fig. 5A). The Pdx1 promoter is abundantly expressed in the dorsal part of the primitive gut from E8.5 (Ohlsson et al 1993) and at lower levels in the distal foregut region, including the posterior stomach and duodenum E10 and onwards (Madsen et al., 2001; Miller et al., 1994). Importantly, this non-pancreatic expression is functionally relevant, as a hypermorphic allelic series of Pdx1 promoter mutants gradually deleting Pdx1 activity provided evidence of a role of Pdx1 in both pyloric gland and Brunners gland formation (Fujitani et al., 2006). The pPdx1-FGF10Flag construct used here was previously shown to disrupt pancreatic morphogenesis by blocking terminal differentiation of all pancreatic cell types (Norgaard et al., 2003). Previous studies of the exogenous Pdx1 promoter (using a 4.5 kb promoter fragment activity) have revealed expression in posterior stomach and duodenal endoderm, in addition to the pancreas. We confirmed the expression of endogenous Pdx1 in the anterior duodenal epithelium, and posterior stomach epithelium at E12.5, E14.5 and E18.5 (Fig. 5C), which was mimicked by the 4.5kb Pdx1 promoter used here (Fig. 6 and data not shown). We analyzed expression of the FGF10FLAG transgene by virtue of the FLAG-tag present as well as by ectopic expression of FGF10 protein and mRNA. At E12.5 FGF10 was only expressed in a few cells in the most posterior regions of the stomach epithelium (Fig 6A), co-responding with a very limited PDX1 domain here (Fig 5C). No expression was found in Wt littermates. In E14.5 TG stomach, a much stronger expression of FGF10FLAG protein was found (Fig. 6B) in stomach epithelial cells. Similar to the pancreas, strongest staining was observed in the plasma membrane rather than in the cytoplasm. No FLAG or FGF10 staining was observed in WT stomach (data not shown), and no FLAG staining was observed in TG stomach mesenchyme (Fig. 6B). Compared to E14.5, reduced expression was observed in E18.5 stomach – a time point where expression of FGF10FLAG persists in pancreas (Fig 6C). We performed in-situ hybridization for fgf10, which revealed transgenic expression in a few antral epithelial cells at E12.5 (Fig. 6A). In contrast, in-situ hybridization revealed a strong expression at E14.5, overlapping with Fgf10 and FLAG staining (Fig. 6B). At E18.5 the fgf10 mRNA level was reduced, and it was only expressed in cells in forming glands (Fig. 6C). No positive epithelial cells were found in WT littermates. Corroborating the histological results, RT-PCR revealed the presence of exogenous FGF10-Flag mRNA in transgenic stomach at E18.5 (Fig. 6D). Given the transient transgenic nature of the e mbryo generation process, individual TG embryos represent individual lines and therefore displayed degrees of variation of phenotype penetrance; the two transgenic embryos analyzed by RT-PCR were from independent litters and differed considerably in the amount of FGF10-Flag mRNA expressed. We accredit this to mosaicism, TG integration site, copy number insertion, or a mix thereof. Notwithstanding, all TG embryos analyzed displayed similar alterations of stomach development.
Figure 5.
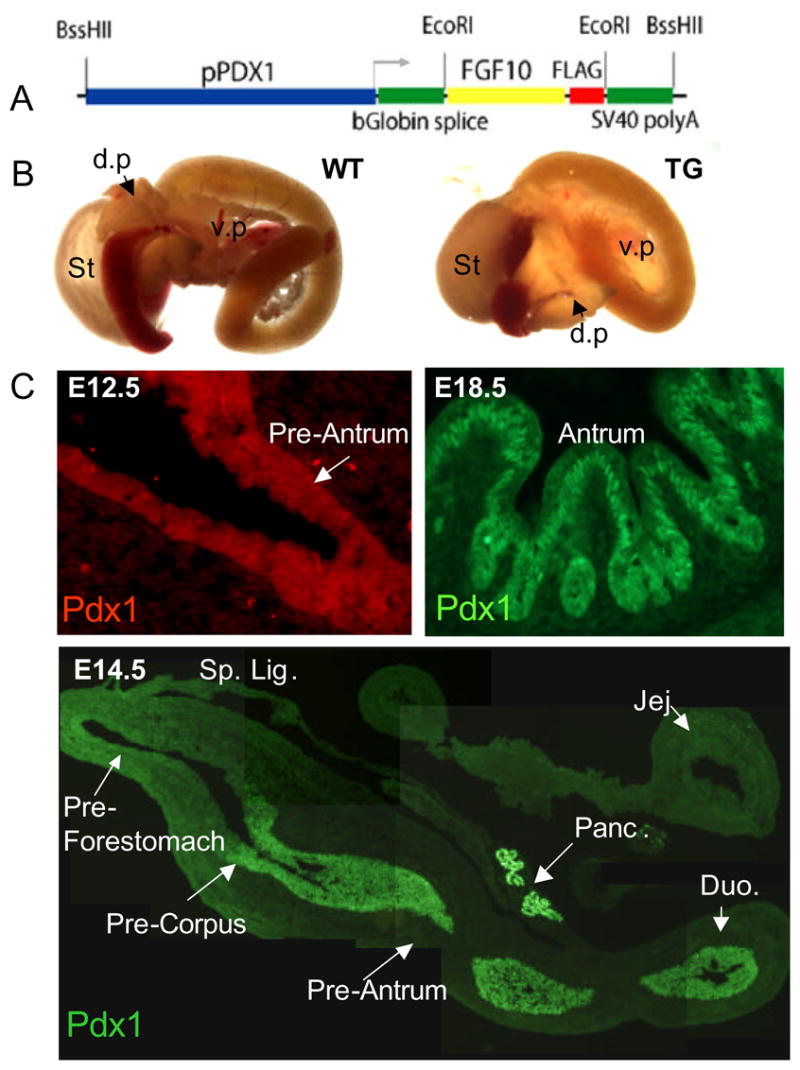
A: pPDX-FGF10-Flag construct. B: Whole gut of E18.5 pPDX-Fgf10FLAG embryo and wildtype littermate. The TG stomach is smaller and appears denser. C: Immunohistochemistry for PDX1 shows expression in E12.5 most posterior stomach, and E14.5, pancreas, anterior duodenum and posterior stomach including antrum and posterior corpus. PDX1 expression is still present in E18.5 antrum.
Figure 6. Gastric expression of exogenous FGF10 mRNA and protein.
