Abstract
The β-barrel proteins of mitochondria are synthesized on cytosolic ribosomes. The proteins are imported by the translocase of the outer membrane (TOM) and the sorting and assembly machinery (SAM). It has been assumed that the SAMcore complex with the subunits Sam35, Sam37 and Sam50 represents the last import stage common to all β-barrel proteins, followed by splitting in a Tom40-specific route and a route for other β-barrel proteins. We have identified new components of the β-barrel assembly machinery and show that the major β-barrel pathway extends beyond SAMcore. Mdm12/Mmm1 function after SAMcore yet before splitting of the major pathway. Mdm12/Mmm1 have been known for their role in maintenance of mitochondrial morphology but we reveal assembly of β-barrel proteins as their primary function. Moreover, Mdm10, which functions in the Tom40-specific route, can associate with SAMcore as well as Mdm12/Mmm1 to form distinct assembly complexes, indicating a dynamic exchange between the machineries governing mitochondrial β-barrel assembly. We conclude that assembly of mitochondrial β-barrel proteins represents a major function of the morphology proteins Mdm12/Mmm1.
Keywords: Mdm10, mitochondria, protein sorting, Saccharomyces cerevisiae , SAM complex
Introduction
All nuclear-encoded mitochondrial precursor proteins have to be transported into or across the mitochondrial outer membrane. This membrane contains the central entry gate of mitochondria, the translocase of the outer membrane (TOM) complex (Neupert, 1997; Endo et al, 2003; Young et al, 2003; Koehler, 2004; Pfanner et al, 2004; Wiedemann et al, 2004a; Rapaport, 2005; Dolezal et al, 2006). The TOM complex consists of receptor proteins that recognize the precursor proteins and a core complex that includes the central pore-forming subunit, Tom40. The TOM complex is able to translocate hundreds of different precursor proteins across the outer membrane to internal mitochondrial compartments; however, the TOM complex is not sufficient to insert a major class of proteins into the outer membrane. Several integral outer membrane proteins possess a β-barrel structure, including Tom40 and the most abundant outer membrane protein, porin. Wiedemann et al (2003) identified a specific machinery in the outer membrane that is responsible for sorting and assembly of β-barrel proteins (SAM complex). The precursors of β-barrel proteins are first translocated via the TOM complex to the trans side of the outer membrane (Model et al, 2001) and are transferred to the SAM complex with the help of chaperone complexes in the intermembrane space, formed by small Tim proteins (Hoppins and Nargang, 2004; Wiedemann et al, 2004b, 2006a).
The SAM complex contains a stable core formed by Sam35 (Tob38/Tom38) (Ishikawa et al, 2004; Milenkovic et al, 2004; Waizenegger et al, 2004), Sam37 (Mas37) (Wiedemann et al, 2003) and Sam50 (Tob55/Omp85) (Kozjak et al, 2003; Paschen et al, 2003; Gentle et al, 2004). Sam50 is an integral β-barrel protein of the outer membrane, conserved from bacteria (Omp85) to man (Voulhoux et al, 2003; Ryan, 2004; Dolezal et al, 2006). Sam35 and Sam37 are peripheral membrane proteins exposed to the mitochondrial surface. The three subunits of the SAMcore complex are required for the biogenesis of all outer membrane β-barrel proteins analyzed.
The mitochondrial distribution and morphology protein Mdm10 was found to associate with the SAMcore complex, forming a SAMholo complex, which plays a specific role in the late steps of assembly of the TOM complex, whereas the assembly pathway of other outer membrane β-barrel proteins does not require Mdm10 (Meisinger et al, 2004). Mdm10 was identified by Sogo and Yaffe (1994) for its role in maintaining mitochondrial morphology. Mdm10 is predicted to form a β-barrel in the outer membrane (Paschen et al, 2003; Wiedemann et al, 2003). The presence of Mdm10 in the SAM complex led to a controversial situation since Boldogh et al (2003) had reported that Mdm10 was associated with two other outer membrane proteins, Mdm12 (Berger et al, 1997) and Mmm1 (Burgess et al, 1994). Mdm12 and Mmm1 were shown to be required for mitochondrial morphology and distribution like Mdm10. Cells lacking Mmm1, Mdm10 or Mdm12 display a similar morphological defect with condensed giant mitochondria (Burgess et al, 1994; Sogo and Yaffe, 1994; Berger et al, 1997; Prokisch et al, 2000; Hobbs et al, 2001; Kondo-Okamoto et al, 2003), supporting the view of a close connection of the three proteins (Boldogh et al, 1998, 2003, 2005). The discussion whether Mdm10 is a subunit of the SAM complex or the Mdm12/Mmm1 complex is controversial (Boldogh et al, 2005; Jensen, 2005; Stojanovski et al, 2006).
Here, we used affinity purification under mild conditions to determine interaction partners of Mdm10. Surprisingly, we found that Mdm10 shows a dual localization in both the SAM complex and the Mdm12/Mmm1 complex, thereby clarifying the controversial debate on its localization. We then tested the hypothesis that also Mdm12/Mmm1 may perform a function in protein assembly. Indeed, mitochondria defective in Mdm12 or Mmm1 were strongly impaired in outer membrane protein assembly. Mdm12/Mmm1 are required after the SAMcore complex but before separation of the Tom40-specific route and thus before Mdm10. Mdm12/Mmm1 are the first mitochondrial morphology proteins that function in the major β-barrel assembly pathway of mitochondria.
Results
Dual localization of Mdm10
To analyze if Mdm10 was present in the SAM complex or the Mdm12/Mmm1 complex, we used yeast cells that expressed chromosomally integrated tagged forms of distinct components of these complexes. Mitochondria were isolated from the cells, lysed with the mild detergent digitonin and subjected to affinity chromatography. Tagged Mdm10 (Meisinger et al, 2004) pulled out the three subunits of the SAMcore complex (Sam50, Sam37, Sam35), as well as Mmm1 and Mdm12 (Figure 1A, lane 2). Control proteins, such as Tom proteins, the mitochondrial intermembrane space assembly protein Mia40 (Chacinska et al, 2004) and the subunit Tim23 of the translocase of the inner membrane, were not copurified with Mdm10 (Figure 1A, lane 2).
Figure 1.
