Abstract
On the basis of kinetic data on ribosome protein synthesis, the mechanical energy for translocation of the mRNA–tRNA complex is thought to be provided by GTP hydrolysis of an elongation factor (eEF2 in eukaryotes, EF-G in bacteria). We have obtained cryo-EM reconstructions of eukaryotic ribosomes complexed with ADP-ribosylated eEF2 (ADPR-eEF2), before and after GTP hydrolysis, providing a structural basis for analyzing the GTPase-coupled mechanism of translocation. Using the ADP-ribosyl group as a distinct marker, we observe conformational changes of ADPR-eEF2 that are due strictly to GTP hydrolysis. These movements are likely representative of native eEF2 motions in a physiological context and are sufficient to uncouple the mRNA–tRNA complex from two universally conserved bases in the ribosomal decoding center (A1492 and A1493 in Escherichia coli) during translocation. Interpretation of these data provides a detailed two-step model of translocation that begins with the eEF2/EF-G binding-induced ratcheting motion of the small ribosomal subunit. GTP hydrolysis then uncouples the mRNA–tRNA complex from the decoding center so translocation of the mRNA–tRNA moiety may be completed by a head rotation of the small subunit.
Keywords: cryo-EM, EF-G, elongation, GTPase, Switch 1
Introduction
The elongation cycle of protein synthesis is conducted by the ribosome with the help of two GTPases named eukaryotic elongation factors 1A and 2 (eEF1A and eEF2, respectively) in eukaryotes (EF-Tu and EF-G in bacteria). eEF1A/EF-Tu delivers an aminoacylated tRNA (aa-tRNA) to the ribosomal A site where it is decoded based on its anticodon match with the codon of messenger RNA (mRNA). Decoding involves three universally conserved bases (G530, A1492, and A1493 in Escherichia coli) in the ribosomal RNA (rRNA) of the small subunit (SSU). When cognate tRNA is delivered to the A site, G530 switches from a syn to an anti conformation to interact with the second position of the anticodon and the third position of the codon, whereas the bases A1492 and A1493 flip out of the internal loop of helix 44 to form A-minor interactions with the first two base pairs between the codon and anticodon (Ogle et al, 2001). GTP hydrolysis allows eEF1A/EF-Tu to leave the ribosome and the aa-tRNA to fully accommodate the A site. The large subunit (LSU) of the ribosome then rapidly catalyzes peptide bond formation by transferring the peptidyl moiety from the peptidyl tRNA in the P site to the aa-tRNA in the A site. Translocation of the mRNA–tRNA complex from the A and P sites to the P and E sites, respectively, must then occur so the next aa-tRNA can be decoded in the vacant A site. This step will necessarily involve the disruption of interactions between the decoding center and the mRNA–tRNA complex, by a mechanism that is poorly understood.
Translocation is catalyzed by the binding and subsequent GTP hydrolysis of eEF2/EF-G. EF-G and eEF2 are composed of six domains, numbered I–V and G′ (Aevarsson et al, 1994; Czworkowski et al, 1994; Jorgensen et al, 2003) (Supplementary Figure S1). Cryo-EM reconstructions of EF-G (Agrawal et al, 1998, 1999; Stark et al, 2000) and eEF2 (Gabashvili et al, 2000; Spahn et al, 2004) bound to the ribosome reveal that the tip of domain IV reaches into the ribosomal decoding center. Mutations of a conserved histidine residue (His583 in E. coli) located at the tip of domain IV of EF-G decrease the rate of translocation by more than two orders of magnitude (Savelsbergh et al, 2000). This conserved histidine is post-translationally modified into a diphthamide in eEF2. Mutations to the diphthamide precursor (His699 in yeast) inhibit cell growth (Phan et al, 1993; Kimata and Kohno, 1994). This inhibition in cell growth is at least partially due to a decreased efficiency in translocation, as shown by sensitivity to translation inhibitors and decreased total translation in vivo in a H699N mutant (Ortiz et al, 2006). Finally, bacterial toxins, such as Pseudomonas aeruginosa exotoxin A and Corynebacterium diphtheriae diphtheria toxin, inhibit translocation by transferring the ADP-ribosyl moiety from NAD+ onto the imidazole ring of the diphthamide in eEF2 (reviewed in Armstrong et al, 2002).
The GTP-binding site in domain I of eEF2/EF-G is surrounded by a mixed, six-stranded sheet and five helices (Aevarsson et al, 1994; Czworkowski et al, 1994; Jorgensen et al, 2003), a structure conserved among guanine nucleotide-binding proteins (reviewed in Vetter and Wittinghofer, 2001). GTP hydrolysis by EF-G has been described as a general GTPase switch mechanism where the factor is in its active form when bound to GTP and, upon GTP hydrolysis, ‘switched' to its inactive form (Kaziro, 1978). The finding that EF-G bound to nonhydrolyzable GTP analogs (GDPCP or GDPNP) still promotes translocation of the mRNA–tRNA complex to a puromycin-reactive state certainly supports this assumption (Kaziro, 1978). From these data, it was suggested that EF-G·GTP binds with high affinity to the ribosome and undergoes the essential conformational changes required to translocate the mRNA–tRNA complex. GTP hydrolysis would then induce additional conformational changes in EF-G after translocation, resulting in a loss of EF-G·GDP affinity for the ribosome and dissociation of the complex (see Spirin, 1985). This model, however, was later challenged by time-resolved pre-steady-state experiments that suggested translocation is actually preceded by GTP hydrolysis (Rodnina et al, 1997). This finding led to the speculation that EF-G is a motor protein and that the mechanical work required for translocation is provided by the chemical energy released from GTP hydrolysis (Rodnina et al, 1997).
A comparison of the X-ray crystal structures of EF-G (Aevarsson et al, 1994; Czworkowski et al, 1994; Hansson et al, 2005) and eEF2 (Jorgensen et al, 2003, 2004, 2005) with the cryo-EM reconstructions of ribosome-bound EF-G (Agrawal et al, 1998, 1999; Stark et al, 2000; Valle et al, 2003b) and eEF2 (Gabashvili et al, 2000; Spahn et al, 2004) indicates gross conformational changes in these factors upon binding to the ribosome. The conformational changes of eEF2/EF-G include a hinge-like motion of domains III, IV, and V with respect to domains I, II, and G′, which induce a ratchet-like rotation of the SSU with respect to the LSU of the ribosome (Frank and Agrawal, 2000; Spahn et al, 2004). The combination of the conformational changes in eEF2/EF-G and ribosomal ratcheting account for an 8 Å shift of the decoding center in the direction of translocation (VanLoock et al, 2000). This observation suggests that ribosomal ratcheting facilitates translocation, but the 8 Å shift falls significantly short of the greater than 20 Å required for full translocation of tRNA from the ribosomal A to P or P to E sites (Yusupov et al, 2001). Even though the entire distance may not be supplied by ribosomal moveable parts, it seems highly likely that another event is required to complete translocation.