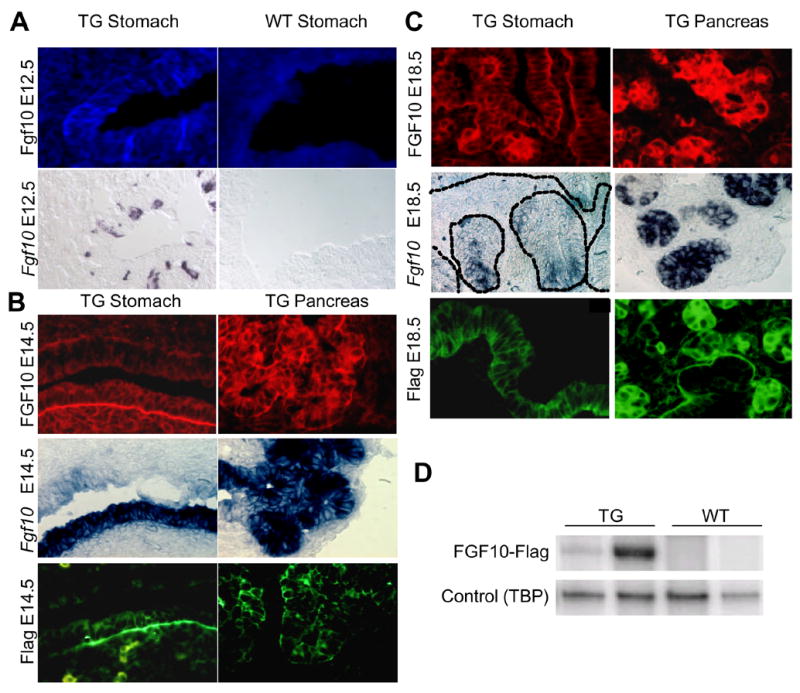
A: E12.5 posterior stomach. Expression of FGF10 was detected with an antibody that detects only high levels of the protein. Fgf10 mRNA was detected by in situ hybridization on an adjacent slide. A few cells in the most posterior stomach of the TG expressed FGF10. No staining was detected in WT littermates. B: E14.5 posterior stomach (lower epithelium). Flag and FGF10 protein detected by immunohistochemistry and Fgf10 mRNA detected by in situ hybridization. No staining was detected in WT littermates (data not shown) n=3. ISH and IHC were performed on consecutive slides. C: E18.5 antrum. Flag and FGF10 protein detected by immunohistochemistry and Fgf10 mRNA detected by in situ hybridization. No staining was detected in WT littermates (data not shown) n=3. Fgf10 mRNA was only detected in glandular structures at this stage although the protein could also be detected in luminal cells. D: Semi-quantitative RT-PCR for the Fgf10-Flag construct in E18.5 stomach of two transgenic mice and their WT littermates. The shown bands are representative examples from three experiments
Disrupted stomach morphogenesis in pPdx1-FGF10FLAG mice
Visual inspection of E12.5 TG embryos (n=3) revealed minor disruptions of stomach morphogenesis. In contrast, at E14.5 (n=3), we detected a major disruption of normal stomach morphogenesis. The size of the stomach appeared slightly reduced, and the ventricular lumen of the stomach appeared filled with tissue. Furthermore, we could not identify the presence of a pyloric sphincter, leaving the attachment zone of the pancreas to the gastrointestinal tract undecided. We first performed a histological analysis of the developing stomach using markers of the epithelium (β-catenin), and the developing circular and longitudinal muscles (smooth muscle actin) (Fig. 7A). In the normal E14.5 stomach, the ventricular space is lined by a homogeneous pseudo-stratified epithelium, with little folding. At a fixed radial distance, smooth muscle (SM) cells differentiate, aligning accordingly to the future position of the contractile muscles of the muscularis (Fig. 7A). In transgenic littermates, almost the entire luminal area was populated by an increasingly folded endodermal epithelium. SM cell differentiation occurred at a similar radial distance to the hyperplastic epithelium, properly following the epithelial folding pattern, suggesting that the irregular muscle condensation is secondary to the morphogenesis of the epithelium (Fig. 7A). Consequently, we interpret these data to signify that the epithelial/mesenchymal signaling regulating SM differentiation is not disturbed. At E18.5 the phenotype was even more striking, with excess folding of the epithelium in the corpus and antral regions, which branched into complex tree-like structures never observed in the WT littermates (Fig. 7A, 7B and Supp. Fig. 3 and 4). The forestomach region developed a stratified squamous epithelium indistinguishable from the WT forestomach region, although in some regions we noted a disturbed differentiation process, which included failure in development of the stratified epithelium (Supp. Fig. 3, Supp. Fig 4C–D). In those regions we also observed that the normal confinement of DNA-incorporating nuclei to the basal cells seen in WT, was disrupted (Supp. Fig. 4E–F). The smooth muscle development appeared normal in the forestomach.
Figure 7. Disturbed gastric morphogenesis in Pdx1-FGF10 embryos.
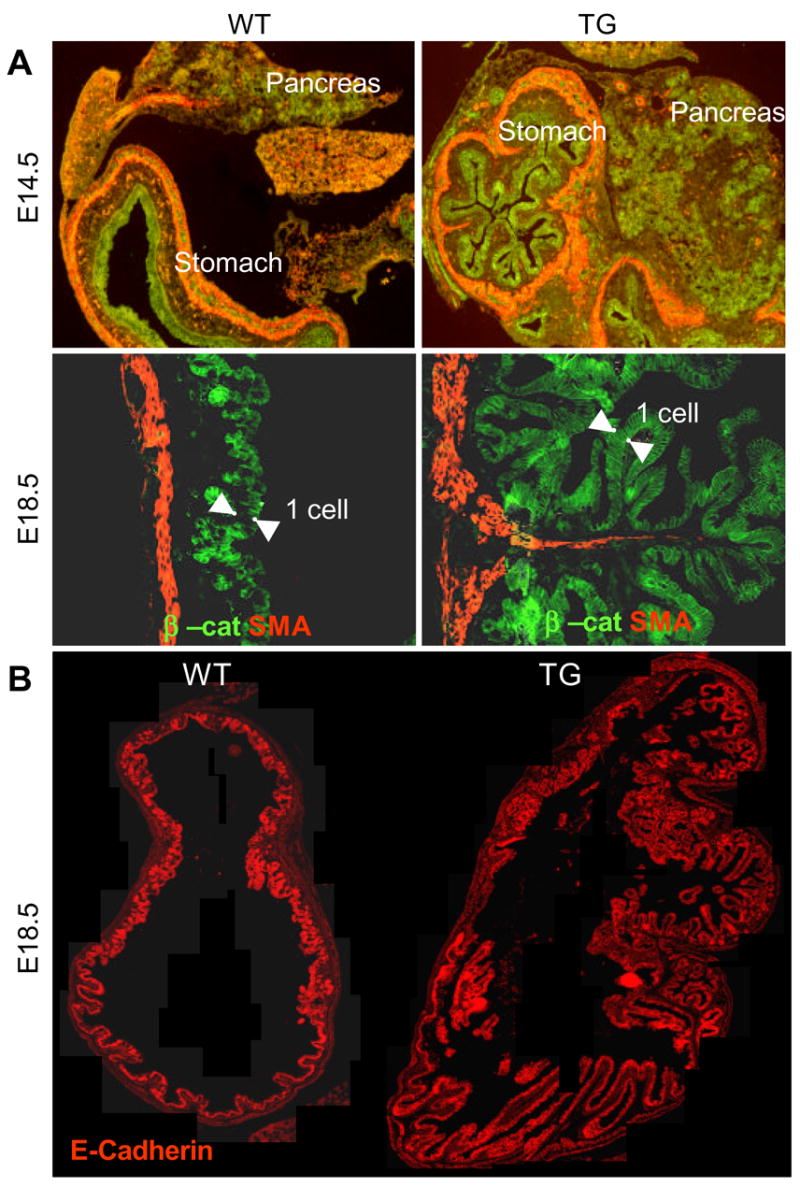
A: Immunohistochemistry for smooth muscle actin and the epithelial marker beta-catenin. B: Immunohistochemistry for E-cadherin on E18.5 TG and WT stomach.
Conservation of distal foregut patterning in pPdx1-FGF10FLAG mice
Due to the difficulty in defining the stomach/duodenal junction morphologically in both E14.5 and E18.5 transgenic embryos, we decided to investigate whether this region had been properly specified using a combination of stomach, pancreatic, and intestine-specific markers. Using the region-specific lectins DBA and UEA which marks the prospective duodenal and gastric regions at E14.5, respectively, we were able to define the attachment zone of the Nkx6.1 -expressing hyperplastic pancreas cells within the prospective duodenal region (Fig. 8A–D), as it normally occurs. However, the regionalization of the pancreas/duodenum connection was not strict as Nkx6.1-expressing pancreatic cells were clearly observed within the duodenal area at E14.5. Given the complete absence of a main pancreatic duct, and the absence of the normal narrowing of the evaginating pancreatic epithelium, this is not surprising. We noted that UEA-positive gastric cells were clearly observed in a more anterior location than the pancreatic/duodenal zone, indicating the conservation of the distal foregut patterning in the FGF10 transgenic embryo. It therefore seems that the distal foregut patterning mechanisms operate even in the absence of a pyloric sphincter and a normal pancreas. These results were corroborated at E18.5, where both pancreatic evaginations, dorsally and ventrally, entered the adjacent endoderm at the level of the Cdx2-expressing duodenum. (Fig. 8E).
Figure 8. Distal foregut patterning.