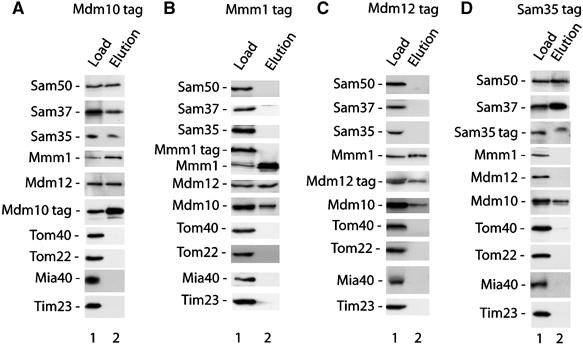
Mdm10 is present in different protein complexes, SAM and Mdm12/Mmm1. Mitochondria from S. cerevisiae strains containing tagged Mdm10 (A), Mmm1 (B), Mdm12 (C) and Sam35 (D) were isolated and lysed with 1% digitonin as described in Materials and methods. After affinity chromatography, protein complexes were eluted and separated by SDS–PAGE. Proteins were analyzed by Western blotting with the indicated antibodies. Load, 16%; elution, 100%.
A different picture, however, was observed when mitochondria containing tagged Mmm1 were used. Only Mdm12 and Mdm10 were copurified whereas none of the three subunits of the SAMcore complex was found in the eluates (Figure 1B, lane 2). Thus, although Mmm1 was copurified with tagged Mdm10, as well as Mdm10 was copurified with tagged Mmm1, the eluates significantly differed with respect to other proteins. Mdm12 was the only additional protein found in both eluates. To further analyze this unusual pull-down behavior, we used mitochondria containing tagged Mdm12. Mmm1 and Mdm10 were copurified but none of the subunits of the SAMcore complex (Figure 1C, lane 2).
These results indicated that Mdm12 and Mmm1 are associated with Mdm10 but not with the SAMcore complex, whereas Mdm10 is associated with Mdm12/Mmm1 as well as the three subunits of the SAMcore complex. A direct explanation for these findings would be the presence of Mdm10 in different outer membrane complexes, the SAM complex and the Mdm12/Mmm1 complex. To obtain further evidence, we used mitochondria carrying tagged Sam35. Indeed, Sam50, Sam37 and Mdm10 were copurified but neither Mdm12 nor Mmm1 (Figure 1D, lane 2).
We conclude that the mitochondrial outer membrane possesses distinct complexes containing Mdm10: the SAM complex with Sam50, Sam37 and Sam35; and the Mdm12/Mmm1 complex. Thus, the apparently controversial observations on the localization of Mdm10 (Boldogh et al, 2003; Meisinger et al, 2004) can now be explained by a dual localization of this protein.
Morphology proteins Mdm12/Mmm1 are required for β-barrel assembly
The interaction of Mdm10 with both SAM and Mdm12/Mmm1 raised the hypothesis that also the morphology proteins Mdm12/Mmm1 may play a role in mitochondrial protein assembly. To test this hypothesis, we analyzed yeast mutants lacking either the MDM12 gene or the MMM1 gene. Mitochondria were isolated from mdm12Δ and mmm1Δ yeast and the corresponding wild-type strains. The steady-state levels of proteins were analyzed by immunodecoration (Figure 2A and B). Mmm1 was largely absent in mdm12Δ mitochondria and the levels of Mdm12 were strongly reduced in mmm1Δ mitochondria in agreement with previous observations on the close interaction of both proteins (Boldogh et al, 2003). In addition, the levels of further outer membrane proteins like Tom40, Mdm10 and SAMcore subunits were moderately reduced in both mutant mitochondria whereas the levels of the inner membrane proteins Tim23 and ADP/ATP carrier were same as those of wild-type mitochondria (Figure 2A and B). For most of the outer membrane proteins analyzed here, a β-barrel fold has been reported (Paschen et al, 2003; Wiedemann et al, 2003; Pfanner et al, 2004), suggesting a possible relation of Mdm12/Mmm1 with the steady-state levels of mitochondrial β-barrel proteins. We thus lysed isolated mitochondria with digitonin and analyzed the TOM complex by blue native electrophoresis. Upon immunodecoration for Tom40, we observed the mature TOM complex of 450 kDa in both mdm12Δ mitochondria and mmm1Δ mitochondria but in reduced amounts compared to wild-type mitochondria (Figure 2C and D). In addition, small amounts of a 100 kDa TOM form were observed in the mutant mitochondria, that is, Tom40 that was not fully assembled into the mature 450 kDa complex. Previous studies had shown that appearance of the small TOM form under steady-state conditions indicated an impaired assembly of the TOM complex (Dekker et al, 1998; Meisinger et al, 2004; Waizenegger et al, 2004) (it should be noted that the 100 kDa form contains one dimer of Tom40 whereas the 450 kDa complex contains three dimers (Dekker et al, 1998; Model et al, 2002) and thus in an immunodecoration for Tom40 the molar ratio of 100 kDa form to 450 kDa form is underestimated by a factor of 3). When porin complexes of digitonin-lysed mitochondria were analyzed by blue native electrophoresis and immunodecoration (Krimmer et al, 2001), a significant reduction in both mdm12Δ and mmm1Δmitochondria was observed(Figure 2E). The possibility that the extraction of porin from mitochondrial membranes by digitonin was less efficient from the mutant mitochondria than from wild-type mitochondria was of concern. We thus compared the digitonin extracts by SDS–polyacrylamide gel electrophoresis (PAGE) and Western blotting and observed a similar extractability of porin from wild-type and mutant mitochondria (Figure 2F), supporting the view that the defect observed by blue native electrophoresis may be caused by an impaired assembly of porin complexes in mitochondria lacking Mdm12/Mmm1.
Figure 2.

Steady-state protein levels in mitochondria lacking Mdm12 or Mmm1. (A) Mitochondria were isolated from wild-type (WT) and mdm12Δ yeast strains. Indicated amounts of mitochondrial protein (μg) were subjected to SDS–PAGE and Western blot analysis. AAC, ADP/ATP carrier. (B) Western blot analysis of mitochondria from WT and mmm1Δ yeast strains after separation by SDS–PAGE. (C) Mitochondria from WT and mdm12Δ strains were lysed by digitonin-containing buffer and subjected to blue native electrophoresis. The TOM complex was detected by Western blotting and immunodecoration with affinity-purified Tom40 antibodies. (D) Western blot analysis of mitochondria from WT and mmm1Δ strains separated by blue native electrophoresis as described for panel C. (E) Porin complexes of mitochondria from WT, mmm1Δ or mdm12Δ yeast strains were analyzed by blue native electrophoresis and Western blotting after solubilization in 0.6% digitonin-containing buffer. (F) Analysis of solubilized mitochondria (0.4 or 1% digitonin) from WT, mmm1Δ or mdm12Δ yeast strains by SDS–PAGE and immunodecoration with porin antiserum.