To understand the molecular mechanism of translocation, the structural changes accompanying eEF2/EF-G binding as well as that of GTP hydrolysis during translocation need to be studied in detail. To this end, we have obtained cryo-EM reconstructions of eEF2 and ADP-ribosylated eEF2 (ADPR-eEF2) bound to the 80S ribosome from the thermophilic fungus Thermomyces lanuginosus. It has been shown that the binding of eEF2 to the ribosome is sufficient to place A site tRNA into a puromycin-reactive state, even in the absence of GTP hydrolysis (i.e., using GDPCP) (Davydova and Ovchinnikov, 1990). Conversely, ADPR-eEF2, with GDPCP or GTP, binds the ribosome as efficiently as eEF2, but is incapable of advancing the A site tRNA to a puromycin-reactive state (Davydova and Ovchinnikov, 1990). In the absence of A-site tRNA, however, ADPR-eEF2 binds the ribosome as efficiently as native eEF2, as shown using binding (Davydova and Ovchinnikov, 1990) and kinetic assays (Nygard and Nilsson, 1990). These data indicate that the interaction between the ADPR moiety and the codon–anticodon of A-site tRNA is insufficient to promote ratcheting of the SSU, and thus translocation. Conversely, the function of native eEF2/EF-G appears independent of the presence of A-site tRNA, because the rates of GTP hydrolysis (Rodnina et al, 1997) and EF-G·GDP dissociation (Wilden et al, 2006), determined using pre-steady-state kinetics and fluorescence titrations, are the same with or without A-site tRNA. Based on these observations, we decided to omit A-site tRNA and use the ADPR moiety of ribosome-bound ADPR-eEF2 as a distinct density marker to identify structural movements in domain IV of ADPR-eEF2 strictly as a function of GTP hydrolysis. For the reasons listed above, we are able to use this system to infer the GTP hydrolysis-induced conformational changes in native eEF2/EF-G and the ribosome in a physiological context and to investigate the structural role of GTP hydrolysis in translocation.
Our data provide the first visualization of the critical Switch 1 loop in the GTP-bound form of eEF2 and ADPR-eEF2, and reveal the structural basis for the conformational signal transduction from the GTP-binding site to the tip of domain IV near the ribosomal decoding center. The interpretation of our cryo-EM maps indicates that GTP hydrolysis results in a movement at the tip of domain IV of eEF2, which likely severs the connection between the mRNA–tRNA complex and the ribosomal decoding center. We present a novel two-step model of translocation, generalized so as to encompass both bacteria and eukaryotes, that describes how the ribosomal binding of eEF2/EF-G creates a ratcheted complex that partially translocates the mRNA–tRNA complex to a puromycin-reactive state. The bulk of translocation, however, is then facilitated by a head rotation of the SSU that can only occur after GTP hydrolysis has severed the connection between the decoding center in the body of the SSU and the mRNA–tRNA complex, which remains bound to the head of the SSU.
Results
Cryo-EM reconstructions of eEF2 and ADPR-eEF2 bound to the 80S ribosome
Cryo-EM and image reconstruction was used to determine the three-dimensional structures of native eEF2 and ADPR-eEF2 bound to the 80S ribosome in the presence of the nonhydrolyzable GTP analog GDPNP. The resolutions of the maps were determined to be 11.3 Å for the eEF2 complex and 9.7 Å for the ADPR-eEF2 complex (FSC=0.5 criterion). The primary difference between the native eEF2 (Figure 1A) and ADPR-eEF2 (Figure 1B) complexes is additional density that can be attributed to the ADPR moiety, at the tip of domain IV of ADPR-eEF2, in the ribosomal A site (circled in Figure 1B).
Figure 1.
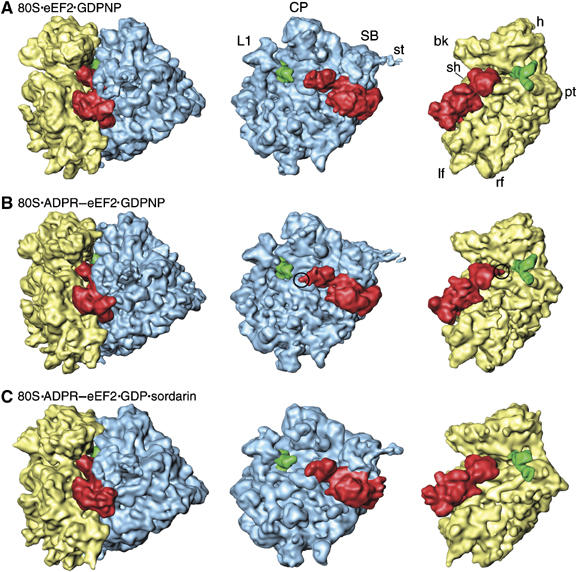
Cryo-EM reconstructions of 80S·eEF2/ADPR-eEF2 complexes. The computationally separated densities from the 80S·eEF2·GDPNP (A), the 80S·ADPR-eEF2·GDPNP (B), and the 80S·ADPR-eEF2·GDP·sordarin (C) complexes. The 60S subunit (LSU) is colored blue, the 40S subunit (SSU) is yellow, the P/E-site tRNA is green, and eEF2 and ADPR-eEF2 are red. Density attributed to the ADPR moiety is circled in (B). Landmarks in (A) are: CP, central protuberance; SB, stalk base; st, stalk; L1, L1 protuberance; h, head; bk, beak; pt, point; sh, shoulder; lf, left foot; rf, right foot.
To trap the GDP state of ADPR-eEF2, we used sordarin, which is highly specific to fungal eEF2 and prevents the dissociation of eEF2·GDP from the ribosome in a manner similar to fusidic acid (Justice et al, 1998). The 80S·ADPR-eEF2·GDP·sordarin complex was resolved to 11.7 Å. The density map (Figure 1C) shows that after GTP hydrolysis, the ADPR moiety has moved substantially, away from the ribosomal intersubunit space and towards the top of helix 44 in the rRNA of the SSU.
In all three reconstructions, the position of the tRNA is similar to that previously observed for P/E-hybrid state tRNA bound to the E. coli 70S ribosome (Valle et al, 2003b). Analogous to the E. coli P/E-state tRNA, the tRNA in our reconstructions has its CCA end positioned in the E site of the LSU, whereas the anticodon stem loop (ASL) is near the P site of the SSU (see Figure 1).
Docking of atomic structures
In the absence of an X-ray structure of any eukaryotic ribosome, we made use of the fact that the ‘common core' of the ribosome responsible for decoding and tRNA movement is conserved among eukaryotes and prokaryotes, suggesting a common mechanism of protein synthesis in all organisms (Gutell et al, 1985). This idea is supported by findings of high-fidelity docking of the atomic structures of the individual ribosomal subunits from bacteria and archaea into the cryo-EM density of the eukaryotic ribosome from Saccharomyces cerevisiae (Spahn et al, 2001). Indeed, the quasi-atomic ribosome model previously prepared for the S. cerevisiae 80S·eEF2·GDP·sordarin complex (Spahn et al, 2004) could be docked directly into our maps (Supplementary Figure S2). The crystal structures of sordarin-bound eEF2 (Jorgensen et al, 2003) and ADPR-eEF2 (Jorgensen et al, 2004) were docked into their respective cryo-EM maps by manual fitting, followed by real-space refinement (Supplementary Figure S1 and Supplementary Table SI, and Materials and methods).