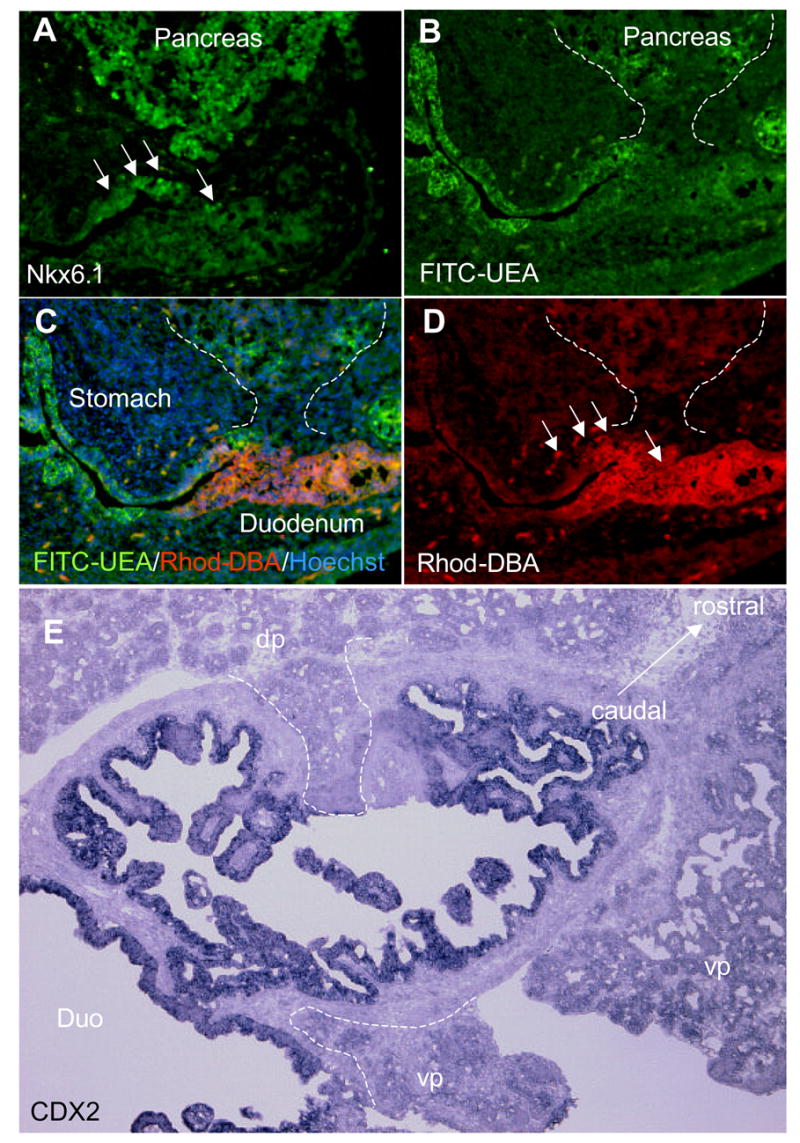
A: Nkx6.1 immunostaining showing the position of the pancreas at E14.5. B: FITC -UEA staining at E14.5. C: FITC-UEA, Rhodamine-DBA and Hoechst staining merge. D: Rhodamine-DBA staining showing the position of the duodenum at E14.5. E: Cdx2 in situ of the duodenum at E18.5
Proliferation analysis of stomach epithelial cells
The increased folding of the epithelium might be suggestive of an increased cellular proliferative rate, or a result of FGF10-mediated morphogenetic effects upon branch-point selection. To analyze for total epithelia area changes, we measured gastric glandular epithelial area at E12.5, E14.5 and E18.5. We also compared cell proliferation and cell death rates within the glandular region (antrum/corpus, forestomach excluded). We did not find any evidence suggesting an increase in total epithelial area (Fig. 9B), nor did we find a reduction in cell death using a combination of Hoechst and TUNEL-staining, as this parameter was negligible in the normal embryonic stomach as well as in the TG stomach (data not shown). To account for any change in the proliferative index, we analyzed for M-phase-activity (using phosphorylated histone H3, pHH3), specifically in the stomach epithelium by co-staining with E-cadherin. Analysis of E12.5, E14.5 and E18.5 TG mice and WT littermates did not reveal a significant increase in the proliferative activity in stomach glandular epithelium (Fig. 9A). Also, the number of cells (counted as Hoechst nuclei) per area was unchanged, signifying that cell size was not affected by ectopic FGF10 (results not shown). We can therefore exclude that the increased folding is caused by an overall increase in epithelial area. Instead, it seems likely that FGF10 is specifically promoting epithelial folding as a chemotaxic factor similar to what is seen in the lung (Park et al., 1998). Therefore, to elucidate whether FGF10 changes the pattern of proliferating cells rather than the overall proliferative index, we analyzed the distribution of cycling cells with PCNA staining. Cycling PCNA positive cells are confined to the glandular compartment in normal E18.5 antrum epithelium, but was distributed throughout the luminal and glandular compartments in the TG antrum (Fig. 9C). This disruption of the proliferative niche was not absolute, as patches of PCNA negative luminal cells were found (Fig 9C, upper right panel). To further pursue the effect of FGF10 on luminal and glandular compartments we next analyzed the distribution of gland and luminal markers.
Figure 9. Analysis of cellular proliferation.
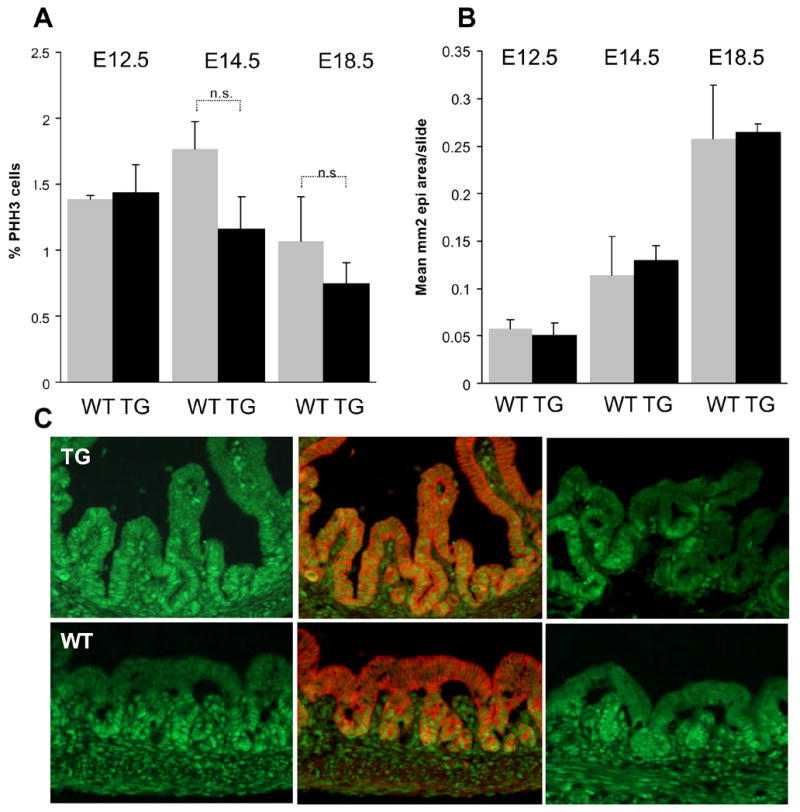
A: Proliferation of epithelial cells in the glandular stomach as detected by pHH3, E-cadherin and Hoechst co-staining is not significantly altered by ectopic FGF10 at E12.5, E14.5 and E18.5 in the stomach (n=3 WT/TG embryos with each n=3 slides distributed across the stomach with no more than 160μm between slides analyzed at each time point.). B: Epithelial area per slide as measured by E-cadherin staining is unaltered by transgene expression at E12.5, E14.5 and E18.5. (n=3 WT/TG embryos with each n=4 (E18.5) or n=3 slides distributed across the stomach with no more than 120 μm between slides analyzed at each time point.) C: PCNA immunostaining shows disruption of the proliferative nice in E18.5 antral stomach. PCNA is shown in green and E-cadherin in red. The two PCNA stainings are from different embryos.
Differential effects of FGF10 on development of stomach secretory cells
Pepsinogen C and H+/K+ ATPase is expressed by chief and parietal cells respectively in the glands of the E18.5 stomach, while Trefoil factor 1 (TFF1, formerly pS2) and Trefoil factor 2 (TFF2, formerly spasmolytic polypeptide, SP) are expressed by mucus producing luminal cells (Otto and Patel, 1999). Immunostaining for Pepsinogen C showed a 68% increase in the number of chief cells (P<0.05) (Fig. 10B and 10C) and an expansion of these cells to nodes of branching epithelium not located within the mesenchyme (Fig. 10B). Interestingly, immunostaining for the C terminal end of the H+,K+-ATPase α-unit showed a 78% decrease in parietal cells (P<0.05). This decrease was corroborated by multiplex-RT-PCR. mRNA expression of the mucin associated Trefoil Factor 2 was unchanged (Fig. 10A), but in-situ hybridization revealed a shift in localization from luminal cells in the WT corpus to glandular cells in the TG (Fig. 10B). It thus seems that the Tff2 expressing mucus cells form unperturbed by FGF10, but that their localization has been shifted. Trefoil factor 1 expression and localization were unchanged (Fig. 10A and data not shown). In-situ hybridization for intestinal fatty acid binding protein (Ifabp), that is expressed in the stomach of Ngn3 null mice (Lee et al., 2002) and in intestinal metaplasia of the stomach, did not show any expression in the stomach of pPDX-FGF10 mice (Fig. 10C and data not shown), and neither did the intestinally expressed TFF3 (data not shown). The analysis of exocrine cells showed that not only are the luminal and glandular cell types displaced by FGF10, certain cell fates are also suppressed while others are accelerated. Previously published results show suppression of terminal endocrine cell fates by FGF10 during ectopic expression in the pancreas (Norgaard et al., 2003). We therefore continued to analyze differentiation of endocrine cell types.
Figure 10. Analysis of Cell Differentiation.