To directly probe for a role of Mdm12/Mmm1 in the assembly of mitochondrial β-barrel proteins, we studied TOM assembly in a kinetically linear range by importing the radiolabeled precursor of Tom40 into isolated mitochondria. Upon lysis with digitonin, the assembly steps were monitored by blue native electrophoresis. In wild-type mitochondria, assembly of Tom40 occurs via two intermediates of about 250 kDa (assembly I) and 100 kDa (assembly II) before the mature 450 kDa complex is formed (Figure 3A, lanes 4–6) (Model et al, 2001; Wiedemann et al, 2003; Meisinger et al, 2004). In mdm12Δ mitochondria as well as mmm1Δ mitochondria, formation of the 450 kDa TOM complex was strongly impaired (Figure 3A, lanes 2 and 3; and B, lanes 7 and 8), demonstrating that Mdm12/Mmm1 are required for assembly of the precursor into the mature TOM complex.
Figure 3.
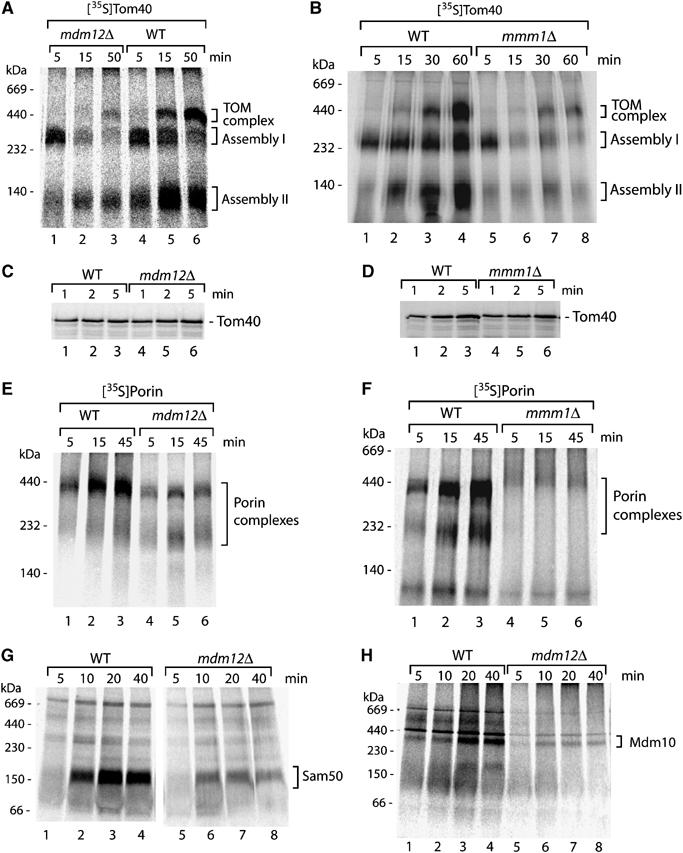
Mitochondria lacking Mdm12/Mmm1 are impaired in assembly of β-barrel proteins. (A, B) Assembly defect of Tom40. 35S-labeled precursor of Tom40 was imported into mitochondria isolated from wild-type (WT), mdm12Δ and mmm1Δ yeast strains as described in Materials and methods. The incubation time for the import reaction is indicated (min). Assembly into complexes was analyzed by digitonin lysis, blue native electrophoresis and digital autoradiography. (C, D) Import of radiolabeled Tom40 precursor into mitochondria from WT, mmm1Δ or mdm12Δ yeast strains. Mutant mitochondria were adjusted to the same amount of TOM complexes and import was performed at 20°C. After treatment with proteinase K (10 μg/ml), the mitochondria were reisolated and analyzed by SDS–PAGE and digital autoradiography. (E, F) Assembly defect of porin. 35S-labeled precursor of porin was imported into isolated yeast mitochondria. Assembly into complexes was analyzed as described for panels (A, B). (G) Assembly of radiolabeled Sam50 precursor in WT and mdm12Δ mitochondria was performed as described for panel A. (H) Assembly of radiolabeled Mdm10 in WT and mdm12Δ mitochondria was performed as described for panel (A).
To determine the assembly stage that depended on Mdm12/Mmm1, we made use of the observation that the 250 kDa intermediate represents the interaction of the precursor protein with the SAMcore complex (Mihara, 2003; Paschen et al, 2003; Wiedemann et al, 2003; Gentle et al, 2004; Ishikawa et al, 2004; Pfanner et al, 2004). Mitochondria deficient in any subunit of the SAMcore complex (Sam35, Sam37 or Sam50) are inhibited from forming of the 250 kDa intermediate (Kozjak et al, 2003; Paschen et al, 2003; Wiedemann et al, 2003; Gentle et al, 2004; Ishikawa et al, 2004; Milenkovic et al, 2004; Waizenegger et al, 2004). In contrast, mitochondria lacking Mdm10 efficiently form the 250 kDa intermediate but are impaired in formation of the mature TOM complex in agreement with the role of Mdm10 in late steps of TOM assembly that take place after the SAMcore complex (Meisinger et al, 2004, 2006b). Both mdm12Δ mitochondria and mmm1Δ mitochondria formed the 250 kDa assembly intermediate of Tom40 after a 5 min import, indicating that the SAMcore complex was functional in these mitochondria (Figure 3A, lane 1; and B, lane 5). Moreover, translocation of the Tom40 precursor to a protease-protected location occurred with similar efficiency in wild-type, mdm12Δ and mmm1Δ mitochondria (Figure 3C and D), indicating that the mutant mitochondria were competent in the early import step of precursor translocation through the TOM complex. (At longer import times, the amount of Tom40 precursor accumulated at the 250 kDa intermediate decreased in mdm12Δ and mmm1Δ mitochondria (Figure 3A and B); as the subsequent assembly steps of Tom40 are impaired in the mutant mitochondria, the turnover of the precursor at the 250 kDa stage is likely increased.) We conclude that Mdm12/Mmm1 are required for Tom40 assembly after the SAMcore complex.
We thus asked if Mdm12/Mmm1 performed a similar function like Mdm10 in outer membrane protein assembly. Mdm10 is selectively required for the biogenesis of Tom40 but not of other β-barrel proteins such as the most abundant outer membrane protein porin (Meisinger et al, 2004, 2006b). We analyzed the assembly of the radiolabeled precursor of porin into mature complexes by blue native electrophoresis (Krimmer et al, 2001; Wiedemann et al, 2003; Meisinger et al, 2004). Surprisingly, the assembly of porin was strongly inhibited in both mdm12Δ mitochondria and mmm1Δ mitochondria (Figure 3E and F). Thus, mitochondria lacking Mdm12 or Mmm1 show a β-barrel assembly defect that is different from mitochondria lacking Mdm10. Additionally, we studied the assembly of two further β-barrel proteins, Sam50 and Mdm10, and observed a significant inhibition in mdm12Δ mitochondria (Figure 3G and H). We conclude that Mdm12/Mmm1 are required for the function of the general β-barrel pathway that is used by all β-barrel proteins analyzed and thus Mdm12/Mmm1 act before the separation of the Tom40-specific route from the general β-barrel pathway.