Conformational signal transduction within eEF2 involves the Switch 1 loop
The ribosomal binding site for GTP-dependent factors for initiation (Allen et al, 2005; Myasnikov et al, 2005), elongation (Stark et al, 1997; Agrawal et al, 1998), and release (Klaholz et al, 2004) in bacteria, as well as for eEF2 (Gomez-Lorenzo et al, 2000; Spahn et al, 2004), is near the universal GTPase-associated center (GAC) of the LSU. The proximity of the GAC to the conserved GTP-binding site in the aforementioned, ribosome-bound G proteins, combined with biochemical data (Skold, 1983; Rodnina et al, 1999), suggests that the GAC is involved with the cyclic function (ribosomal binding, GTP hydrolysis, factor release) in all of the factors. A comparison of our 80S·ADPR-eEF2·GDPNP and 80S·ADPR-eEF2·GDP·sordarin complexes shows rearrangements of ADPR-eEF2 and the GAC that, upon GTP hydrolysis, alter the contacts between the LSU and ADPR-eEF2 (Figure 2A and B). Density assigned exclusively to ADPR-eEF2 in the two structures was isolated from the cryo-EM reconstruction by segmentation, that is, by removing density attributed to components of the ribosome (see Materials and methods). Examination of the segmented volumes of ADPR-eEF2 revealed that GTP hydrolysis causes movements of domains G′, I, and II toward the GAC, whereas domains III, IV, and V move in the opposite direction (Figure 2C).
Figure 2.
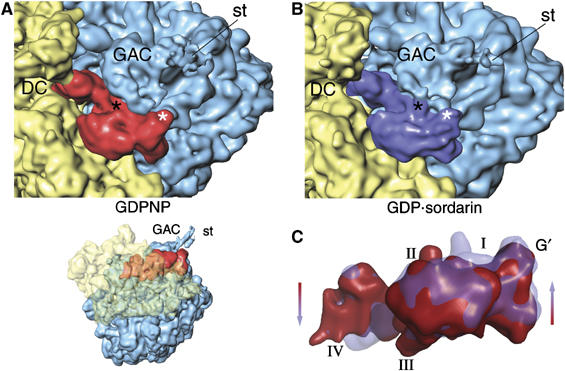
GTP hydrolysis causes a shift of domains I, II, and G′ of ADPR-eEF2 toward the GAC. Both ADPR-eEF2 and the GAC in the LSU undergo significant rearrangements due to GTP hydrolysis, as seen by comparing densities in the GTP-bound (GDPNP, A) and GDP-bound (GDP:sordarin, B) complexes. These conformational changes induce a reorganization of the contacts between the GAC and ADPR-eEF2, which are highlighted in black and white asterisks in the two panels. (C) Overlaying the densities attributed to ADPR-eEF2 from before (red) and after (blue) GTP hydrolysis exposes the conformational changes induced in ADPR-eEF2. This overlay reveals a shift in domains I, II, and G′ of ADPR-eEF2 toward the GAC upon GTP hydrolysis, while domains III, IV, and V are seen to shift in the opposite direction, away from the LSU. DC, decoding center; GAC, GTPase associated center; st, stalk.
For all available GTPase structures in both the GDP- and GTP-bound states, significant conformational changes in the GTP-binding domain are confined primarily to two conserved loops, named Switch 1 and Switch 2. The Switch 1 loop is disordered in all crystal structures of nucleotide-free, GDP-bound, and GDPNP-bound EF-G (Aevarsson et al, 1994; Czworkowski et al, 1994; Hansson et al, 2005) and eEF2 (Jorgensen et al, 2003, 2004, 2005), but its proximity to the GTP-binding site suggests that, like other GTPases, it coordinates the two terminal phosphate groups of bound GTP through binding of a Mg2+ ion. In the aa-tRNA·EF-Tu·GDPNP ternary complex of EF-Tu, the Switch 1 motif is composed of two α-helices, one of which makes a close contact to the tRNA (Nissen et al, 1995). The Switch 1 region from the crystal structure of the EF-Tu ternary complex was superimposed onto our quasi-atomic models by simply aligning the conserved helix A in domain I of the two structures. By doing so, it becomes obvious that unassigned density in the 80S·eEF2·GDPNP (Figure 3A) and 80S·ADPR-eEF2·GDPNP (Figure 3B) complexes is well accounted for by the EF-Tu Switch 1 structure. Although the resolution of our maps is insufficient to confidently determine the secondary structure of the Switch 1 loop in eEF2, the occupation of this superimposed model from EF-Tu in the Switch 1 density of our reconstructions implies a similar location, and potentially similar structure, of this polypeptide in the two elongation factors. This first localization of a ribosome-bound, ordered Switch 1 loop demonstrates that prior to GTP hydrolysis, this region in eEF2, and probably in all ribosomal GTPases, adopts an ordered conformation, similar to that of EF-Tu in the GTP-bound state. Indeed, the recent crystal structure of an EF-G homolog, EF-G-2, with bound GTP (PDB entry 1WDT) displays a Switch 1 conformation similar to that of EF-Tu in the GTP-bound state.
Figure 3.
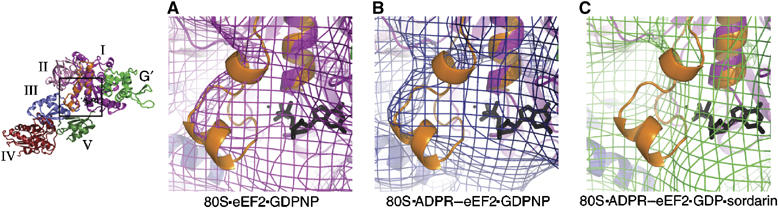
Superimposition of the Switch 1 loop from the EF–Tu ternary complex. The Switch 1 loop from the crystal structure of the EF–Tu ternary complex was superimposed onto our quasi-atomic models of eEF2 bound to the ribosome by least-squares alignment of the conserved helix A in domain I of the two structures. The Switch 1 loop from EF–Tu is accommodated by density in the cryo-EM structures of eEF2 in the GTP state (A) and ADPR-eEF2 in the GTP state (B), but the density of ADPR-eEF2 in the GDP state (C) shows that the Switch 1 loop has become disordered after GTP hydrolysis has occurred. Domains of eEF2 and ADPR-eEF2 are color-coded, the Switch 1 loop is colored orange, and GDP is colored black in all three panels. This figure was generated using PyMOL (DeLano, 2002).