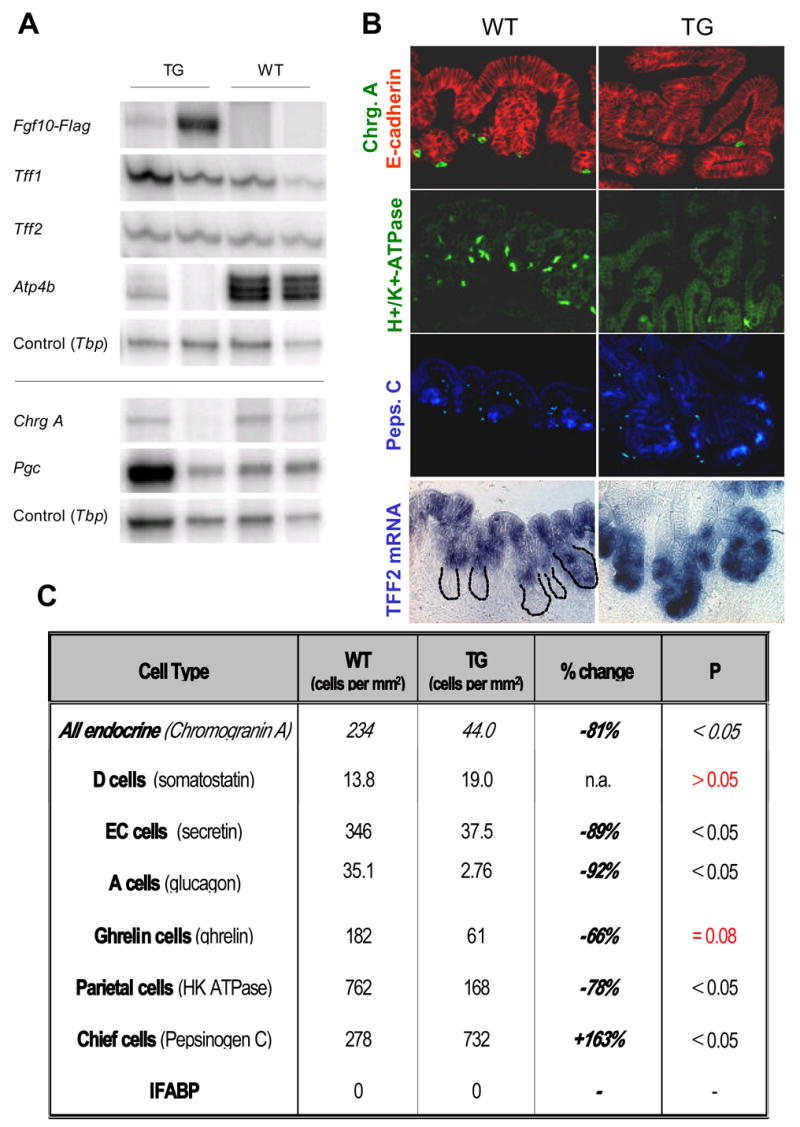
A: Semi-quantitative multiplex RT-PCR of the differentiation markers Trefoil Factor 1 (Tff1), Trefoil Factor 2 (Tff2), H+/K+-ATPase (Atp4b), Chromogranin A (Chrg A), and pepsinogen C (Pgc). The shown bands are representative examples from two experiments. B: Immunostainings of Chromogranin A with E-Cadherin, H+/K+-ATPase and pepsinogen and in-situ hybridization of Tff2 in E18.5 pPDX-FGF10 and WT littermates (n=3). C: Quantification of immunostainings shown in B and immunostainings for somatostatin, secretin, glucagon and ghrelin. Positive cells per mm2 epithelium as marked by E-cadherin was quantified on three slides distributed throughout the stomach for each of three different WT or TG mice and the mean calculated. Statistical significance was tested using the Student’s T-test. No Ifabp was detected in the stomach by in situ hybridization of either WT or TG.
FGF10 attenuates stomach endocrine terminal differentiation
We described the onset of gastric endocrine differentiation at E15.5–E16.5 using the pan-endocrine marker Chromogranin A. More specifically, the gastric endocrine cell complement includes secretin, glucagon, somatostatin, ghrelin, and gastrin producing cells, predominantly. The majority of the endocrine cells are formed based on the activity of bHLH components, including the Ngn3 gene (Jenny et al., 2002; Jensen et al., 2000; Lee et al., 2002), and are repressed by the Notch system, acting through Hes1 (Jensen et al., 2000). We compared E18.5 WT and TG littermates for effects on stomach cell differentiation by semi-quantitative multiplex RT-PCR (Fig. 10A) and immunohistochemical evaluation of markers for differentiated stomach cell types (Fig. 10B and 10C). Multiplex RT-PCR showed a down-regulation of mRNA for Chromogranin A (with relative ratios of Chgr.A/TBP of TG1: 0.07, TG2: 0.02, WT1: 0.21, WT2: 0.24; Fig 10A). This effect was most pronounced in the TG embryo with the highest expression of ectopic FGF10. Immunohistochemistry for Chromogranin A positive cells likewise showed a significant reduction (81% reduced compared to WT (P<0.05)) However, a few Chromogranin A positive cells were still present and we went on to discern the identity of those cells. Glucagon staining revealed an almost complete loss of A cells (92% reduced compared to WT (P<0.05)) and staining against secretin showed a significant decrease in enterochromaffin cells (89% reduced compared to WT (P<0.05)), while analyzing for somatostatin producing D cells showed no significant change. This is a surprising result, since a lack of somatostatin expression was reported in the Ngn3−/− stomach (Lee et al., 2002). Quantitation of Grelin cells did not reveal a statistically significant change, although ghrelin cells were significantly reduced in two out of three transgenic mice. Stomach gastrin expressing cells (G-cells) where not detected at this time in development in WT or TG embryos.
FGF10 leads to nuclear translocation of FGFR2
As shown by multiplex-RT-PCR, mRNA for FGFR2b is present in the embryonic stomach, and based on immunohistochemistry the protein is present in the cytoplasm/membrane of the stomach epithelium at E14.5 and E18.5. Upon expression of FGF10 in the TG mice the levels of FGFR2b are unchanged (Fig. 12), but immunohistochemical detections reveals a change in FGFR2 localization from the cytoplasm/membrane to the nucleus in the forming glands (Fig. 11B). This localization event was never detected in stomach epithelium of wildtype littermates (Fig. 11B and 4A and B). Given the observed increase in pepsinogen-expressing cells, we speculated whether nuclear localization of FGFR2 was associated with chief cell differentiation. Co-staining with pepsinogen and FGFR2 showed no co-localization (Fig. 11B). In contrast, the widespread nuclear presence of FGFR2 in E-cadherin expressing and terminal marker negative stomach epithelial cells suggests that FGFR2 nuclear entry is a result of ongoing FGF10 signaling in the arrested progenitors.
Figure 12. Changes in cell signaling in the pPDX-FGF10 stomach.
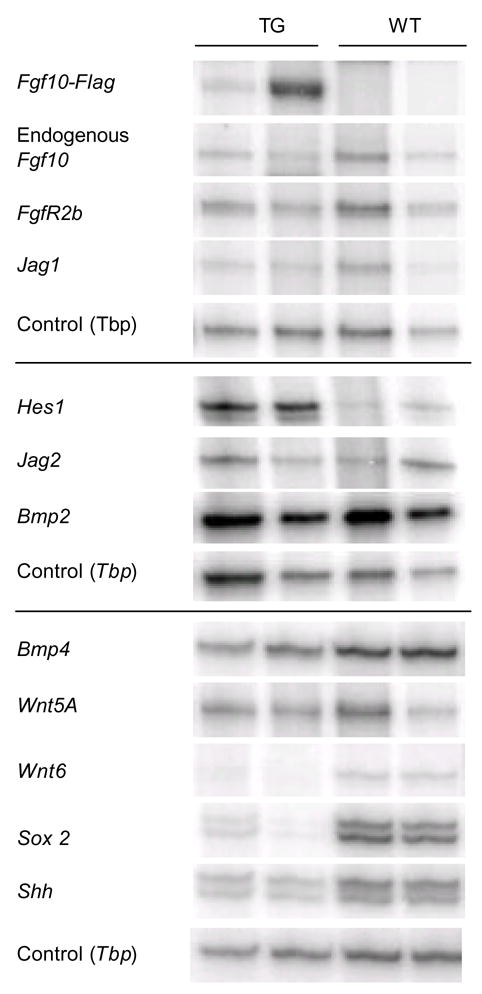
Semi quantitative multiplex RT-PCR of the expression of ectopic FGF10-Flag, endogenous Fgf10, the notch targets Hes1, notch ligands Jagged1 and Jagged2, Bmp2 and 4, Wnt5a and 6, Sox2, Sonic hedgehog (Shh), and the FGF10 receptor FGFR2b. Tbp was used as control. The shown bands are representative examples from two experiments
Figure 11. Changes in Hes expression and FGFR2 intracellular distribution.