Specificity of β-barrel assembly defect in mitochondria lacking Mdm12/Mmm1
We performed several control experiments to demonstrate that the defect of mitochondria lacking Mdm12 or Mmm1 is specific for the β-barrel pathway. First, we studied the import of preproteins destined for internal mitochondrial compartments in order to exclude a general defect of mdm12Δ or mmm1Δ mitochondria in protein import. We used the β-subunit of the F1-ATPase (F1β) and a matrix-targeted model protein consisting of the presequence of Fo-ATPase subunit 9 and dihydrofolate reductase (Su9-DHFR) (Pfanner et al, 1987). Import of the proteins was determined by processing to the mature-sized form and the membrane potential (Δψ)-dependent translocation to a protease-protected location (Wiedemann et al, 2006b). mdm12Δ and mmm1Δ mitochondria imported the preproteins like wild-type mitochondria (Figure 4A and B), indicating that the mutant mitochondria were competent in protein translocation across the outer membrane to internal compartments.
Figure 4.
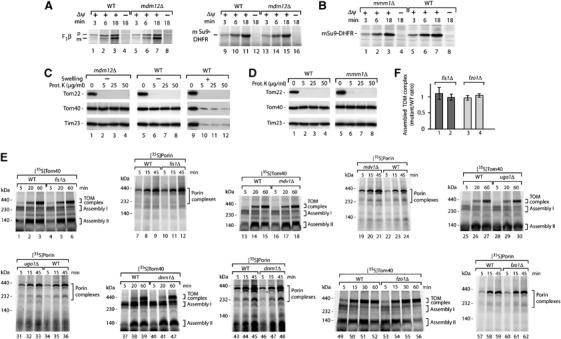
Mitochondrial fusion and fission components are not required for assembly of β-barrel proteins. (A) 35S-labeled precursors of Su9-DHFR and F1β were imported into isolated mitochondria from wild-type (WT) and mdm12Δ yeast in the presence or absence of a membrane potential (Δψ). The incubation time for the import reaction is indicated (min). After treatment with proteinase K, the samples were analyzed by SDS–PAGE and digital autoradiography. p, precursor; m, mature. (B) Import of 35S-labeled precursor of Su9-DHFR into WT and mmm1Δ mitochondria. Proteinase K-treated samples were separated by SDS–PAGE. (C) mdm12Δ mitochondria possess an intact outer membrane. Mitochondria, isolated from WT and mdm12Δ strains, were subjected to swelling where indicated, treated with proteinase K and subjected to SDS–PAGE and Western blotting. (D) mmm1Δ mitochondria possess an intact outer membrane. Isolated mitochondria were treated with proteinase K and analyzed by SDS–PAGE and Western blotting. (E) 35S-labeled precursors of Tom40 and porin were imported into mitochondria isolated from WT yeast and the indicated mitochondrial fission and fusion mutant strains (derived from the Euroscarf collection). Mitochondria were lysed by digitonin, protein complexes were separated by blue native electrophoresis and analyzed by digital autoradiography. (F) Quantification of assembled TOM complex in fis1Δ and fzo1Δ mitochondria using ImageQuant software (GE Healthcare). Import of the radiolabeled precursor of Tom40 was performed as described for panel (E). For each time point (20 and 60 min for fis1Δ, 30 and 60 min for fzo1Δ), the ratio of mutant to wild-type mitochondria is shown. Bars indicate s.e.m. from at least three independent experiments.
Second, the assembly of β-barrel proteins of the outer membrane has been shown to depend on an intact intermembrane space. Opening of the outer membrane, which leads to a release of intermembrane space proteins like the small Tim proteins, strongly inhibits assembly of Tom40 and porin (Smith et al, 1994; Wiedemann et al, 2004b). We thus tested the integrity of the outer membrane by treating isolated mitochondria with increasing concentrations of proteinase K. The outer membrane receptor Tom22 was degraded whereas Tom40 as well as Tim23, which is exposed to the intermembrane space, was protected against the protease in both mdm12Δ and mmm1Δ mitochondria (Figure 4C, lanes 2–4; Figure 4D, lanes 6–8). For comparison, we subjected mitochondria to swelling in order to open the outer membrane (Wiedemann et al, 2004b). Now both Tom40 and Tim23 were accessible to proteinase K (Figure 4C, lanes 10–12). We conclude that the outer membrane barrier of mdm12Δ and mmm1Δ mitochondria is intact.
Mitochondrial fusion and fission components are not required for β-barrel assembly
As mutants lacking Mdm10, Mdm12 or Mmm1 are defective in both mitochondrial morphology and assembly of outer membrane proteins, we asked whether alteration of mitochondrial morphology leads to defects in β-barrel assembly. We therefore analyzed deletion mutants of mitochondrial outer membrane proteins, which have been reported to be required for mitochondrial fission (Otsuga et al, 1998; Bleazard et al, 1999; Fekkes et al, 2000; Mozdy et al, 2000; Tieu and Nunnari, 2000; Cerveny et al, 2001; Jofuku et al, 2005) or fusion (Hermann et al, 1998; Rapaport et al, 1998; Sesaki and Jensen, 2001; Ishihara et al, 2004; Meeusen et al, 2004). We used isolated mitochondria and the radiolabeled precursors of Tom40 and porin. As some of the fusion mutants have a tendency to lose mitochondrial DNA, wild-type mitochondria were isolated from both rho+ and rho− strains and the assembly reactions in mutant mitochondria were compared to both types of wild-type mitochondria. None of the mutants of the outer membrane components known to be required for mitochondrial fission (Fis1, Mdv1, Dnm1) or fusion (Fzo1, Ugo1) showed a significant defect in assembly of β-barrel proteins (Figure 4E and F). We conclude that alterations of mitochondrial fusion or fission do not lead to defects in protein assembly of the mitochondrial outer membrane.