GTP hydrolysis causes conformational changes in eEF2
Conformational changes within Switch 2, upon GTP hydrolysis, have been implicated as a major source of domain movements for eEF2 (Jorgensen et al, 2003), eIF5B (Roll-Mecak et al, 2000), and EF-Tu (Berchtold et al, 1993; Kjeldgaard et al, 1993). After GTP hydrolysis, the Switch 1 loop of eEF2 becomes disordered at the resolution of our reconstructions (Figure 3C), as observed in the 70S·EF-Tu·Phe-tRNAPhe·kirromycin complex also trapped in a post-GTP hydrolysis state (Valle et al, 2003a). The disordering of the switch indicates that GTP hydrolysis has indeed taken place in the 80S·ADPR-eEF2·GDP·sordarin complex. Our quasi-atomic model of ADPR-eEF2 shows the proximity of the Switch 1 loop in eEF2 with the sarcin-ricin loop (SRL) of the LSU (Figure 4), which has been implicated to play a role in catalyzing GTP hydrolysis (Hausner et al, 1987; Wool et al, 1992). After GTP hydrolysis, domains III and V move away from Switch 2 in domain I, with domain III rotating by approximately 5o with respect to domains I and II. Domains IV and V rotate, as one rigid body, by about 9o with respect to domains I and II. This movement results in a shift of the tip of domain IV, where the diphthamide resides in eEF2, by approximately 6 Å toward the top of helix 44 of the SSU (Figure 4).
Figure 4.
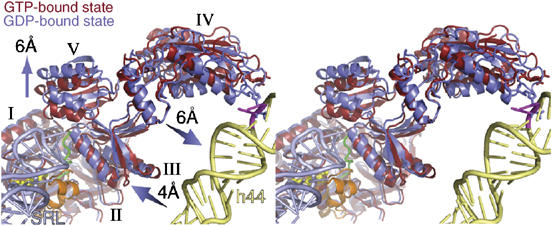
Stereo-view of domain movements in ADPR-eEF2 caused by GTP hydrolysis. The real-space refined, quasi-atomic models of ADPR-eEF2 before (red) and after (blue) GTP hydrolysis are shown together. GTP hydrolysis causes a shift in domains III and V of eEF2 away from domain I. Domain IV and V rotate slightly, as a single rigid body, resulting in a 6 Å shift of the tip of domain IV toward helix 44 in the rRNA of the SSU. The proximity of the SRL (light blue) to the Switch 1 loop (orange) in the GTP-bound state of the quasi-atomic model supports the role of the SRL in catalyzing GTP hydrolysis. GDP is shown in yellow stick representation, the Switch 2 loop is colored green, and the Switch 1 loop in the GTP-bound state is orange. Helix 44 from the 18S rRNA of the SSU is colored light yellow and the SRL from the 25S rRNA of the LSU is colored light blue. The location of two universally conserved adenine bases, A1492 and A1493 (E. coli numbering) in helix 44, that compose part of the ribosomal decoding center, is represented as magenta sticks. This figure was generated with PyMOL (DeLano, 2002).
Despite the small mass, density assigned to the ADPR moiety is well resolved in the map of the 80S·ADPR-eEF2·GDPNP complex, most likely due to the presence of two electron-dense phosphates. The ADPR moiety from the X-ray crystal structure of ADPR-eEF2 (Jorgensen et al, 2004) can be docked into its attributed density in the cryo-EM map as a rigid body (Figure 5A and Supplementary Figure S3). Fitting of the ADPR moiety into the map of the 80S·ADPR-eEF2·GDP·sordarin complex was facilitated by superimposition of the X-ray structure from the ADPR-eEF2·ETA complex (Jorgensen et al, 2005) onto the tip of domain IV of ADPR-eEF2 as determined by real-space refinement (Figure 5B). By comparing the densities of the 80S·ADPR-eEF2·GDPNP and 80S·ADPR-eEF2·GDP·sordarin complexes, it becomes obvious that GTP hydrolysis triggers a movement of the ADPR moiety into the major groove of helix 44, which comprises part of the ribosomal decoding center (Figure 5C). This movement is independently corroborated, in both magnitude and direction, by comparisons of the GTP- and GDP-bound structures of ADPR-eEF2 using (i) real-space refinement of domain IV (Figure 4), (ii) visual inspection of the segmented ADPR-eEF2 densities (Figure 2C), and (iii) analysis of density attributed exclusively to the ADPR moiety at the tip of domain IV (Figure 5). This movement, which—as we show below—is due exclusively to GTP hydrolysis, likely reflects the natural motion of the diphthamide residue in domain IV of eEF2 from the GTP- to the GDP-bound state and, in a translocating ribosome, would physically disrupt the interaction between the decoding center and the mRNA–tRNA duplex (Supplementary Movie S1).
Figure 5.
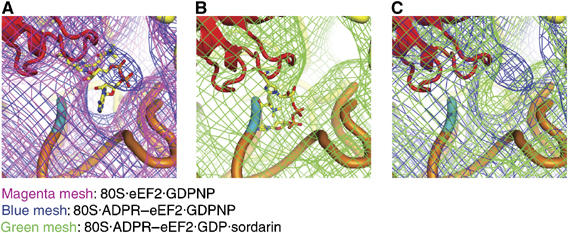
Docking of the X-ray structure of the ADPR moiety into the cryo-EM density map. (A) Comparison of the density obtained from the eEF2 (magenta mesh) and ADPR-eEF2 (blue mesh) maps, both with GDPNP, shows that density attributed to the ADPR occupies the ribosomal A site before GTP hydrolysis. (B) Fitting of the ADPR moiety after GTP hydrolysis in the 80S·ADPR-eEF2·GDP·sordarin cryo-EM density was facilitated by superimposition of domain IV of the ADPR-eEF2·exotoxin A crystal structure (Jorgensen et al, 2005). The docked ADPR moiety is well accounted for by extra density in this map. (C) A comparison of density obtained from the 80S·ADPR-eEF2·GDPNP (blue mesh) and 80S·ADPR-eEF2·GDP·sordarin (green mesh) complexes indicates that the ADPR moiety moves out of the A site, and into helix 44, after GTP hydrolysis. In all panels, ADPR-eEF2 is red, the ADPR moiety is colored according to atom composition, SSU rRNA is yellow, helix 44 of the SSU is orange, with the phosphate backbone of the conserved adenines in helix 44 (A1492 and A1493 in E. coli) colored cyan.
Conformational changes in ADPR-eEF2 are a direct result of GTP hydrolysis
To determine whether the movements at the tip of domain IV are a result of GTP hydrolysis or due to the binding of sordarin, we prepared a ‘control' complex containing both GDPNP and sordarin (80S·ADPR-eEF2·GDPNP·sordarin). The reconstruction of the complex at 8.9 Å resolution reveals that the ADPR moiety remains in the ribosomal intersubunit space, similar to the 80S·ADPR-eEF2·GDPNP complex, which is also in the GTP-bound state (Figure 6A). A comparison of the cryo-EM maps confirms that the segmented densities attributed to ADPR-eEF2 are virtually superimposable in the GTP-bound state (i.e. with GDPNP), regardless of the presence of sordarin (Figure 6B). The observation that density of the size expected for ADPR resides in precisely the same location in the two independent reconstructions, in a location moreover that agrees with its location in the X-ray map of ADPR-eEF2, justifies the conclusion that the mass is due to ADPR and unlikely due to random noise fluctuations in the separate reconstructions. Furthermore, density attributed to sordarin can be resolved in the 80S·ADPR-eEF2·GDPNP·sordarin map, and it appears at the location known from the X-ray map of sordarin-bound eEF2, as indicated in Figure 6B. As the mass of sordarin is comparable to that of the ADPR moiety, this finding further validates the assignment of density to the ADPR moiety in the cryo-EM reconstructions presented here.