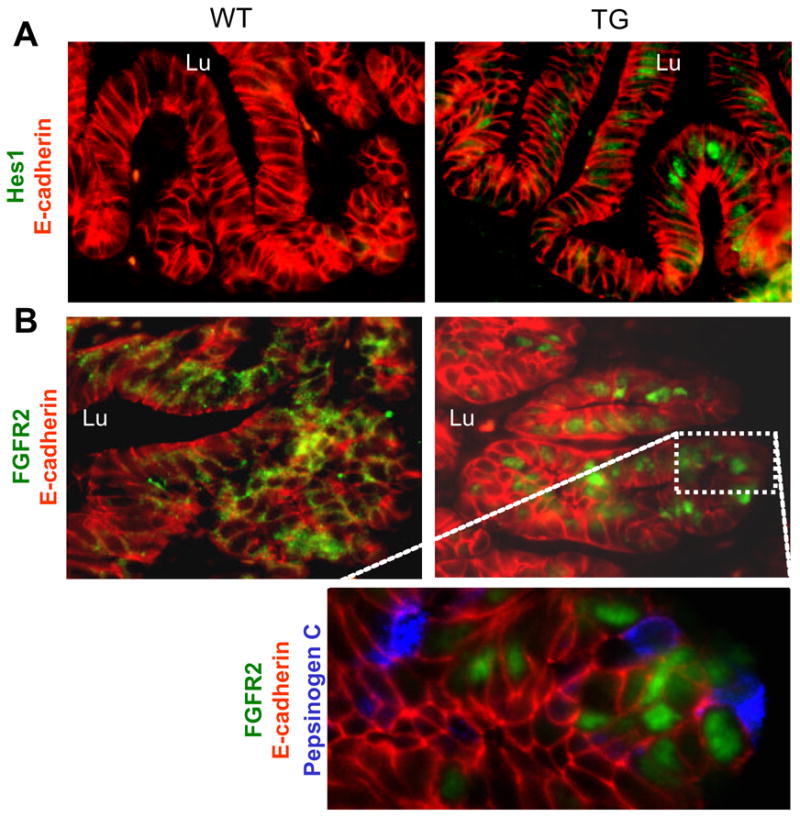
A: Immunohistochemistry of Hes1 and E-cadherin. B: Immunohistochemistry of FGFR2 and E-cadherin. Enlargement of area in the TG also shows pepsinogen staining. Lu: towards the lumen of the stomach.
FGF10 induces Hes1 expression in embryonic stomach
We next analyzed expression of notch pathway components in the pPdx-FGF10FLAG stomach by multiplex-RT-PCR (Fig. 12). Hes1 expression is very low in wildtype mice, but it was considerably increased in the TG mice, while levels of Jagged1 and 2 were unchanged (Fig. 12). Immunohistochemistry for Hes1 using an antibody (Lee et al., 2005) that in our hands only allows detection of high Hes1 levels, showed upregulation of Hes1 in the epithelium of the TG mouse (Fig. 11A). Hes1 was detected in the forming glands in the antrum, consistent with the area where we find the strongest expression of FGFR2 and ectopic FGF10 at E18.5.
Effects of FGF10 on gastric morphogenetic signaling networks
Sonic Hedgehog (Shh) has been shown to be important for stomach gland development in both chicken and mouse (Fukuda et al., 2003; van den Brink et al., 2001) and is downstream of FGFR2b in limb development (Revest et al., 2001). Shh is expressed in gastric epithelium and has been localized to parietal cells in human adult stomach and in parietal and zymogenic cells in murine adult stomach (van den Brink et al., 2001). Our expression ana lysis showed a 2-fold downregulation of Shh in E18.5 TG stomach (Fig. 12). We also analyzed the expression of BMP4, as this mesenchymal expressed signaling molecule has been shown to be a downstream target of Shh in mouse adult stomach (van den Brink et al., 2001). No change in BMP4 expression was noted. Similarly, BMP2 expressed by gastric mesenchymal cells (Kaestner et al., 1997), was unaltered (Fig. 12). Wnt-signaling is critical for intestinal progenitor/stem cell maintenance, and multiple studies indicate a role in both normal and transformed growth of gastric cells (Perreault et al., 2005; Tsukamoto et al., 2003; Kim et al., 2003). Wnt5A is highly expressed in the gastric and intestinal mesenchyme (Lickert et al., 2001), but we did not observe any changes in overall gastric expression in the Pdx1-FGF10 transgenic model. In contrast, Wnt6 is expressed in the normal embryonic stomach, but we were unable to detect any expression of Wnt6 in transgenic stomach (Fig. 12). In-situ hybridization for Wnt6 in gastric development showed that Wnt6 is exclusively expressed by the gastric epithelium (Supp. Fig. 5). The transcription factor Sox2 is expressed in gastric and esophageal epithelium with a more anterior expression domain early in gut development, that expands into the posterior stomach just prior to the secondary transition (Fig 4 and results not shown) and is also one of a few transcription factors expressed in embryonic stem cells that is lost upon differentiation (Carlin et al., 2006). Sox2 expression was lowered in the pPDX-FGF10 mouse in a manner correlating with levels of ectopic FGF10 (Fig. 12).
Discussion
Although the stomach is amongst the largest organs, relatively little is known of its morphogenesis and development, particularly in relation to which genetic networks may coordinate the process of gastric cell differentiation. To better characterize the temporal onset, and local patterning of gastric cell differentiation, we employed staining techniques for the terminal gastric products allowing the visualization of chief, parietal, mucous and gastric endocrine cells, combined with an RT-PCR procedure. We conclude that each of the three major gastric regions; forestomach, corpus and antrum, initiates terminal differentiation concomitantly at E15.5. We define this strict temporal onset of terminal differentiation as the “secondary transition”, reserving the temporal specification of pro-gastric endodermal cells towards the gastric fate at early somite stages as the “primary transition” – a process involving early splanchnic mesoderm signaling controlled by Hox-gene family patterning (Pitera et al., 1999; Aubin et al., 2002) and the homeobox gene Barx1 (Kim et al., 2005). No terminal differentiation events appear associated with the gastric primary transition, in contrast to that of the pancreas. The gastric secondary transition is characterized by the initial presence of terminal cell types, and coincides with the time when the caudal gastric epithelium is first starting to form glandular structures. At E15.5 the invagination of epithelium and subsequent glandular formation is initiated, but is far from completed when the terminal cellular fates are induced (E16.5). Thus, we can conclude that gastric cytodifferentiation during the embryonic development, operates irrespectively of the presence of a mature gastric pit-gland unit.
In comparison to the pancreas, where the “pancreatic secondary transition” occurs at E13.5–E14.5 and signifies the development of mature exocrine, ductal, and insulin-producing cells, the stomach is two days delayed. This delay argues against sharing of any systemic factors that might help trigger the secondary transition in these adjacent organs. It is currently believed that organ-intrinsic factors acting in mesenchymal-to-epithelial signaling are the major determinants in orchestrating the onset of the secondary transition in the pancreas. FGF10 is a critical part in this and helps to maintain the undifferentiated pancreatic progenitor state prior to the secondary transition, in part through maintenance of Notch signaling (Norgaard et al., 2003). These observations motivated us to address if FGF signaling similarly guide stomach progenitor cell maintenance and if this may be similarly involving the control of Notch signaling. To address the identity of FGF-family proteins expressed in gastric development, we screened all members by RT-PCR performed on a kinetic series of embryonic stomach cDNA. These results highlighted FGF10 as a plausible FGF-family member playing a role prior to the secondary transition in the stomach. FGF10 is expressed abundantly in the adjacent gastric mesenchyme, in a decreasing gradient from the distal mesenchymal cells in the posterior stomach (Fig. 3 and Supp. Fig. 1A and 2). It is thus positioned as a component that might influence gastric epithelial cells, prior to the secondary transition. The predominant receptor for FGF10, FGFR2IIIb, was in turn expressed mainly in the gastric epithelium, arguing for a mesenchymal/epithelial interaction, similar to what is observed in lung and pancreas. We noted that FGFR2 expression was also not uniform, as epithelium in the corpus expressed only minor amounts of FGFR2 prior to the secondary transition, which could indicate that corpus might differentiate between FGF10 availability as compared to the antrum. As FGF10 and FGFR2IIIb expression is maintained after the secondary transition, FGF-signaling appears to have a later role in the more mature gastric epithelium as well. We find expression of multiple Notch signaling components in gastric progenitor cells. Notch1, Notch2, the downstream target Hes1, and the ligand Jagged2, are all expressed at high levels in the stomach prior to the secondary transition. Following the transition, expression can still be observed in the basal population of the forestomach and in epithelial cells of the antrum and corpus, but not in differentiated squamous cells. It is thus likely that notch signaling also plays a role in murine gastric development.