Rapid induction of protein assembly defects in mmm1-1 mutant yeast
As mutants of the SAMcore complex as well as mutants of MDM12/MMM1 show defects in β-barrel assembly and in mitochondrial morphology, the primary function of these components is of major interest. In the case of components of the SAMcore complex, a primary function in protein assembly is evident (Stojanovski et al, 2006). Mutants of SAMcore subunits grown at low temperature show kinetic defects in assembly of β-barrel proteins whereas mitochondrial morphology is not affected. Only upon shift of the cells to elevated temperature for longer time spans, defects in mitochondrial morphology are observed; under these conditions, the steady-state levels of outer membrane protein complexes like the TOM complex are significantly reduced (Wiedemann et al, 2003; Meisinger et al, 2004, 2006b). These findings show that the SAMcore complex plays a primary role in the assembly of outer membrane proteins such as the TOM complex, which is required for import of various morphology factors (Stojanovski et al, 2006).
In the case of Mdm12/Mmm1, the selective defect of deletion mutants in the β-barrel pathway at a post-SAMcore stage (Figure 3) and the normal β-barrel assembly in mutants of the various fusion and fission components (Figure 4E) similarly suggest a primary function of Mdm12/Mmm1 in mitochondrial protein assembly. To obtain further evidence, we used a temperature-conditional mutant of MMM1 (Burgess et al, 1994) and monitored the time dependence of induction of phenotypes upon shift to nonpermissive temperature. We observed that 33°C was a lethal temperature for the mmm1-1 cells, whereas the growth was impaired at 30°C and was comparable to wild-type cells at 23°C (Figure 5A). Mitochondrial morphology was analyzed by confocal laser scanning microscopy. Upon growth of mmm1-1 cells at 24°C, as well as after a shift to 33°C for 30 min , the cells preferentially showed the typical tubular mitochondrial morphology like wild-type cells (Figure 5B). After a 90 min shift to 33°C, mmm1-1 cells contained condensed giant mitochondria whereas wild-type mitochondria still showed the tubular morphology (Figure 5B). A quantification revealed that after the 30 min shift, 5% of the mmm1-1 cells displayed an altered mitochondrial morphology, whereas after the 90 min shift, 99% of mmm1-1 cells contained altered mitochondria (Figure 5C). We conclude that after a 90 min shift to 33°C, the morphological phenotype of mmm1-1 cells is expressed. We isolated mitochondria from the cell cultures used for the morphological analysis. The steady-state levels of the mutant protein Mmm1-1 as well as of Mdm12 decreased upon a 30 min shift to 33°C whereas the steady-state levels of other proteins analyzed, including outer membrane proteins and control proteins of internal mitochondrial compartments, were not affected (Figure 5D). After a shift of 90 min to 33°C, the steady-state level of Mdm10 was moderately reduced (5D). Upon analysis by blue native electrophoresis, it became evident that assembly of the mature 450 kDa TOM complex was significantly impaired after the 90 min shift to 33°C (Figure 5E).
Figure 5.

Early induction of protein assembly defect in mmm1-1 mutant cells. (A) Growth phenotype of the conditional mmm1-1 yeast strain. WT and mmm1-1 cells were spotted via serial dilution onto YPG agar plates and incubated at 23, 30 and 33°C. (B) Mitochondrial morphology defect of mmm1-1 cells. Precultures (YPG, 24°C) of WT and mmm1-1 cells were diluted with fresh medium of appropriate temperature to maintain 24°C or reach 33°C by mixing. The cultures were further incubated at 24 or 33°C. After 30 and 90 min, cells were immediately stained with DiOC6 and analyzed by laser scanning confocal microscopy. DIC, differential interference contrast; scale bar, 2 μm. (C) Statistics of the mitochondrial morphology of WT and mmm1-1 cells grown at 24°C or after shift to 33°C for 30 and 90 min. Number of cells analyzed for each condition >500. (D) Analysis of steady-state protein levels of WT and mmm1-1 mitochondria isolated from yeast strains that were grown at 24°C and shifted to 33°C for 30 and 90 min where indicated as described for panel (B). (E) The endogenous TOM complex of mitochondria isolated as described for panel (D) was analyzed by blue native electrophoresis and immunodecoration with Tom40-specific antibodies. (F) 35S-labeled precursor of Tom40 was imported into WT and mmm1-1 mitochondria isolated as described for panel (D) for the indicated periods (min). TOM complex assembly was analyzed by blue native electrophoresis and digital autoradiography. (G) Quantification of assembled TOM complex in mmm1-1 mitochondria using ImageQuant software (GE Healthcare). The experiments were performed as described for panel (F). Bars indicate s.e.m. from four independent experiments. (H) Import of the radiolabeled precursors of Sam50 and porin was performed and analyzed as described for panel (F).
We then studied the kinetics of TOM assembly in the mutant mitochondria by using the radiolabeled precursor of Tom40 and observed an efficient assembly process in mitochondria isolated from non-shifted mmm1-1 cells (Figure 5F, lanes 4–6; and G). After the 30 min shift, a significant defect in formation of the mature TOM complex was observed compared to wild-type mitochondria (Figure 5F, lane 12 versus lane 9; and G). Similarly, after the 90 min shift, the formation of mature TOM complex was strongly inhibited (Figure 5F, lane 18 versus lane 15; and G). In both cases, the formation of assembly intermediate I, which represents the SAMcore complex, was not impaired, thus confirming the results obtained with mmm1Δ mitochondria (Figure 3B). The assembly of further β-barrel proteins, like Sam50 and porin, was also impaired in the mmm1-1 mutant after the 30 min shift to nonpermissive conditions (Figure 5H).
As the β-barrel assembly defect is observed very early (after a 30 min shift), it is obviously not a consequence of morphological alterations of the mutant mitochondria. The morphology defect is observed under conditions (90 min shift) where the steady-state levels of mature assembled TOM complex are significantly reduced. These findings support the view of a primary role of Mmm1 in β-barrel assembly.
Overexpression of Mmm1, Mdm10 or Mdm12 partially suppresses the growth defect of sam37Δ cells
To obtain additional in vivo evidence for a relation of Mmm1, Mdm12 and Mdm10 with the SAMcore complex, we used sam37Δ cells that cannot grow at 37°C on fermentable and non-fermentable media (Gratzer et al, 1995; Wiedemann et al, 2003). We asked if overexpression of these three so-called morphology components was able to suppress the growth defect of sam37Δ cells. As control, we overexpressed Sam37 as well as Sam35 and Sam50 in the deletion mutant. Whereas Sam37 rescued growth on both media as expected, the two other subunits of the SAMcore complex, Sam35 and Sam50, were only able to suppress the growth defect on fermentable medium (Figure 6). Transformation of the mutant cells with the control plasmid (no insert) did not lead to suppression. Each of the three morphology components partially suppressed the growth defect of sam37Δ cells on fermentable medium (Figure 6). Thus, Mdm10, Mmm1 and Mdm12 can partially substitute for Sam37, supporting the view of a functional relation of the Mdm/Mmm proteins to the SAMcore complex.