Figure 6.
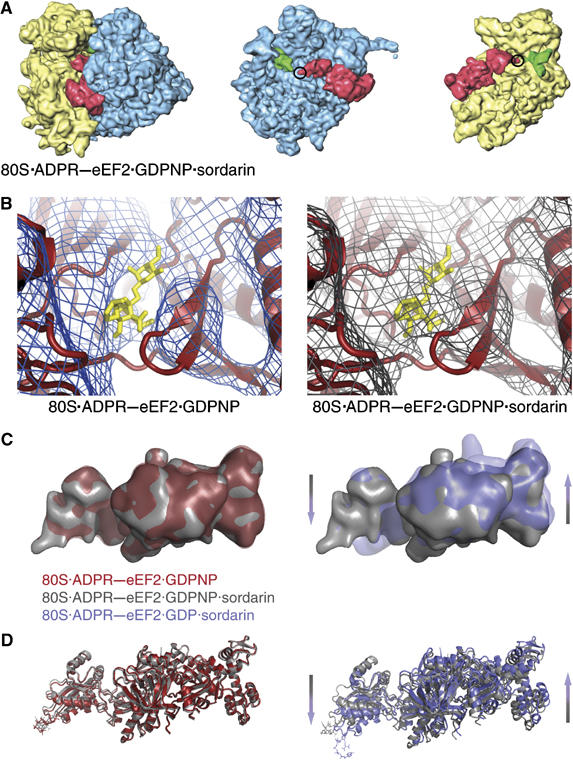
Comparison of ADPR-eEF2 conformational changes due to sordarin binding versus GTP hydrolysis. (A) The computationally separated densities from the 80S·ADPR-eEF2·GDPNP·sordarin complex reveals that the ADPR moiety occupies the intersubunit space (circled), as it does in the 80S·ADPR-eEF2·GDPNP complex (Figure 1B). This observation indicates that the binding of sordarin does not induce conformational changes in eEF2, when bound to the ribosome and GDPNP. (B) The primary difference in density between the 80S·ADPR-eEF2·GDPNP and 80S·ADPR-eEF2·GDPNP·sordarin complexes is density that can be attributed to the presence of sordarin, which is represented as yellow sticks in both panels. (C) Density attributed to ADPR-eEF2 from the 80S·ADPR-eEF2·GDPNP map (red) is virtually superimposable with that from the 80S·ADPR-eEF2·GDPNP·sordarin map (gray). Conversely, superimposition of density attributed to ADPR-eEF2 from the 80S·ADPR-eEF2·GDP·sordarin map (blue) with that from the 80S·ADPR-eEF2·GDPNP·sordarin map (gray) demonstrates significant conformational changes in the factor that are due exclusively to GTP hydrolysis. The direction and magnitude of these conformational changes are in agreement with those in the quasi-atomic models obtained after real-space refinement (D). The quasi-atomic models from the 80S·ADPR-eEF2·GDPNP (red) and 80S·ADPR-eEF2·GDPNP·sordarin (gray) complexes are very similar, whereas a comparison of the 80S·ADPR-eEF2·GDP·sordarin (blue) and 80S·ADPR-eEF2·GDPNP·sordarin (gray) quasi-atomic models shows conformational changes that are a result of GTP hydrolysis.
The differences when comparing the conformation of ADPR-eEF2 in the 80S·ADPR-eEF2·GDPNP·sordarin map to ADPR-eEF2 in the GDP-bound complex (Figure 6C) are the same as noted earlier (i.e. domains I, II, and G′ move toward the GAC, whereas domains III, IV, and V move away from the LSU, upon GTP hydrolysis). As another important control, the two nearly identical maps of GTP-bound states (80S·ADPR-eEF2·GDPNP and 80S·ADPR-eEF2·GDPNP·sordarin complexes) also resulted in nearly identical quasi-atomic models after real-space refinement (Figure 6D), underscoring the reliability of the model fitting procedures used in this study. The combination of these data confirms that (i) sordarin is able to bind to eEF2 in both GDP- and GTP-bound states and (ii) the conformational changes of ADPR-eEF2 reported here are due solely to GTP hydrolysis and not to sordarin binding.
Discussion
In contrast to native eEF2, it was shown that ADPR-eEF2 is unable to promote translocation of A-site tRNA to a puromycin-reactive state (Davydova and Ovchinnikov, 1990). The puromycin-reactive state was credited to binding of either eEF2 (Davydova and Ovchinnikov, 1990) or EF-G (Kaziro, 1978) to the ribosome, even without GTP hydrolysis. Later, it was shown that the binding of EF-G/eEF2 to the ribosome, again before GTP hydrolysis, is sufficient to promote or stabilize a conformation of the ribosome with the SSU ratcheted with respect to the LSU (Frank and Agrawal, 2000; Spahn et al, 2004). The combination of these data suggests that ADPR-eEF2 is unable to induce ratcheting, and therefore unable to produce puromycin-reactive tRNAs in a functioning ribosome.
In the absence of A-site tRNA, the conformational changes in eEF2 as a function of GTP hydrolysis presented here using the ADP-ribosylated factor would likely pertain to native eEF2 as well. This statement is supported by analysis of the cryo-EM reconstructions of ADPR-eEF2 and eEF2, both bound with GDPNP and a vacant A site, showing that the only discernible difference between the two structures is density attributed to the ADPR moiety (Figure 1A and B). Both structures reveal an ordered Switch 1 loop (Figure 3A and B) in the GTP-bound state, which becomes disordered in the ADPR-eEF2·GDP complex (Figure 3C), indicating that GTP has indeed been hydrolyzed (Figure 3C). This observation reveals that the rearrangement of the Switch 1 loop, as well as individual domain movements presented here, are a direct result of GTP hydrolysis and unrelated to the ADP-ribosylation. Even though our reconstructions lack A-site tRNA and therefore represent a non-functional complex, the observed conformational changes of eEF2/EF-G induced by GTP hydrolysis likely mimic those in translocation. Support for this statement is provided by studies showing similar rates for GTP hydrolysis by EF-G (Rodnina et al, 1997) and for EF-G·GDP dissociation (Wilden et al, 2006) using either translocating or vacant ribosomes. In a physiological context, the shift in domain IV of eEF2 would potentially sever the connection between the ribosomal decoding center and the codon–anticodon of the mRNA–tRNA duplex during translocation.
There is a wealth of kinetic data depicting a series of steps throughout the translocation process in E. coli (Rodnina et al, 1997, 1999; Savelsbergh et al, 2003; Wintermeyer et al, 2004; Wilden et al, 2006). These steps, which are likely conserved in all organisms, are (1) binding of EF-G·GTP to the pre-translocation ribosome, (2) GTP hydrolysis, (3) ‘unlocking' of the ribosome, (4) conformational rearrangement of the ribosome, Pi release, and tRNA translocation (5) release of EF-G·GDP, along with rearrangement of the ribosome to its post-translocational state. Our data are compatible with this kinetic model and provide several details that have been missing until now. The structures inferred from our density maps indicate that the ‘unlocking' event is the severing of the mRNA–tRNA complex from the decoding center and that the ribosomal rearrangement that drives translocation reported by kinetic studies can be explained by the head rotation, about the neck region, of the SSU.