In order to investigate the role of FGF10/FGFR2 signaling in embryonic stomach, we forced epithelial expression of FGF10, disrupting the FGF10 gradient from the mesenchyme. The results provide evidence for a role of FGF10 in gastric gland formation and progenitor maintenance and the resulting suppression of differentiation bears some resemblance to that observed during pancreatic development (Norgaard et al., 2003).
The most apparent phenotypic disruption is a luminal branching of the epithelium, contrasting with the folding into the mesenchyme observed during gland formation in the normal stomach. We show that this luminal b ranching is not due to an increase in overall epithelial area or proliferation, but rather a change from gland niche restricted to diffuse proliferation. This phenotype reflects what has been seen in the lung, where FGF10 regulates directional outgrowth as a chemoattractant, but only acts as a modest proliferation factor (Park et al., 1998). Very recently, two studies describing the gastric phenotype of Fgf10 −/−, Ffgr2IIb −/− mice (Spencer-Dene et al., 2006), and virus-mediated overexpression of Fgf10 and a secreted FGFR2b in chicken (Shin et al., 2006) observed a similar role of Fgf10 to the one reported here in regulation of gland formation. Both knockout mouse models show a decrease in the formation of complex glands, as does the model of secreted FGFR2b in chicken, while the virally mediated fgf10-overexpression model shows hypermorphic epithelial structures similar to what we report here. In contrast to our over-expression model, the chicken study ascribes the role of Fgf10 in gland formation to a direct regulation of cell proliferation. However, only a modest increase in the percentage of proliferating cells upon over-expression of fgf10 (Shin et al., 2006) is observed. This difference may be due to species differences in regulation of proliferation, also supported by the observation that the chicken gastric epithelium at late stage 36/day 10 does not have a defined proliferative zone (Shin et al., 2006). Also, the level of Fgf10 expression in the chicken study likely exceeds that obtained through the ectopic expression in the mouse, possibly leading to the observed difference in mitogenic activity. This argument would also help explain the difference in mitogenic FGF10 signaling in the pancreas as compared to the stomach in the pPDX-FGF10FLAG model; Activity of the Pdx1-promoter in the pancreas is higher than in the gut (Fig. 5), and expression of TG fgf10 protein is indeed lower in the stomach (this paper and Norgaard et al). Thus, we believe it is entirely possible that fgf10 operates both as a chemotaxic factor in gland development at lower levels, and at higher levels through induction of epithelial growth.
We hypothesized that the increased branching would associate with exogenous gland formation, and indeed expression of the glandular marker pepsinogen C was increased and chief cells had spread to branching nodes in the lumen. In contrast, the parietal cell type was almost absent, while the luminal marker TFF1 was preferentially expressed in forming glandular structures, suggesting that luminal cells ectopically differentiate in the glandular domain. This reversal of localization of pepsinogen and TFF1 is similar to what is seen in the chicken Fgf10 overexpression model (Shin et al., 2006). We are curious as to why the chief cell fate was promoted while parietal cell fate was repressed. Both cell types are glandular, altho ugh chief cells are located deeper in the gland. We speculate that early gastric A-P patterning might have been affected. We show that at the initial time of their development, a spatial separation of parietal and chief cells in the corpus and a ntrum exist. Therefore, the observed loss of parietal cells and gain of chief cells in the transgenic model would reflect that the corpus acquires antral identity, as we also found no data to support expansion of the antrum by increased proliferation. The hypothesis that Fgf10 regulates AP-patterning is reinforced by the fact that Shh, which is normally expressed mostly in the anterior stomach (Aubin et al., 2002), is downregulated by overexpression of FGF10 (this study) and upregulated in the fgf10 knockout (Spencer-Dene et al., 2006). It is worthwhile to note that for both the fgf10 and Fgfr2IIIb knockout mice a reduced antral stomach epithelium is observed (Spencer-Dene et al., 2006), consistent with a role of FGF-signaling in promoting antral regionalization. Interestingly, “antralization” is observed in certain gastric metaplasias, also known as “SPEM” (spasmolytic peptide expressing metaplasia), characterized by oxyntic atrophy and a shift in localization of TFF2 mRNA from the mucous neck cells in the normal adult fundus to the lower two thirds of the glands in the H. felis–infected fundus and in SPEM (Nomura et al., 2004; Wang et al., 1998), similar to what we see. Obviously the two phenotypes cannot be directly compared, as the embryonic stomach described here does not posses the mature glands described in SPEM, but these results nonetheless suggest that ectopic expression of FGF10 in the embryonic stomach results in a SPEM-like embryonic phenotype and consequently indicates a role of FGF signaling in gastric metaplastic development (Schmidt et al., 1999). We speculate that the normal gradient of FGF10 and FGFR2-expression in the stomach may help specify antrum versus corpus/fundus fate choices. The lower levels of FGF10 and FGFR2 in corpus (this study and (Aubin et al., 2002)) suggest that progenitors here normally experience lower FGFR2 signaling as compared to antrum. In TG embryos, on the other hand, the FGF10 availability is much increased, possibly reaching up to, or above, normal antral levels, and presumably causing the antralization of the corpus. Further experiments are needed to validate if this gradient of FGF signaling helps specify stomach region identity.
Similar to what is seen in the pancreas of pPDX1-FGF10FLAG mice (Norgaard et al., 2003), we saw almost complete elimination of endocrine cell fates. Taken together with the attenuation of parietal cell differentiation, we conclude that Fgf10 inhibits differentiation of the gastric progenitors towards the majority of cell fates, thus maintaining the progenitor cells. In the pancreas, this Fgf10 effect is mediated through Notch signaling (Norgaard et al., 2003). When we analyzed downstream changes in transcription of notch signaling components, we noted that Fgf10 elicits an upregulation of Hes1 expression; supporting our hypothesis that Fgf-signaling is upstream of Notch signaling in the stomach. This does not seem to occur through increased levels of jagged-ligand production, as we previously suggested for the pancreas. It has previously been shown that Hes1 regulates endocrine differentiation in the murine stomach (Jensen et al., 2000), and active Notch signaling inhibit terminal cell fates in the stomach of the chicken (Matsuda et al., 2005). Interestingly, activated Notch also promote glandular specification in the chicken (Matsuda et al., 2005), suggesting that not only the progenitor maintenance effect of FGF10, but also the increased formation of glandular structures seen, could be mediated through notch signaling. Further research is needed to elucidate this aspect in the mouse.
Fgfr signaling affects several downstream signaling pathways, and so we also analyzed changes in alternative pathways. An immediate molecular effect of ectopic expression of FGF10 was the occurrence of nuclear localization of FGFR2. The biological relevance of nuclear localization of FGFR2 is unknown, but in this case it seems to be a response to hyper-stimulation of the receptor, as it was never observed in wildtype stomach epithelium. However, nuclear localization of FGFR2 as a response to FGF can not be dismissed as just an artifact of over-expression of ligand in all cases, as it has previously been described in normal Sertoli cells, where it disappears in FGF9 null mice (Schmahl et al., 2004). Nuclear localization of FGFR2 also occurs in osteoblasts in association with prostaglandin induction (Sabbieti et al., 2005), and thus seems to have functional relevance. Interestingly, nuclear FGFR1 acts as a transcriptional regulator that stimulates growth and differentiation responses (reviewed in (Stachowiak et al., 2003)), and nuclear FGFR2 has been associated with differentiation of Sertoli cells (Schmahl et al., 2004). A direct association with terminal differentiation in the gastric epithelium is not evident, as nuclear FGFR2 did not co-localize with pepsinogen in the pPdx1-FGF10 TG mice.
Stomach development and homeostasis has been shown to be regulated in part by Wnt, Shh and BMP–signaling and we therefore analyzed whether ectopic FGF10 expression had any influence on the transcription of components within these pathways. Our data suggest negative effects of FGF10 on Wnt6 and Shh expression, but not on BMP2, BMP4 and Wnt5A. It has previously been shown that Shh helps to induce BMP4 expression in adjacent submucosal cells of the hindgut in chicken, but this induction was not found in midgut (Roberts et al., 1995). Our results indicate that in the mouse, gastric BMP4 expression is likewise independent of Shh in the embryonic stomach. We also analyzed the expression level of the transcription factor Sox2, which is selectively expressed in the stomach and esophagus regions of the endoderm, and found a marked downregulation. This was surprising, given that Sox2 is induced in FGF treated osteoblasts (Mansukhani et al., 2005), but reinforces the hypothesis that Fgf10 regulates AP-patterning. It is noteworthy that Hes1, Wnt6, Shh and Sox2 all are expressed by gastric epithelium, whereas the BMP2, 4 and Wnt5a are mesenchymal. This supports the fact that we did not see any changes in morphology of the mesenchyme, and shows that FGF10 ectopically expressed by epithelial cells predominantly influences the epithelium itself.