Figure 6.
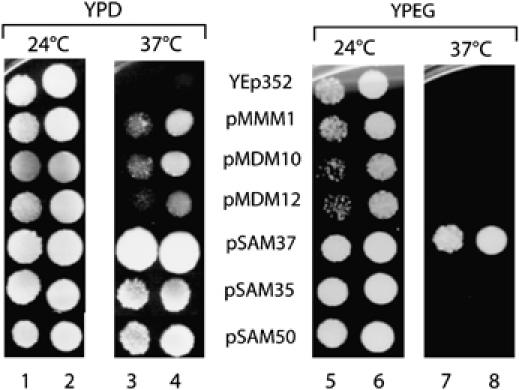
Partial suppression of sam37Δ mutant cells by overexpression of Mmm1, Mdm10 or Mdm12. The sam37Δ yeast strain was transformed with the control plasmid YEp352 or with plasmids containing the endogenous promoter, open reading frame and terminator region of MMM1, MDM10, MDM12, SAM37, SAM35 and SAM50. Growth was analyzed on agar plates at fermentable (YPD) or non-fermentable (YPEG) conditions at 24 and 37°C.
Discussion
We have identified two new components of the mitochondrial pathway for assembly of β-barrel proteins. Mdm12 and Mmm1 were originally identified in genetic screens for their involvement in maintenance of mitochondrial morphology and distribution (Burgess et al, 1994; Berger et al, 1997). Mdm12 and Mmm1 are closely connected, as the deletion of either one component leads to a nearly complete loss of mitochondrial localization of the other partner (Boldogh et al, 2003; this study). We found that Mdm12/Mmm1 function in the major β-barrel pathway that is responsible for biogenesis of outer membrane proteins like porin, Tom40, Sam50 and Mdm10. Mdm12/Mmm1 shift the branching point of the β-barrel pathway to a stage after the SAMcore complex.
To date, it had been assumed that splitting of β-barrel assembly into the Tom40-specific route and the general route for the other β-barrel proteins occurs upon exiting from the SAMcore complex. Mdm10 and Mim1 (Tom13) then selectively direct the final steps of assembly of Tom40 with the other subunits of the TOM complex (Ishikawa et al, 2004; Meisinger et al, 2004, 2006b; Waizenegger et al, 2005). We show that mitochondria deficient in Mdm12 or Mmm1 are competent in forming the SAMcore intermediate of β-barrel proteins, demonstrating that Mdm12/Mmm1 operate after the SAMcore complex. The separation of the Tom40-specific pathway occurs after the action of Mdm12/Mmm1.
Interestingly, Mdm10 of the Tom40-specific pathway can associate with either complex of the major β-barrel pathway, the SAMcore complex and the Mdm12/Mmm1 complex, forming the larger SAMholo complex and the Mdm12/Mmm1/Mdm10 complex, respectively. This dual localization of Mdm10 solves the previous controversy on its localization (Boldogh et al, 2003; Meisinger et al, 2004) and indicates a dynamic interplay between the components of the major β-barrel pathway and those of the Tom40-specific pathway. The association of Mdm10 with a fraction of SAMcore and Mdm12/Mmm1 complexes may help to regulate the substrate flow in the β-barrel pathways. It is conceivable that Mdm10 acts as a switch that shuttles between both complexes and channels precursor proteins into the TOM-specific pathway, that is, for association with other TOM proteins. Indeed, Meisinger et al (2006b) showed that Tom7 controls the association of Mdm10 with the SAM complex and thus differentially regulates the assembly efficiency of porin and Tom40. We did not find Mim1 in association with SAM components or Mdm/Mmm components (data not shown), supporting the view that Mim1 is located in a distinct outer membrane complex dedicated to Tom40 assembly (Ishikawa et al, 2004; Waizenegger et al, 2005).
Mutants of MDM10, MDM12 or MMM1 show a similar alteration of mitochondrial morphology, that is, the formation of condensed giant mitochondria (Burgess et al, 1994; Sogo and Yaffe, 1994; Berger et al, 1997; Dimmer et al, 2002; Meisinger et al, 2004, 2006b). Our findings that mutants of these proteins are impaired in mitochondrial β-barrel assembly raise the question whether the primary function of this group of Mdm/Mmm proteins is assembly of proteins or maintenance of mitochondrial morphology. First, the requirement for these Mdm/Mmm proteins at defined stages of β-barrel assembly argues against an unspecific/indirect impairment of protein assembly in the mutant mitochondria. Second, we asked if alteration of mitochondrial morphology causes defects in β-barrel assembly and analyzed Saccharomyces cerevisiae mutant strains with defined defects in mitochondrial morphology, that is, the mitochondrial outer membrane components that are directly required for mitochondrial fission and fusion. Mutants of these fission and fusion components (mdv1Δ, fis1Δ, dnm1Δ, fzo1Δ, ugo1Δ) did not display any significant defect in β-barrel assembly, demonstrating that the defects in mitochondrial morphology do not cause an impairment of outer membrane protein assembly. Third, we used a temperature-conditional mutant of MMM1 and found a sequential induction of phenotypes with the β-barrel assembly defect occurring significantly before the morphological alteration. These findings indicate that defects in the β-barrel pathway lead to impaired assembly of protein complexes of the mitochondrial outer membrane, including protein complexes that are required to maintain a proper mitochondrial morphology. Indeed, mutants of SAMcore components as well as TOM subunits show similar morphological alterations (giant mitochondria) like mutants of Mdm10, Mdm12 and Mmm1 (Dimmer et al, 2002; Altmann and Westermann, 2005; Meisinger et al, 2004, 2006b) and overexpression of Mdm10, Mdm12 and Mmm1 can partially suppress the growth defect of mutants impaired in the SAMcore complex.
In summary, we have identified a new stage of the major β-barrel assembly pathway of mitochondria that requires the morphology proteins Mdm12/Mmm1. The SAMcore complex as well as Mdm12/Mmm1 can associate with Mdm10 that functions in the Tom40-specific route, indicating a dynamic cooperation of the machineries required for mitochondrial β-barrel assembly. Our findings suggest that these Mdm/Mmm proteins play a primary role in assembly of protein complexes of the mitochondrial outer membrane whereas the maintenance of mitochondrial morphology is a secondary consequence of the assembly function.