This head rotation about the neck region of the SSU can occur only after eEF2/EF-G has disconnected the tether that links the mRNA–tRNA complex, still bound to the head, to the decoding center in the body of the SSU. In all three of our maps, eEF2 is bound and the P/E-site tRNA is interacting strongly with the head of the SSU (Figure 1). Interestingly, the head of the SSU is rotated significantly when eEF2 is bound as compared to its position in the 80S ribosome with tRNA in the P/P site and no factors bound (Spahn et al, 2004). This suggests that in a translating ribosome, the head rotation drags the mRNA and ASL of tRNAs from the A and P sites toward the P and E sites, respectively. Indeed, alignment of the bodies of the SSU of quasi-atomic models determined for the pre-translocational (Spahn et al, 2001) and the eEF2-bound (Spahn et al, 2004) 80S ribosomes from yeast reveals that head rotation alone could advance the mRNA–tRNA complex by approximately 12–13 Å (Figure 7). Summation of this movement with the ∼8 Å movement induced by eEF2/EF-G binding (VanLoock et al, 2000), gives a distance that is consistent with the ∼20 Å separating A and P, or P and E sites on the SSU (Yusupov et al, 2001). Disruptions of the interactions between the head of the SSU and tRNAs must be induced by a combination of reverse-ratcheting of the SSU and back-rotation of the head, movements that both occur upon release of Pi (Wilden et al, 2006) and eEF2/EF-G·GDP (Supplementary Movie S2).
Figure 7.
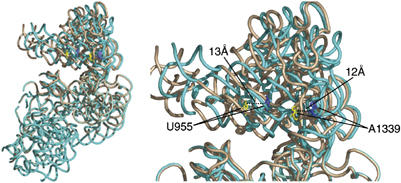
Head rotation of the 40S subunit of the yeast 80S ribosome. Binding of eEF2 induces a subunit rearrangement that involves ratcheting of the SSU with respect to the LSU, as well as head rotations in the SSU. To discriminate the difference induced exclusively by the head rotation, we superimposed the bodies of the SSU from the quasi-atomic models of the pre-translocational (Spahn et al, 2001) and the ratcheted, eEF2-bound (Spahn et al, 2004) 80S ribosomes, by explicit least-squares fitting using the program O (Jones et al, 1991). The SSU of the pre-translocational ribosome with P-site tRNA (PDB ID: 1K5X) is shown in tan, and that of the eEF2-bound ribosome (PDB ID: 1S1H) is shown in blue. Highlighted residues U955 and A1339 are in regions known to interact with ribosome-bound tRNA (Yusupov et al, 2001). This alignment demonstrates that head rotation alone can account for 12–13 Å movements of tRNA during translocation. Similar ratcheting and head rotations have been noted for the SSU of the bacterial ribosome (Schuwirth et al, 2005), suggesting that the mechanism of translocation is conserved.
In contrast to GTP hydrolysis, the mere binding of eEF2/EF-G induces ratcheting of the SSU with respect to the LSU (Frank and Agrawal, 2000; Spahn et al, 2004), which primes the ribosome for translocation. The ratcheting moves the ASL of A-site tRNA by 8 Å toward the P site (VanLoock et al, 2000), which is apparently adequate for puromycin reactivity, even in the presence of nonhydrolyzable GTP analogs (Kaziro, 1978; Davydova and Ovchinnikov, 1990). But for translocation to proceed, this ‘metastable' complex requires GTP hydrolysis to sever a critical connection between the moiety formed by mRNA, tRNA, and the head of the SSU with the decoding center in the body. Uncoupling of this link allows head rotation of the SSU about the neck region in the direction of translocation, as shown here and in previous ribosome structures from eukaryotes and bacteria (Spahn et al, 2004; Schuwirth et al, 2005). If GTP cannot be hydrolyzed after eEF2/EF-G has induced the ratcheting movement in the presence of A-site tRNA (i.e., when GDPNP is present and the decoding center is not reset), then reverse-ratcheting is bound to occur, and the mRNA and tRNAs will return to their original positions. This proposed mechanism explains why the puromycin reaction is reversible before the hydrolysis of GTP (Tanaka et al, 1977), why GTP hydrolysis increases the rate of translocation 50-fold (Rodnina et al, 1997), and why the cryo-EM reconstruction of the 70S·(tRNA)2·EF-G·GDPCP complex shows low occupancy and fragmented density for EF-G (Agrawal et al, 1999). Conversely, eEF2/EF-G binds to, and induces ratcheting of, the ribosome with nonhydrolyzable GTP analogs as effectively as it does with GTP in the absence of A-site tRNA, as shown here in our complexes, as well as in equivalent complexes in E. coli (Agrawal et al, 1999; Valle et al, 2003b). As these systems are starting without tRNA in the A site, bases A1492 and A1493 of the ribosomal decoding center remain stacked in helix 44. Consequently, there is no rigid link at the decoding center between the head and the body of the SSU, so head rotation occurs immediately after eEF2/EF-G binding and ribosomal ratcheting, regardless of whether or not GTP hydrolysis has occurred.
GTP hydrolysis, in summary, produces only subtle conformational changes in eEF2/EF-G that, in turn, trigger the large conformational changes in the ribosome necessary for completion of translocation. By this rationale, GTP hydrolysis would indeed precede the bulk of translocation, yet without requiring EF-G to act as a direct motor protein, as suggested (Rodnina et al, 1997). Rather, GTP hydrolysis of EF-G acts as a switch releasing the translocative potential of the ribosome that is established by peptide bond formation and ribosomal ratcheting upon binding of EF-G·GTP.