We here provide evidence to support a role of FGF10 in gland formation and gastric patterning, in addition to demonstrating a functional role for FGF10 upstream of Hes1 in gastric progenitor maintenance, extending the knowledge of FGF10 as a mesenchymal-derived factor involved in epithelial cell differentiation also to that of the stomach. FGF10-mediated mesenchymal-epithelial signaling thus appears to be conserved from anterior endoderm (lung) (Sekine et al., 1999), over distal foregut (stomach and pancreas (Bhushan et al., 2001)). The role of FGF10 in more posterior regions of the gut needs to be clarified, although data obtained in parallel with this study indicate a similarly important role for FGF10 in duodenal progenitor maintenance (will be described elsewhere). Therefore, similar to the conserved nature of epithelial Notch signaling along the gut tract, FGF10 signaling operates to help maintain the epithelial progenitor niche. Further studies should be aimed at defining how these signaling systems may interact, as such knowledge is likely to be important in understanding both the development of gastric cancer, as well as programs of stomach regeneration, and how these processes may be modulated.
Supplementary Material
Supplementary figure 1: FGF10 and FGFR2, 3 and 4 expression in stomach. Images of in situ hybridiza tions on E14.5 whole embryos obtained from Genepaint (www.genepaint.org) (Visel A et al., Nucleic Acids Res. 2004, D552-6). A: FGF10 probe. A’: enlargement of A. B: FGFR2 probe. A Blast-analysis revealed that this probe is specific for FGFR2IIIb. B’: enlargement of B. C: FGFR3 probe. C’: enlargement of C. D: FGFR4 probe. D’: enlargement of D.
Supplementary figure 2: FGF10 expression is highest towards the posterior end of the stomach. Images of in situ hybridizations on E14.5 whole embryos obtained from Genepaint (www.genepaint.org) (Visel A et al., Nucleic Acids Res. 2004, D552–6). 1–11 shows consecutive sagittal sections from anterior end of an E14.5 stomach towards the posterior end. Diagram illustrates the position of the sections shown.
Supplementary figure 3: Stomach morphology is severely disturbed in the pPDX-FGF10 mouse. A: Composite image showing E-cadherin immunostaining of E18.5 wildtype stomach. B: Composite image showing E-cadherin immunostaining of E18.5 pPDX-FGF10 mouse. White boxes outline regions shown in higher power in Fig. 4 (antral/forestomach regions). Transgenic forestomach represented in a heterogeneous fashion in TG embryos, where certain regions were indistinguishable from WT squamous epithelium. However, other regions (Box) displayed an almost complete elimination of squamous cell differentiation.
Supplementary figure 4: A–D: Higher-magnification of antral and forestomach regions shown in Supp. Fig 3. E,F: BrdU (red) and β-catenin double immunofluorescence of E18.5 forestomach. Proliferating cells are normally observed only at the basal layer in the normal forestomach; however, in select regions of the TG forestomach the distribution of BrdU-incorporating nuclei was prominent throughout the cell layers. This was accompanied by the absence of cell shredding seen in C.
Supplementary figure 5: Wnt6 is expressed by the stomach epithelium. In-situ hybridization for Wnt6 on E18.5 CD1 stomach (A and B), intestine (C) and kidney (D).
Supplementary Table 1: Multiplex primers and FGF10-Flag primers used for genotyping.
Acknowledgments
This work was supported by NIH-R01 DK070636(J.J.) and the American Diabetes Association (Career Development Award, J.J). Further support was obtained from NIH grant P30 DK57516 (Diabetes and Endocrinology Research Center). We thank Dr. Nadean Brown for supplying us with the rabbit α-Hes1 antibody, Dr. L. Sussel for supplying the protocol for secretin and ghrelin immunohistochemistry, Dr. R. Kageyama for the Hes1 ISH plasmid, Dr J. Hald for the Notch1 and Notch2 plasmid, Dr. G. Weinmaster for the Jagged2 plasmid and Dr. A.P. McMahon for the Shh plasmid. The U. Colorado Cancer Center transgenic facility is thanked for helping generate the transgenic embryos used in this study and Maureen A. Reddan is thanked for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialized mesoderm differentiation in the intestine and pancreas. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, de Angelis MH, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Aubin J, Dery U, Lemieux M, Chailler P, Jeannotte L. Stomach regional specification requires Hoxa5-driven mesenchymal-epithelial signaling. Development. 2002;129:4075–4087. doi: 10.1242/dev.129.17.4075. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Itoh N, Kato S, Thiery JP, Czernichow P, Bellusci S, Scharfmann R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128:5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- Carlin R, Davis D, Weiss M, Schultz B, Troyer D. Expression of early transcription factors Oct4, Sox2 and Nanog by porcine umbilical cord (PUC) matrix cells. Reprod Biol Endocrinol. 2006;4:8. doi: 10.1186/1477-7827-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BJ, Kleeberger W, Ball DW. Notch in lung development and lung cancer. Semin Cancer Biol. 2004;14:357–364. doi: 10.1016/j.semcancer.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Lim L. Transcription factors in mouse lung development and function. Am J Physiol Lung Cell Mol Physiol. 2001;280:L823–L838. doi: 10.1152/ajplung.2001.280.5.L823. [DOI] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukamachi H, Mizuno T, Takayama S. Epithelial-mesenchymal interactions in differentiation of stomach epithelium in fetal mice. Anat Embryol (Berl) 1979;157:151–160. doi: 10.1007/BF00305155. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Kameda T, Saitoh K, Iba H, Yasugi S. Down-regulation of endodermal Shh is required for gland formation in chicken stomach. Mech Dev. 2003;120:801–809. doi: 10.1016/s0925-4773(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, Guillemot F. neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc Natl Acad Sci U S A. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald J, Hjorth JP, German MS, Madsen OD, Serup P, Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Hart A, Papadopoulou S, Edlund H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev Dyn. 2003;228:185–193. doi: 10.1002/dvdy.10368. [DOI] [PubMed] [Google Scholar]
- Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development. 2000;127:3913–3921. doi: 10.1242/dev.127.18.3913. [DOI] [PubMed] [Google Scholar]
- Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, Gradwohl G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. Embo J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. Gene regulatory factors in pancreatic development. Dev Dyn. 2004;229:176–200. doi: 10.1002/dvdy.10460. [DOI] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, Madsen OD. Control of endodermal endocrine development by Hes-1. Nat Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jensen J, Serup P, Karlsen C, Nielsen TF, Madsen OD. mRNA profiling of rat islet tumors reveals nkx 6.1 as a beta- cell-specific homeodomain transcription factor. J Biol Chem. 1996;271:18749–18758. doi: 10.1074/jbc.271.31.18749. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Silberg DG, Traber PG, Schutz G. The mesenchymal winged helix transcription factor fkh6 is required for the control of gastrointestinal proliferation and differentiation. Gene Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- Kim BM, Buchner G, Miletich I, Sharpe PT, Shivdasani RA. The stomach mesenchymal transcription factor Barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev Cell. 2005;8:611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Kim HS, Hong EK, Park SY, Kim WH, Lee HS. Expression of beta-catenin and E-cadherin in the adenoma-carcinoma sequence of the stomach. Anticancer Res. 2003;23:2863–2868. [PubMed] [Google Scholar]
- Koike T, Yasugi S. In vitro analysis of mesenchymal influences on the differentiation of stomach epithelial cells of the chicken embryo. Differentiation. 1999;65:13–25. doi: 10.1046/j.1432-0436.1999.6510013.x. [DOI] [PubMed] [Google Scholar]
- Lee CS, Perreault N, Brestelli JE, Kaestner KH. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002;16:1488–1497. doi: 10.1101/gad.985002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaigre F, Zaret KS. Liver development update: new embryo models, cell lineage control, and morphogenesis. Curr Opin Genet Dev. 2004;14:582–590. doi: 10.1016/j.gde.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Li H, Edlund H. Persistent expression of Hlxb9 in the pancreatic epithelium impairs pancreatic development. Dev Biol. 2001;240:247–253. doi: 10.1006/dbio.2001.0440. [DOI] [PubMed] [Google Scholar]