Materials and methods
Yeast growth and isolation of mitochondria
The S. cerevisiae strains mmm1-1 (Burgess et al, 1994), sam37Δ (tom37Δ) (Ryan et al, 1999), Sam35-His10 (Milenkovic et al, 2004) and the strain expressing Mdm10-His10 (Meisinger et al, 2004) have been described before. The strains Mdm12-His10 (background YPH499, C-terminal deca-histidine tag) and ProtA-Mmm1 (BY4741, N-terminal protein A-tag) were generated by homologous recombination with PCR products from affinity tag replacement cassettes (Meisinger et al, 2001; Kozjak et al, 2003). Other yeast deletion strains were obtained from Euroscarf (BY4741). To avoid adaptation/suppression, the strains were used directly from the stock. For overexpression, the open reading frames of MMM1, MDM10, MDM12, SAM37, SAM35 and SAM50 under control of the endogenous promoter and terminator were cloned into the vector YEp352 (Hill et al., 1986). The resulting plasmids were able to complement the growth phenotype of the respective deletion strains. After transformation of the sam37Δ strain and selection on medium without uracil, yeast clones were tested as described in the legend of Figure 6.
Yeast cells were grown in medium (1% (w/v) yeast extract, 2% (w/v) bactopeptone) with 2% (w/v) glucose (YPD), 2% (w/v) sucrose (YPS), 3% (w/v) glycerol (YPG) or 2% (v/v) ethanol and 2% (w/v) glycerol (YPEG) at 20–33°C. For isolation of mitochondria used in import experiments, mdm12Δ yeast and the corresponding wild-type strain were grown at 20°C, and mmm1Δ yeast and the corresponding wild-type strain were grown at 23°C. Cells of mmm1-1 yeast and the corresponding wild-type strain were cultured at 24°C in YPG medium to an optical density (OD) of 1 (mid-log-phase). The cultures were split and diluted with fresh medium to an OD of 0.33. The fresh medium had a temperature of either 24 or 38°C in order to instantly set permissive (24°C) and non-permissive (33°C) growth conditions for the mmm1-1 mutant. Yeast cells were further incubated for 30 and 90 min. Mitochondria were stained by direct application of 150 nM of the lipophilic dye 3,3′dihexyloxacarbocyanine iodide (DiOC6) (Pringle et al, 1989) to the growth medium. Mitochondrial morphology was analyzed immediately with an epifluorescence or confocal microscope. Images of mitochondria from cells with a representative phenotype were taken and processed as described (Meisinger et al, 2004). Mitochondria were isolated by differential centrifugation, resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS/KOH, pH 7.2) and aliquots were frozen at −80°C (Meisinger et al, 2006a).
In vitro import and rupturing of the outer membrane
pGEM vectors carrying open reading frames of mitochondrial precursor proteins were used for in vitro transcription with SP6 RNA polymerase (Stratagene). Radiolabeled precursor proteins were synthesized in vitro in rabbit reticulocyte lysate in the presence of [35S]methionine/cysteine (GE Healthcare). For in vitro import, isolated mitochondria (25–50 μg protein) were incubated with radiolabeled precursor proteins in 100 μl import buffer (3% (w/v) BSA, 80 mM KCl, 5 mM MgCl2, 2 mM NADH, 5 mM creatine phosphate, 100 μg/ml creatine kinase, 10 mM MOPS/KOH, pH 7.2, 2–4 mM ATP) at 25°C. Import reactions were stopped on ice by addition of 8 μM antimycin A, 20 μM oligomycin and 1 μM valinomycin (Sigma). Mitochondria were reisolated and washed with SEM buffer.
To selectively rupture the outer membrane and generate mitoplasts by swelling, mitochondria were resuspended in SEM buffer and the suspension was diluted with 9 volumes of EM buffer (1 mM EDTA, 10 mM MOPS/KOH, pH 7.2) and incubated on ice for 15 min. Mitochondria or mitoplasts were treated with proteinase K (5–50 μg/ml) for 15 min on ice. The protease was inactivated by addition of 2 mM PMSF and incubation for 5 min on ice. Further details of the methods are described in Wiedemann et al (2006b).
Isolation of tagged protein complexes, SDS–PAGE and blue native electrophoresis
Protein A- or decahistidine-tagged protein complexes were isolated essentially as described (Meisinger et al, 2001, 2004). In brief, mitochondria were solubilized at 4°C in 1% (w/v) digitonin buffer (20 mM Tris–HCl, pH 7.4, 0.1 mM EDTA, 50 mM NaCl, 10% (w/v) glycerol), centrifuged at 12 000 g at 4°C for 15 min and the supernatant was subjected to Ni–NTA or IgG chromatography. The IgG column was washed with digitonin buffer and the protein A tag was cleaved with TEV protease for elution. The Ni–NTA column was washed with digitonin buffer with 200 mM NaCl and 50–70 mM imidazole whereas decahistidine-tagged proteins were eluted with 200 mM imidazole. Eluates or isolated mitochondria (25–50 μg protein) were resuspended in SDS sample buffer (2% (w/v) SDS, 10% (w/v) glycerol, 60 mM Tris–HCl, pH 6.8, 0.2–1% (v/v) β-mercaptoethanol, 0.01% bromophenol blue), heated to 95°C for 5 min and subjected to SDS-PAGE. For separation of protein complexes, mitochondria were resuspended in digitonin buffer (0.5–1% (w/v) digitonin for porin, 1% (w/v) digitonin for TOM) and incubated for 15 min on ice. A 5 μl portion of blue native-sample buffer (5% (w/v) Coomassie brilliant blue G-250, 100 mM Bis–Tris/HCl, pH 7.0, 500 mM ɛ-amino-n-caproic acid) was added, samples were centrifuged at 12 000 g and 4°C for 15 min and the supernatants were applied to a 6–16.5% blue native electrophoresis gradient gel at 4°C. Radiolabeled proteins were detected by digital autoradiography. Non-relevant gel lanes were excised digitally. Endogenous proteins or protein complexes were analyzed by Western blotting on PVDF membranes, decoration with specific antibodies and detection by ECL (GE Healthcare).
Acknowledgments
We thank Drs J Nunnari, JM Shaw, LA Pon, D Rapaport, T Endo and MP Yaffe for discussion. We are grateful to B Schönfisch for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388 and 746, Gottfried Wilhelm Leibniz Program, Max Planck Research Award, Bundesministerium für Bildung und Forschung and the Fonds der Chemischen Industrie. DS is a recipient of an Alexander von Humboldt Research Fellowship.