Our structural data, combined with previously published biochemical and kinetic data, provide a basis for explaining the stepwise mechanism of translocation. First eEF2/EF-G binds the pre-translocational ribosome. Whether tRNAs are in classical, A/A and P/P positions (Yusupov et al, 2001; Valle et al, 2003b), or they are sampling A/P and P/E hybrid states (Sharma et al, 2004) in the pre-translocational ribosome, binding of eEF2/EF-G will stabilize the tRNAs in the hybrid states. The proximity of the SRL to the Switch 1 loop of the ribosome-bound eEF2/EF-G supports the crucial role of the SRL in the GTPase function of the translocase (Hausner et al, 1987; Wool et al, 1992), although the exact interaction is not apparent at the resolution of our reconstructions. Binding of eEF2/EF-G to the ribosome causes the factors to undergo a conformational change, previously observed in cryo-EM reconstructions (Agrawal et al, 1999; Spahn et al, 2004), which induces ratcheting of the SSU with respect to the LSU (Frank and Agrawal, 2000; Spahn et al, 2004). The ratcheting moves the ASL of the tRNAs, along with the decoding center of the SSU, by approximately 8 Å in the direction of translocation (VanLoock et al, 2000). Ratcheting and coincident movement of tRNAs allows domain IV of eEF2/EF-G to occupy the ribosomal A site while changing the contacts between ribosomal subunits (Gao et al, 2003; Spahn et al, 2004) and inducing puromycin reactivity (Kaziro, 1978; Davydova and Ovchinnikov, 1990). Upon binding of eEF2/EF-G and ratcheting of the SSU, GTP is hydrolyzed, potentially due to interactions that alter the conformations of the Switch 1 and 2 loops of eEF2/EF-G. Conformational changes of eEF2/EF-G, upon GTP hydrolysis, include a 6 Å shift of domain IV that uncouples the rigid connection between the codon-ASL of the A-site tRNA and the decoding center. This event severs the connection between the mRNA–tRNA complex and the decoding center, allowing the head of the SSU to rotate about its neck region. Head rotation of the SSU translocates the ASL of tRNAs by the remaining distance required for full translocation. Finally, eEF2/EF-G leaves the ribosome, allowing the head to back-rotate and the SSU to back-ratchet to their original positions. The final outcome is the post-translocational ribosome with tRNAs in the P/P and E/E positions (Supplementary Movie S2).
Materials and methods
Preparation of ribosomes, eEF2, and ADPR-eEF2
80S ribosomes were purified as described previously (Sengupta et al, 2004). Briefly, ribosomes were pelleted through a sucrose cushion and further purified by hydrophobic interaction chromatography.
Purification of eEF2 from S. cerevisiae was performed as described (Jorgensen et al, 2004). The catalytic fragment of ETA with a C-terminal 6 × His tag was overexpressed and purified as reported previously (Armstrong et al, 2002). eEF2 was labeled with ADP-ribose and purified by anion-exchange chromatography as described previously (Jorgensen et al, 2004).
Cryo-EM and image processing
Ribosomes (30 nM 80S) were incubated on ice with eEF2 or ADPR-eEF2, nucleotide (GDPNP or GTP) and, if applicable, sordarin for 20 min. Carbon-coated holey grids were prepared following standard procedures, and micrographs were taken under low-dose conditions on a Philips Tecnai F30 at 300 kV and a × 37 642 magnification. Micrographs were scanned at a pixel size of 7 μm on a Zeiss/Imaging scanner (Z/I Imaging), corresponding to a 1.86 Å pixel size in the digitized image. Particles were chosen via automated particle picking followed by visual inspection, and data were processed with the SPIDER software package using standard procedures (Frank et al, 1996). Briefly, each data set was subdivided into defocus groups ranging from 0.8 to 4.2 μm underfocus in increments of 375 Å. Table I summarizes the number of particles, defocus range, and resolution attained for each of the three reconstructions. The reconstructed volumes were generated using three-dimensional projection alignment procedures (Frank et al, 2000) and CTF correction (Zhu et al, 1997), followed by multiple rounds of angular refinement with angular increments ranging from 2.0 to 0.5. X-ray solution scattering data for the 70S ribosome from E. coli (Gabashvili et al, 2000) were used to correct the Fourier amplitudes of the refined volumes.
Table 1.
Project details for image processing of cryo-EM data sets for each ribosomal complex reconstructed
| Complex | No. of particles | Defocus range (μm) | Resolution (Å)a |
|---|---|---|---|
| 80S·eEF2·GDPNP | 93 807 | 1.0–4.2 | 11.3 |
| 80S·ADPR-eEF2·GDPNP | 193 547 | 0.8–3.5 | 9.7 |
| 80S·ADPR-eEF2·GDP·sordarin | 70 510 | 0.8–3.7 | 11.7 |
| 80S·ADPR-eEF2·GDPNP·sordarin | 102 689 | 1.4–4.0 | 8.9 |
| aFinal resolution, as determined by FSC=0.5 cut-off. | |||
By comparing density attributed to eEF2/ADPR-eEF2 to that of the ribosome at various thresholds, the occupancy of eEF2/ADPR-eEF2 was estimated to be greater than 80% in all three complexes. Computationally separated volumes of eEF2 or ADPR-eEF2 were segmented from the 80S complex volumes by density clustering procedures using SPIDER (Frank et al, 1996). Briefly, clustered densities were isolated at various threshold values and manually assigned as either ribosomal or eEF2/ADPR-eEF2 components. Assignment of individual clusters was facilitated by the use of masks of the separated 40S and 60S subunit volumes from the yeast 80S ribosome (Spahn et al, 2004).
Docking and refining of atomic structures into cryo-EM density
The quasi-atomic model of the SSU from the S. cerevisiae 80S·eEF2·sordarin complex (Spahn et al, 2004) (PDB ID: 1S1H) was docked directly into our cryo-EM maps after alignment of the reconstructed volumes. The X-ray crystal structure of ADPR-eEF2 (PDB ID: 1U2R) was first docked manually into its attributed density in the 80S·eEF2·GDPNP cryo-EM map. This was done by independent fitting of the N-terminal domains (I, II, and G′) and the C-terminal domains (III, IV, and V) using the program O (Jones et al, 1991). This crudely fitted model was used as the input for real-space refinement against the cryo-EM density obtained for each of the four complexes presented here. This step prevented bias, as all eEF2/ADPR-eEF2 structures are in exactly the same orientation before refinement. The quasi-atomic model of each complex was then refined against the respective cryo-EM density of the entire complex, by using real-space refinement in the program RSRef2.0 (Chapman, 1995; Fabiola and Chapman, 2005). First, the model was refined as two individual rigid bodies: domains I, II, and G′ made up one body, and the second rigid body was composed of domains III, IV, and V. The six domains of eEF2/ADPR-eEF2 (I–V and G′) were further refined as individual rigid bodies (Supplementary Figure S1 and Supplementary Table SI).
For the 80S·ADPR-eEF2·GDPNP complex, the ADPR moiety from the X-ray crystal structure of ADPR-eEF2 (residues 699 and 1699 in 1U2R.pdb) was fitted manually using adjustment of threshold values of the density (Supplementary Figure S3). Docking of the ADPR moiety into the 80S·ADPR-eEF2·GDP·sordarin complex involved superimposition of the crystal structure of the ETA catalytic fragment in complex with eEF2 (PDB ID: 1ZM2) and our real space refined model. The docking was facilitated by use of an explicit least-squares fitting algorithm in the program O (Jones et al, 1991) to align domain IV of ADPR-eEF2 from the two structures.
The Switch 1 loop from the X-ray crystal structure of the EF–Tu ternary complex (Nissen et al, 1995) was superimposed in the quasi-atomic models presented here by alignment of the conserved helix A from the EF-Tu crystal structure (residues 25–35, pdb id: 1EFT) with that of the eEF2/ADPR-eEF2 quasi-atomic models (residues 33–43).