- Lickert H, Kispert A, Kutsch S, Kemler R. Expression patterns of Wnt genes in mouse gut development. Mech Dev. 2001;105:181–184. doi: 10.1016/s0925-4773(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Madsen OD, Serup P, Jensen J, Petersen HV, Heller RS. An historical and phylogenetic perspective on islet development. In: Habener JF, Hussain MA, editors. In Molecular basis of pancreas development and function. Norvell, Ma, USA: Kluwer academic press; 2001. pp. 1–17. [Google Scholar]
- Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Wakamatsu Y, Kohyama J, Okano H, Fukuda K, Yasugi S. Notch signaling functions as a binary switch for the determination of glandular and luminal fates of endodermal epithelium during chicken stomach development. Development. 2005;132:2783–2793. doi: 10.1242/dev.01853. [DOI] [PubMed] [Google Scholar]
- Milano J, McKay J, Dagenais C, Foster-Brown L, Pognan F, Gadient R, Jacobs RT, Zacco A, Greenberg B, Ciaccio PJ. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82:341–358. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- Miller CP, McGehee RE, Jr, Habener JF. IDX-1: a new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. Embo J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Lamotte L, Couton D, Joshi RL. Interplay between FGF10 and Notch signalling is required for the self-renewal of pancreatic progenitors. Int J Dev Biol. 2006;50:17–26. doi: 10.1387/ijdb.052080fm. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Saitoh K, Kameda T, Kuroiwa A, Mizutani M, Koike C, Iba H, Yasugi S. BMPs are necessary for stomach gland formation in the chicken embryo: a study using virally induced BMP-2 and Noggin expression. Development. 2000;127:981–988. doi: 10.1242/dev.127.5.981. [DOI] [PubMed] [Google Scholar]
- Nomura S, Baxter T, Yamaguchi H, Leys C, Vartapetian AB, Fox JG, Lee JR, Wang TC, Goldenring JR. Spasmolytic polypeptide expressing metaplasia to preneoplasia in H. felis-infected mice. Gastroenterology. 2004;127:582–594. doi: 10.1053/j.gastro.2004.05.029. [DOI] [PubMed] [Google Scholar]
- Norgaard GA, Jensen JN, Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev Biol. 2003;264:323–338. doi: 10.1016/j.ydbio.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Otto WR, Patel K. Trefoil factor family (TFF)-domain peptides in the mouse: embryonic gastrointestinal expression and wounding response. Anat Embryol (Berl) 1999;199:499–508. doi: 10.1007/s004290050247. [DOI] [PubMed] [Google Scholar]
- Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. FGF-10 is a chemotactic factor for distal epithelial buds during lung development. Dev Biol. 1998;201:125–134. doi: 10.1006/dbio.1998.8994. [DOI] [PubMed] [Google Scholar]
- Perreault N, Sackett SD, Katz JP, Furth EE, Kaestner KH. Foxl1 is a mesenchymal Modifier of Min in carcinogenesis of stomach and colon. Genes Dev. 2005;19:311–315. doi: 10.1101/gad.1260605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitera JE, Smith VV, Thorogood P, Milla PJ. Coordinated expression of 3′ hox genes during murine embryonal gut development: an enteric Hox code. Gastroenterology. 1999;117:1339–1351. doi: 10.1016/s0016-5085(99)70284-2. [DOI] [PubMed] [Google Scholar]
- Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, JOHNSON RL, BURKE AC, NELSON CE, MORGAN BA, Tabin C. Sonic hedgehog is an endodermal signal inducing BMP4 and Hox genes during induction and regionalization of the chick hindgut. Development. 1995;121:3163–3174. doi: 10.1242/dev.121.10.3163. [DOI] [PubMed] [Google Scholar]
- Sabbieti MG, Marchetti L, Gabrielli MG, Menghi M, Materazzi S, Menghi G, Raisz LG, Hurley MM. Prostaglandins differently regulate FGF-2 and FGF receptor expression and induce nuclear translocation in osteoblasts via MAPK kinase. Cell Tissue Res. 2005;319:267–278. doi: 10.1007/s00441-004-0981-8. [DOI] [PubMed] [Google Scholar]
- Schmahl J, Kim Y, Colvin JS, Ornitz DM, Capel B. Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development. 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- Schmidt PH, Lee JR, Joshi V, Playford RJ, Poulsom R, Wright NA, Goldenring JR. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest. 1999;79:639–646. [PMC free article] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Shin M, Noji S, Neubuser A, Yasugi S. FGF10 is required for cell proliferation and gland formation in the stomach epithelium of the chicken embryo. Dev Biol. 2006;294:11–23. doi: 10.1016/j.ydbio.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Spencer-Dene B, Sala FG, Bellusci S, Gschmeissner S, Stamp G, Dickson C. Stomach development is dependent on fibroblast growth factor 10/fibroblast growth factor receptor 2b-mediated signaling. Gastroenterology. 2006;130:1233–1244. doi: 10.1053/j.gastro.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Fang X, Myers JM, Dunham SM, Berezney R, Maher PA, Stachowiak EK. Integrative nuclear FGFR1 signaling (INFS) as a part of a universal “feed-forward-and-gate” signaling module that controls cell growth and differentiation. J Cell Biochem. 2003;90:662–691. doi: 10.1002/jcb.10606. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Heller RS, Miller CP, Habener JF. Developmental expression of the homeodomain protein IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional reporter. Endocrinology. 1999;140:5374–5381. doi: 10.1210/endo.140.11.7122. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Inada K, Tanaka H, Mizoshita T, Mihara M, Ushijima T, Yamamura Y, Nakamura S, Tatematsu M. Down-regulation of a gastric transcription factor, Sox2, and ectopic expression of intestinal homeobox genes, Cdx1 and Cdx2: inverse correlation during progression from gastric/intestinal-mixed to complete intestinal metaplasia. J Cancer Res Clin Oncol. 2004;130:135–145. doi: 10.1007/s00432-003-0519-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Yamamoto M, Ogasawara N, Ushijima T, Nomoto T, Fujita H, Matsushima T, Nozaki K, Cao X, Tatematsu M. beta-Catenin mutations and nuclear accumulation during progression of rat stomach adenocarcinomas. Cancer Sci. 2003;94:1046–1051. doi: 10.1111/j.1349-7006.2003.tb01399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink GR, Hardwick JC, Tytgat GN, Brink MA, ten Kate FJ, van Deventer SJ, Peppelenbosch MP. Sonic hedgehog regulates gastric gland morphogenesis in man and mouse. Gastroenterology. 2001;121:317–328. doi: 10.1053/gast.2001.26261. [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den BM, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Wang TC, Goldenring JR, Dangler C, Ito S, Mueller A, Jeon WK, Koh TJ, Fox JG. Mice lacking secretory phospholipase A2 show altered apoptosis and differentiation with Helicobacter felis infection. Gastroenterology. 1998;114:675–689. doi: 10.1016/s0016-5085(98)70581-5. [DOI] [PubMed] [Google Scholar]
- Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev. 2005;19:1686–1691. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1: FGF10 and FGFR2, 3 and 4 expression in stomach. Images of in situ hybridiza tions on E14.5 whole embryos obtained from Genepaint (www.genepaint.org) (Visel A et al., Nucleic Acids Res. 2004, D552-6). A: FGF10 probe. A’: enlargement of A. B: FGFR2 probe. A Blast-analysis revealed that this probe is specific for FGFR2IIIb. B’: enlargement of B. C: FGFR3 probe. C’: enlargement of C. D: FGFR4 probe. D’: enlargement of D.
Supplementary figure 2: FGF10 expression is highest towards the posterior end of the stomach. Images of in situ hybridizations on E14.5 whole embryos obtained from Genepaint (www.genepaint.org) (Visel A et al., Nucleic Acids Res. 2004, D552–6). 1–11 shows consecutive sagittal sections from anterior end of an E14.5 stomach towards the posterior end. Diagram illustrates the position of the sections shown.
Supplementary figure 3: Stomach morphology is severely disturbed in the pPDX-FGF10 mouse. A: Composite image showing E-cadherin immunostaining of E18.5 wildtype stomach. B: Composite image showing E-cadherin immunostaining of E18.5 pPDX-FGF10 mouse. White boxes outline regions shown in higher power in Fig. 4 (antral/forestomach regions). Transgenic forestomach represented in a heterogeneous fashion in TG embryos, where certain regions were indistinguishable from WT squamous epithelium. However, other regions (Box) displayed an almost complete elimination of squamous cell differentiation.
Supplementary figure 4: A–D: Higher-magnification of antral and forestomach regions shown in Supp. Fig 3. E,F: BrdU (red) and β-catenin double immunofluorescence of E18.5 forestomach. Proliferating cells are normally observed only at the basal layer in the normal forestomach; however, in select regions of the TG forestomach the distribution of BrdU-incorporating nuclei was prominent throughout the cell layers. This was accompanied by the absence of cell shredding seen in C.
Supplementary figure 5: Wnt6 is expressed by the stomach epithelium. In-situ hybridization for Wnt6 on E18.5 CD1 stomach (A and B), intestine (C) and kidney (D).
Supplementary Table 1: Multiplex primers and FGF10-Flag primers used for genotyping.