References
- Altmann K, Westermann B (2005) Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell 16: 5410–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP (1997) Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol 136: 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1: 298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh I, Vojtov N, Karmon S, Pon LA (1998) Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol 141: 1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Fehrenbacher KL, Yang HC, Pon LA (2005) Mitochondrial movement and inheritance in budding yeast. Gene 354: 28–36 [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA (2003) A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell 14: 4618–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Delannoy M, Jensen RE (1994) MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol 126: 1375–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny KL, McCaffery JM, Jensen RE (2001) Division of mitochondria requires a novel DNM1-interacting protein, Net2p. Mol Biol Cell 12: 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuán Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N (2004) Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 23: 3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJT, Ryan MT, Brix J, Müller H, Hönlinger A, Pfanner N (1998) Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol Cell Biol 18: 6515–6524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B (2002) Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell 13: 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313: 314–318 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci 116: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP (2000) Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol 151: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T (2004) The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol 164: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratzer S, Lithgow T, Bauer RE, Lamping E, Paltauf F, Kohlwein SD, Haucke V, Junne T, Schatz G, Horst M (1995) Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol 129: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol 143: 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A (1986) Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2: 163–167 [DOI] [PubMed] [Google Scholar]
- Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE (2001) Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol 152: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins SC, Nargang FE (2004) The Tim8–Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J Biol Chem 279: 12396–12405 [DOI] [PubMed] [Google Scholar]
- Ishihara N, Eura Y, Mihara K (2004) Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 117: 6535–6546 [DOI] [PubMed] [Google Scholar]
- Ishikawa D, Yamamoto H, Tamura Y, Moritoh K, Endo T (2004) Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J Cell Biol 166: 621–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RE (2005) Control of mitochondrial shape. Curr Opin Cell Biol 17: 384–388 [DOI] [PubMed] [Google Scholar]
- Jofuku A, Ishihara N, Mihara K (2005) Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem Biophys Res Commun 333: 650–659 [DOI] [PubMed] [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Kondo-Okamoto N, Shaw JM, Okamoto K (2003) Mmm1p spans both the outer and inner mitochondrial membranes and contains distinct domains for targeting and foci formation. J Biol Chem 278: 48997–49005 [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N (2003) An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem 278: 48520–48523 [DOI] [PubMed] [Google Scholar]
- Krimmer T, Rapaport D, Ryan MT, Meisinger C, Kassenbrock CK, Blachly-Dyson E, Forte M, Douglas MG, Neupert W, Nargang FE, Pfanner N (2001) Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J Cell Biol 152: 289–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, McCaffery JM, Nunnari J (2004) Mitochondrial fusion intermediates revealed in vitro. Science 305: 1747–1752 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006a) Isolation of yeast mitochondria. Methods Mol Biol 313: 33–39 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Rissler M, Chacinska A, Sanjuán Szklarz LK, Milenkovic D, Kozjak V, Schönfisch B, Lohaus C, Meyer HE, Yaffe MP, Guiard B, Wiedemann N, Pfanner N (2004) The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev Cell 7: 61–71 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N (2001) Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol Cell Biol 21: 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Wiedemann N, Rissler M, Strub A, Milenkovic D, Schönfisch B, Müller H, Kozjak V, Pfanner N (2006b) Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J Biol Chem 281: 22819–22826 [DOI] [PubMed] [Google Scholar]
- Mihara K (2003) Cell biology: moving inside membranes. Nature 424: 505–506 [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Kozjak V, Wiedemann N, Lohaus C, Meyer HE, Guiard B, Pfanner N, Meisinger C (2004) Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J Biol Chem 279: 22781–22785 [DOI] [PubMed] [Google Scholar]
- Model K, Meisinger C, Prinz T, Wiedemann N, Truscott KN, Pfanner N, Ryan MT (2001) Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat Struct Biol 8: 361–370 [DOI] [PubMed] [Google Scholar]
- Model K, Prinz T, Ruiz T, Radermacher M, Krimmer T, Kühlbrandt W, Pfanner N, Meisinger C (2002) Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J Mol Biol 316: 657–666 [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol 151: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, Shaw JM (1998) The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol 143: 333–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W (2003) Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature 426: 862–866 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Tropschug M, Neupert W (1987) Mitochondrial protein import: nucleoside triphosphates are involved in conferring import-competence to precursors. Cell 49: 815–823 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N, Meisinger C, Lithgow T (2004) Assembling the mitochondrial outer membrane. Nat Struct Mol Biol 11: 1044–1048 [DOI] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW (1989) Fluorescence microscopy methods for yeast. Methods Cell Biol 31: 357–435 [DOI] [PubMed] [Google Scholar]
- Prokisch H, Neupert W, Westermann B (2000) Role of MMM1 in maintaining mitochondrial morphology in Neurospora crassa. Mol Biol Cell 11: 2961–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D (2005) How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J Cell Biol 171: 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D, Brunner M, Neupert W, Westermann B (1998) Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem 273: 20150–20155 [DOI] [PubMed] [Google Scholar]
- Ryan MT, Müller H, Pfanner N (1999) Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J Biol Chem 274: 20619–20627 [DOI] [PubMed] [Google Scholar]
- Ryan MT (2004) Chaperones: inserting beta barrels into membranes. Curr Biol 14: R207–R209 [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE (2001) UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol 152: 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Hicks S, Baker K, McCauley R (1994) Rupture of the mitochondrial outer membrane impairs porin assembly. J Biol Chem 269: 28460–284664 [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP (1994) Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol 126: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Rissler M, Pfanner N, Meisinger C (2006) Mitochondrial morphology and protein import—a tight connection? Biochim Biophys Acta 1763: 414–421 [DOI] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol 151: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J (2003) Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299: 262–265 [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Habib SJ, Lech M, Mokranjac D, Paschen SA, Hell K, Neupert W, Rapaport D (2004) Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep 5: 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T, Schmitt S, Zivkovic J, Neupert W, Rapaport D (2005) Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep 6: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, Kozjak V, Chacinska A, Schönfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C (2003) Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424: 565–571 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Frazier AE, Pfanner N (2004a) The protein import machinery of mitochondria. J Biol Chem 279: 14473–14476 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Truscott KN, Pfannschmidt S, Guiard B, Meisinger C, Pfanner N (2004b) Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J Biol Chem 279: 18188–18194 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Chacinska A (2006a) Chaperoning through the mitochondrial intermembrane space. Mol Cell 21: 145–148 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Rehling P (2006b) Import of precursor proteins into isolated yeast mitochondria. Methods Mol Biol 313: 373–383 [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU (2003) Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112: 41–50 [DOI] [PubMed] [Google Scholar]