Accession Number
PDB IDs: the coordinates for the docked and refined eEF2 and ADPR-eEF2 have been deposited in the Protein Data Bank with accession numbers: 2P8W for eEF2•GDPNP, 2P8X for ADPR-eEF2•GDPNP, 2P8Y for ADPR-eEF2•GDP•sordarin, and 2P8Z for ADPR-eEF2•GDPNP•sordarin. The cryo-EM maps have been deposited at the 3D-EM database, EMBL-EBI with accession numbers: EMD-1342 for the 80S•eEF2•GDPNP complex, EMD-1343 for the 80S•ADPR-eEF2-GDPNP complex, EMD-1344 for the 80S•ADPR-eEF2•GDP•sordarin complex, and EMD-1345 for the 80S•ADPR-eEF2•GDPNP•sordarin complex.
Supplementary Material
Supplementary Information
Supplementary Movie 1
Supplementary Movie 2
Acknowledgments
We are grateful to R Grassucci for assistance with data collection, T Shaikh for assistance with data processing, M Watters for assistance in generating figures, Y Chen for assistance in generating the animation, J Nyborg (1943–2005) and R Jorgensen for valuable discussions, and J Sengupta for discussion of the manuscript. Sordarin was a kind gift from M Justice, Merck Research Laboratories. This work was supported by the Howard Hughes Medical Institute and National Institute of Health grants to JF, Human Frontier Science Program and Danish Natural Science Research Council grants to GRA and PN, a Hallas-Møller fellowship from the Novo Nordisk Foundation to PN, and a Canadian Institutes of Health Research Grant to ARM.
References
- Aevarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J 13: 3669–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6: 643–647 [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Frank J (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci USA 95: 6134–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 [DOI] [PubMed] [Google Scholar]
- Armstrong S, Yates SP, Merrill AR (2002) Insight into the catalytic mechanism of Pseudomonas aeruginosa exotoxin A. Studies of toxin interaction with eukaryotic elongation factor-2. J Biol Chem 277: 46669–46675 [DOI] [PubMed] [Google Scholar]
- Berchtold H, Reshetnikova L, Reiser CO, Schirmer NK, Sprinzl M, Hilgenfeld R (1993) Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365: 126–132 [DOI] [PubMed] [Google Scholar]
- Chapman MS (1995) Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron density function. Acta Crystallogr A A51: 69–80 [Google Scholar]
- Czworkowski J, Wang J, Steitz TA, Moore PB (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J 13: 3661–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydova EK, Ovchinnikov LP (1990) ADP-ribosylated elongation factor 2 (ADP-ribosyl-EF-2) is unable to promote translocation within the ribosome. FEBS Lett 261: 350–352 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Fabiola F, Chapman MS (2005) Fitting of high-resolution structures into electron microscopy reconstruction images. Structure 13: 389–400 [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 318–322 [DOI] [PubMed] [Google Scholar]
- Frank J, Penczek P, Agrawal RK, Grassucci RA, Heagle AB (2000) Three-dimensional cryoelectron microscopy of ribosomes. Methods Enzymol 317: 276–291 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P (2000) Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100: 537–549 [DOI] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J (2003) Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801 [DOI] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J (2000) Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J 19: 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF (1985) Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acids Res Mol Biol 32: 155–216 [DOI] [PubMed] [Google Scholar]
- Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT (2005) Crystal structure of a mutant elongation factor G trapped with a GTP analogue. FEBS Lett 579: 4492–4497 [DOI] [PubMed] [Google Scholar]
- Hausner TP, Atmadja J, Nierhaus KH (1987) Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie 69: 911–923 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Merrill AR, Yates SP, Marquez VE, Schwan AL, Boesen T, Andersen GR (2005) Exotoxin A–eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436: 979–984 [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR (2003) Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol 10: 379–385 [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Yates SP, Teal DJ, Nilsson J, Prentice GA, Merrill AR, Andersen GR (2004) Crystal structure of ADP-ribosylated ribosomal translocase from Saccharomyces cerevisiae. J Biol Chem 279: 45919–45925 [DOI] [PubMed] [Google Scholar]
- Justice MC, Hsu MJ, Tse B, Ku T, Balkovec J, Schmatz D, Nielsen J (1998) Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J Biol Chem 273: 3148–3151 [DOI] [PubMed] [Google Scholar]
- Kaziro Y (1978) The role of guanosine 5′-triphosphate in polypeptide chain elongation. Biochim Biophys Acta 505: 95–127 [DOI] [PubMed] [Google Scholar]
- Kimata Y, Kohno K (1994) Elongation factor 2 mutants deficient in diphthamide formation show temperature-sensitive cell growth. J Biol Chem 269: 13497–13501 [PubMed] [Google Scholar]
- Kjeldgaard M, Nissen P, Thirup S, Nyborg J (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1: 35–50 [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, Van Heel M (2004) Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427: 862–865 [DOI] [PubMed] [Google Scholar]
- Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP (2005) Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol 12: 1145–1149 [DOI] [PubMed] [Google Scholar]
- Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270: 1464–1472 [DOI] [PubMed] [Google Scholar]
- Nygard O, Nilsson L (1990) Kinetic determination of the effects of ADP-ribosylation on the interaction of eukaryotic elongation factor 2 with ribosomes. J Biol Chem 265: 6030–6034 [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG (2006) Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem 281: 32639–32648 [DOI] [PubMed] [Google Scholar]
- Phan LD, Perentesis JP, Bodley JW (1993) Saccharomyces cerevisiae elongation factor 2. Mutagenesis of the histidine precursor of diphthamide yields a functional protein that is resistant to diphtheria toxin. J Biol Chem 268: 8665–8668 [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Matassova NB, Katunin VI, Semenkov YP, Wintermeyer W (1999) Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc Natl Acad Sci USA 96: 9586–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Cao C, Dever TE, Burley SK (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103: 781–792 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W (2003) An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 11: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W (2000) Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol 300: 951–961 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J (2004) Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11: 957–962 [DOI] [PubMed] [Google Scholar]
- Sharma D, Southworth DR, Green R (2004) EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA 10: 102–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold SE (1983) Chemical crosslinking of elongation factor G to the 23S RNA in 70S ribosomes from Escherichia coli. Nucleic Acids Res 11: 4923–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirin AS (1985) Ribosomal translocation: facts and models. Prog Nucleic Acid Res Mol Biol 32: 75–114 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M (1997) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389: 403–406 [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W (2000) Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100: 301–309 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Iwasaki K, Kaziro Y (1977) Translocation reaction promoted by polypeptide chain elongation factor-2 from pig liver. J Biochem (Tokyo) 82: 1035–1043 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003a) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003b) Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- VanLoock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC (2000) Movement of the decoding region of the 16 S ribosomal RNA accompanies tRNA translocation. J Mol Biol 304: 507–515 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
- Wilden B, Savelsbergh A, Rodnina MV, Wintermeyer W (2006) Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc Natl Acad Sci USA 103: 13670–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV (2004) Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans 32: 733–737 [DOI] [PubMed] [Google Scholar]
- Wool IG, Gluck A, Endo Y (1992) Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem Sci 17: 266–269 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zhu J, Penczek PA, Schroder R, Frank J (1997) Three-dimensional reconstruction with contrast transfer function correction from energy-filtered cryoelectron micrographs: procedure and application to the 70S Escherichia coli ribosome. J Struct Biol 118: 197–219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Movie 1
Supplementary Movie 2


